We narrowed to 2,963 results for: PHI-1
-
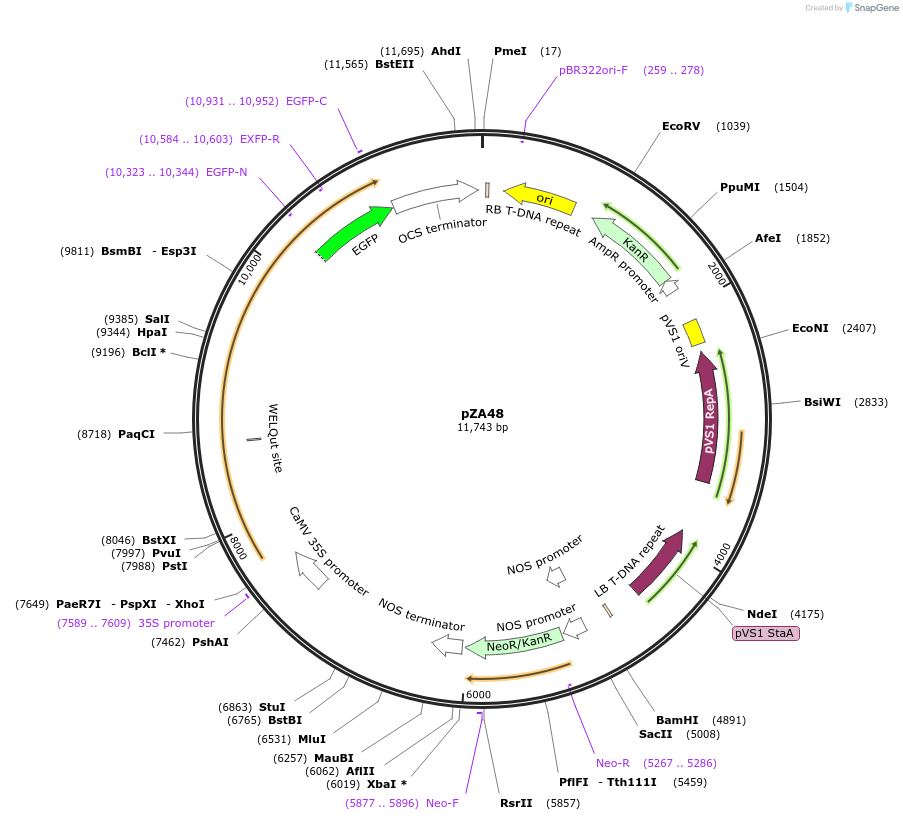
Plasmid#158526PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V/L17E-eGFP. Km plant selection marker.DepositorInsertZAR1-eGFP
TagseGFPExpressionPlantMutationL17E, D481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
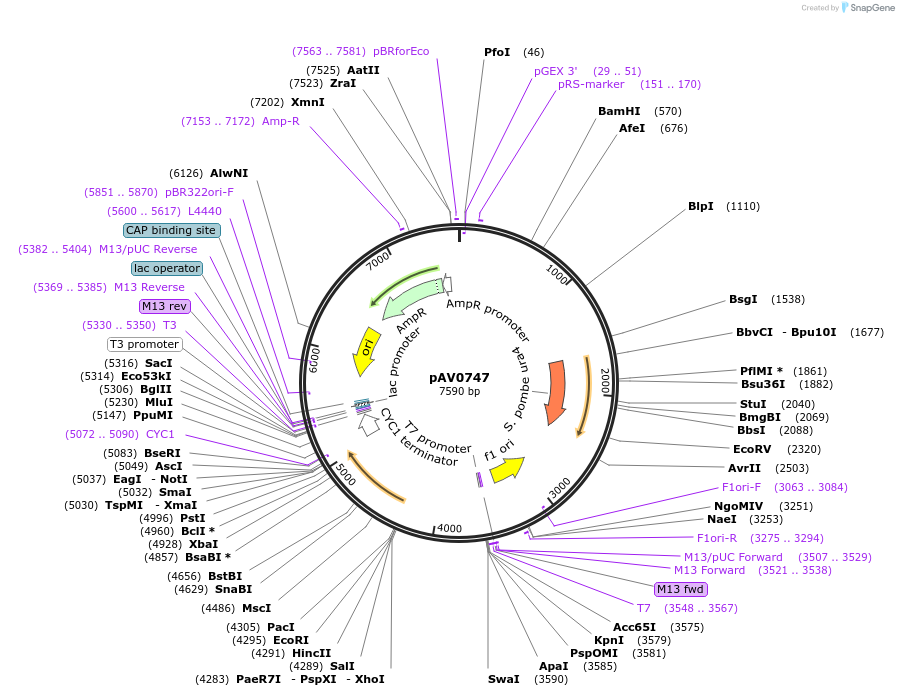
pAV0747
Plasmid#133481PurposeExpression in fission yeast from promoter ppom1 , integrated at ura4 locusDepositorInsertpUra4(AfeI)-p(pom1)-sfGFP-terminator(ScCYC1)
ExpressionYeastAvailable SinceNov. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
