8,194 results
-
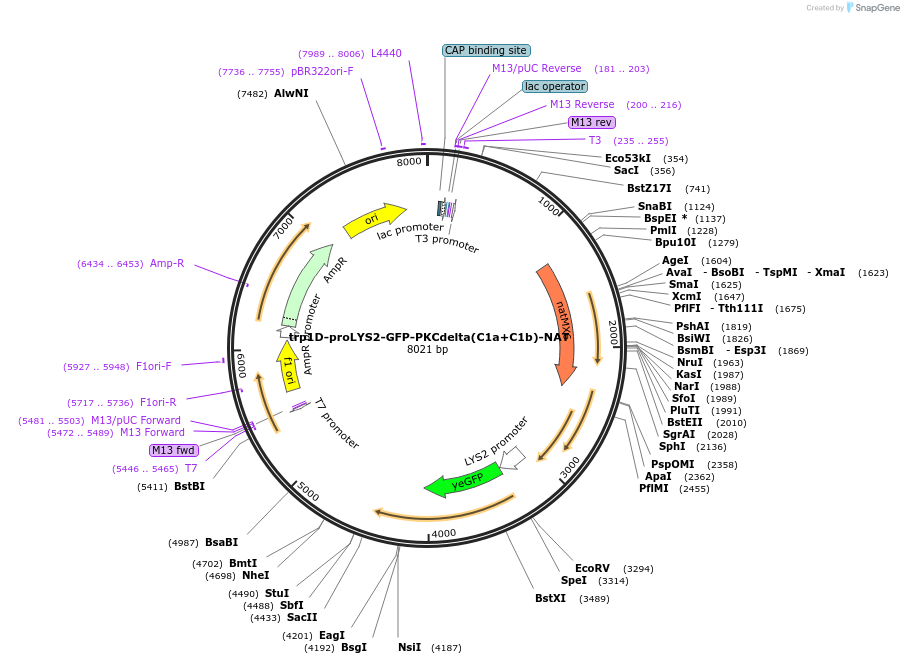
Plasmid#184774PurposeMarker for DAG. Yeast expression of mouse PKCdelta under LSY2 promoter. Uses antibiotic resistance marker natMX6. Replaces endogenous TRP1 upon genome integration, leading to trp1D.DepositorAvailable SinceFeb. 6, 2023AvailabilityAcademic Institutions and Nonprofits only
-
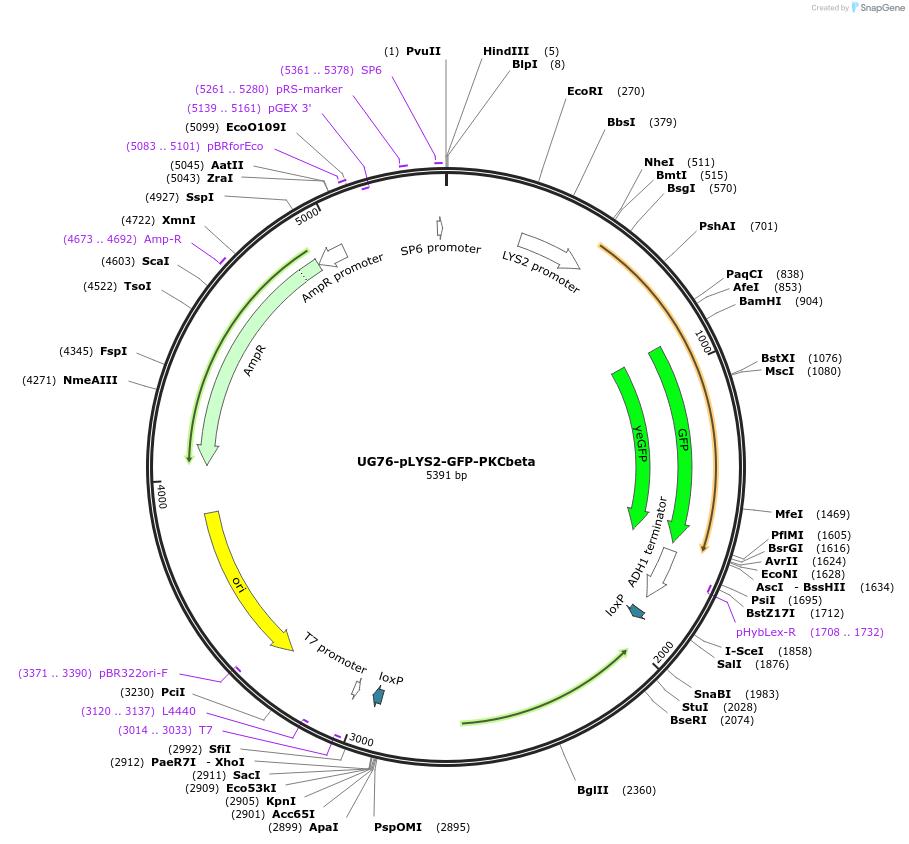
UG76-pLYS2-GFP-PKCbeta
Plasmid#184762PurposeMarker for DAG. Yeast expression of human PKC beta under LSY2 promoter. Uses auxotrophic marker TRP1(Kluyveromyces lactis).DepositorAvailable SinceFeb. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
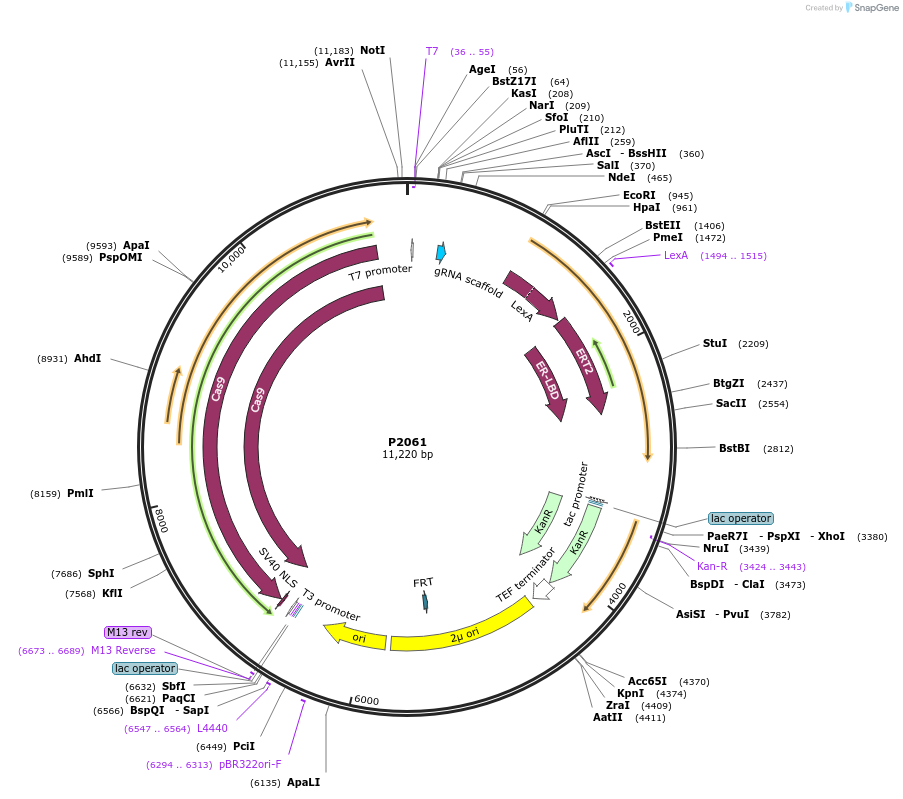
P2061
Plasmid#186299PurposeExpresses LexA-HBD-B112 under the control of pAct1 and Cas9 under the control of LexA-HBD-B112+Beta-estradiol inducible promoterDepositorInsertsLexA-HBD-B112
