We narrowed to 44,705 results for: cha;
-
Plasmid#226131PurposeExpression of Ufd1R64C-3C-His6DepositorInsertUfd1R64C-3C-His6
ExpressionBacterialMutationR64CAvailable SinceSept. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
pAG416-GAL-TDP-43-M337V
Plasmid#181721PurposeGalactose inducible expression of TDP-43-M337VDepositorAvailable SinceApril 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
pAG416-GAL-TDP-43-N267S
Plasmid#181720PurposeGalactose inducible expression of TDP-43-N267SDepositorAvailable SinceApril 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
pAG416-GAL-TDP-43-Q331K
Plasmid#181719PurposeGalactose inducible expression of TDP-43-Q331KDepositorAvailable SinceApril 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
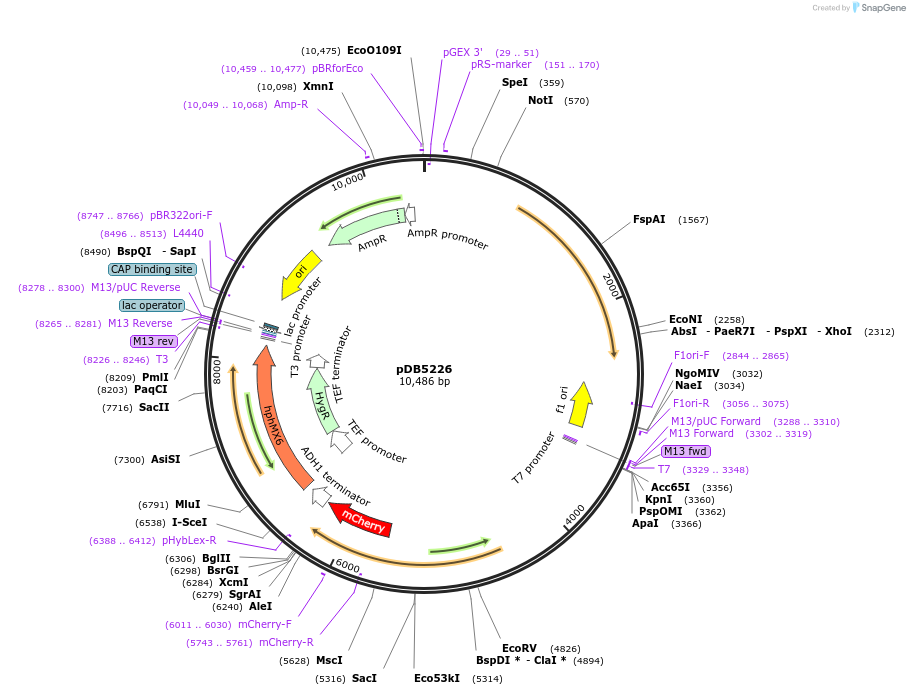
pDB5226
Plasmid#215493PurposeA pAde6 backbone integrating plasmid expressing Pil1-mCherry under the inducible promoter 41nmt1DepositorInsertPil1-mCherry
ExpressionYeastPromoterP41nmt1Available SinceMarch 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
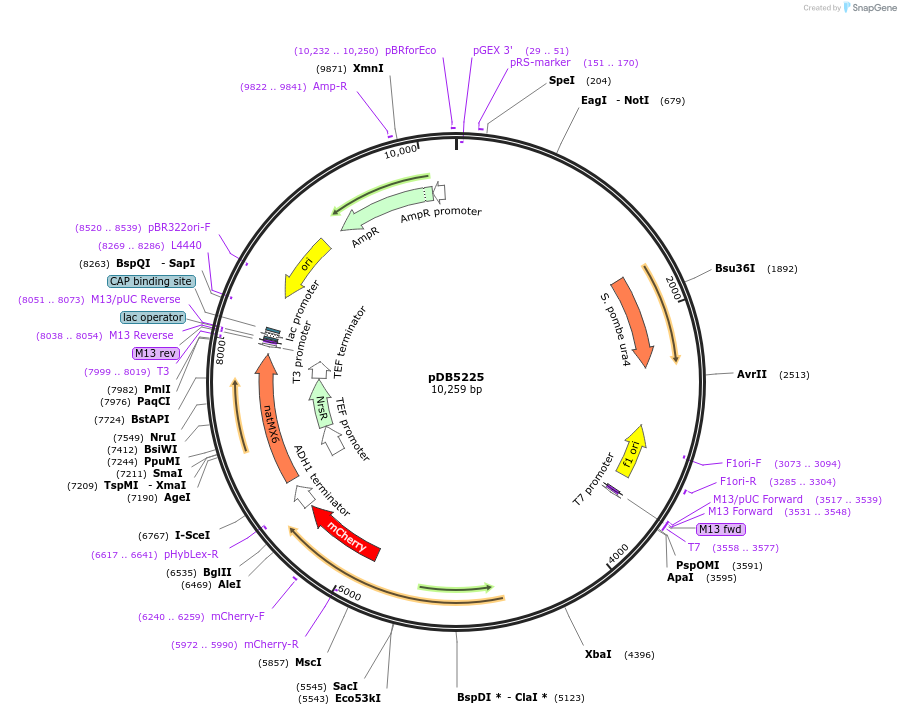
pDB5225
Plasmid#215492PurposeA pUra4 backbone integrating plasmid expressing Pil1-mCherry under the inducible promoter 41nmt1DepositorInsertPil1-mCherry
