We narrowed to 10,222 results for: ADA
-
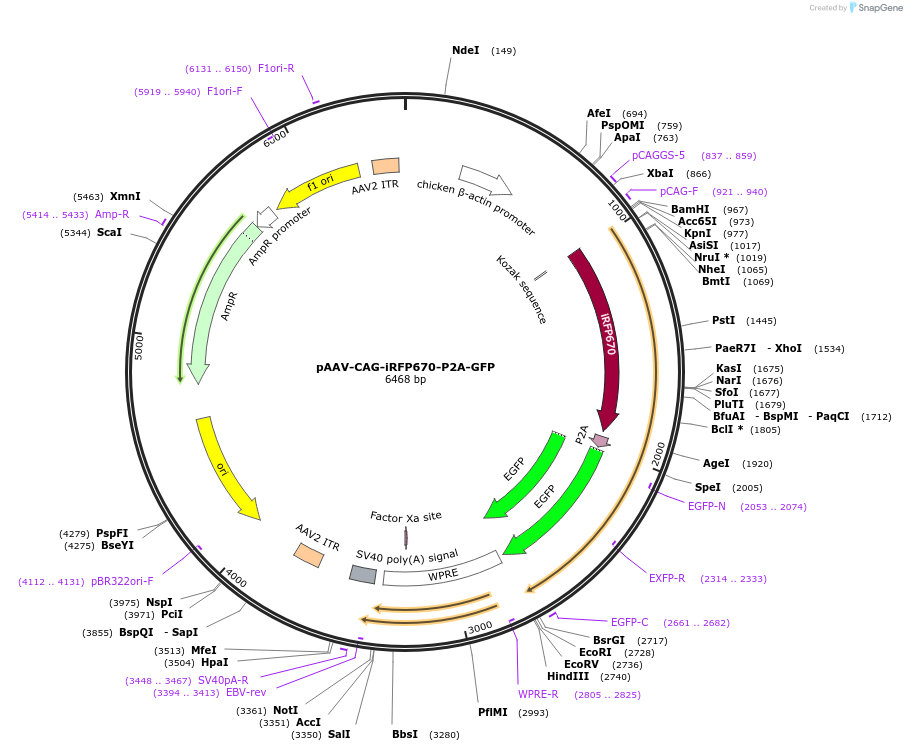
Plasmid#197169PurposeExpresses the protein of iRFP670-P2A-GFP in mammalian cellsDepositorInsertiRFP670-P2A-GFP
UseAAVAvailable SinceMarch 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
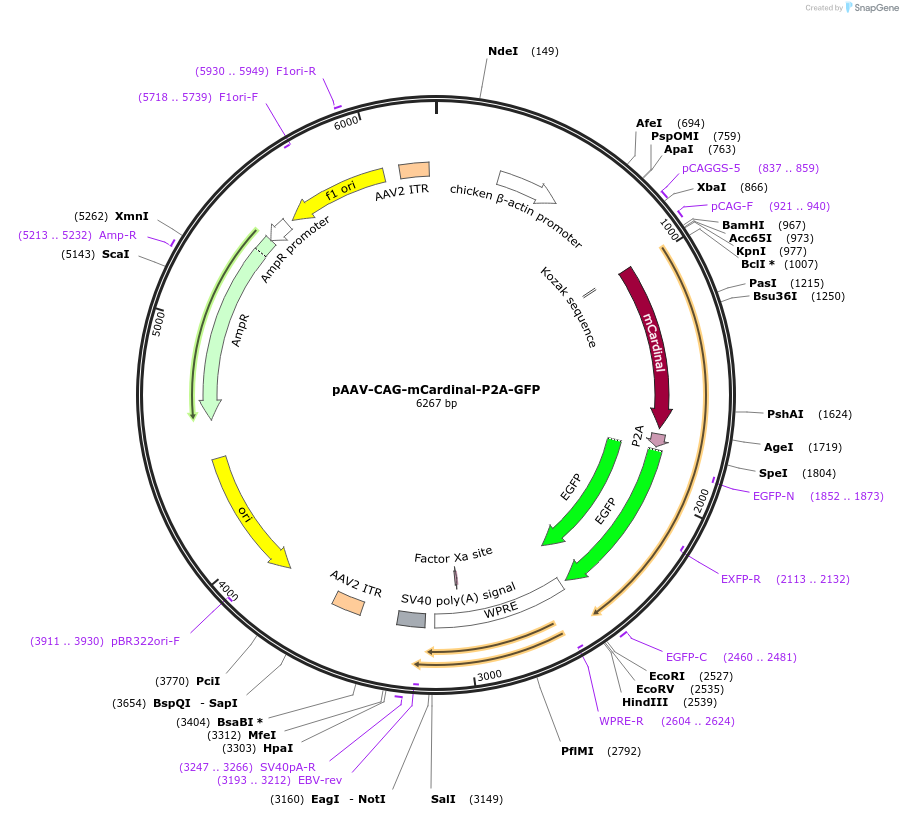
pAAV-CAG-mCardinal-P2A-GFP
Plasmid#197167PurposeExpresses the protein of mCardinal-P2A-GFP in mammalian cellsDepositorInsertmCardinal-P2A-GFP
UseAAVAvailable SinceMarch 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