pAV0748
Plasmid#133482PurposeExpression in fission yeast from promoter ppak1 , integrated at ura4 locusDepositorInsertpUra4(AfeI)-p(pak1)-sfGFP-terminator(ScCYC1)
ExpressionYeastAvailable SinceNov. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
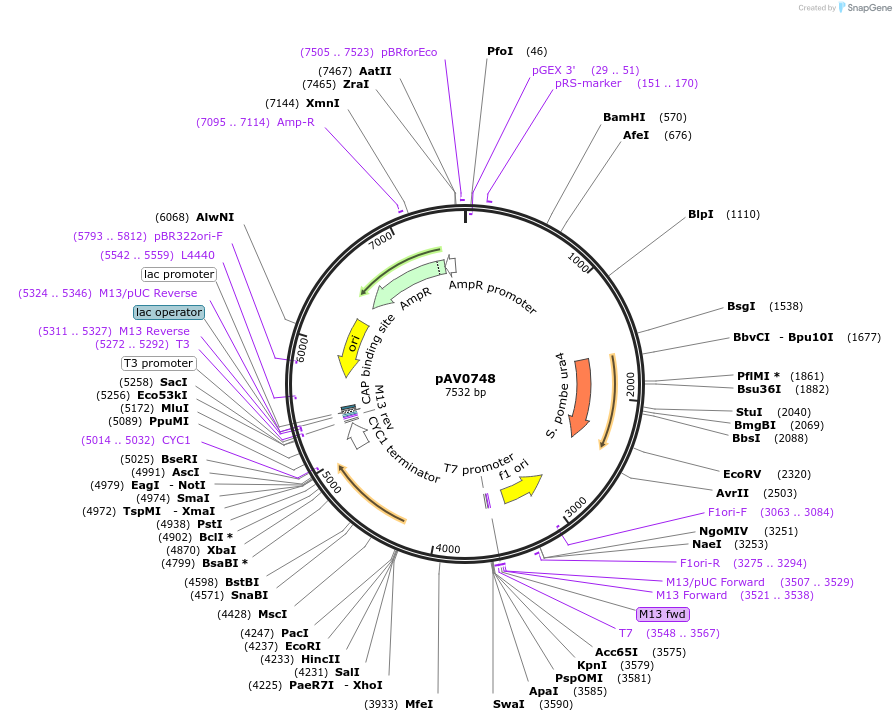
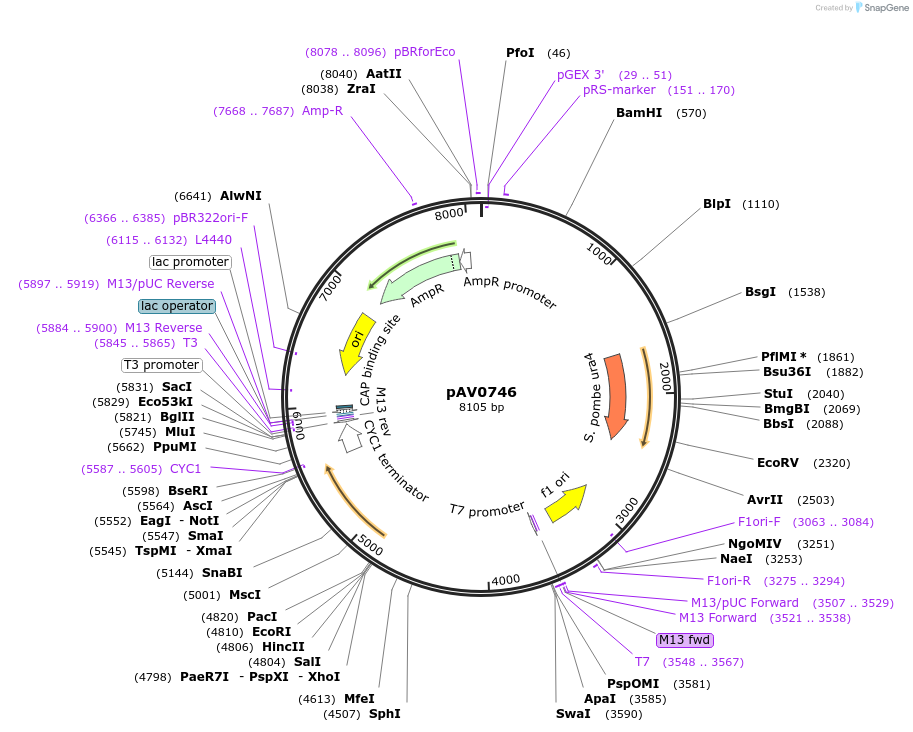
pAV0746
Plasmid#133480PurposeExpression in fission yeast from promoter prga3 , integrated at ura4 locusDepositorInsertpUra4(AfeI)-p(rga3)-sfGFP-terminator(ScCYC1)
ExpressionYeastAvailable SinceNov. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