ExpressionYeastPromoterP41nmt1Available SinceMarch 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
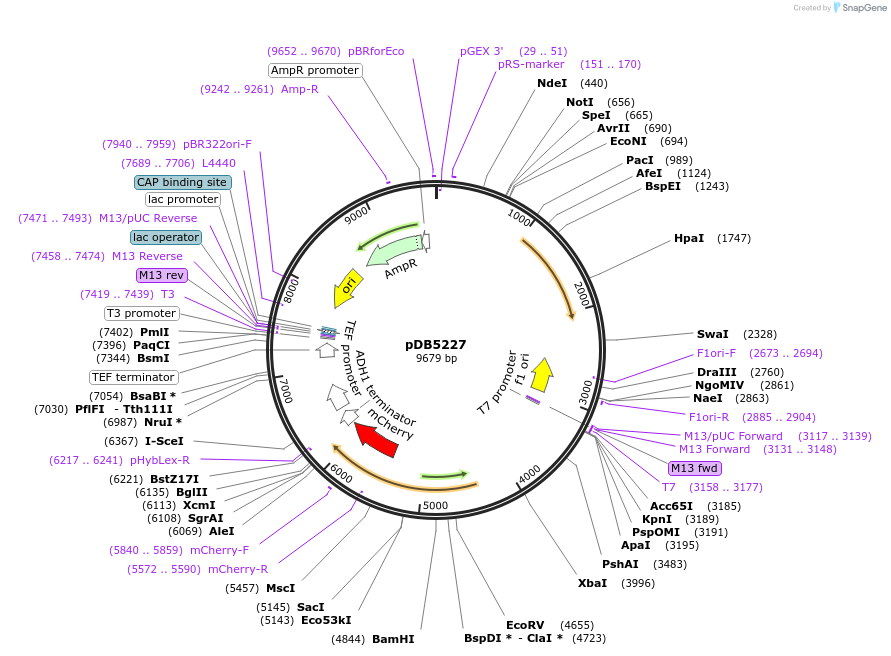
pDB5227
Plasmid#215494PurposeA pLys3 backbone integrating plasmid expressing Pil1-mCherry under the inducible promoter 41nmt1DepositorInsertPil1-mCherry
ExpressionYeastPromoterP41nmt1Available SinceMarch 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
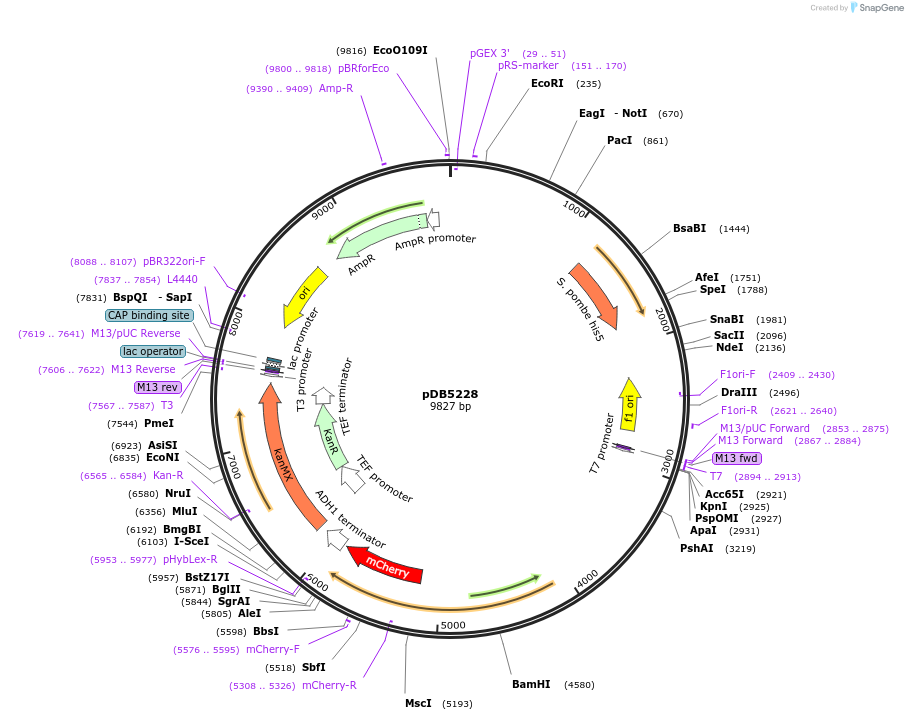
pDB5228
Plasmid#215495PurposeA pHis5 backbone integrating plasmid expressing Pil1-mCherry under the inducible promoter 41nmt1DepositorInsertPil1-mCherry
ExpressionYeastPromoterP41nmt1Available SinceMarch 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
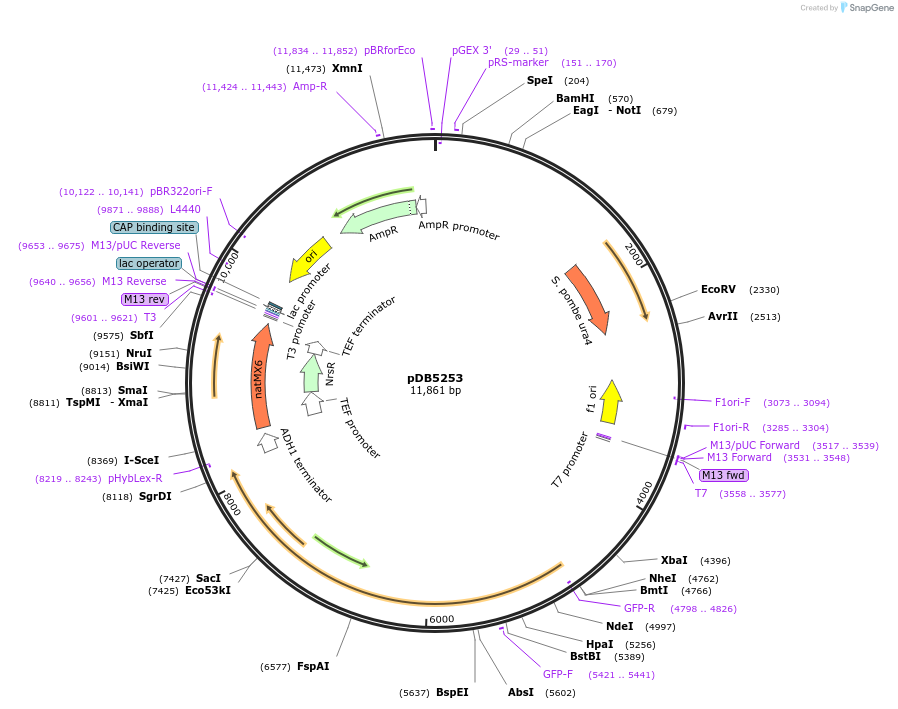
pDB5253
Plasmid#215501PurposeA pUra4 backbone integrating plasmid expressing mECitrine-CtNbr1 under the inducible promoter 41nmt1DepositorInsertmECitrine-CtNbr1
ExpressionYeastPromoterP41nmt1Available SinceMarch 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
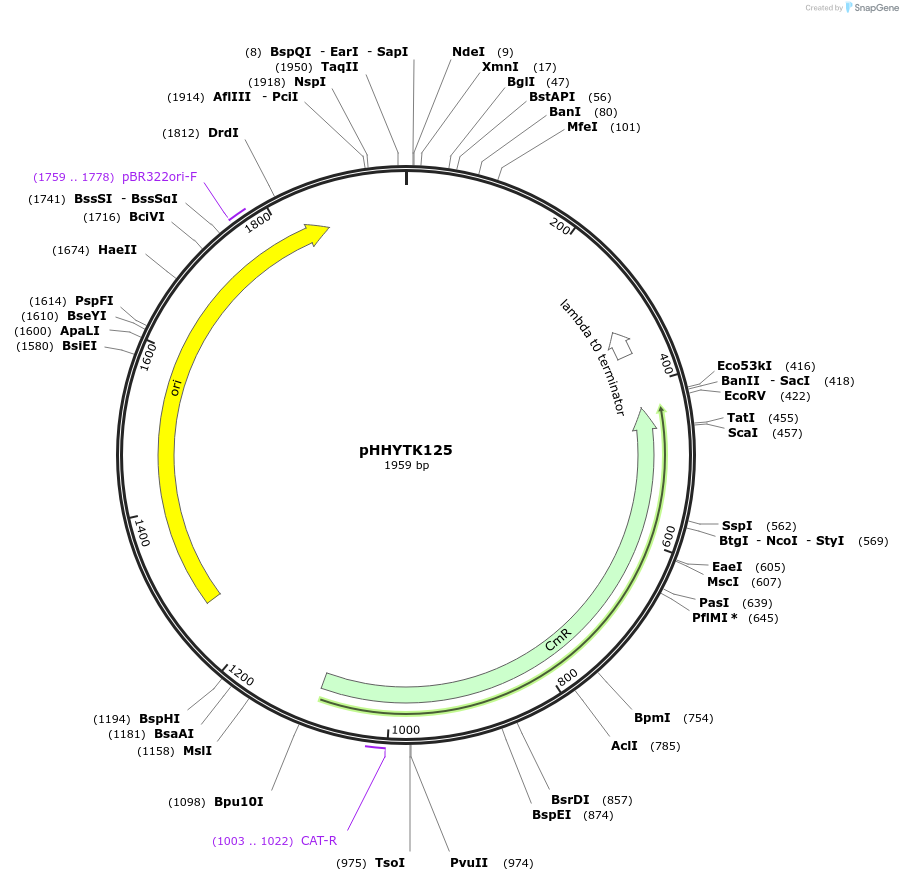
pHHYTK125
Plasmid#200137PurposeType 3a part for Modular Cloning in YeastDepositorInsertOM-Tom70 (2-98)