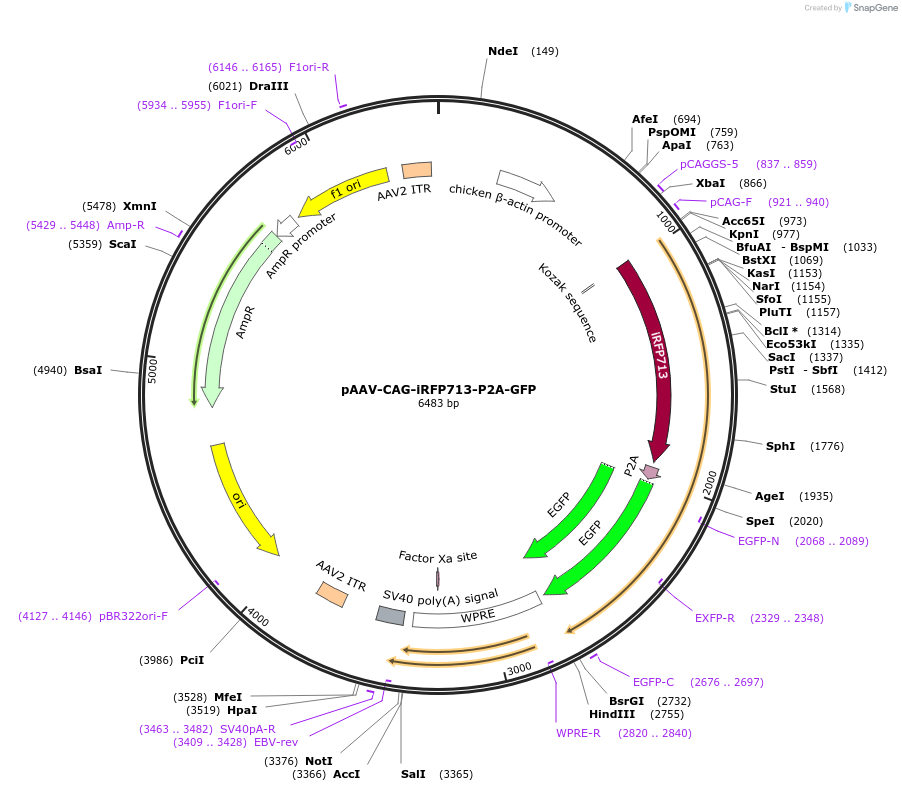
pAAV-CAG-iRFP713-P2A-GFP
Plasmid#197166PurposeExpresses the protein of iRFP713-P2A-GFP in mammalian cellsDepositorInsertiRFP713-P2A-GFP
UseAAVAvailable SinceMarch 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
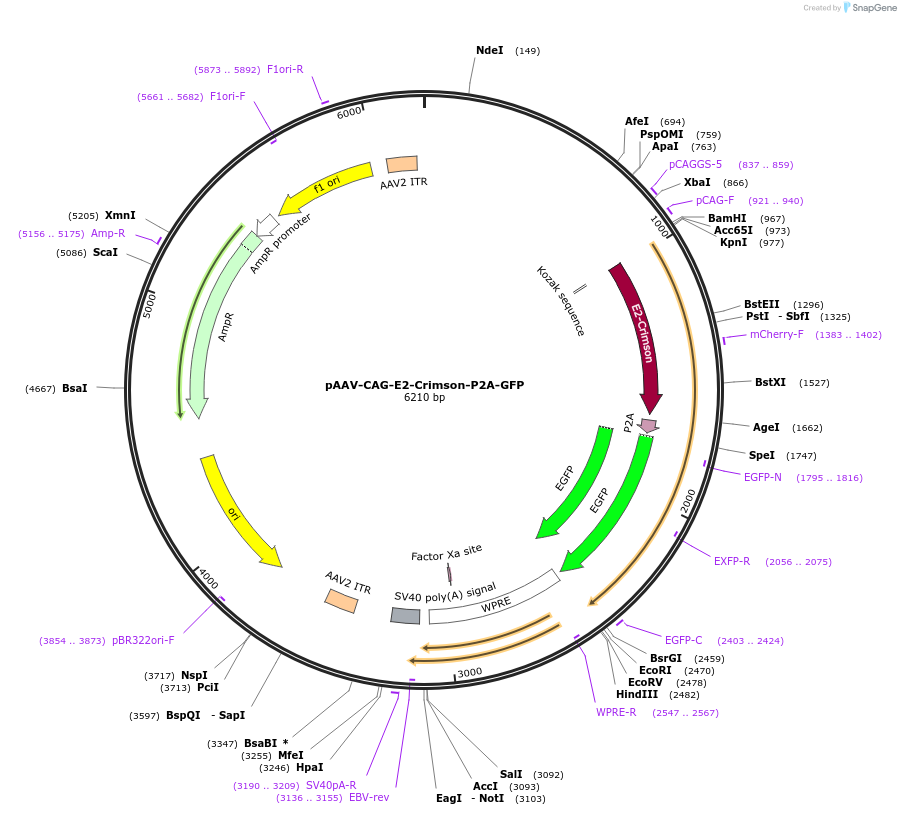
pAAV-CAG-E2-Crimson-P2A-GFP
Plasmid#197162PurposeExpresses the protein of E2-Crimson-P2A-GFP in mammalian cellsDepositorInsertE2-Crimson-P2A-GFP
UseAAVAvailable SinceMarch 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