-
-
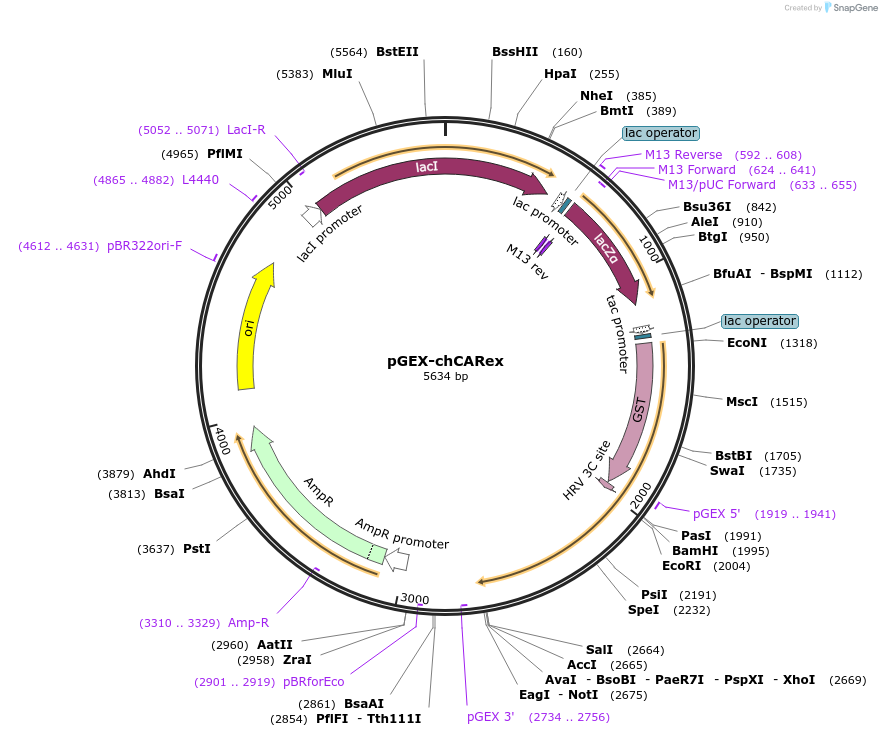
pGEX-chCARex
Plasmid#87233Purposebacterial expression of GST-CAR extracellular domainDepositorAvailable SinceFeb. 28, 2017AvailabilityAcademic Institutions and Nonprofits only -
KHBD00517
Plasmid#39618DepositorAvailable SinceSept. 18, 2012AvailabilityAcademic Institutions and Nonprofits only -
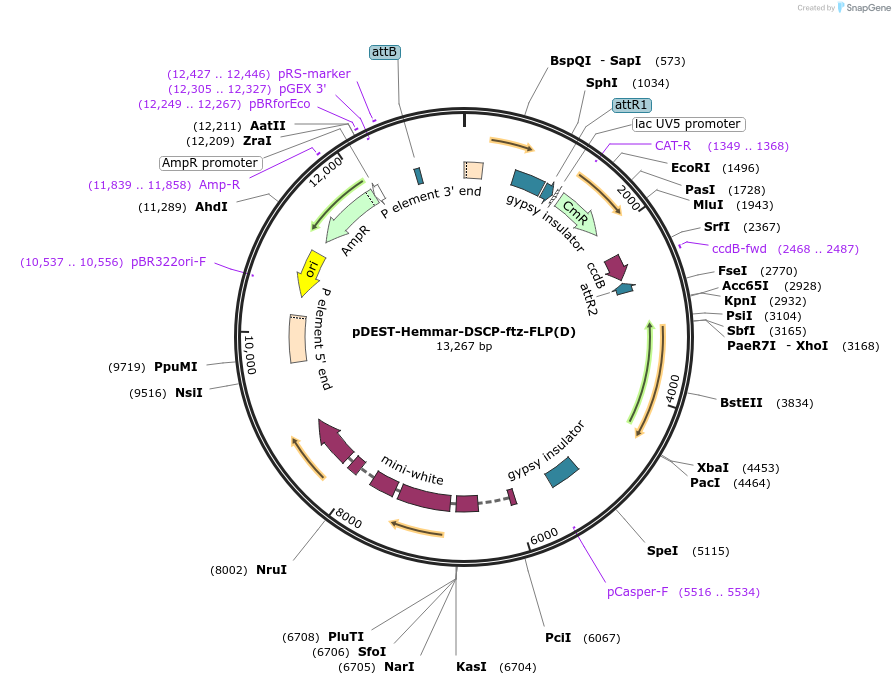
pDEST-Hemmar-DSCP-ftz-FLP(D)
Plasmid#119894PurposeFLP(D) with a Drosophila Synthetic Core Promoter (DSCP) and ftz intronDepositorInsertsUseDrosophila expressionMutationa mutated form of FLP, which at position 5 contai…Available SinceFeb. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
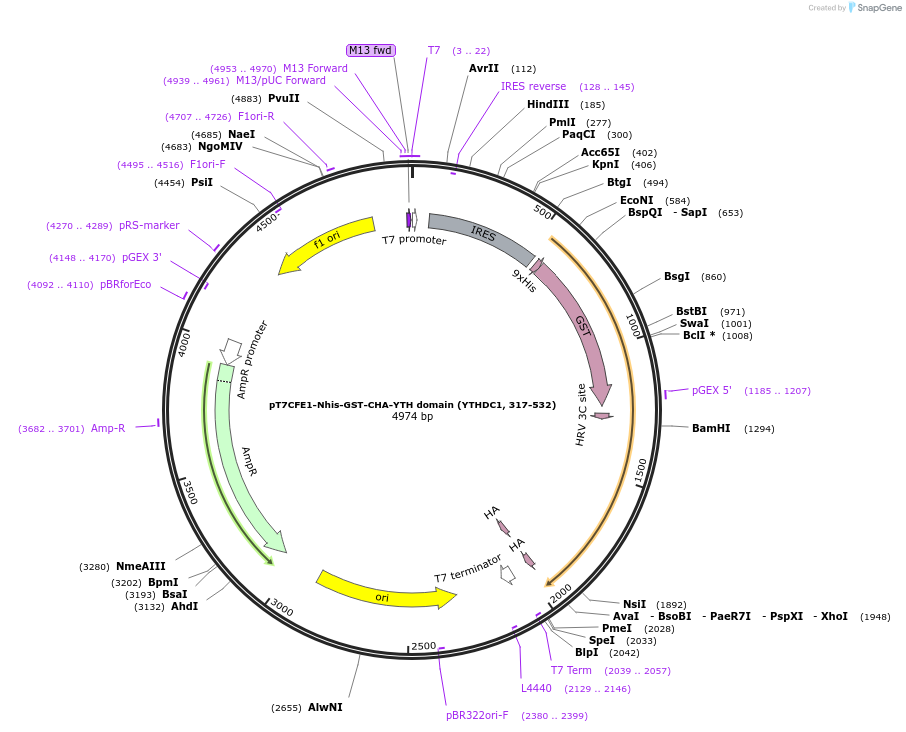
pT7CFE1-Nhis-GST-CHA-YTH domain (YTHDC1, 317-532)
Plasmid#102277PurposeFor expression of His-GST-tagged human YTHDC1 YTH domain using mammalian cell-free protein expression system (1-step human high-yield mini IVT kit, cat. #88890, Thermo Fisher Scientific)DepositorInsertYTH domain of human YTHDC1 (317-532) (YTHDC1 Human)
UseFor mammalian cell-free protein expression system…TagsGST, HA, and HISPromoterT7Available SinceJuly 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