UseSynthetic BiologyExpressionBacterialAvailable SinceMay 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
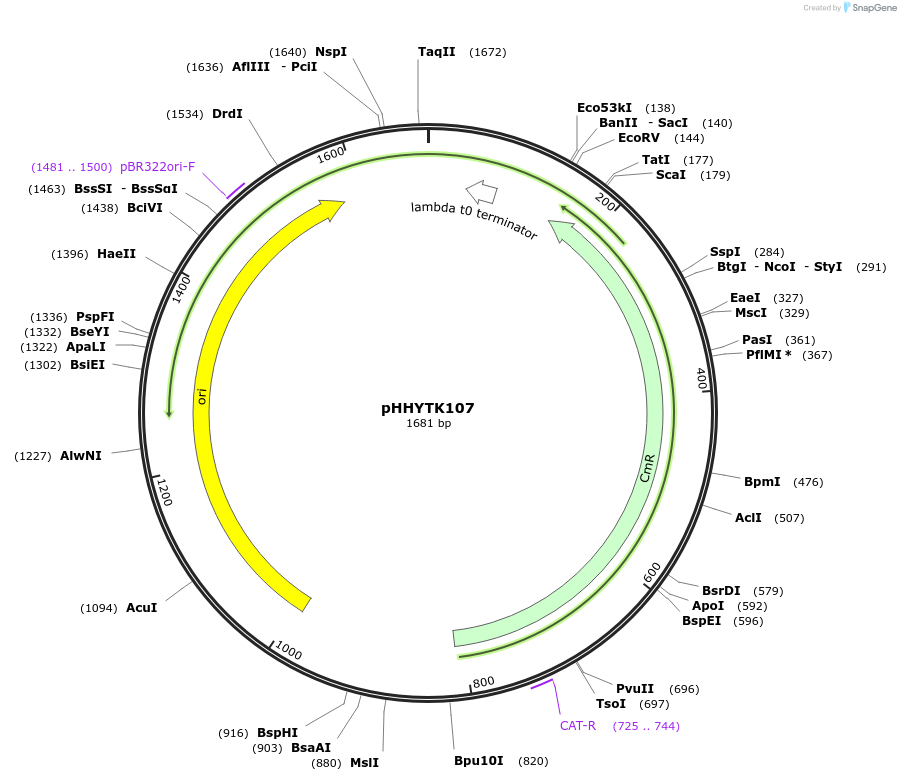
pHHYTK107
Plasmid#200133PurposeType 4a part for Modular Cloning in YeastDepositorInsertKar2-HDEL
UseSynthetic BiologyExpressionBacterialAvailable SinceMay 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
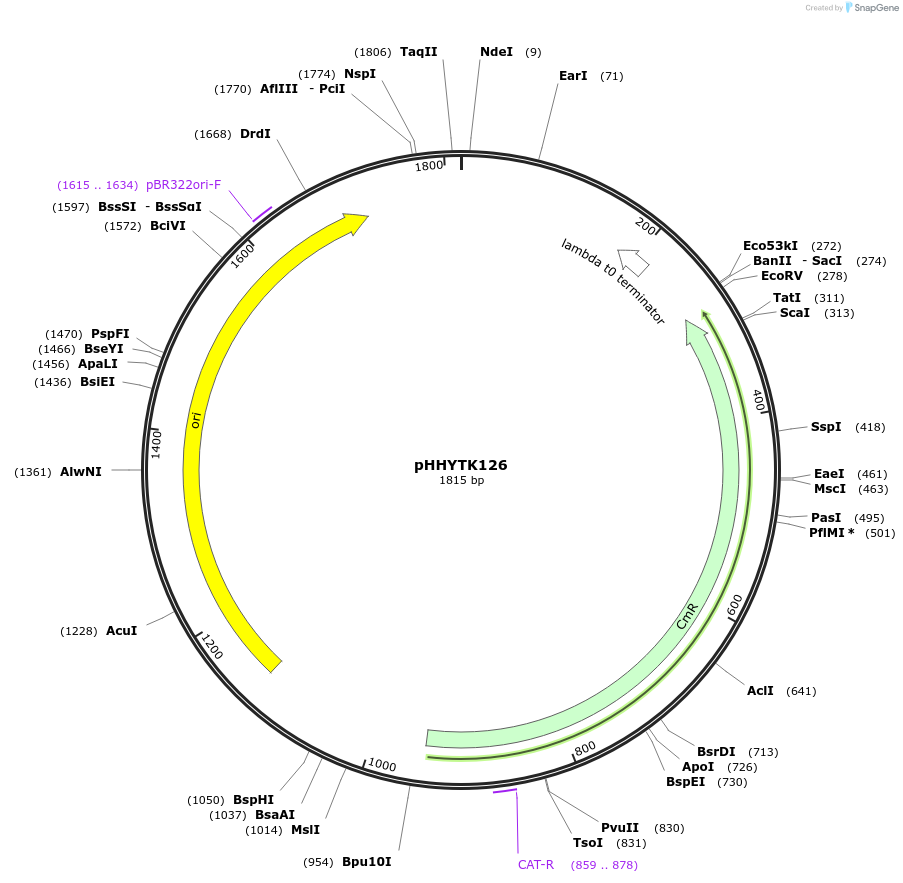
pHHYTK126
Plasmid#200138PurposeType 3a part for Modular Cloning in YeastDepositorInsertPot1-PTS2 (2-50)
UseSynthetic BiologyExpressionBacterialAvailable SinceMay 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
pHHYTK106
Plasmid#200132PurposeType 3a part for Modular Cloning in YeastDepositorInsertKar2-SS
UseSynthetic BiologyExpressionBacterialAvailable SinceMay 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
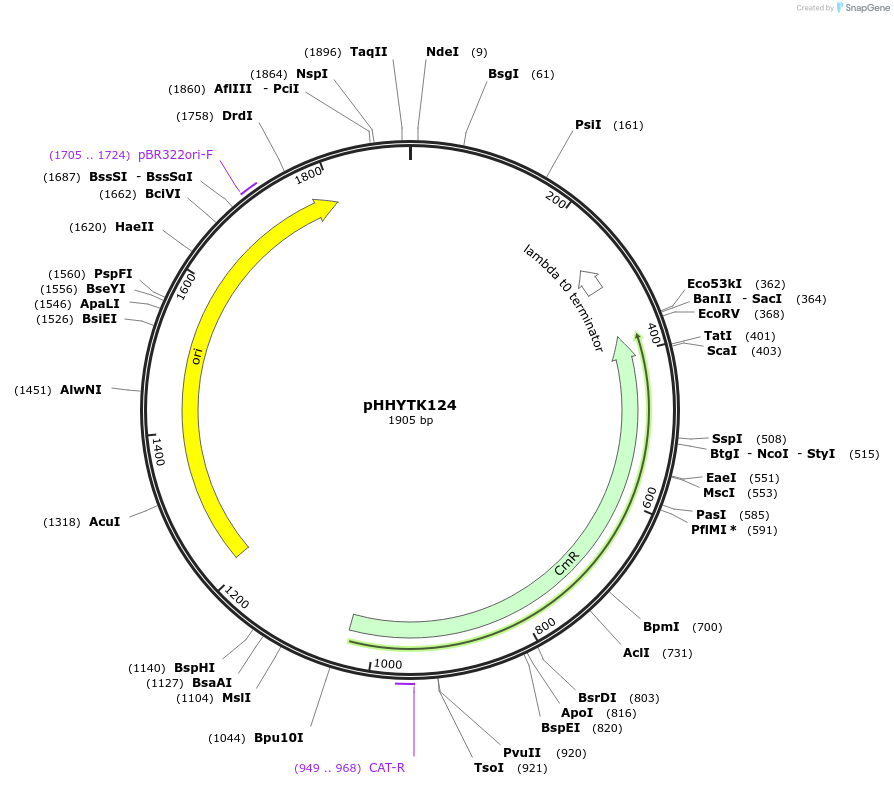
pHHYTK124
Plasmid#200136PurposeType 3a part for Modular Cloning in YeastDepositorInsertIMS-Mia40 (2-70)
UseSynthetic BiologyExpressionBacterialAvailable SinceMay 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
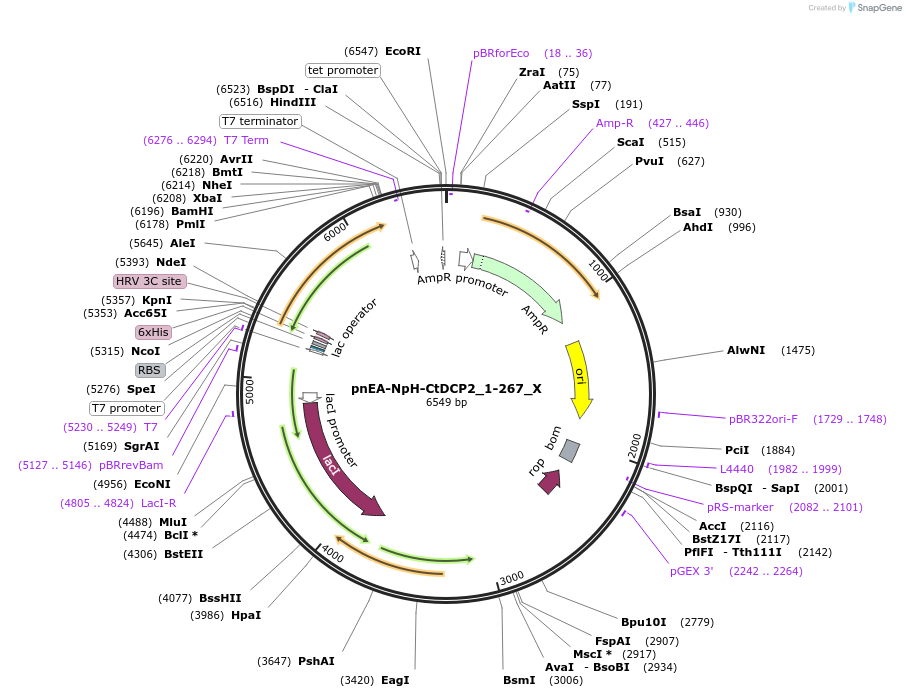
pnEA-NpH-CtDCP2_1-267_X