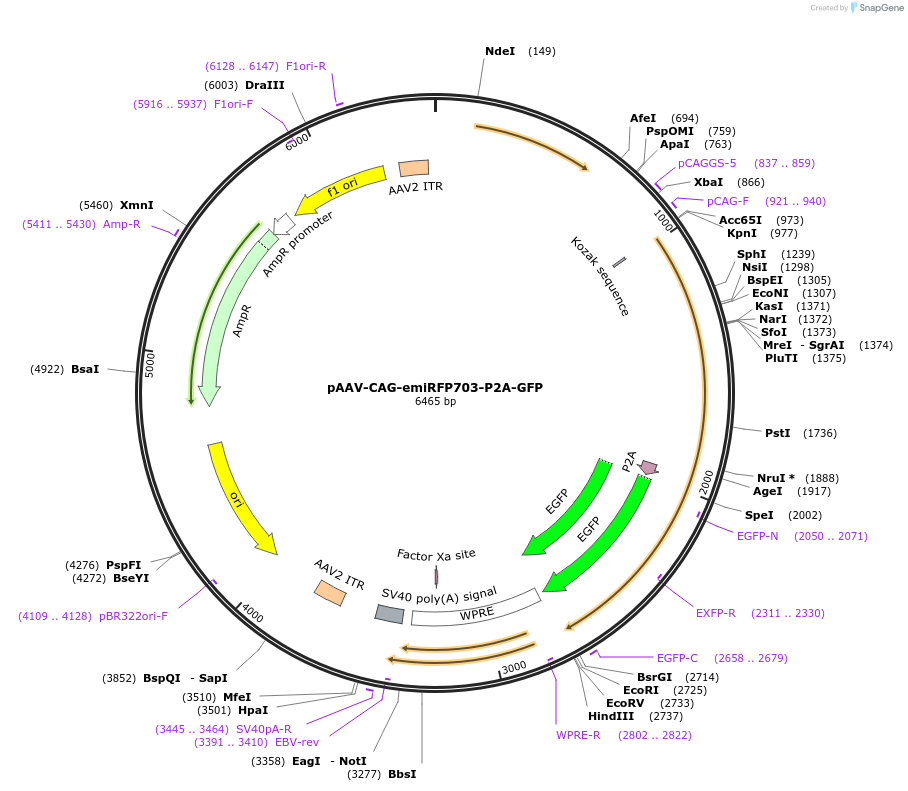
pAAV-CAG-emiRFP703-P2A-GFP
Plasmid#197158PurposeExpresses the protein of emiRFP703-P2A-GFP in mammalian cellsDepositorInsertemiRFP703-P2A-GFP
UseAAVAvailable SinceMarch 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
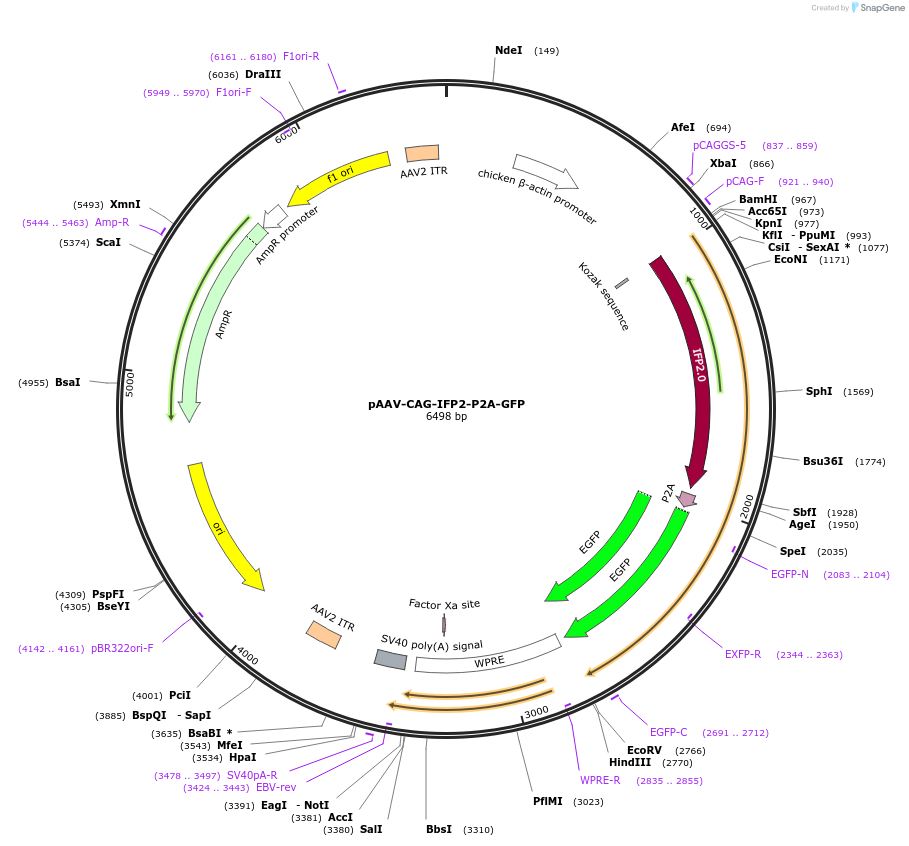
pAAV-CAG-IFP2-P2A-GFP
Plasmid#197161PurposeExpresses the protein of IFP2-P2A-GFP in mammalian cellsDepositorInsertIFP2-P2A-GFP
UseAAVAvailable SinceMarch 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
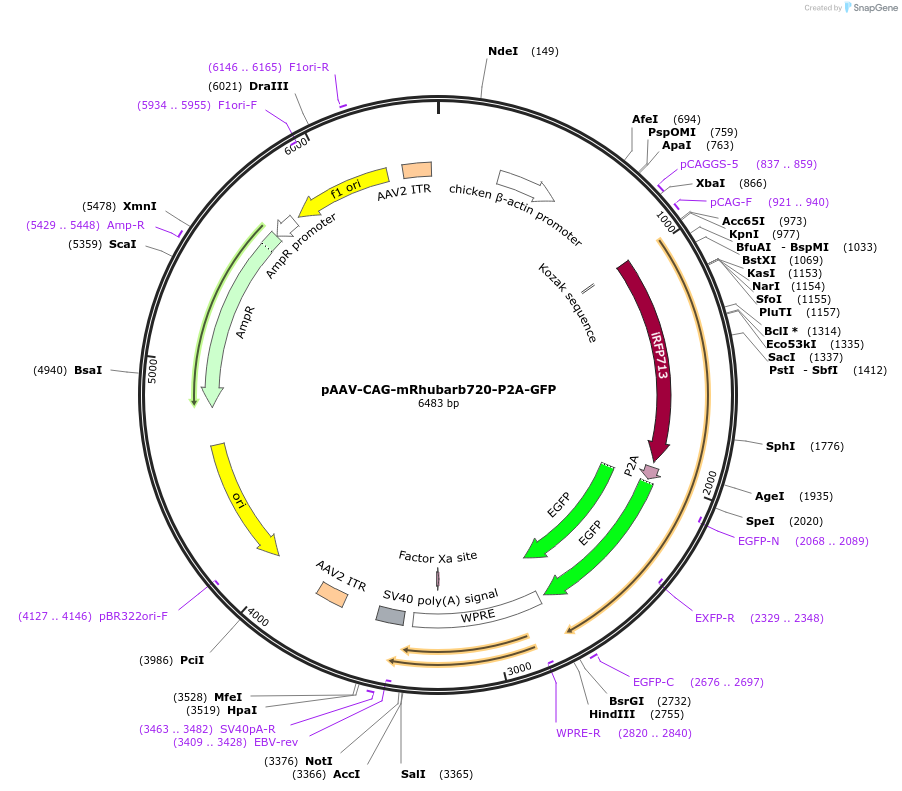
