We narrowed to 9,659 results for: SUB;
-
Plasmid#10982DepositorInsertrpb1-C67Y (RPO21 Budding Yeast)
ExpressionBacterial and YeastMutationMutation 200 G to A in the RPB1 ORF resulting i…Available SinceDec. 16, 2005AvailabilityAcademic Institutions and Nonprofits only -
pL-rpb1-C70Y
Plasmid#10983DepositorInsertrpb1-C70Y (RPO21 Budding Yeast)
ExpressionBacterial and YeastMutationMutation 209 G to A in the RPB1 ORF resulting i…Available SinceDec. 16, 2005AvailabilityAcademic Institutions and Nonprofits only -
pL-rpb1-H80Y
Plasmid#10984DepositorInsertrpb1-H80Y (RPO21 Budding Yeast)
ExpressionBacterial and YeastMutationMutation 238 C to T in the RPB1 ORF resulting i…Available SinceDec. 16, 2005AvailabilityAcademic Institutions and Nonprofits only -
pDHJS3 AN-
Plasmid#78241PurposeOne of four plasmids needed to create the mismatched double Holliday junction substrate (MM-DHJS).DepositorInsertsloxP
sequence "A" with NheI site
loxP2
Available SinceSept. 26, 2016AvailabilityAcademic Institutions and Nonprofits only -
pDHJS3 AS-
Plasmid#78237PurposeOne of four plasmids needed to create the double Holliday junction substrate (DHJS).DepositorInsertsloxP
sequence "B"
loxP2
UsePhagemidAvailable SinceSept. 26, 2016AvailabilityAcademic Institutions and Nonprofits only -
pDNR-hAkt1
Plasmid#49387Purposedonor vector for cloning human Akt1 with 6xHN tagDepositorInsertAkt1 (AKT1 Human)
UseCre/Lox; Donor vectorTags6xHN (6 repeating His-Asn subunits)Mutationtga to cca at nt 1725 with respect to GenBank ID …PromoternoneAvailable SinceDec. 2, 2013AvailabilityAcademic Institutions and Nonprofits only -
pJRL2 PHO5(UASm1)pr VYFP (EB#1549)
Plasmid#14841DepositorInsertPHO5 promoter UASm1 (PHO5 Budding Yeast)
TagsVYFPExpressionYeastMutationUASm1 (AAGCTT was substituted for CACGTT). Unindu…Available SinceMay 25, 2007AvailabilityAcademic Institutions and Nonprofits only -
pL-rpb1-D260N
Plasmid#10985DepositorInsertrpb1-D260N (RPO21 Budding Yeast)
ExpressionBacterial and YeastMutationMutation 778 G to A in the RPB1 ORF resulting i…Available SinceDec. 16, 2005AvailabilityAcademic Institutions and Nonprofits only -
pL-rpb1-D261N
Plasmid#10986DepositorInsertrpb1-D261N (RPO21 Budding Yeast)