Plasmid#148004PurposeBacterial Expression of CtDCP2_1-267DepositorInsertCtDCP2_1-267
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
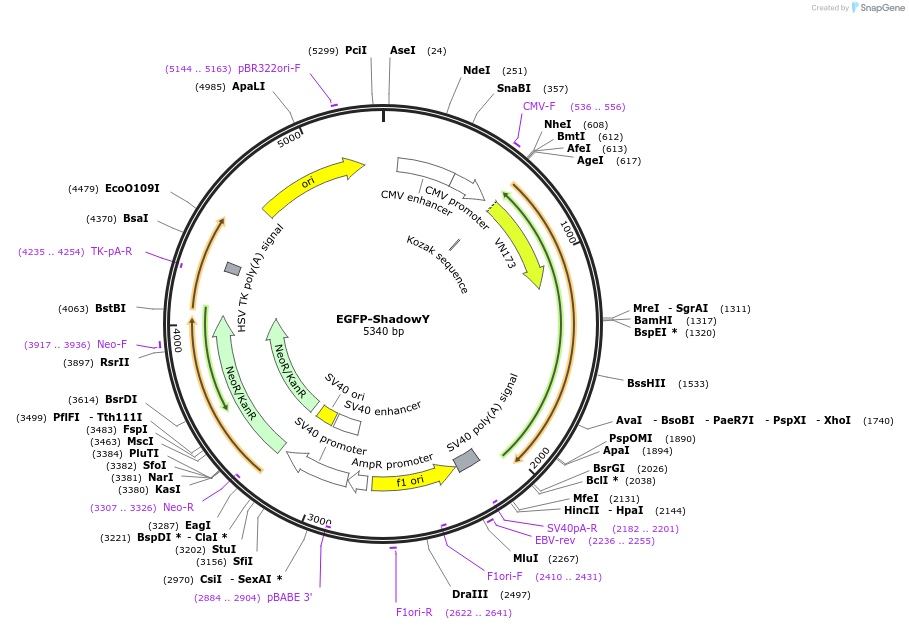
EGFP-ShadowY
Plasmid#193555PurposeExpresses mEGFP-ShadowY in mammalian cells.DepositorInsertShadowY
TagsmEGFPExpressionMammalianPromoterCMVAvailable SinceFeb. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
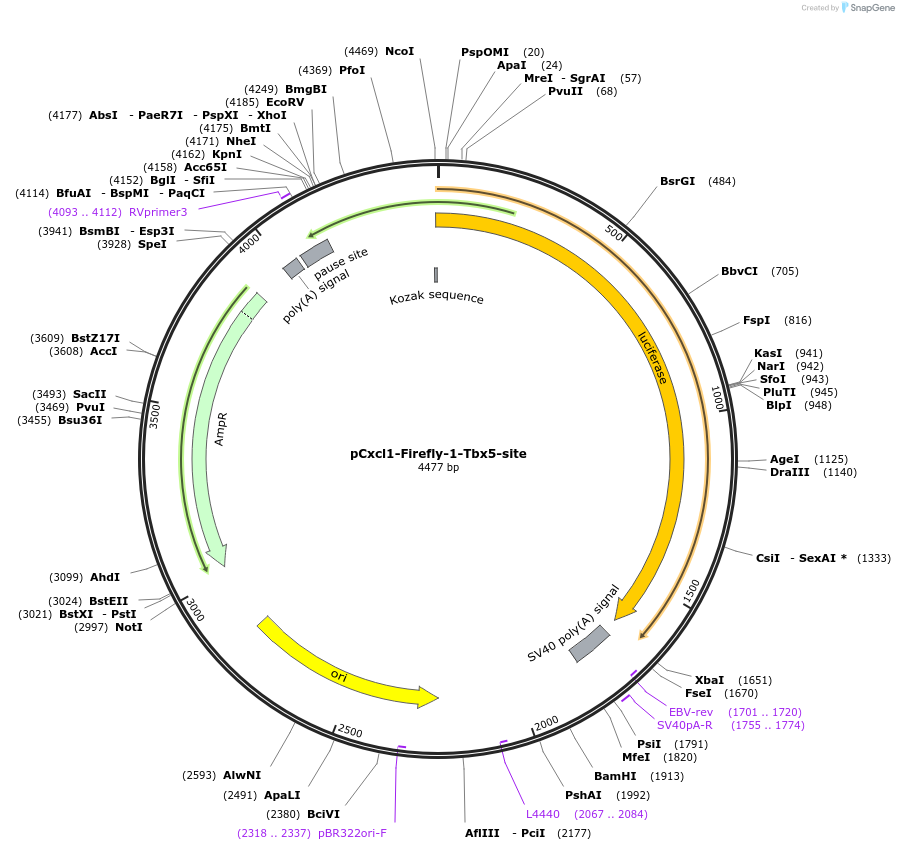
pCxcl1-Firefly-1-Tbx5-site
Plasmid#177832PurposeLuciferase reporter for mouse Cxcl1 with one Tbx5 consensus binding siteDepositorInsertLuciferase
UseLuciferasePromoterCxcl1Available SinceAug. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
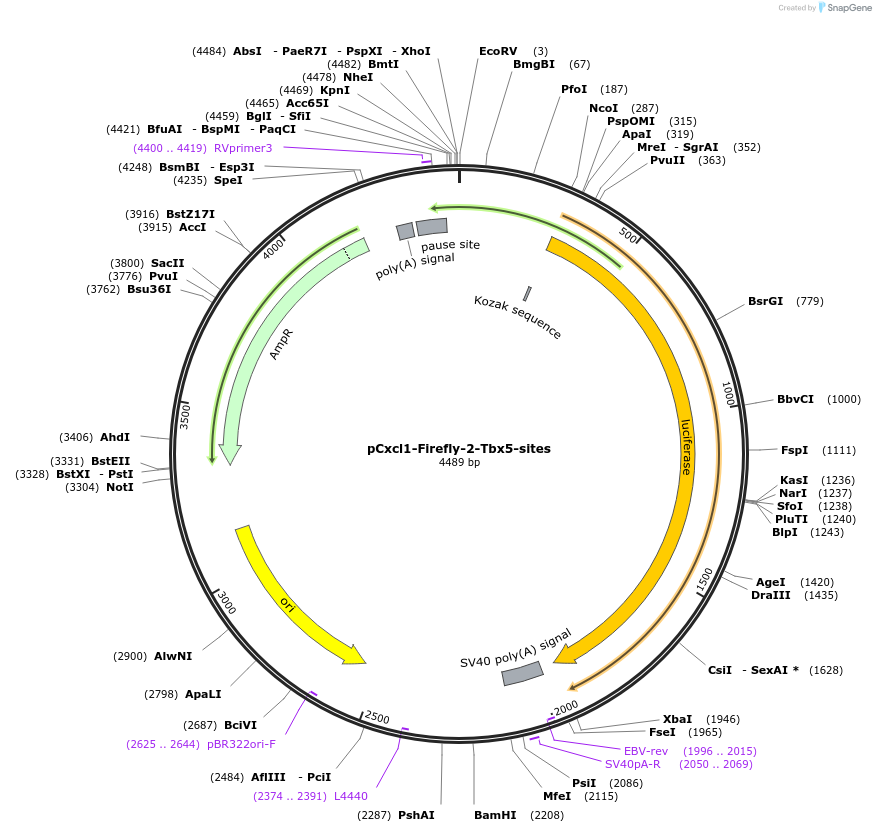
pCxcl1-Firefly-2-Tbx5-sites
Plasmid#177833PurposeLuciferase reporter for mouse Cxcl1 with two Tbx5 consensus binding sitesDepositorInsertLuciferase
UseLuciferasePromoterCxcl1Available SinceAug. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
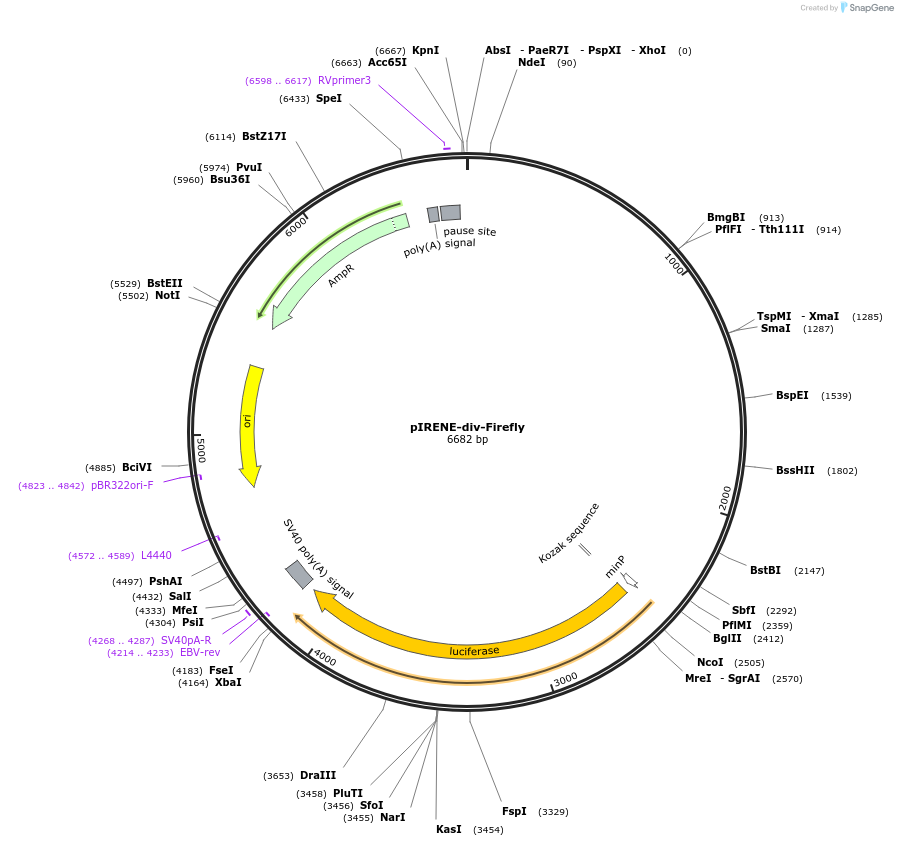
pIRENE-div-Firefly
Plasmid#170661PurposeLuciferase reporter for mouse IRENE-div lncRNADepositorInsertIRENE-div
UseLuciferasePromoterIRENE-divAvailable SinceJan. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