Cas9
UseCRISPR and Synthetic BiologyExpressionBacterial and YeastPromoter2xLexop-minimalCyc1 and PACT1Available SinceFeb. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
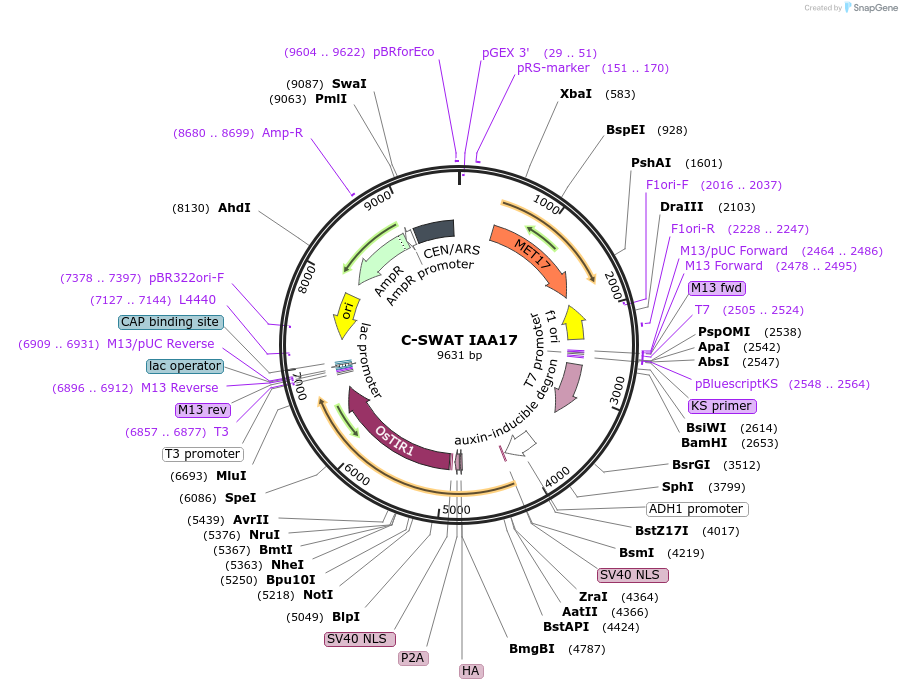
C-SWAT IAA17
Plasmid#193319PurposeC-SWAT compatible vector for C-terminal tagging with IAA17 (Auxin-Inducible Degron). ARF16(PB1)-P2A-osTIR1 has SIR4 homology arms for genome integration.DepositorInsertIAA17
TagsIAA17ExpressionYeastAvailable SinceFeb. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
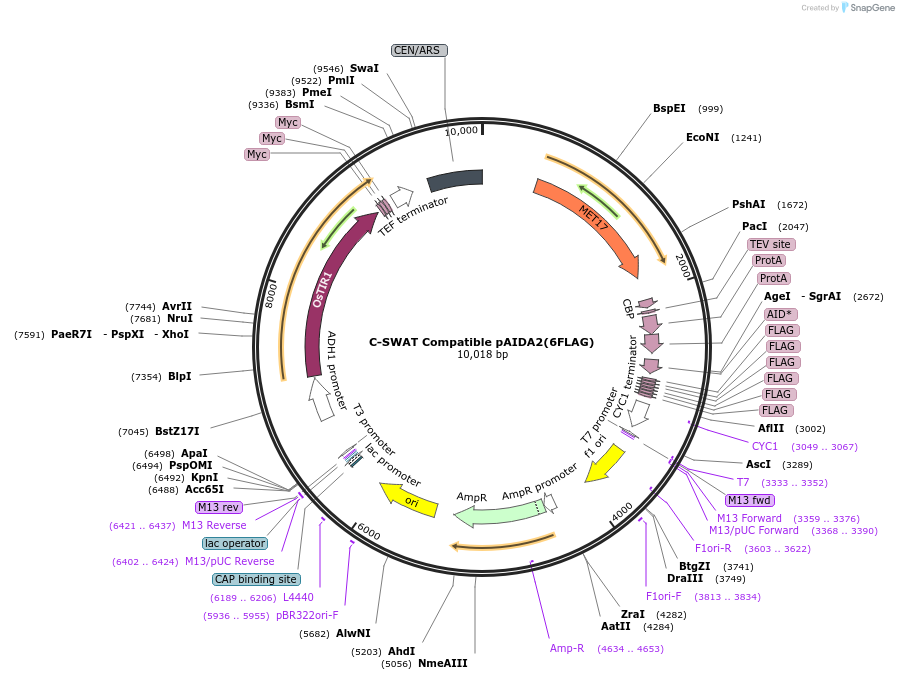
C-SWAT Compatible pAIDA2(6FLAG)
Plasmid#193321PurposeC-SWAT compatible vector for C-terminal tagging with TAP-AID*-6FLAG. osTIR1 is carried downstream of the C-terminal tag.DepositorInsertTAP-AID*-6FLAG