ExpressionBacterial and YeastMutationMutation 781 G to A in the RPB1 ORF resulting i…Available SinceDec. 16, 2005AvailabilityAcademic Institutions and Nonprofits only -
pL-rpb1-R320C
Plasmid#10987DepositorInsertrpb1-R320C (RPO21 Budding Yeast)
ExpressionBacterial and YeastMutationMutation 958 C to T in the RPB1 ORF resulting i…Available SinceDec. 16, 2005AvailabilityAcademic Institutions and Nonprofits only -
pL-rpb1-E1230K
Plasmid#10990DepositorInsertrpb1-E1230K (RPO21 Budding Yeast)
ExpressionBacterial and YeastMutationMutation 3688 G to A in the RPB1 ORF resulting in…Available SinceDec. 16, 2005AvailabilityAcademic Institutions and Nonprofits only -
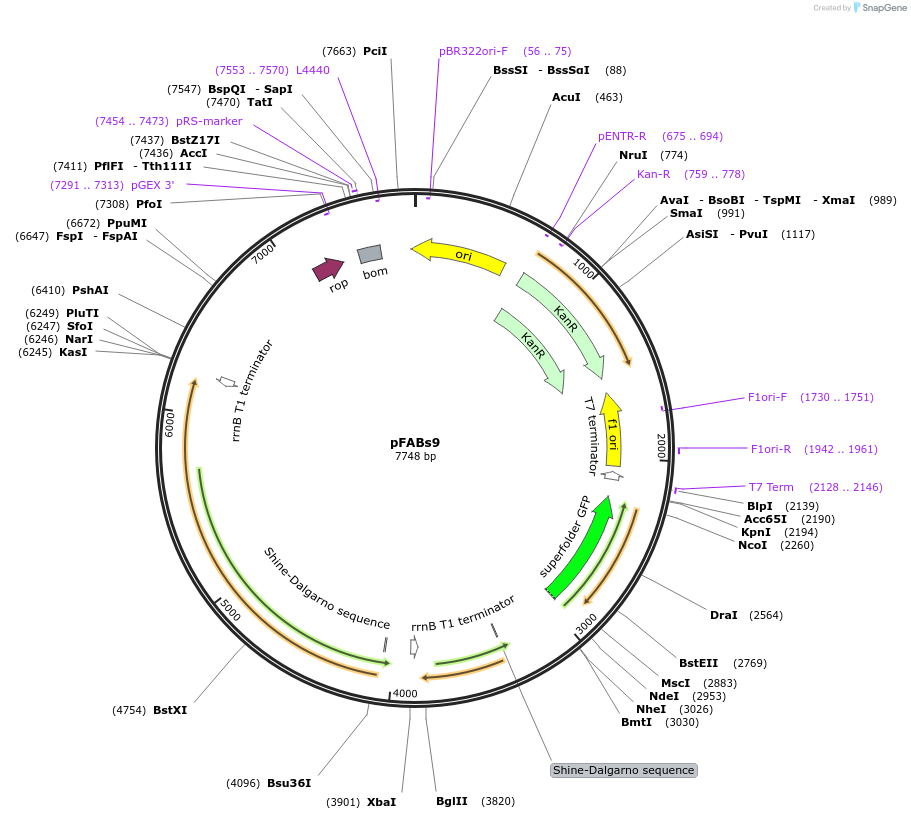
pFABs9
Plasmid#241905PurposeFerrulic acid biosensor - expresses sfGFP in response to substituted cinnamic acidDepositorInsertsfGFP, ferC, ferA
UseSynthetic BiologyMutationnoneAvailable SinceSept. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
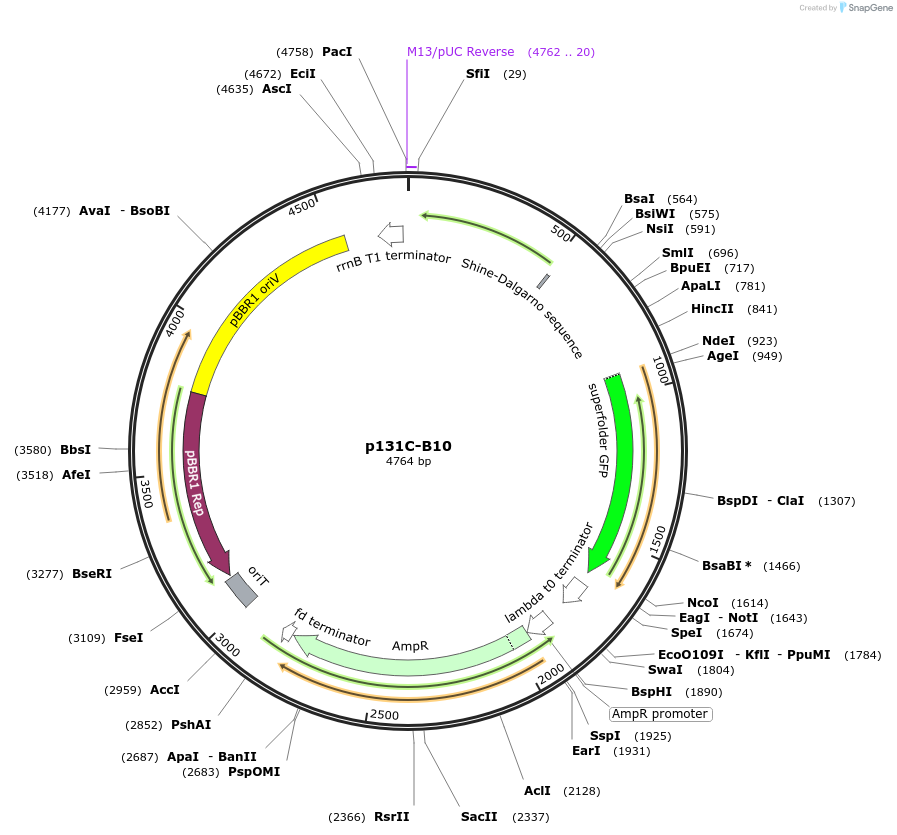
p131C-B10
Plasmid#241908PurposeOptimised PCA biosensor produced sfGFP in response to hydroxyl-substituted benzoic acids, including protocatechuic acid (PCA)DepositorInsertpcaV, sfGFP
UseSynthetic BiologyMutationnoneAvailable SinceAug. 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
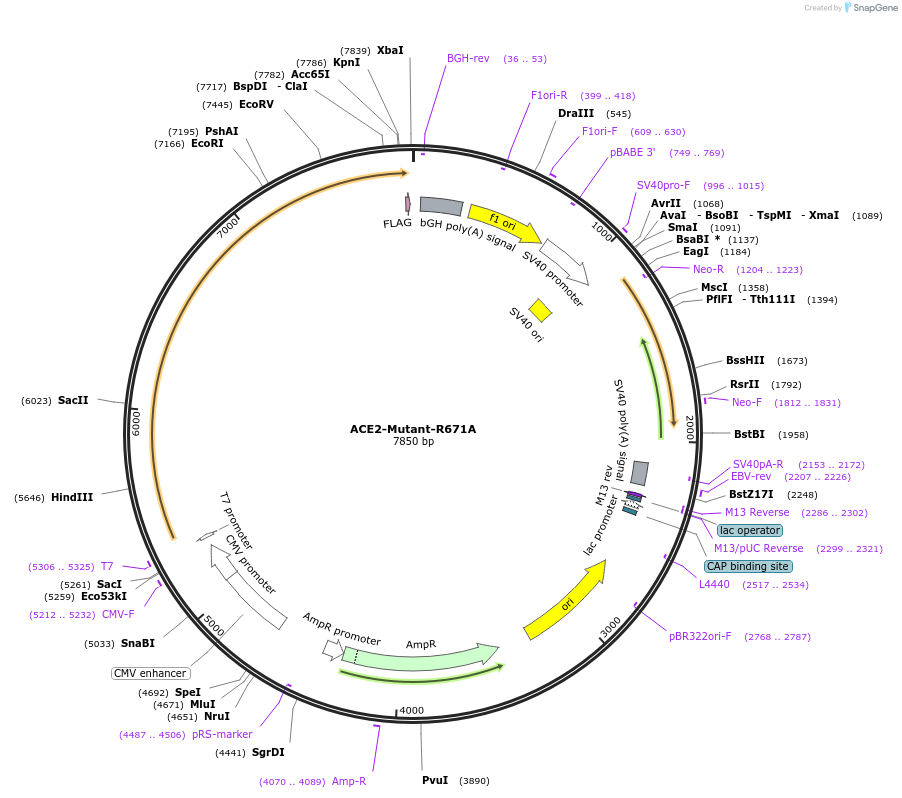
ACE2-Mutant-R671A
Plasmid#240606PurposeTo test ACE2 cleavage by TMPRSS2. This protease targets R and K amino acids of ACE2; R was substituted by A at Residue 671 of ACE2 in the collectrin-like siteDepositorInsertAngiotensin-converting enzyme 2 (ACE2 Human)
TagsFLAG tagExpressionMammalianMutationArginine (R) at position 671 on ACE2 is replaced …PromoterCMVAvailable SinceAug. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
pRSF_AaNHis_RbcL
Plasmid#229517PurposeExpresses Anthoceros agrestis Rubisco large (N terminus hexahistidine)DepositorInsertRubisco large subunit
TagsHexahistidine tagExpressionBacterialPromoterT7Available SinceJune 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
pTYB1-T4gp47
Plasmid#228212PurposeExpression in BL21(DE3) of T4 SbcD-like subunit of palindrome specific endonucleaseDepositorInsert47 (47 Escherichia phage T4)