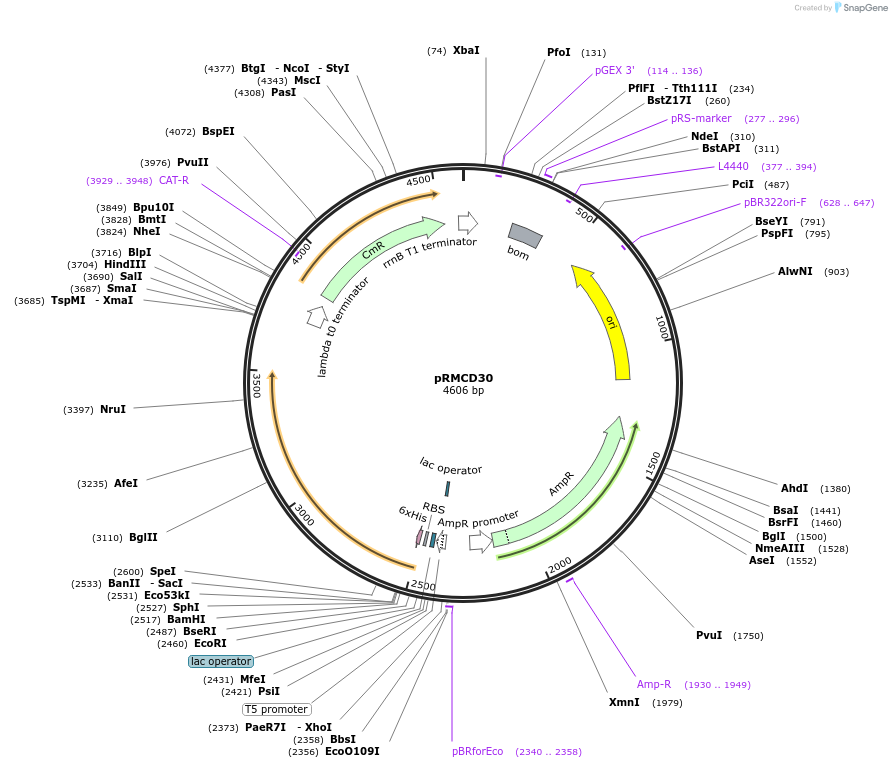
pRMCD30
Plasmid#173033PurposepQE30 with S. flexneri 2a wzzBSF geneDepositorInsertS. flexneri 2a wzzBSF gene
Tags6xHisExpressionBacterialAvailable SinceOct. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
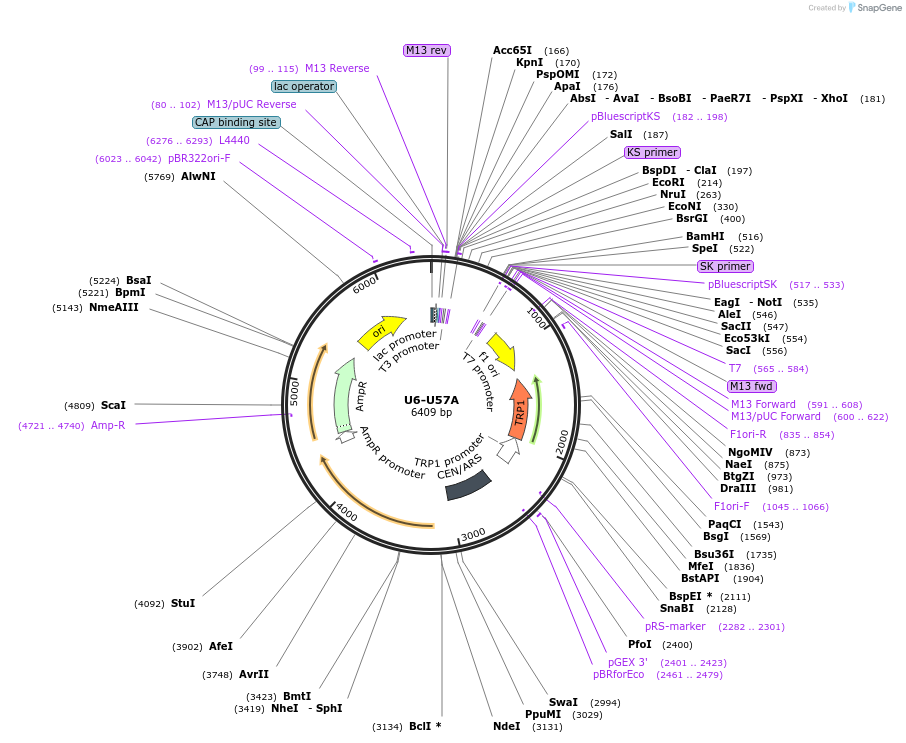
U6-U57A
Plasmid#169927PurposeWT U6 with a U57A mutationDepositorInsertU6 U57A
ExpressionBacterial and YeastAvailable SinceAug. 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
pVG50
Plasmid#164708PurposeyEVenus under the control of tetOpr with ADH1t upstream of the reporterDepositorInsertADH1t - tetOpr - yEVenus - CLN2 PEST - ADH1t
ExpressionYeastAvailable SinceMarch 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
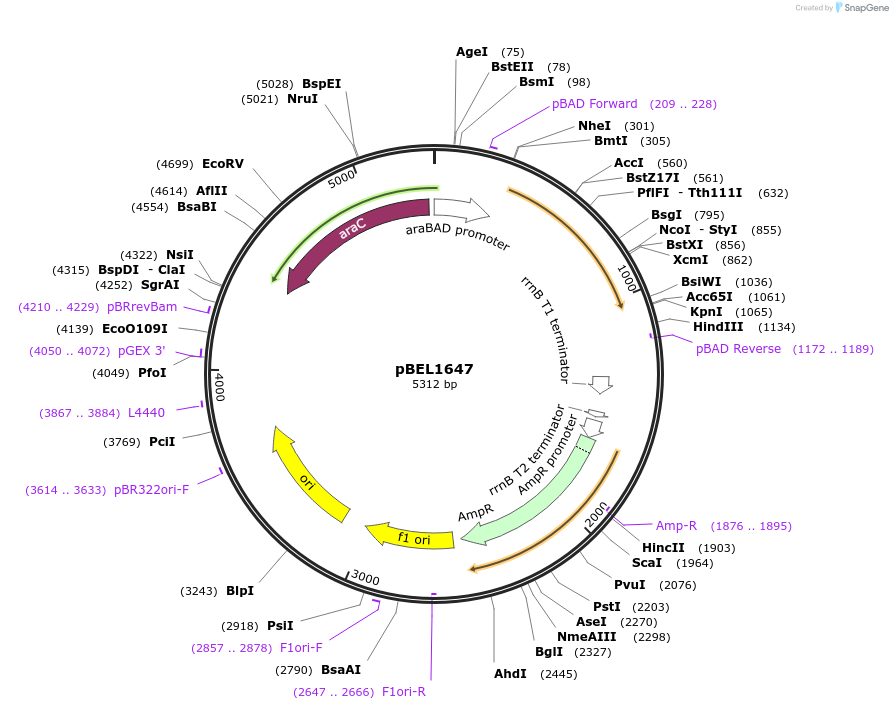
pBEL1647
Plasmid#155148PurposeExpresses MlaF 1-246, taglessDepositorInsertmlaF(1-246)
PromoteraraBADAvailable SinceDec. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
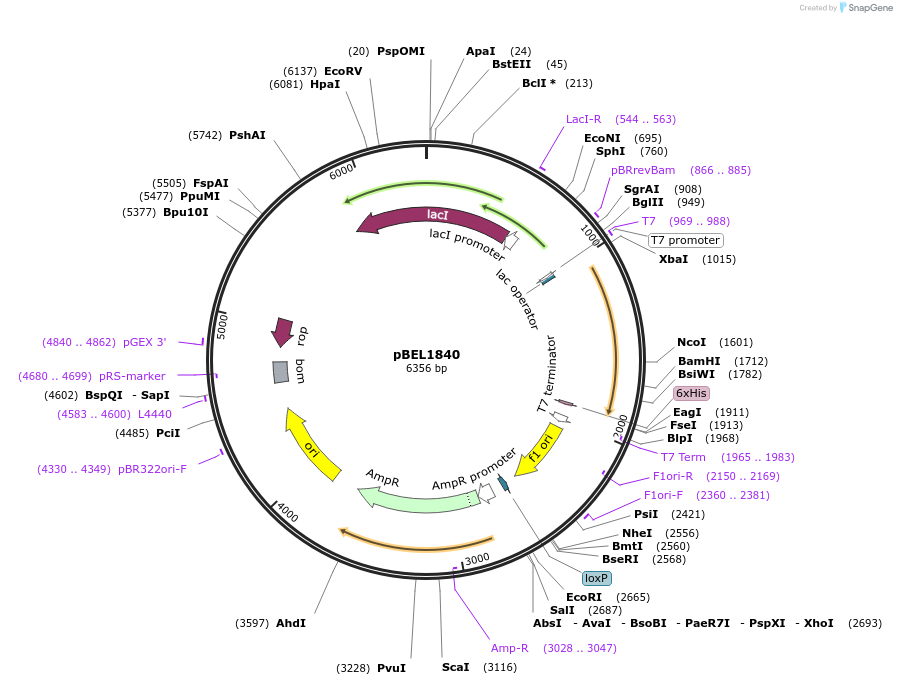
pBEL1840
Plasmid#155150PurposeExpresses MlaF-6xHisDepositorInsertMlaF-6xHis
PromoterT7 promoterAvailable SinceDec. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
