We narrowed to 4,238 results for: pes;
-
Plasmid#131076PurposeActivation of Mafa via SAM systemDepositorAvailable SinceOct. 3, 2025AvailabilityAcademic Institutions and Nonprofits only
-
AAV2 for Split-PE, gRNAs + MMLV-RT(dRH)-P2A-eGFP (PKC1002)
Plasmid#190112PurposeAAV2 for expression of pegRNA and ngRNA (HEKs3, CTT insertion) + MMLV-RT(dRH)-P2A-eGFPDepositorInsertsMMLV-RT(dRH)
U6-pegRNA(HEKs3, CTT ins)-H1-ngRNA(HEK s3)
UseAAVTagsbpNLS and bpNLS-P2A-eGFPMutationmutations from RT in PE2 and truncation of RNAse …PromoterEFS and U6, H1Available SinceSept. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLN193 (SSX1p-[mK-Ex1]-[miR1-Mv3 intron]-[mK-Ex2]-BS(Pe))
Plasmid#105210PurposeModule 1 - SSX1 promoter drives self-inhibiting mKate2 expression (Version 3 intron - see PMID: 29056342 for detailed information)DepositorInsertSSX1p-[mK-Ex1]-[miR1-Mv3 intron]-[mK-Ex2]-BS(Pe)
UseSynthetic BiologyExpressionMammalianAvailable SinceMarch 17, 2018AvailabilityAcademic Institutions and Nonprofits only -
pLN520 (SSX1p-[mK-Ex1]-[miR1-Mv1 intron]-[mK-Ex2]-BS(Pe))
Plasmid#105207PurposeModule 1 - SSX1 promoter drives self-inhibiting mKate2 expression (Version 1 intron - see PMID: 29056342 for detailed information)DepositorInsertSSX1p-[mK-Ex1]-[miR1-Mv1 intron]-[mK-Ex2]-BS(Pe)
UseSynthetic BiologyExpressionMammalianAvailable SinceNov. 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
pLN189 (SSX1p-[mK-Ex1]-[miR1-Mv2 intron]-[mK-Ex2]-BS(Pe))
Plasmid#105209PurposeModule 1 - SSX1 promoter drives self-inhibiting mKate2 expression (Version 2 intron - see PMID: 29056342 for detailed information)DepositorInsertSSX1p-[mK-Ex1]-[miR1-Mv2 intron]-[mK-Ex2]-BS(Pe)
UseSynthetic BiologyExpressionMammalianAvailable SinceNov. 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
TC358
Plasmid#122613PurposeTRE-delta-pes-10-GFPDepositorInsert7XTRE-delta-pes-10-GFP
ExpressionWormAvailable SinceMay 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
pBN100
Plasmid#35106DepositorInsertcaURA3PR