pAAV-CAG-mRhubarb720-P2A-GFP
Plasmid#197159PurposeExpresses the protein of mRhubarb720-P2A-GFP in mammalian cellsDepositorInsertmRhubarb720-P2A-GFP
UseAAVAvailable SinceMarch 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAAV-CAG-BDFP1.6-P2A-GFP
Plasmid#197154PurposeExpresses the protein of BDFP1.6-P2A-GFP in mammalian cellsDepositorInsertBDFP1.6-P2A-GFP
UseAAVAvailable SinceMarch 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
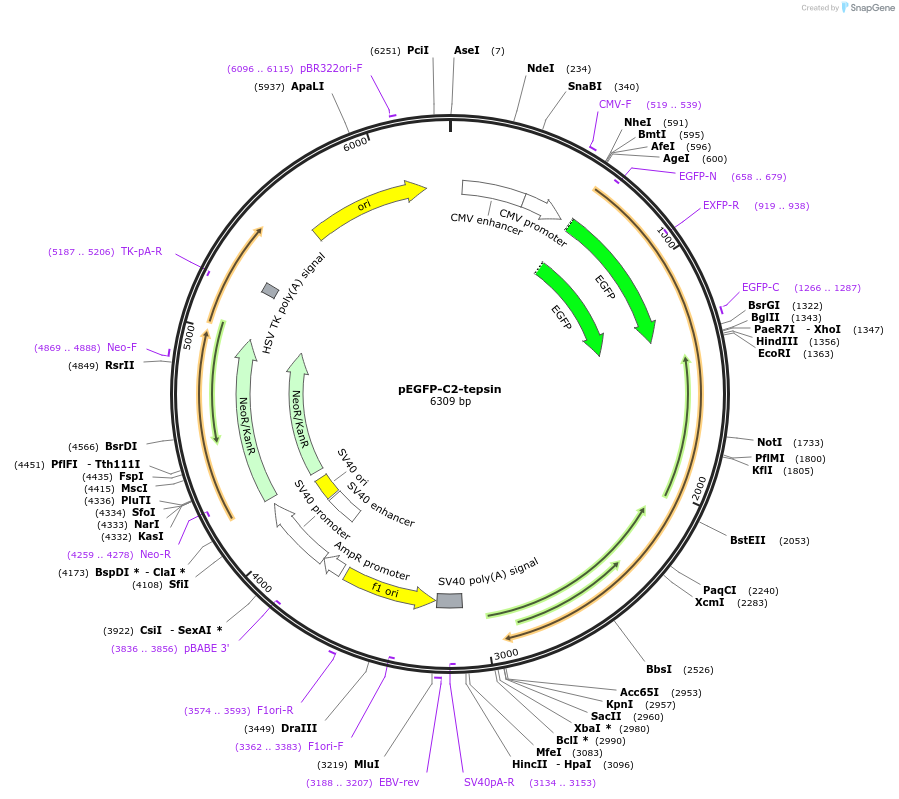
pEGFP-C2-tepsin