TagsTAP-AID*-6FLAGExpressionYeastAvailable SinceJan. 26, 2023AvailabilityAcademic Institutions and Nonprofits only -
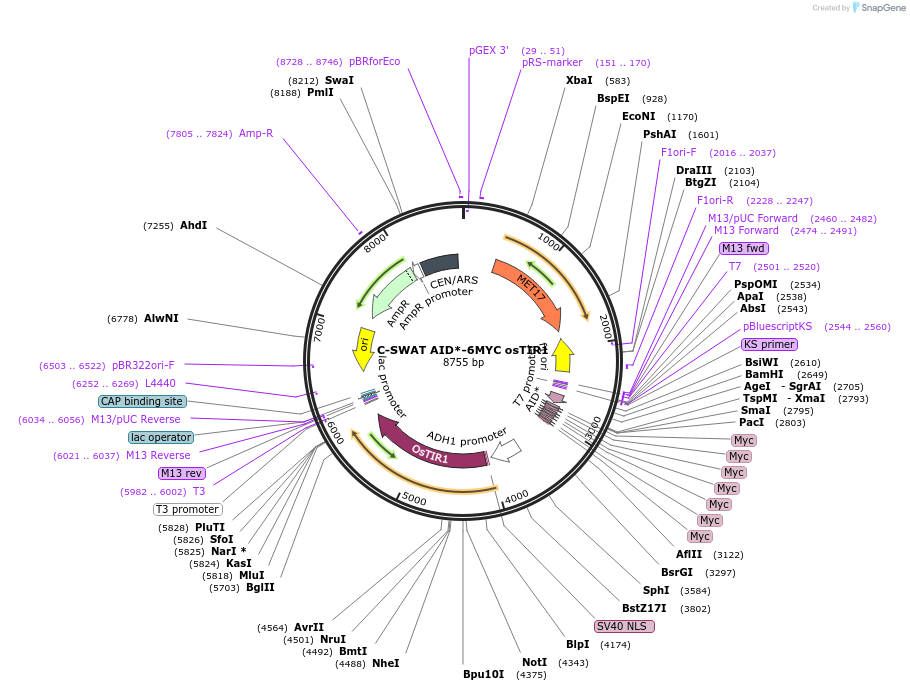
C-SWAT AID*-6MYC osTIR1
Plasmid#193320PurposeC-SWAT compatible vector for C-terminal tagging with AID*-6MYC for auxin induced gene knockdown. osTIR1 is flanked by SIR4 homology arms for integration into the genome.DepositorInsertAID*
Tags6MYCExpressionYeastAvailable SinceJan. 26, 2023AvailabilityAcademic Institutions and Nonprofits only -
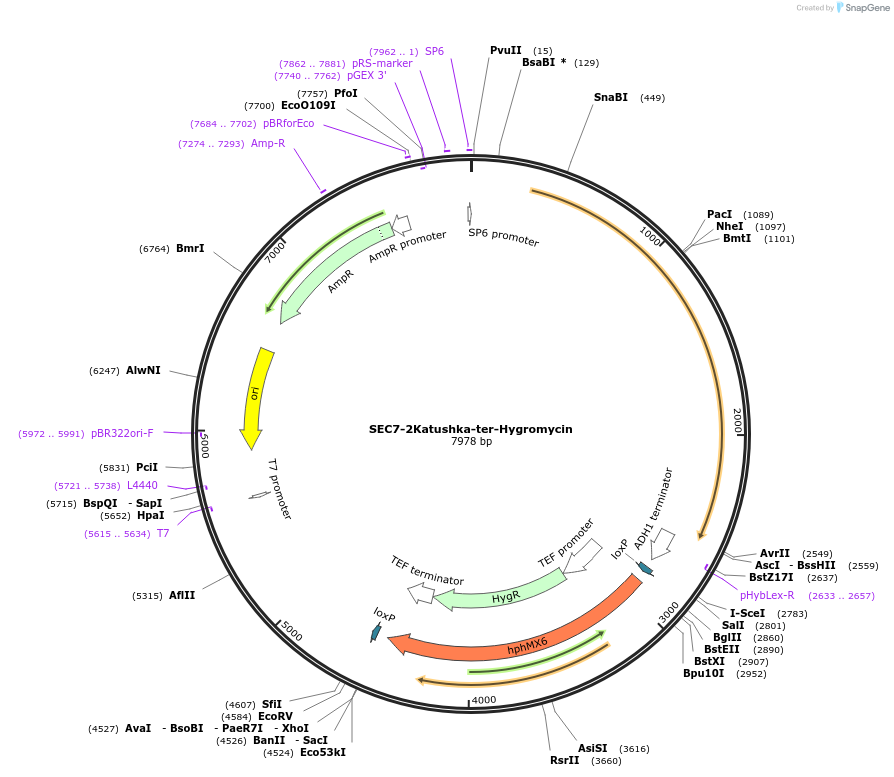
SEC7-2Katushka-ter-Hygromycin
Plasmid#184772PurposeRed marker for yeast late Golgi/early endosome. Integration of 2xKatushka tag at SEC7 C terminus. Uses antibiotic resistance marker hphMX6.DepositorAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