TagsSce VMA intein-chitin binding domainExpressionBacterialPromoterT7Available SinceDec. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
pCMV-EGFP-nonstop
Plasmid#226406PurposeExpresses EGFP without a stop codon that produces a GFP-tagged readthrough RQC substrate.DepositorInsertEGFP without stop codon
ExpressionMammalianPromoterCMVAvailable SinceNov. 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
pUC18-LucFF
Plasmid#220496PurposeTo generate substrates to monitor Ku overloading and its impact on transcription at the DNA ends vicinity.DepositorInsertFirefly Luciferase
ExpressionMammalianPromoterSV40Available SinceJuly 3, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
pRegev_S1.34C>A_46G>U
Plasmid#204161PurposepRegev_S1 with double mutations that restore secondary structure.DepositorInsertS1 exon with 34C>A and 46G>U substitutions in beta globin splicing minigene
UseSynthetic BiologyExpressionMammalianMutation34C>A and 46G>U mutationsPromoterTruncated CAGAvailable SinceAug. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
pCI-neo-μ3A-(HA)3
Plasmid#198179PurposeExpression of HA-tagged AP-3 μ3A in mammalian cellsDepositorInsertAP-3 μ3A
Tagstriple HA tagExpressionMammalianMutationsilent substitution in codon 225PromoterCMVAvailable SinceApril 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
pRSF-1b-S10
Plasmid#128594PurposeExpresses tagless S10 ribosomal protein in E.coliDepositorAvailable SinceMay 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
Lmajor_gp63_10_0460_D463N
Plasmid#171644PurposeExpression of L. major glcyoprotein-63 (D463N) with substrate binding site mutation from chromosome 10 in mammalian cellsDepositorInsertLmjF.10.0460 D463N
TagsMyc-HisExpressionMammalianMutationD463NAvailable SinceDec. 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
pJH223
Plasmid#172151PurposepylBCD variant in pCDF backboneDepositorInsertPT7/lac (pylBCD 34B_sub-pop5); LacI
ExpressionBacterialMutationPylB(N61S, E178K, E251D) PylD(A252V)Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
pJH224
Plasmid#172152PurposepylBCD variant in pCDF backboneDepositorInsertPT7/lac (pylBCD 40C_sub-pop6); LacI