UseYeast replicatingAvailable SinceSept. 2, 2012AvailabilityAcademic Institutions and Nonprofits only -
pBN42
Plasmid#35103DepositorInsertkanPR
UseYeast replicatingAvailable SinceAug. 29, 2012AvailabilityAcademic Institutions and Nonprofits only -
pPH37
Plasmid#35102DepositorInsertfcy1MX4
UseYeast replicatingAvailable SinceApril 19, 2012AvailabilityAcademic Institutions and Nonprofits only -
pPH36
Plasmid#35101DepositorInsertfca1MX4
UseYeast replicatingAvailable SinceApril 19, 2012AvailabilityAcademic Institutions and Nonprofits only -
pBN63
Plasmid#35104DepositorInsertfcy1PR
UseYeast replicatingAvailable SinceAug. 29, 2012AvailabilityAcademic Institutions and Nonprofits only -
pBN62
Plasmid#35105DepositorInsertfcy1PR
UseYeast replicatingAvailable SinceAug. 29, 2012AvailabilityAcademic Institutions and Nonprofits only -
pPH 34
Plasmid#35098DepositorInsertfca1MX3
UseYeast replicatingAvailable SinceSept. 27, 2012AvailabilityAcademic Institutions and Nonprofits only -
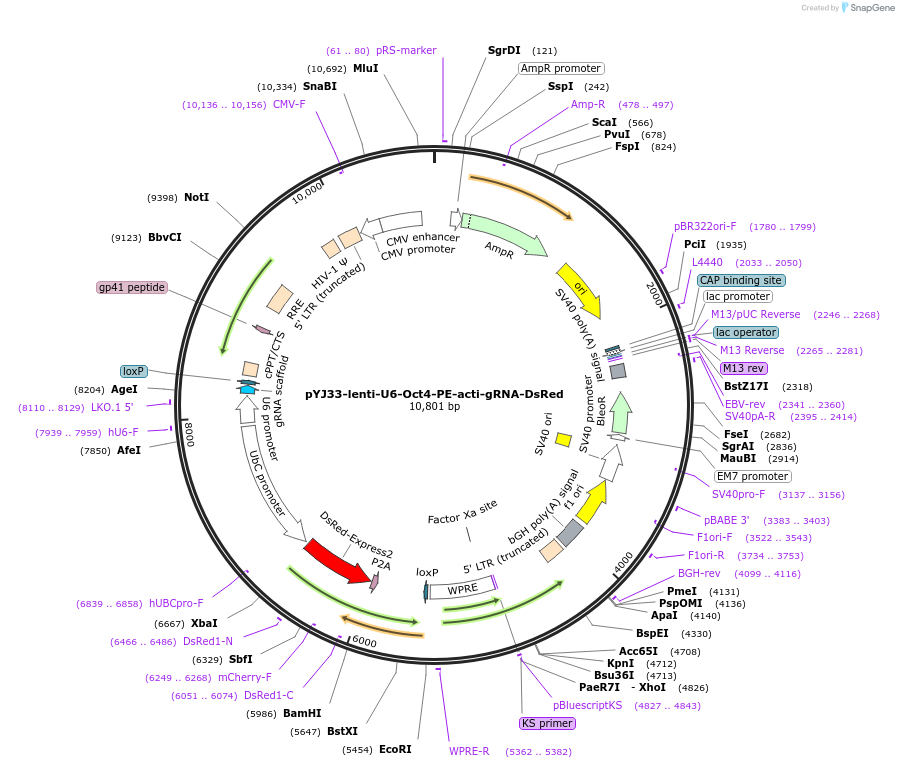
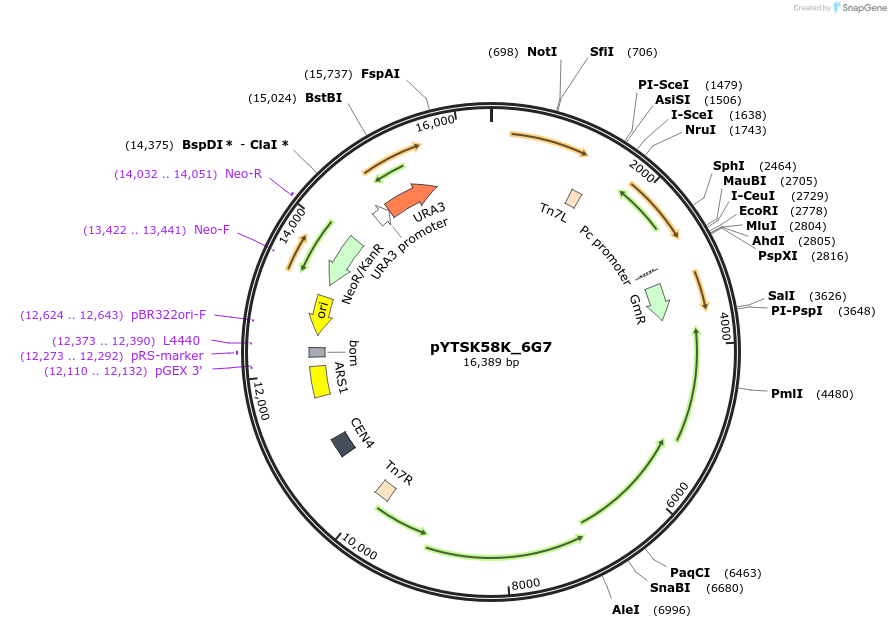
pYTSK58K_6G7
Plasmid#177302Purposegenomic integration of genes (inserted in I-SceI-site) via transposon Tn7 with GmR and PE-HDepositorInsertpe-h
UseSynthetic Biology; Yeast expression, tn7 genomic …ExpressionBacterialAvailable SinceAug. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
pYTRW17K_6G5
Plasmid#177285Purposegenomic integration of genes (inserted in I-SceI-site) via transposon Tn5 with GmR and PE-HDepositorInsertpe-h
UseSynthetic Biology; Yeast expression, tn5 genomic …ExpressionBacterialAvailable SinceAug. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
pRSET Nucleolin
Plasmid#13037DepositorAvailable SinceOct. 5, 2006AvailabilityAcademic Institutions and Nonprofits only -
pPEE7
Plasmid#128357PurposeRecycable marker for use in Cryptococcus neoformans. Contains the amdS1 counterselectable marker flanked by direct repeats.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceOct. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
pPEE8