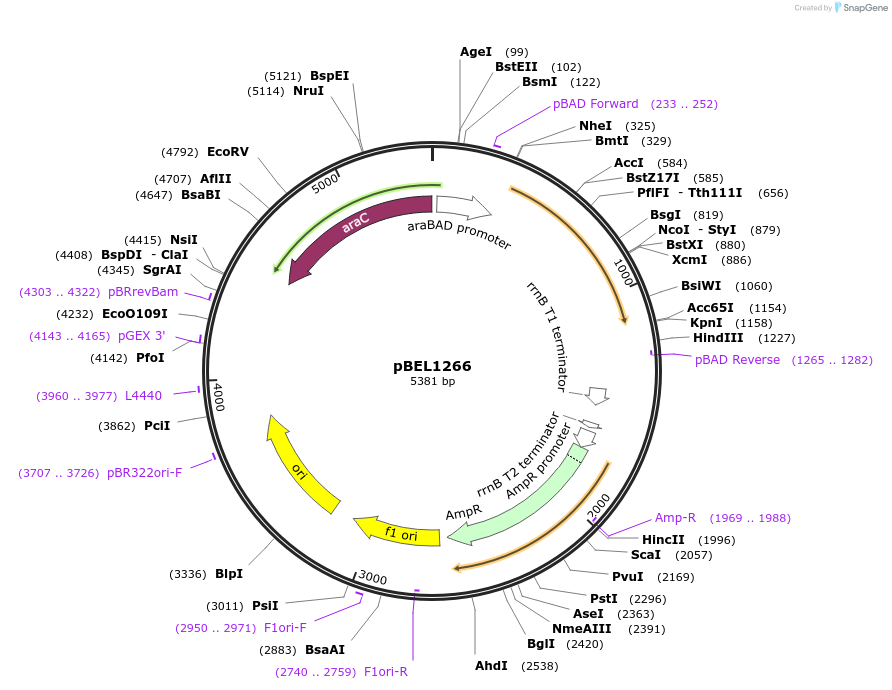
pBEL1266
Plasmid#155142PurposeExpresses MlaF, tagless, araBAD promoterDepositorInsertMlaF
PromoteraraBADAvailable SinceDec. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
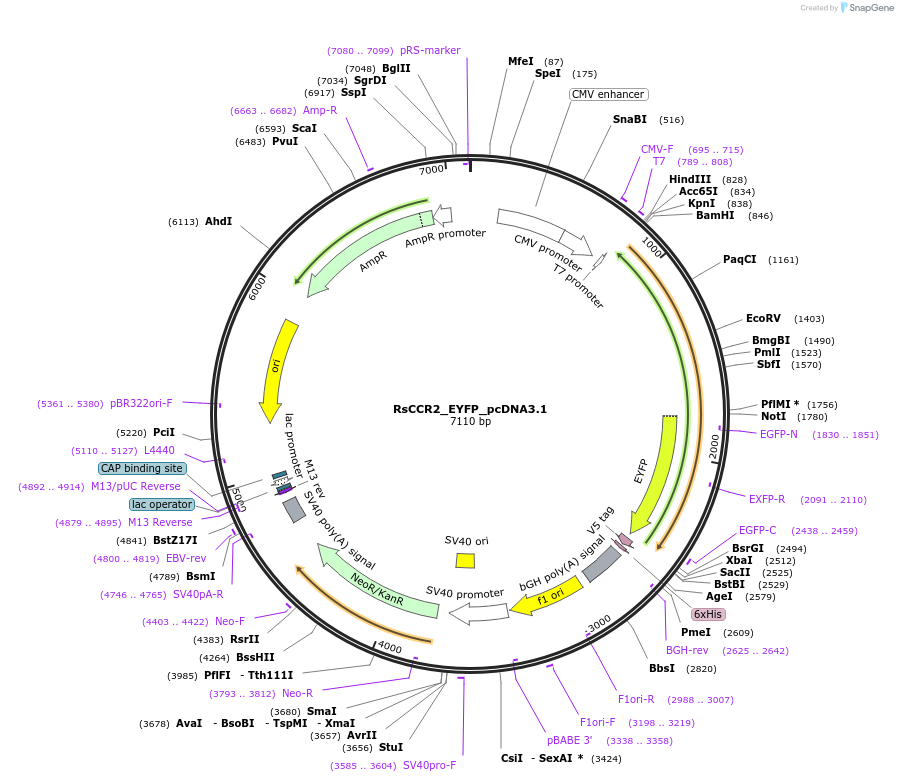
RsCCR2_EYFP_pcDNA3.1
Plasmid#159125PurposeThis plasmid encodes RsCCR2 rhodopsin domainDepositorInsertRsCCR2
TagsEYFPExpressionMammalianAvailable SinceSept. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
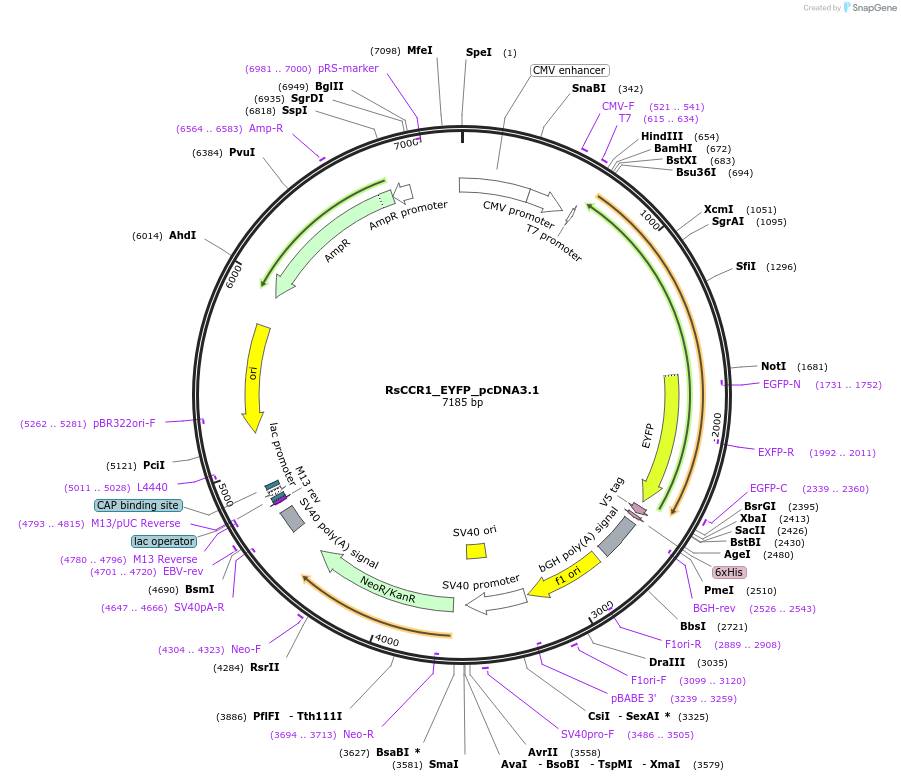
RsCCR1_EYFP_pcDNA3.1
Plasmid#159124PurposeThis plasmid encodes RsCCR1 rhodopsin domainDepositorInsertRsCCR1
TagsEYFPExpressionMammalianAvailable SinceSept. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
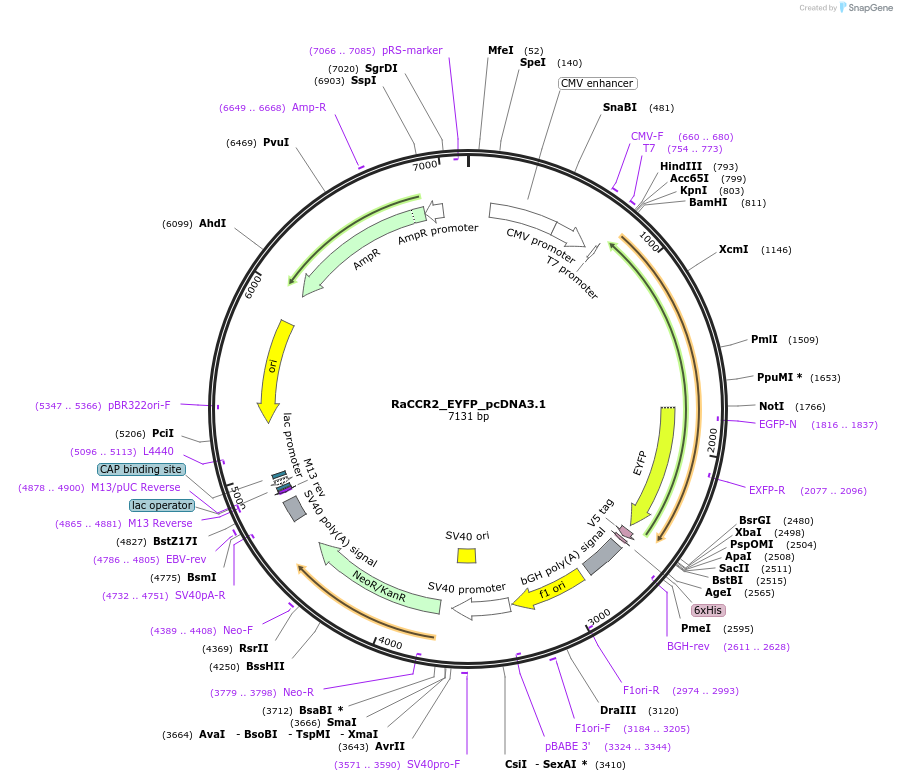
RaCCR2_EYFP_pcDNA3.1
Plasmid#159123PurposeThis plasmid encodes RaCCR2 rhodopsin domainDepositorInsertRaCCR2
TagsEYFPExpressionMammalianAvailable SinceSept. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
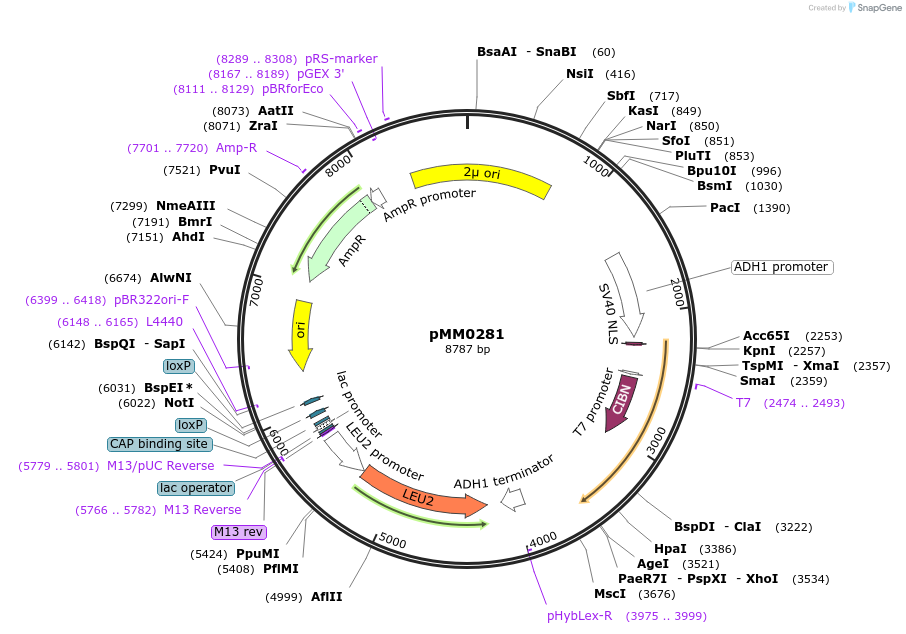
pMM0281
Plasmid#128972PurposeEncodes VP16-CIB1 under pADH1 promoterDepositorInsertpscADH1-SV40NLS-VP16-CIB1-tscADH1
ExpressionYeastAvailable SinceOct. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
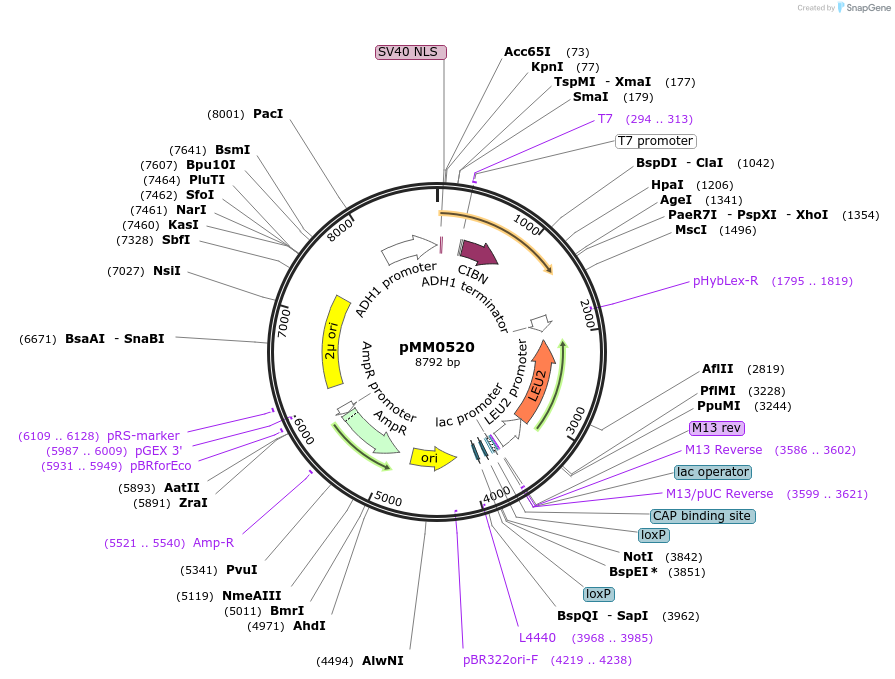
pMM0520
Plasmid#128983PurposeOverexpression plasmid for VP16-CIB1DepositorInsertSV40NLS-VP16-CIB1 LEU2 2_
ExpressionYeastAvailable SinceOct. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
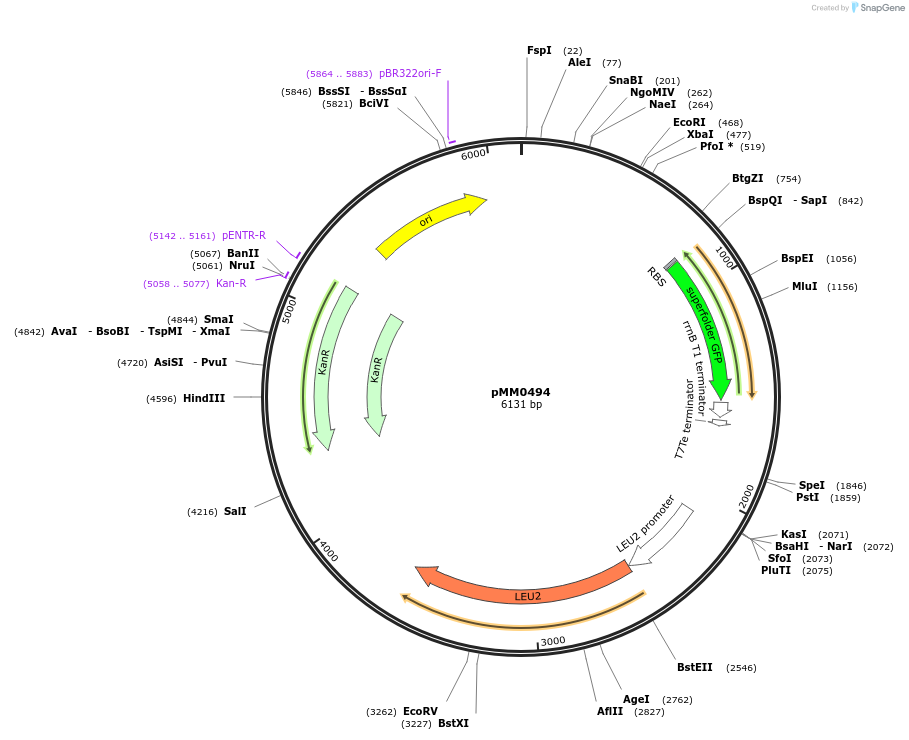
pMM0494
Plasmid#128981PurposeLEU2 preassembled entry vectorDepositorInsertLEU2 5' homology-ConLS-sfGFP dropout-ConR1-LEU2 3' homology
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
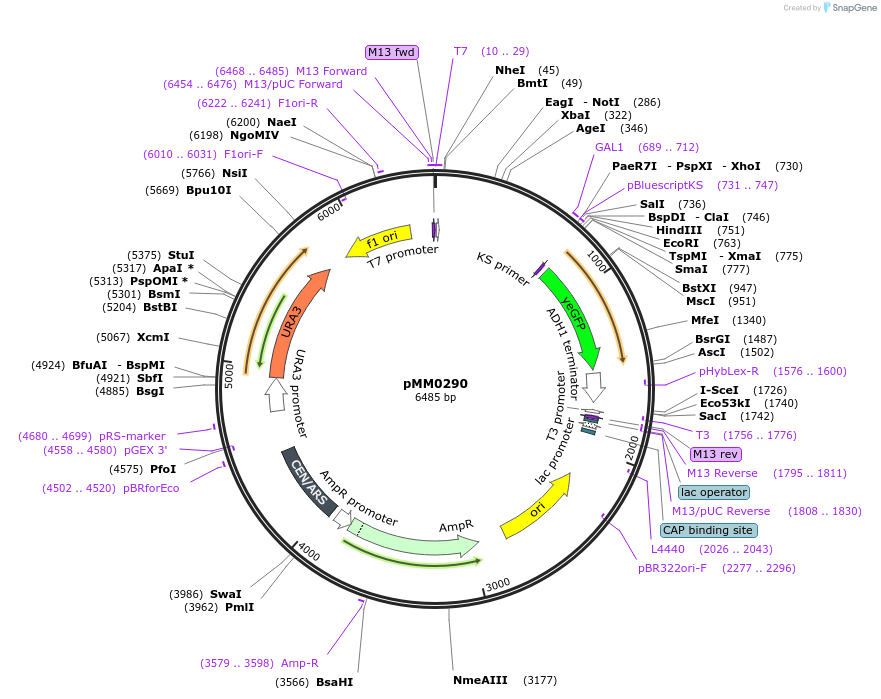
pMM0290
Plasmid#128978PurposeReporter plasmid, pZF(3BSopp)-yEVENUSDepositorInsertpZF(3BSop)-yEVENUS
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
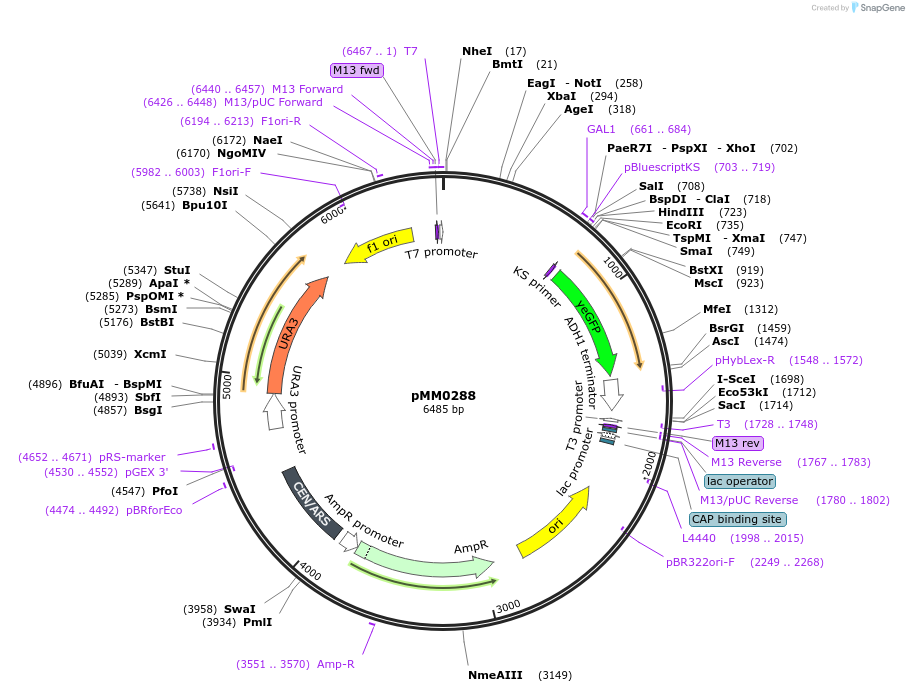
pMM0288
Plasmid#128976PurposeReporter plasmid, pZF(3BSc)-yEVENUSDepositorInsertpZF(3BSc)-yEVENUS
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
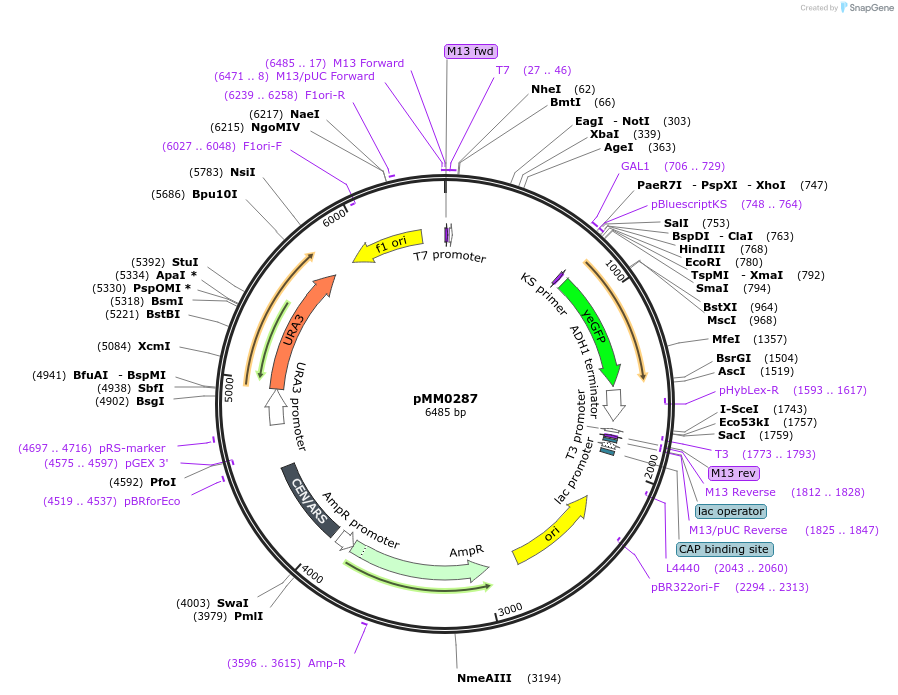
pMM0287
Plasmid#128975PurposeReporter plasmid, pZF(3BS)-yEVENUSDepositorInsertpZF(3BS)-yEVENUS
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
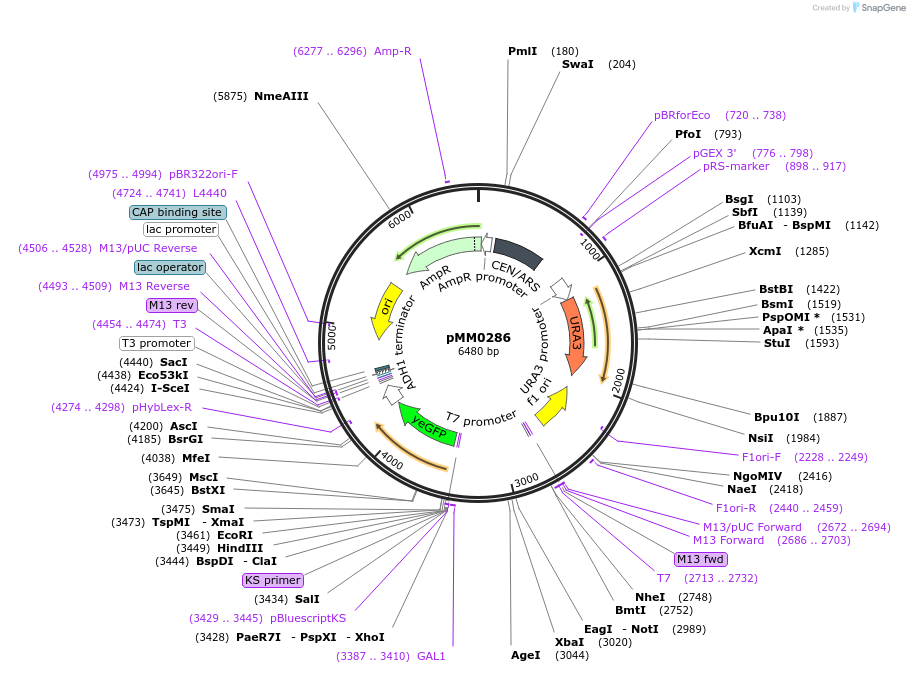
pMM0286
Plasmid#128974PurposeReporter plasmid, pZF(3oBS)-yEVENUSDepositorInsertpZF(3oBS)-yEVENUS