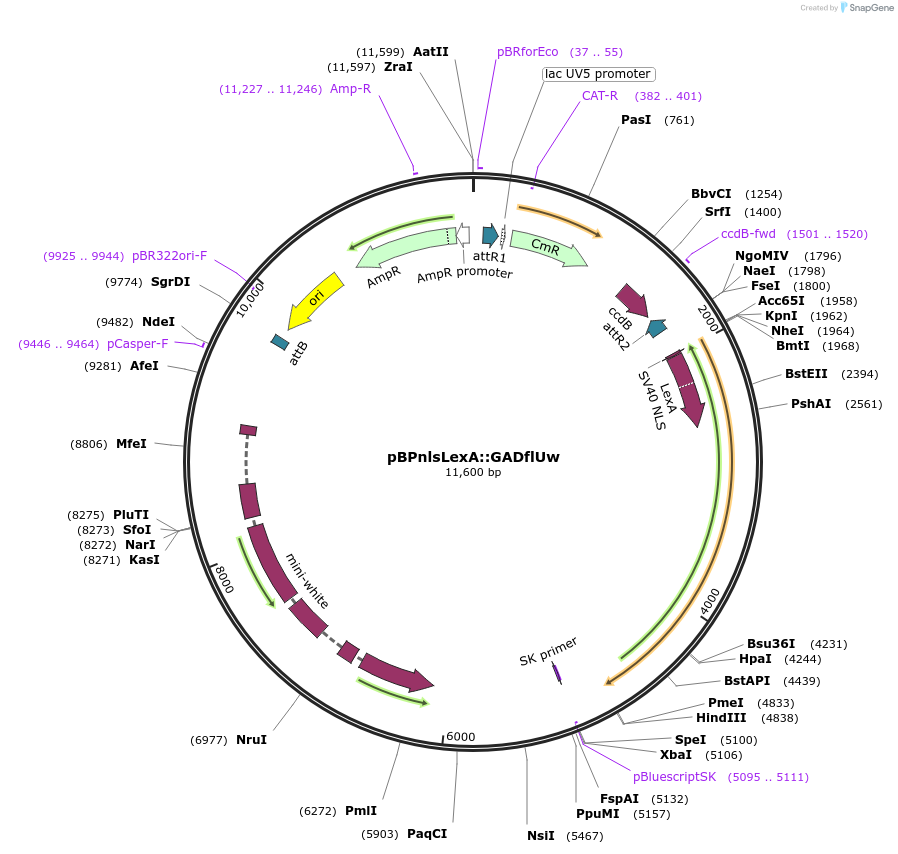
pBPnlsLexA::GADflUw
Plasmid#26232DepositorInsertnlsLexA::GADfl
ExpressionInsectAvailable SinceOct. 7, 2010AvailabilityAcademic Institutions and Nonprofits only -
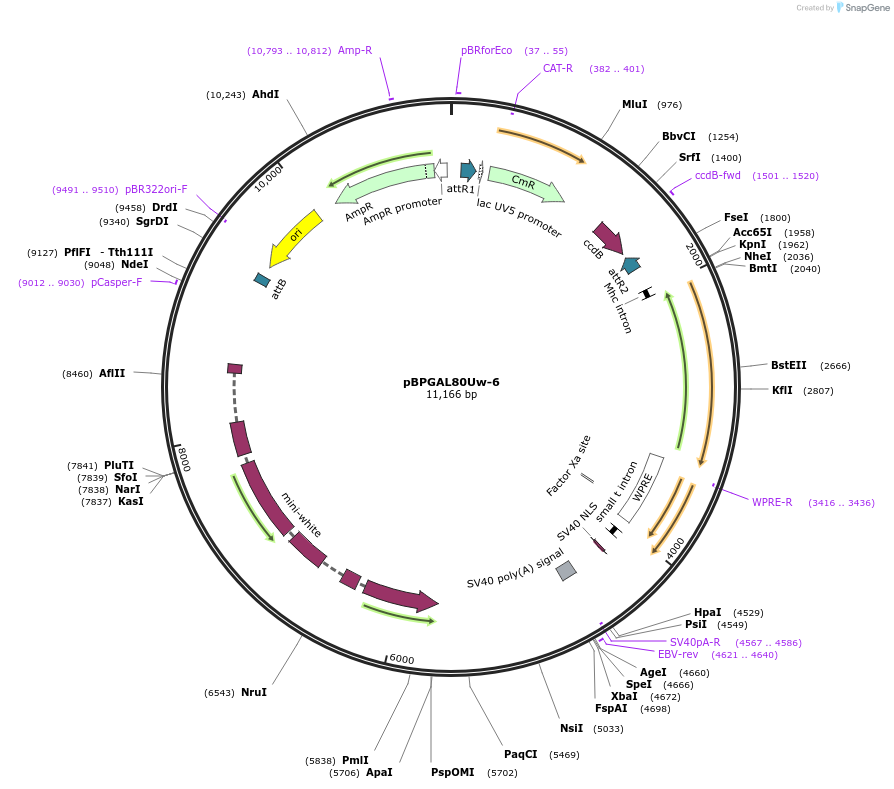
pBPGAL80Uw-6
Plasmid#26236DepositorInsertGAL80 (GAL80 Budding Yeast)
ExpressionInsectAvailable SinceOct. 4, 2010AvailabilityAcademic Institutions and Nonprofits only -
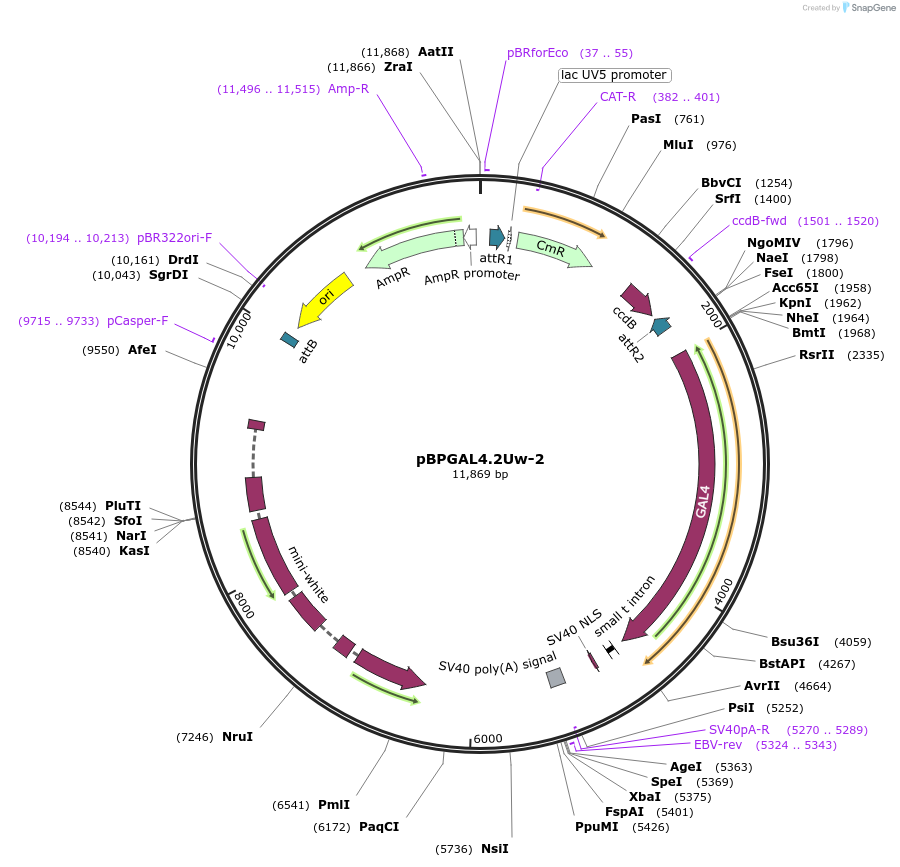
pBPGAL4.2Uw-2
Plasmid#26227DepositorInsertGAL4 (GAL4 Budding Yeast)