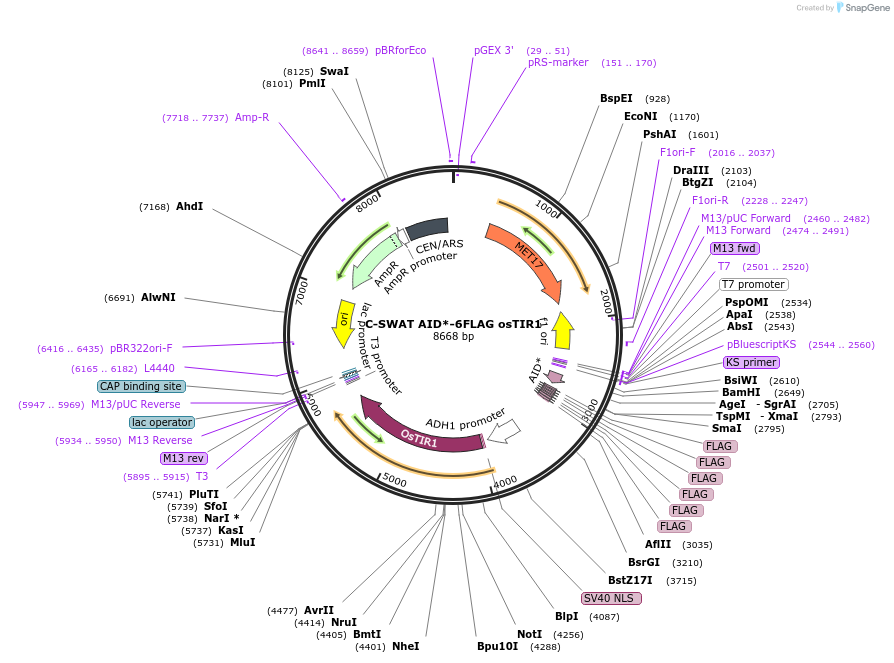
C-SWAT AID*-6FLAG osTIR1
Plasmid#182515PurposeC-SWAT compatible vector for C-terminal tagging with AID*-6FLAG for auxin induced gene knockdown. osTIR1 is flanked by SIR4 homology arms for integration into the genome.DepositorInsertAID*
Tags6FLAGExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
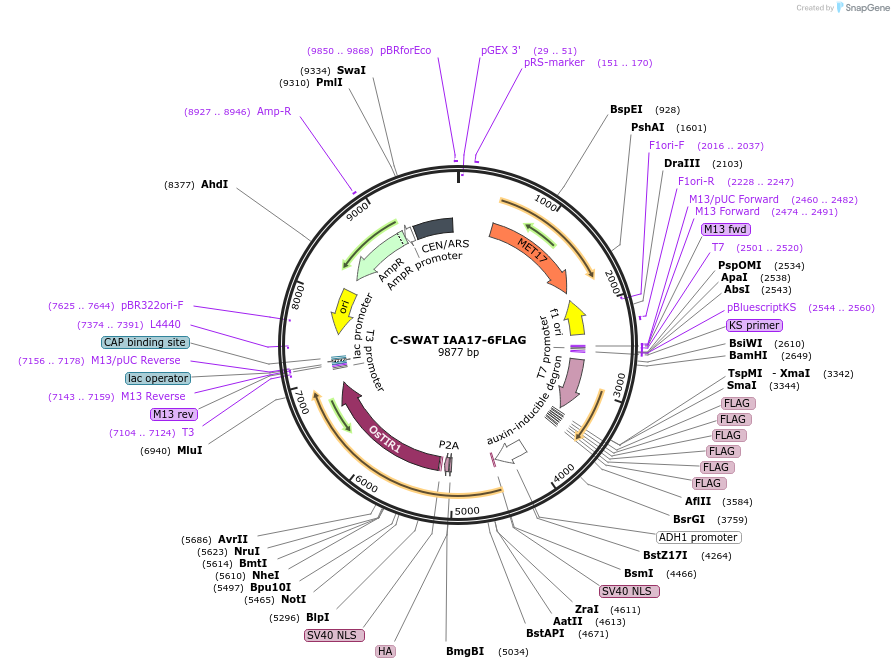
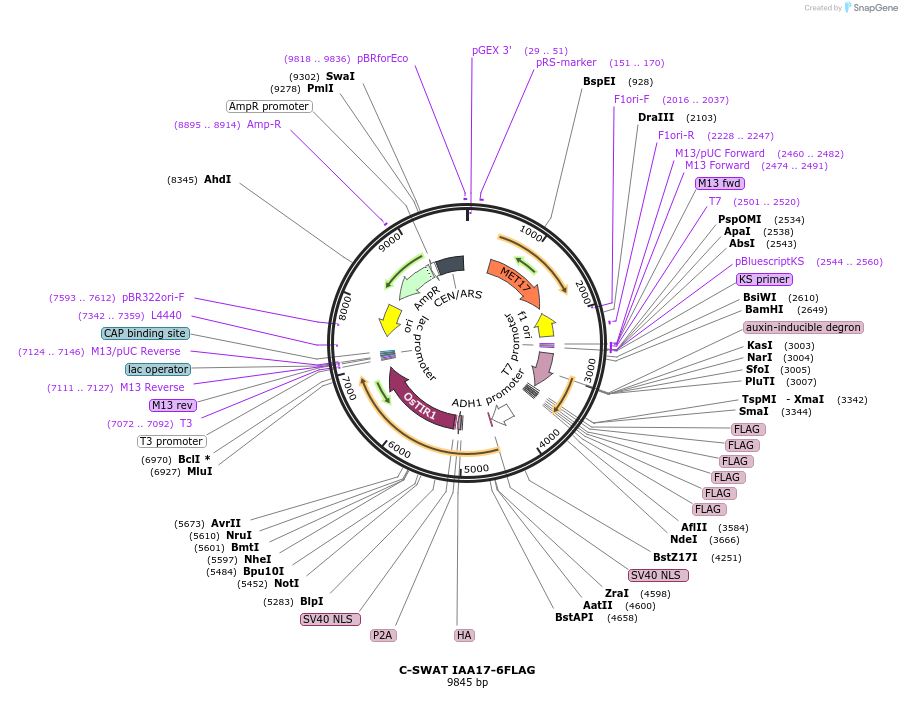
C-SWAT IAA17-6FLAG
Plasmid#182516PurposeC-SWAT compatible vector for C-terminal tagging with IAA17 (Auxin-Inducible Degron) and 6FLAG. ARF16(PB1)-P2A-osTIR1 has SIR4 homology arms for genome integration.DepositorInsertIAA17
Tags6FLAGExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