ExpressionBacterialMutationPylB(N61S, E175K, E178K)Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
pJH463
Plasmid#172155PurposepylBCD variant in pBAD backboneDepositorInsertPara (pylBCD 36A_sub-pop1); AraC
ExpressionBacterialMutationPylB(N61S, E122K, E178K, G188R)Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
pJH574
Plasmid#172158PurposepylBCD variant in pBAD backboneDepositorInsertPara (pylBCD 36A_sub-pop2); AraC
ExpressionBacterialMutationPylB(N61S, E84K, E122K) PylC(E316D)Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
pJH575
Plasmid#172159PurposepylBCD variant in pBAD backboneDepositorInsertPara (pylBCD 34B_sub-pop3); AraC
ExpressionBacterialMutationSumo(S6L) PylB(N61S, E178K, G188R, E251D) PylD(A2…Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
pJH576
Plasmid#172160PurposepylBCD variant in pBAD backboneDepositorInsertPara (pylBCD 34B_sub-pop4); AraC
ExpressionBacterialMutationPylB(N61S, E175K, E178K, E251D) PylD(A252V)Available SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
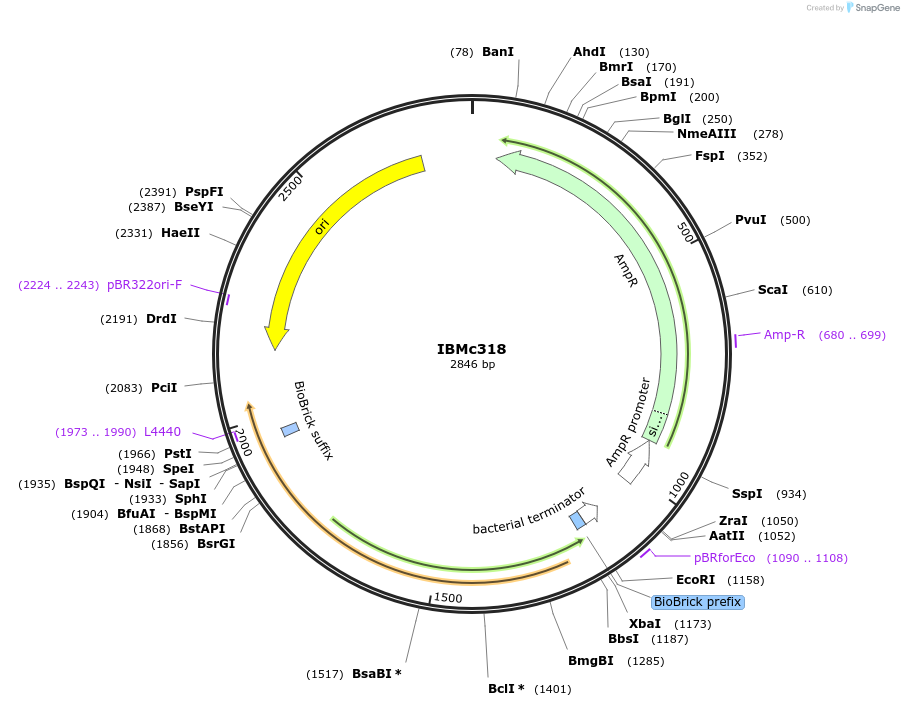
IBMc318
Plasmid#161941PurposeCarries Golden Gate substitution insert for ER-LBD domain-insertion mappingDepositorInsertpSB1A3-BbsI-G-ER-LBD-S-SapI
ExpressionBacterialAvailable SinceMarch 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
pSM002
Plasmid#122936PurposeExpression of the marked mRNA under the control of substitutional promoters.DepositorInsertYML043C (Marked)
ExpressionYeastMutationYML043C: bp 52-63 AAGTTAAAGTAT to AAATTGAAATACPromoter[tetO2]4inPgal1Available SinceApril 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
pSM001
Plasmid#122935PurposeExpression of the marked mRNA under the control of substitutional promoters.DepositorInsertYML029W (Marked)
ExpressionYeastMutationYML029W: bp 52-63 GAGATTGATCTT to GAAATAGACCTAPromoter[tetO2]4inPgal1Available SinceApril 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
pEC005
Plasmid#122939PurposeExpression of the marked mRNA under the control of substitutional promoters.DepositorInsertYML071C (Marked)
ExpressionYeastMutationYML071C: bp 52-63 AAACGGTTAAGT to AAGCGCTTGAGCPromoter[tetO2]4inPgal1Available SinceApril 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