ExpressionInsectAvailable SinceOct. 7, 2010AvailabilityAcademic Institutions and Nonprofits only -
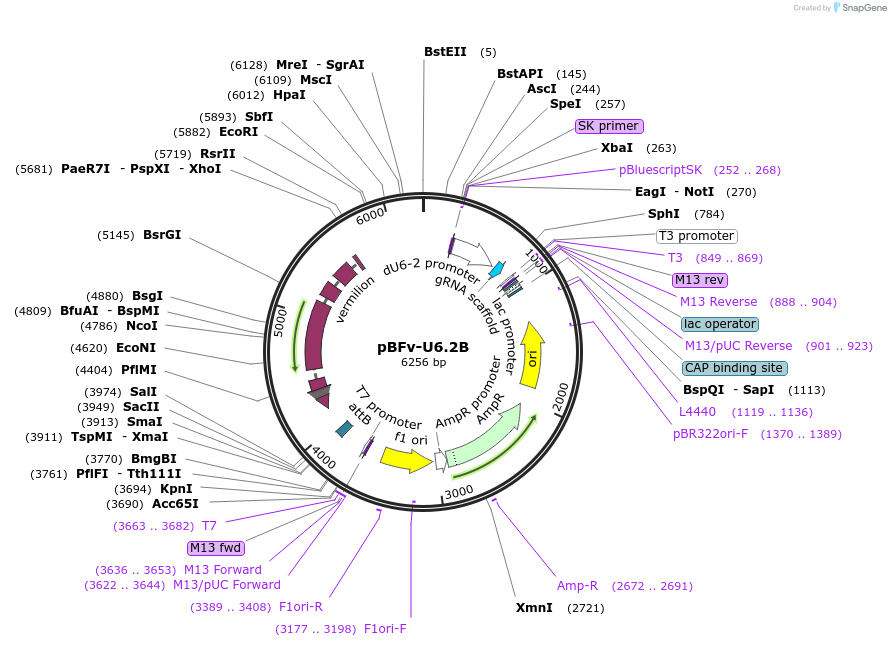
pBFv-U6.2B
Plasmid#138401PurposegRNA expression vector; Can be combined with pBFv-U6.2 to generate double-gRNA expression vectorDepositorInsertU6-2
ExpressionInsectPromoterU6-2Available SinceMarch 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
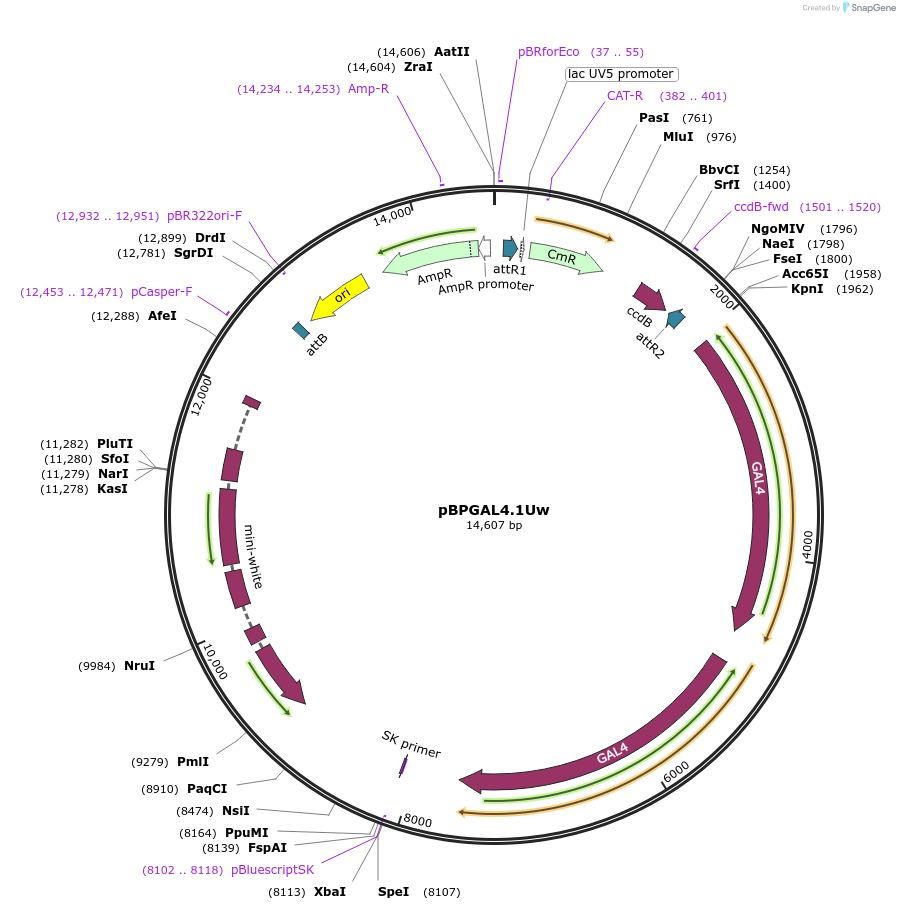
pBPGAL4.1Uw
Plasmid#26226DepositorInsertGAL4 (GAL4 Budding Yeast)
ExpressionInsectAvailable SinceOct. 7, 2010AvailabilityAcademic Institutions and Nonprofits only -