Plasmid#196707PurposeExpresses GFP-tepsin in mammalian cellsDepositorInserttepsin (TEPSIN Human)
TagsEGFPExpressionMammalianMutationsilent substitution in codon 199 to eliminate int…PromoterCMVAvailable SinceMarch 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
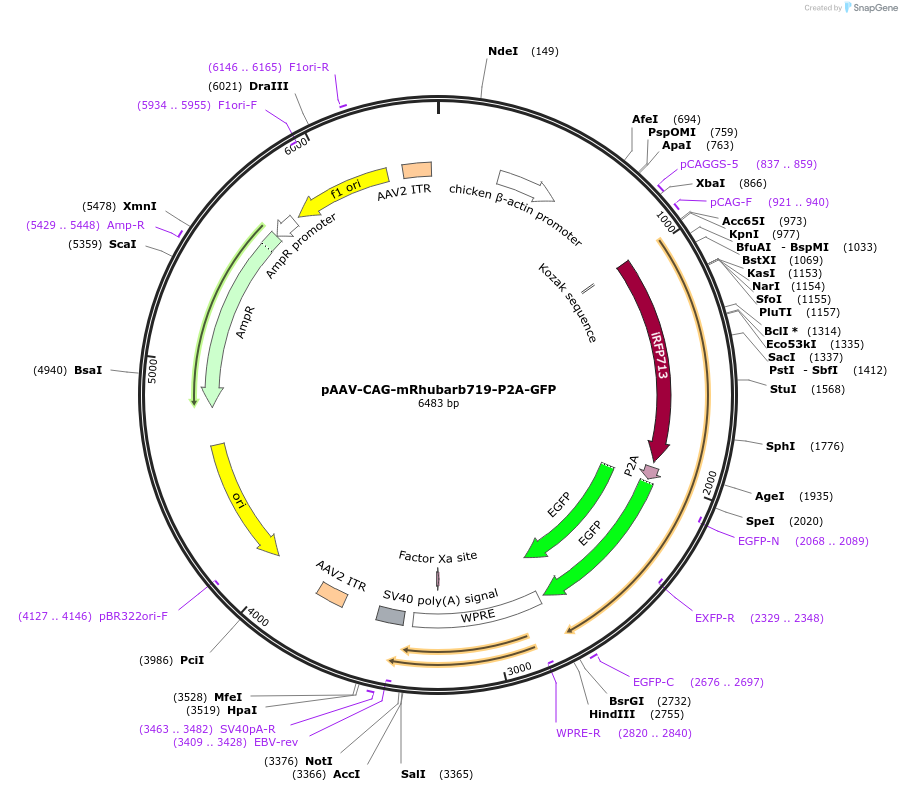
pAAV-CAG-mRhubarb719-P2A-GFP
Plasmid#197152PurposeExpresses the protein of mRhubarb719-P2A-GFP in mammalian cellsDepositorInsertmRhubarb719-P2A-GFP
UseAAVAvailable SinceMarch 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
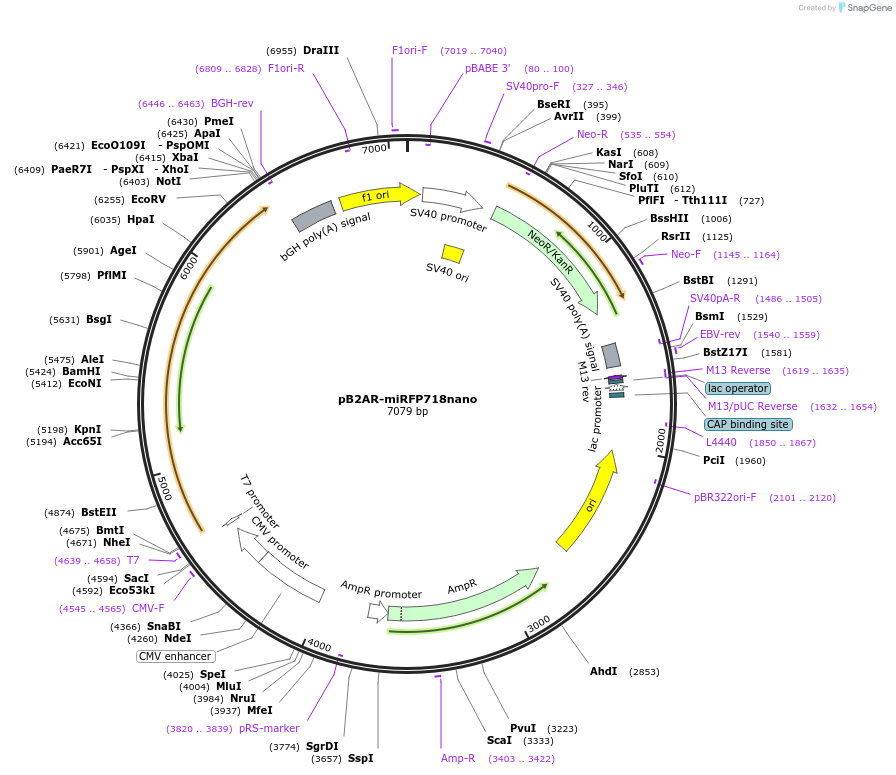