ExpressionYeastAvailable SinceOct. 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
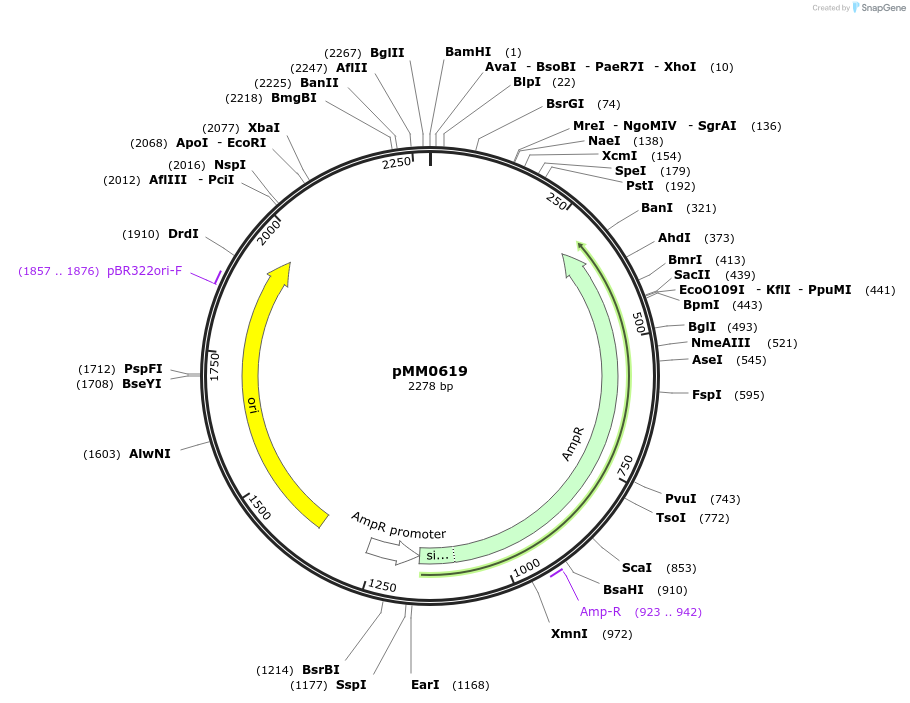
pMM0619
Plasmid#128994PurposeSpacer cassette plasmid flanked by ConL2 and ConREDepositorInsertConL2-Spacer-ConRE-AmpR-Col-E1
ExpressionYeastAvailable SinceOct. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
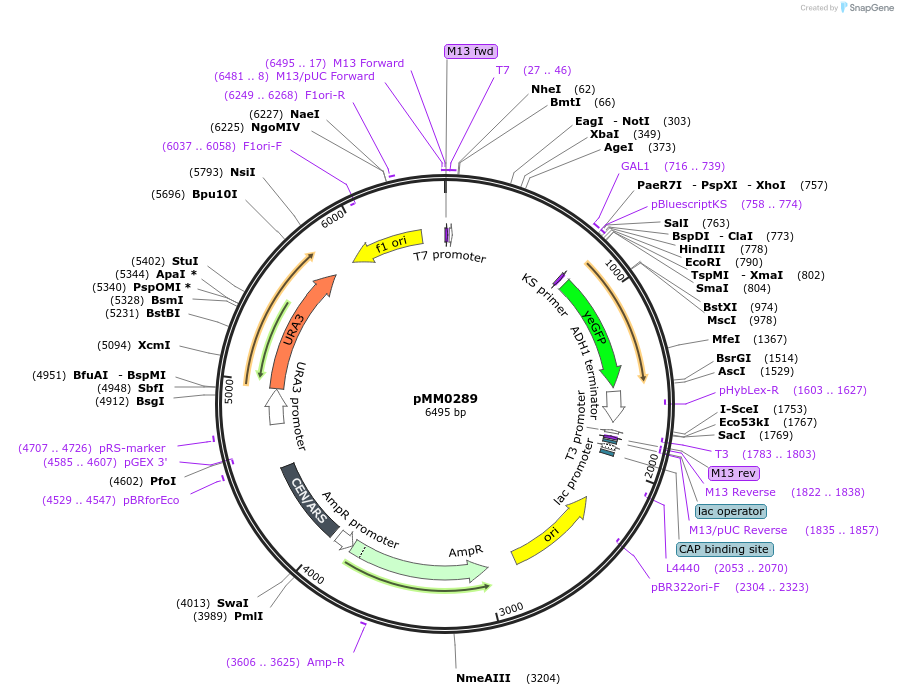
pMM0289
Plasmid#128977PurposeReporter plasmid, pZF(4BSc)-yEVENUSDepositorInsertpZF(4BSc)-yEVENUS
ExpressionYeastAvailable SinceOct. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
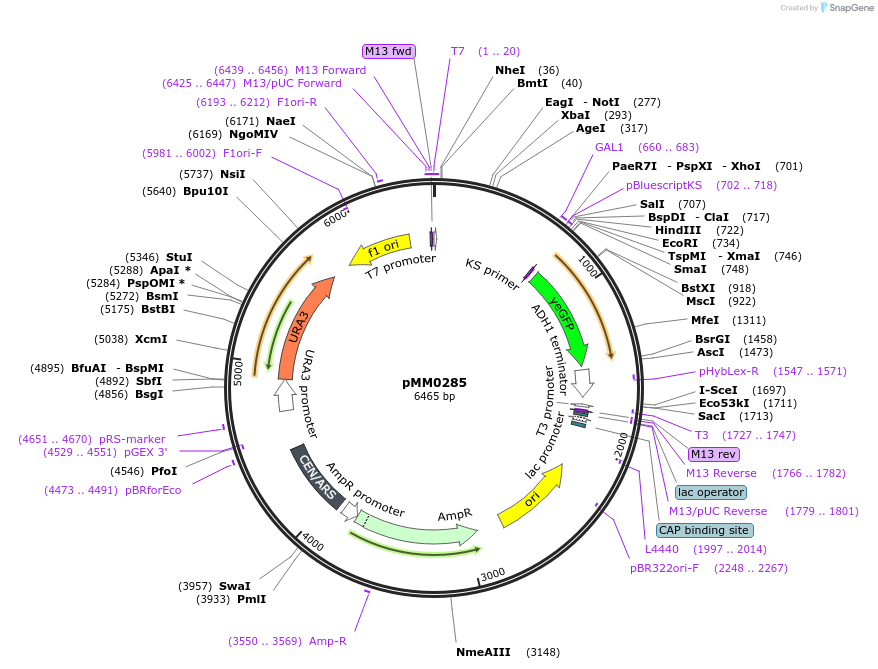
pMM0285
Plasmid#128973PurposeReporter plasmid, pZF(1BS)-yEVENUSDepositorInsertpZF(1BS)-yEVENUS
ExpressionYeastAvailable SinceOct. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
pRS314 snu114-del(IV)
Plasmid#111321Purposesnu114-del(IV)DepositorInsertSNU114
ExpressionYeastMutationdeletion of domain IVaAvailable SinceAug. 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
p413TEF-NQYFP
Plasmid#99556PurposeExpresses Swi1 NQ-YFP to monitor Swi1 state or induce [SWI+].DepositorAvailable SinceAug. 30, 2017AvailabilityAcademic Institutions and Nonprofits only