pSV_207
Plasmid#122938PurposeExpression of the marked mRNA under the control of substitutional promoters.DepositorInsertYML070W (Marked)
ExpressionYeastMutationYML070W: bp 52-63 GGGTTTGCCCTT to GGCTTCGCGCTAPromoter[tetO2]4inPgal1Available SinceApril 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
pSM004
Plasmid#122937PurposeExpression of the marked mRNA under the control of substitutional promoters.DepositorInsertYML065W (Marked)
ExpressionYeastMutationYML065W: bp 52-63 GAGCAGGGAAAT to GAACAAGGTAACPromoter[tetO2]4inPgal1Available SinceApril 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
pSV_228
Plasmid#122933PurposeExpression of the marked mRNA under the control of substitutional promoters.DepositorInsertYBR025C (Marked)
ExpressionYeastMutationYBR025C: bp 52-63 GGTAATAACTTG to GGAAACAATTTAPromoter[tetO2]4inPgal1Available SinceApril 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
pSV_229
Plasmid#122934PurposeExpression of the marked mRNA under the control of substitutional promoters.DepositorInsertYBR026C (Marked)
ExpressionYeastMutationYBR026C: bp 52-63 AAGCACTTCAAA to AAACATTTTAAGPromoter[tetO2]4inPgal1Available SinceApril 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
-
DeltaNIRD KVS-1
Plasmid#113069PurposeExpresses worm truncated KVS1.2 in mammalian cellsDepositorInsertK+ voltage sensitive channel subunit 1.2 (kvs-1 Nematode)
ExpressionMammalianMutationfirst 40 AA deletionAvailable SinceAug. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
C113S KVS-1-HA
Plasmid#113070PurposeExpresses worm KVS-1 C113S in mammalian cellsDepositorInsertC113S K+ voltage sensitive channel subunit 1 (kvs-1 Nematode)
TagsHAExpressionMammalianMutationC113SAvailable SinceJuly 31, 2018AvailabilityAcademic Institutions and Nonprofits only -
-
pUPD-MTS2 (12)
Plasmid#101698PurposeMTS2 in domestication vectorDepositorInsertCCAT-[MTS from β subunit of Nicotiana plumbaginifolia F1 ATPase]-AATG
UseYeast expressionAvailable SinceJuly 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
pUPD2-ATPAMTS (12)
Plasmid#101696PurposeATPAMTS in domestication vectorDepositorInsertCCAT-[MTS from S. cerevisiae ATP synthase subunit alpha (YBL099W)]-AATG