pB2AR-miRFP718nano
Plasmid#195751PurposeBeta-2-adrenergic receptor internally tagged with miRFP718nanoDepositorInsertB2AR-miRFP718nano
ExpressionMammalianPromoterCMVAvailable SinceFeb. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
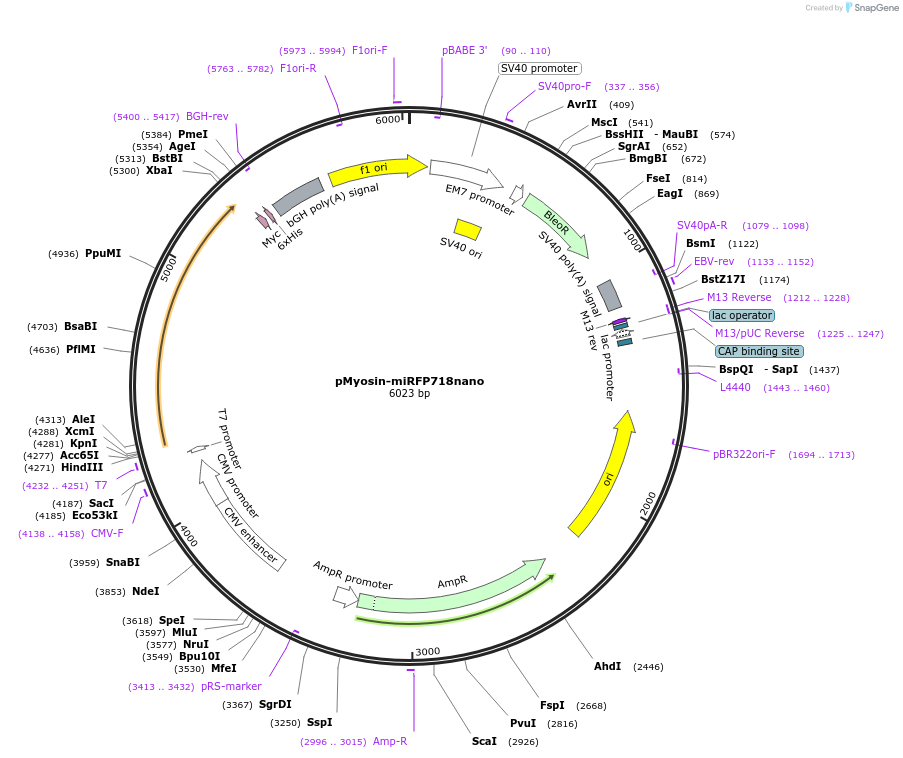
pMyosin-miRFP718nano
Plasmid#195748PurposeLC-Myosin labelled with near-infrared fluorescent protein miRFP718nanoDepositorInsertmiRFP718nano-LC-Myosin
TagsmiRFP718nanoExpressionMammalianPromoterCMVAvailable SinceFeb. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