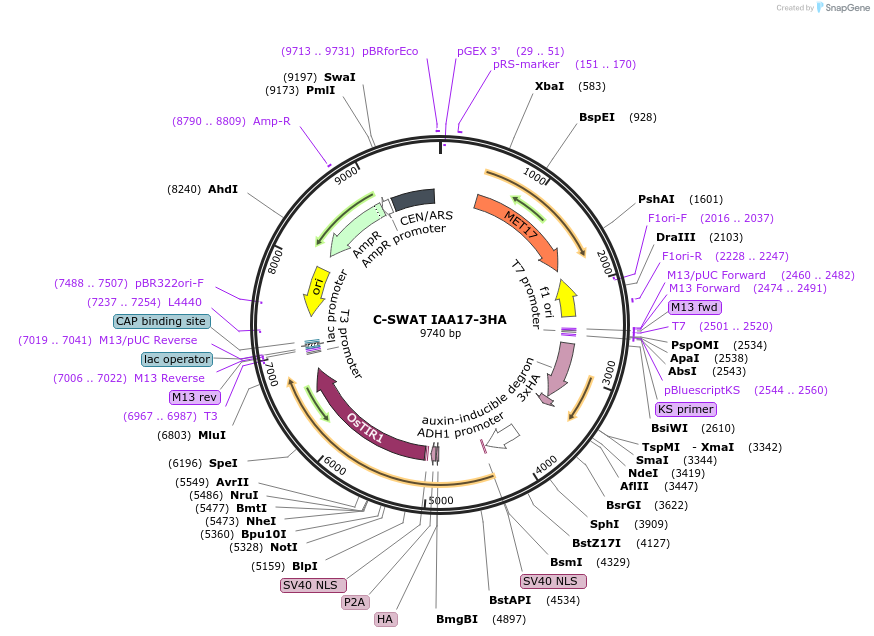
C-SWAT IAA17-3HA
Plasmid#182517PurposeC-SWAT compatible vector for C-terminal tagging with IAA17 (Auxin inducible degron) and 3HA. ARF16(PB1)-P2A-osTIR1 with SIR4 homology arms for genome integration.DepositorInsertIAA17
Tags3HAExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
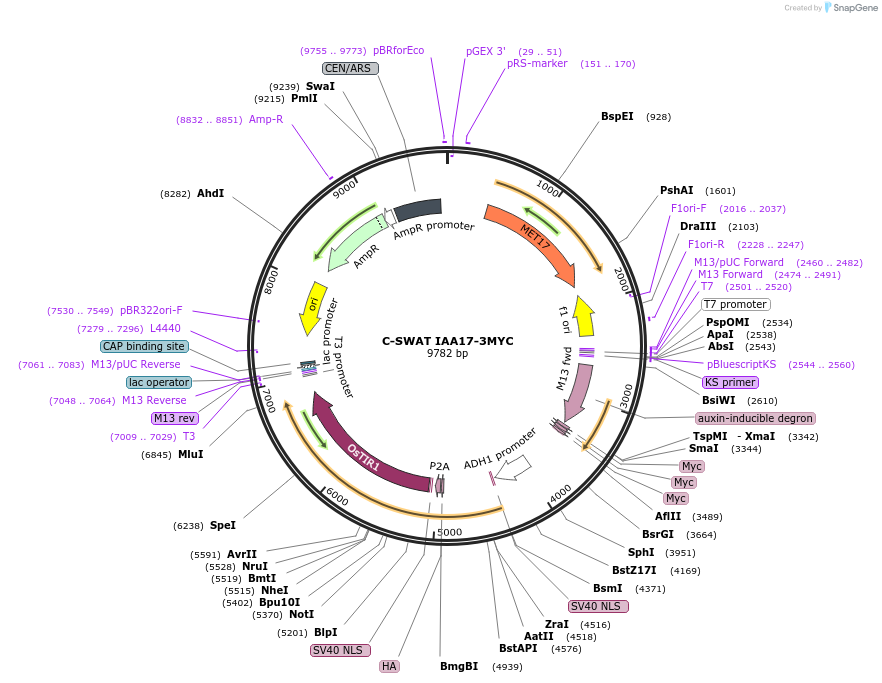
C-SWAT IAA17-3MYC
Plasmid#182518PurposeC-SWAT compatible vector for C-terminal tagging with IAA17 (Auxin inducible degron) and 3MYC. ARF16(PB1)-P2A-osTIR1 with SIR4 homology arms for genome integration.DepositorInsertIAA17
Tags3MYCExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
SIR4 gRNA HIS
Plasmid#182520PurposeExpression vector for gRNA directed against SIR4 ORF.DepositorInsertSIR4 gRNA (SIR4 Budding Yeast)
ExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
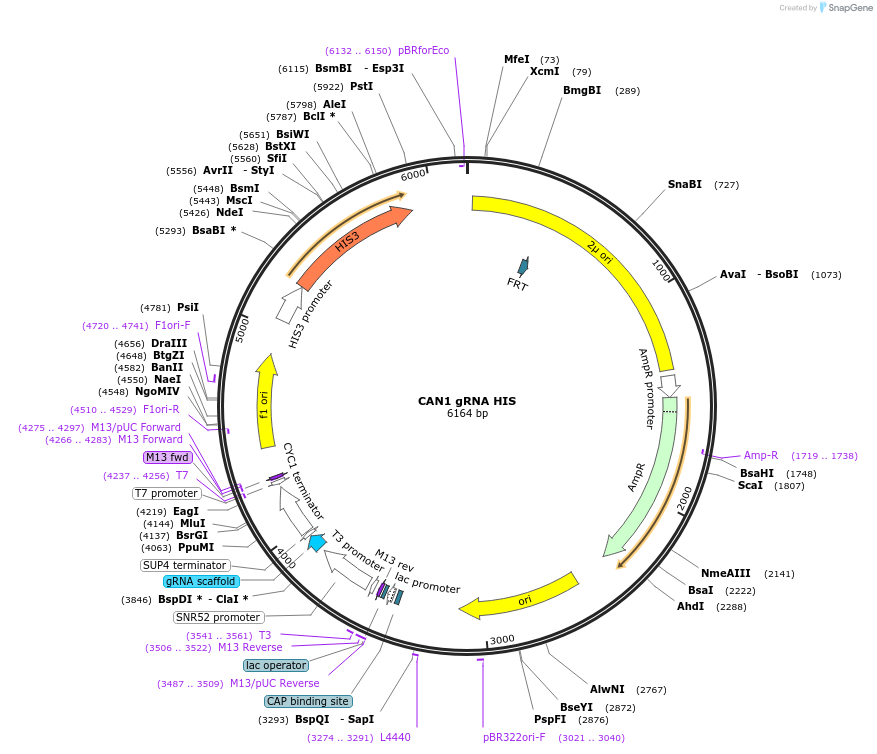
CAN1 gRNA HIS
Plasmid#182521PurposeExpression vector for gRNA directed against CAN1 ORF.DepositorInsertCAN1 gRNA (CAN1 Budding Yeast)
ExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
C-SWAT IAA17-6FLAG
Plasmid#182522PurposeC-SWAT compatible vector for C-terminal tagging with IAA17 (Auxin Inducible Degron) and 6FLAG. ARF16(PB1)-P2A-osTIR1 with CAN1 homology arms for genome integration.DepositorInsertIAA17
Tags6FLAGExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
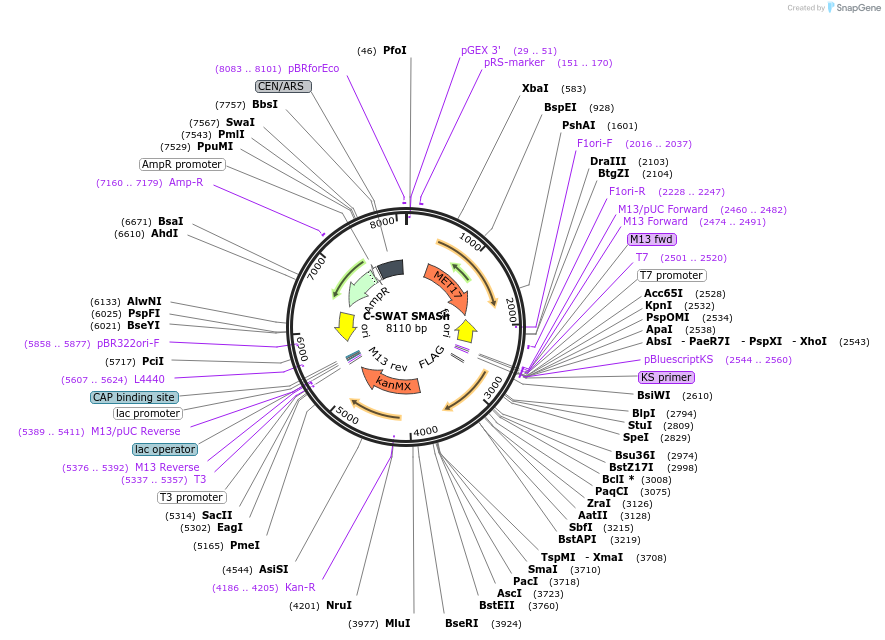
C-SWAT SMASh
Plasmid#182523PurposeC-SWAT compatible vector for C-terminal tagging with SMASh Degron. (Protease Cleavage Site: DEMEECSKHL).DepositorInsertSMASh Tag
ExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
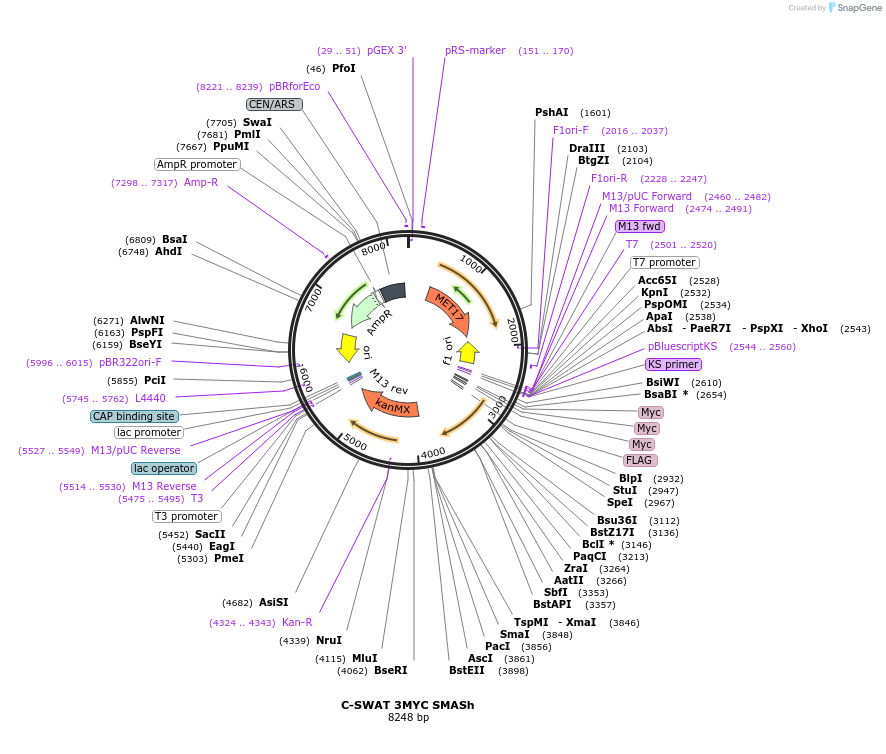
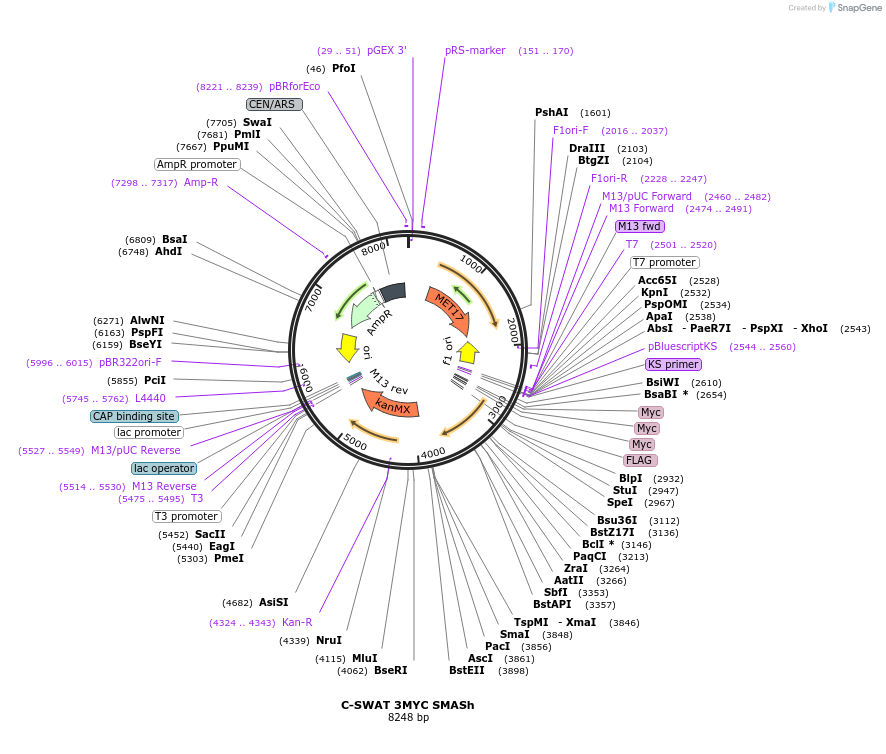
C-SWAT 3MYC SMASh
Plasmid#182524PurposeC-SWAT compatible vector for C-terminal tagging with 3MYC followed by SMASh degron. (Protease Cleavage Site: DEMEECSKHL).DepositorInsertSMASh Tag
Tags3MYCExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
C-SWAT 3MYC SMASh
Plasmid#182525PurposeC-SWAT compatible vector for C-terminal tagging with 3MYC followed by SMASh degron. (Protease Cleavage Site: DEMEECSQHL).DepositorInsertSMASh Tag
Tags3MYCExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
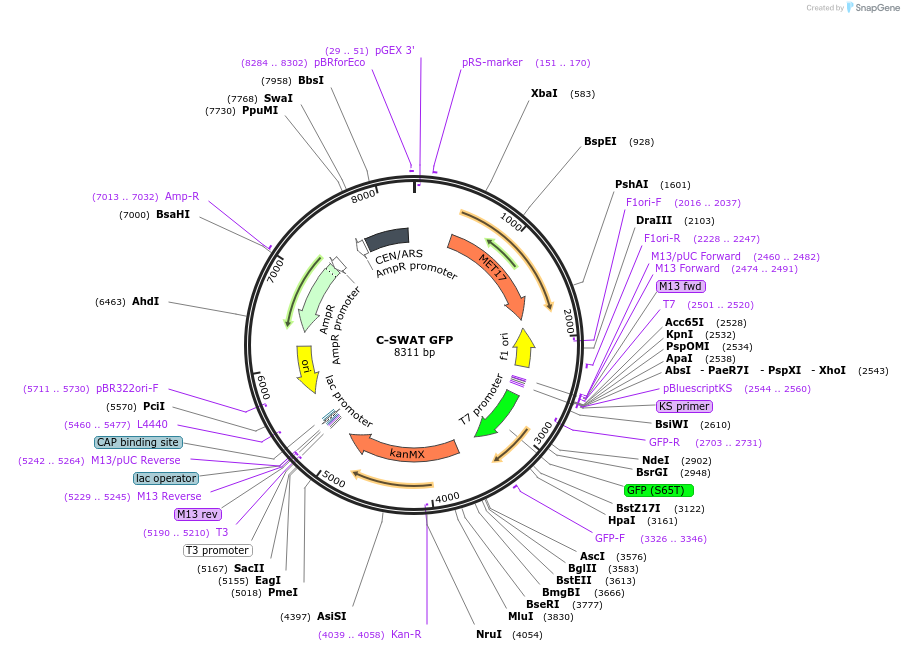
C-SWAT GFP
Plasmid#182510PurposeC-SWAT compatible vector for C-terminal tagging with GFPDepositorInsertGFP
TagsGFPExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
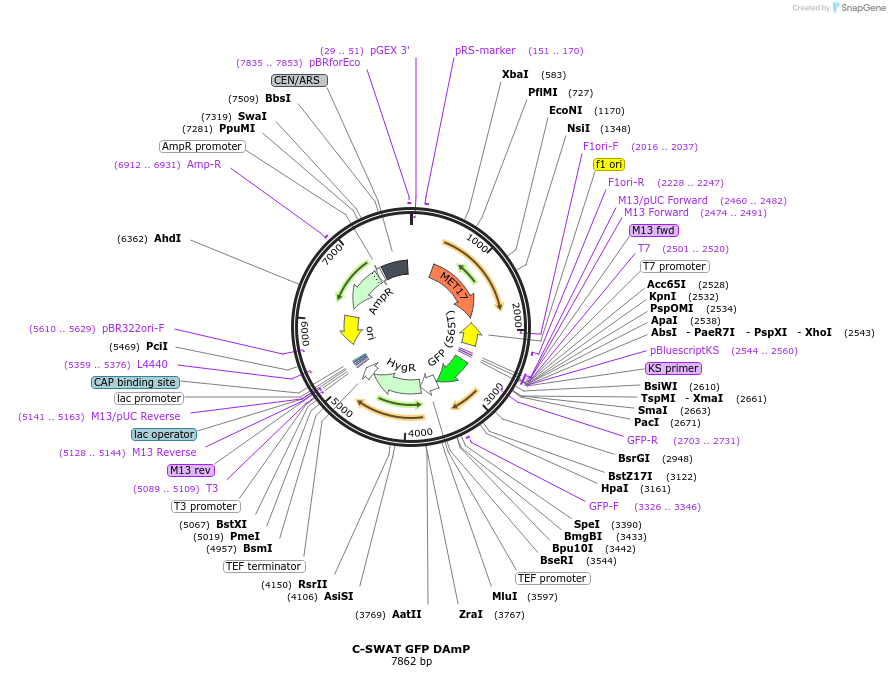
C-SWAT GFP DAmP
Plasmid#182511PurposeC-SWAT compatible vector for C-terminal tagging with GFP DAmP. GFP is immediately followed by the Hygromycin B (Hph) resistance cassette.DepositorInsertGFP
TagsGFPExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
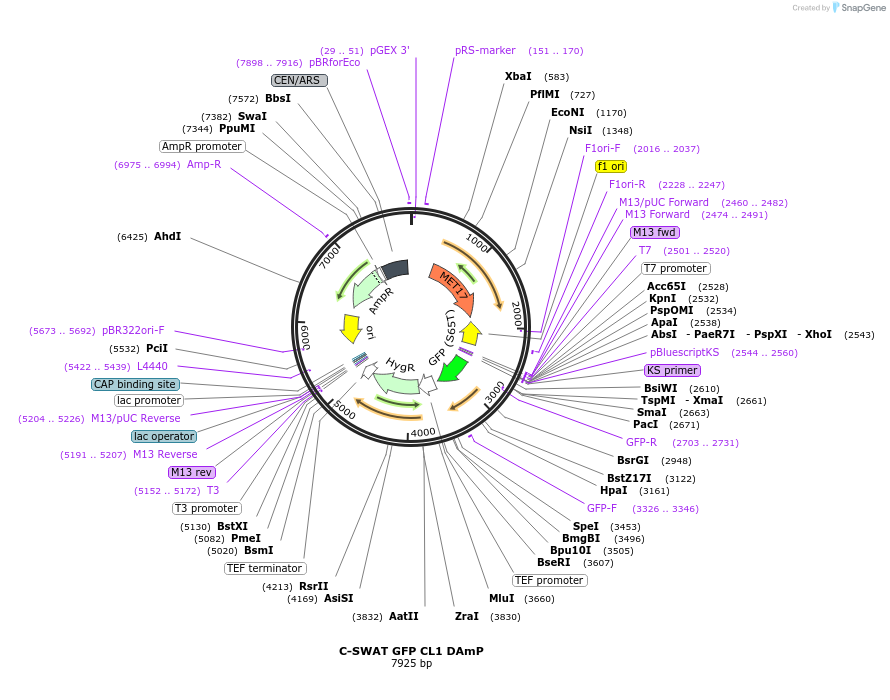
C-SWAT GFP CL1 DAmP
Plasmid#182513PurposeC-SWAT compatible vector for C-terminal tagging with GFP CL1 followed by the DAmP cassette. The stop codon after the CL1 degron is immediately followed by the Hygromycin B (Hph) resistance cassette.DepositorInsertCL1 and DAmP
TagsGFPExpressionYeastAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only