pJFRC14-10XUAS-IVS-GFP-WPRE
Plasmid#26223DepositorInsertGFP
ExpressionInsectAvailable SinceOct. 19, 2010AvailabilityAcademic Institutions and Nonprofits only -
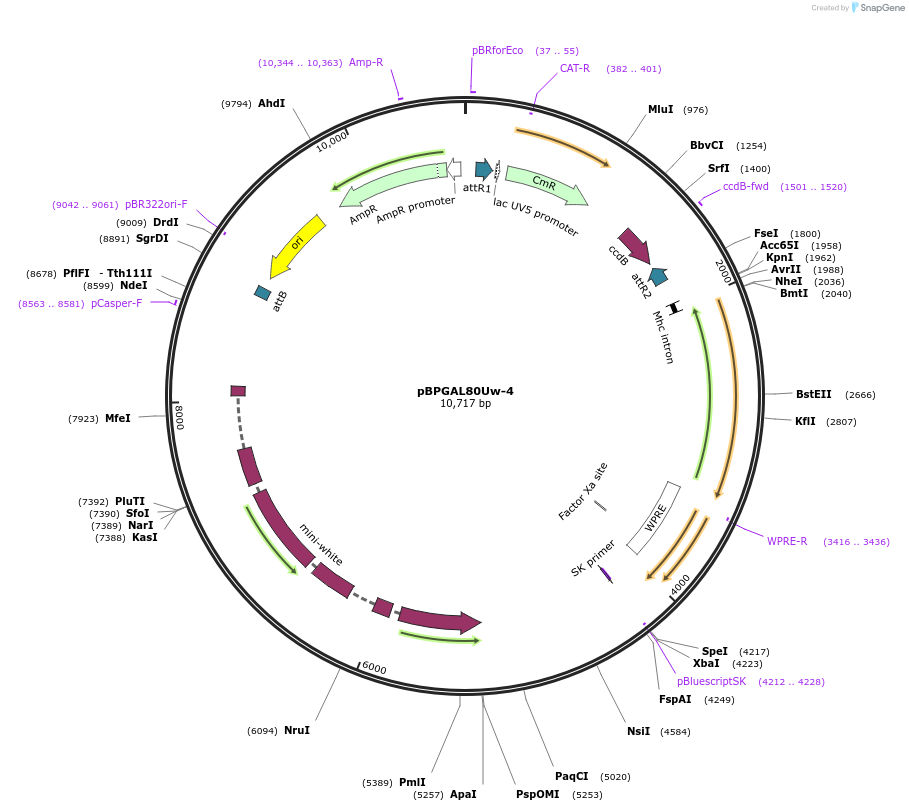
pBPGAL80Uw-4
Plasmid#26235DepositorInsertGAL80 (GAL80 Budding Yeast)
ExpressionInsectAvailable SinceOct. 22, 2010AvailabilityAcademic Institutions and Nonprofits only -
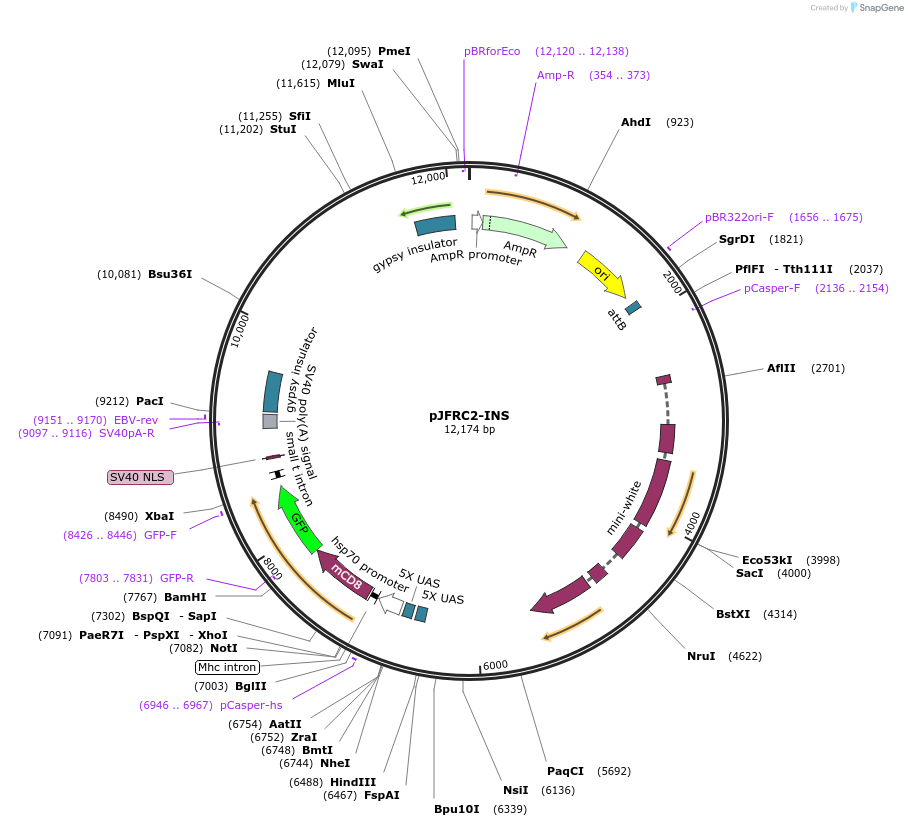
pJFRC2-INS
Plasmid#26215DepositorInsertmCD8::GFP
ExpressionInsectAvailable SinceOct. 19, 2010AvailabilityAcademic Institutions and Nonprofits only -