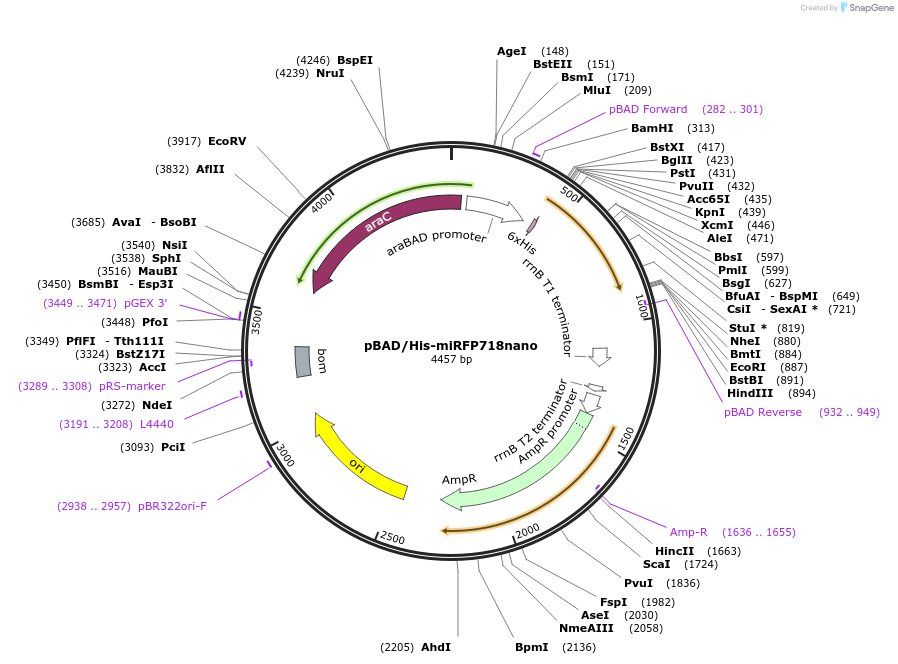
pBAD/His-miRFP718nano
Plasmid#195743PurposeMonomeric near-infrared fluorescent protein miRFP718nano in bacterial plasmidDepositorInsertmiRFP718nano
TagsHisExpressionBacterialPromoteraraCAvailable SinceFeb. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
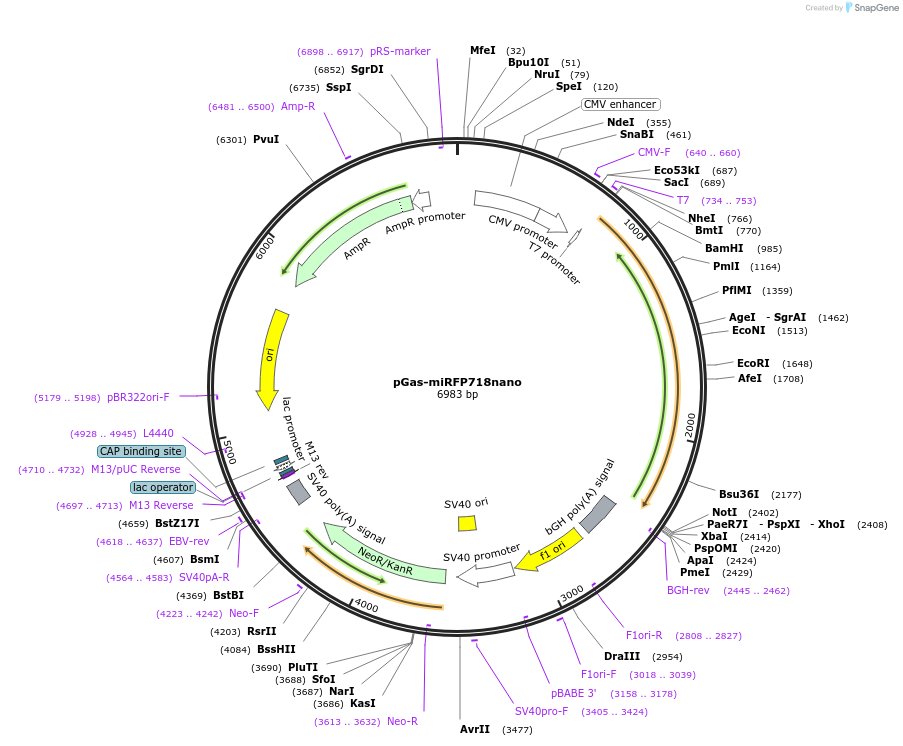
pGas-miRFP718nano
Plasmid#195752PurposeG protein alpha-subunit internally tagged with miRFP718nanoDepositorInsertG protein alpha-s-miRFP718nano