Plasmid#128358PurposeRecycable marker for use in Cryptococcus neoformans. Contains the amdS2 counterselectable marker flanked by direct repeats.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceOct. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
pWXC078
Plasmid#222961PurposeC. elegans codon-optimized mScarlet-I3 with 3 introns cloned into pPD95.75 backboneDepositorInsertmScarlet-I3
ExpressionWormAvailable SinceJuly 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
pPEE16
Plasmid#128360PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 1, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceOct. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
pWXC077
Plasmid#222960PurposeC. elegans codon-optimized mScarlet3 with 3 introns cloned into pPD95.75 backboneDepositorInsertmScarlet3
ExpressionWormAvailable SinceJuly 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
pPEE13
Plasmid#128359PurposeRecycable marker for use in Cryptococcus neoformans. Contains the amdS2 counterselectable marker flanked by the mCherry ORF and a flexable linker at the 5' end.DepositorInsertAcetamidase (amdS)
UseUnspecifiedPromoterTEF1Available SinceSept. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
pPEE27
Plasmid#128367PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
pPJ-1
Plasmid#38144DepositorInsertmRFP
ExpressionWormPromoterpromoterlessAvailable SinceOct. 25, 2012AvailabilityAcademic Institutions and Nonprofits only -
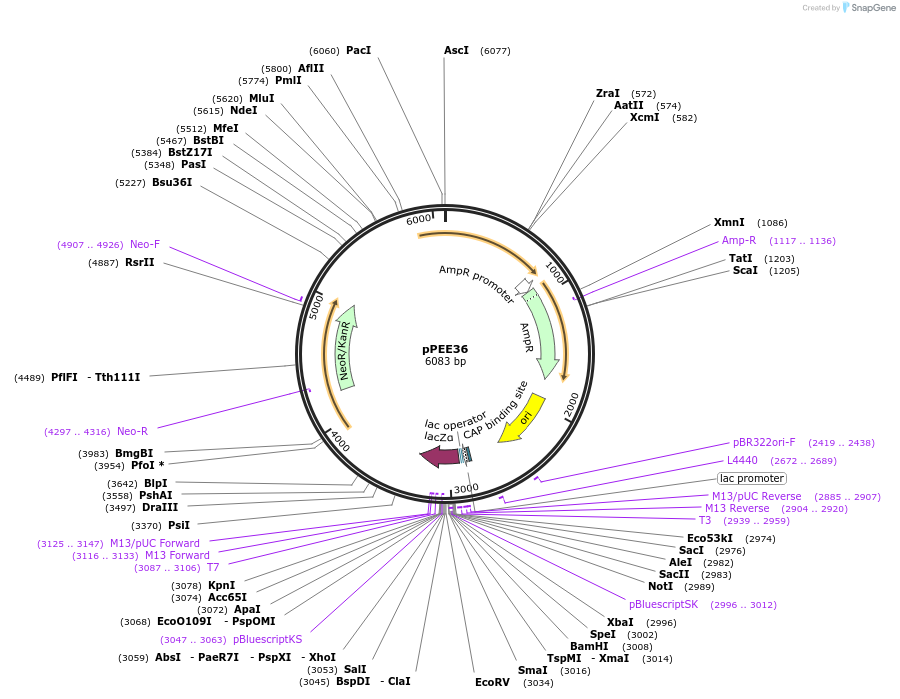
pPEE36
Plasmid#128376PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceApril 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
pPEE38
Plasmid#128378PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
pPEE32
Plasmid#128372PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
pPEE20
Plasmid#128363PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceOct. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
pPEE35
Plasmid#128375PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
pPEE41
Plasmid#128381PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only