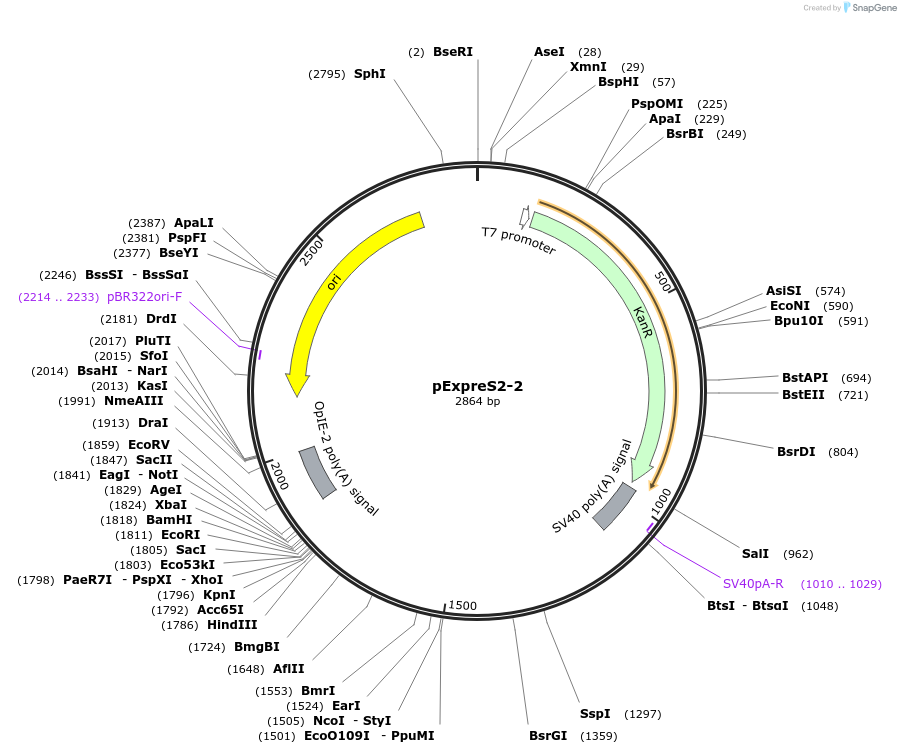
pExpreS2-2
Plasmid#175362PurposeG418R backbone for recombinant protein overexpression with Drosophila ExpreS2 PlatformDepositorTypeEmpty backboneExpressionInsectPromoterFused Actin-HSP70Available SinceMarch 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
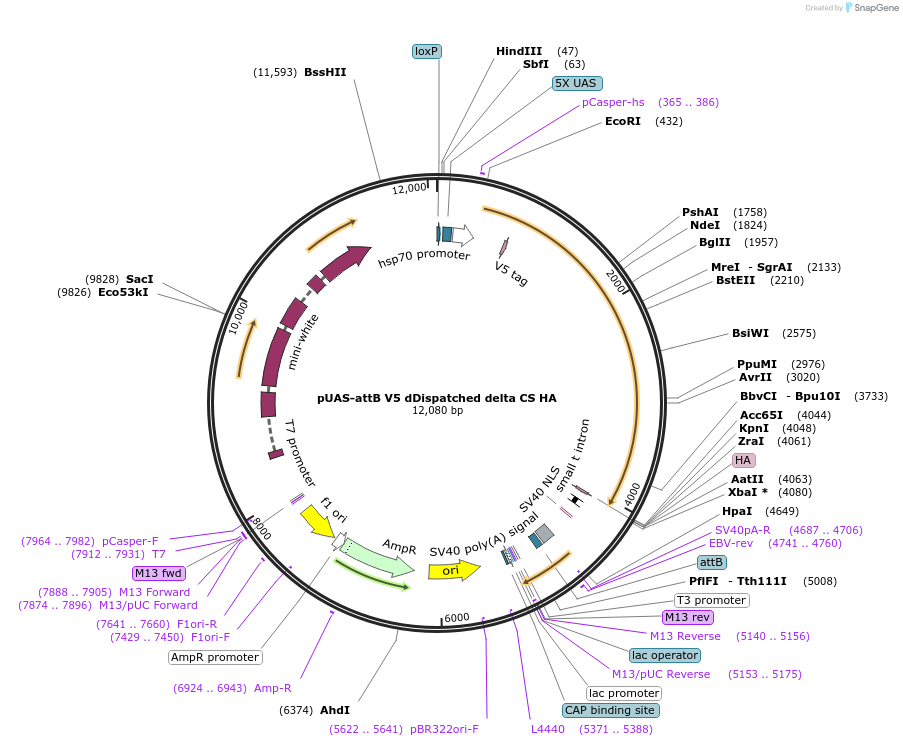
pUAS-attB V5 dDispatched delta CS HA
Plasmid#126412PurposeExpression of V5 dDispatched Delta CS HA in Drosophila cells linesDepositorAvailable SinceJune 14, 2019AvailabilityAcademic Institutions and Nonprofits only