ExpressionYeastAvailable SinceJuly 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
KG#244
Plasmid#110927PurposeVector for expressing proteins in a subset of 9 DA/ DB cholingergic motor neurons in the C. elegans ventral nerve cordDepositorInsertsunc-129 promoter
unc-54 3' control region
ExpressionBacterialAvailable SinceJune 11, 2018AvailabilityAcademic Institutions and Nonprofits only -
YCe1897 HC_Kan_C-KDEL_8a
Plasmid#100681Purposepart designed to occupy position 8a of EMMA. Functional category: Signal peptide for subcellular targetingDepositorInsertC-KDEL
UseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
YCe1903 HC_Kan_SV40-NLS_p6
Plasmid#100684Purposepart designed to occupy position 6 of EMMA. Functional category: Signal peptide for subcellular targetingDepositorInsertSV40-NLS
UseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
YCe2058 HC_Kan_Kozak_ATG_Palm_p6
Plasmid#100694Purposepart designed to occupy position 6 of EMMA. Functional category: Signal peptide for subcellular targetingDepositorInsertKozak_ATG_Palm
UseSynthetic BiologyAvailable SinceNov. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
pDHJS3 AN+
Plasmid#78240PurposeOne of four plasmids needed to create the mismatched double Holliday junction substrate (MM-DHJS).DepositorInsertsloxP
sequence "A" with NheI site
loxP2
UsePhagemidAvailable SinceJune 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
pDHJS3 BS-
Plasmid#78239PurposeOne of four plasmids needed to create the double Holliday junction substrate (DHJS).DepositorInsertsloxP
sequence "A"
loxP2
UsePhagemidAvailable SinceJune 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
pDHJS3 AS+
Plasmid#78236PurposeOne of four plasmids needed to create the double Holliday junction substrate (DHJS).DepositorInsertsloxP
sequence "A"
loxP2
UsePhagemidAvailable SinceJune 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
pDHJS3 BS+
Plasmid#78252PurposeOne of four plasmids needed to create the double Holliday junction substrate (DHJS).DepositorInsertsloxP
sequence "B"
loxP2
UsePhagemidAvailable SinceJune 7, 2016AvailabilityAcademic Institutions and Nonprofits only