ExpressionMammalianPromoterCMVAvailable SinceFeb. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
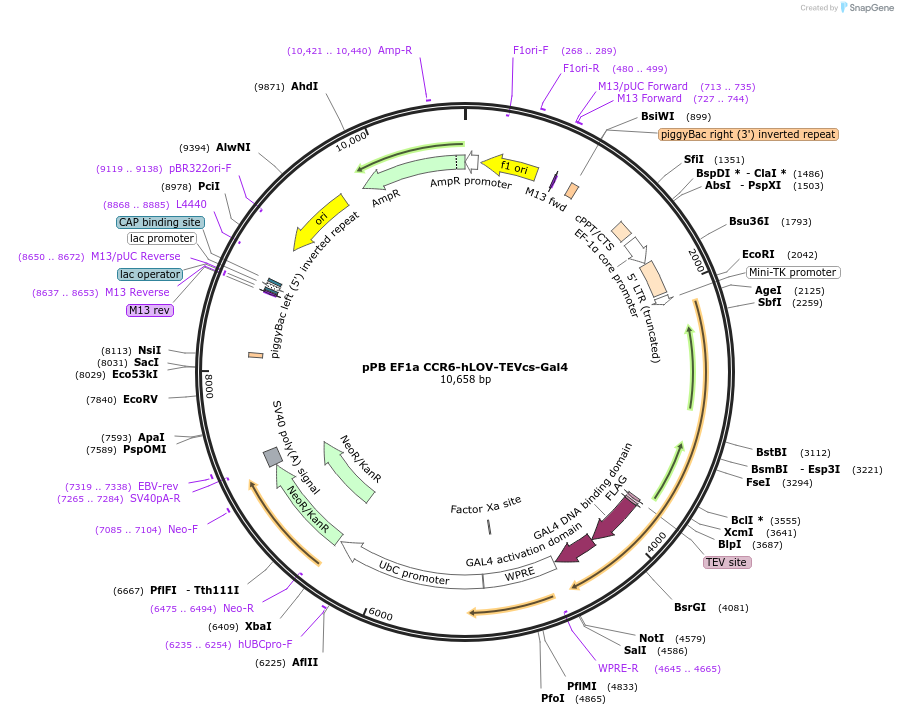
pPB EF1a CCR6-hLOV-TEVcs-Gal4
Plasmid#173120PurposeExpresses CCR6 hLOV receiver construct in DIPGDepositorInsertCCR6-hLOV-TEVcs-Gal4
ExpressionMammalianPromoterEF1aAvailable SinceDec. 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
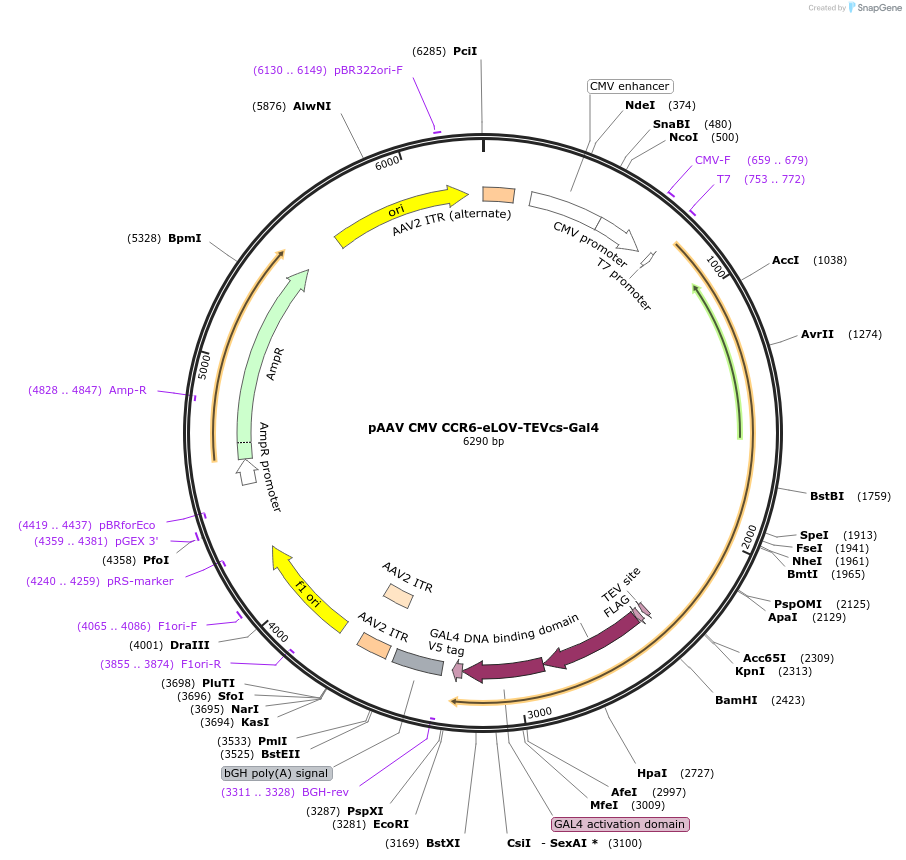
pAAV CMV CCR6-eLOV-TEVcs-Gal4
Plasmid#173113PurposeExpresses CCR6 receiver construct in mammalian cellsDepositorInsertCCR6-eLOV-TEVcs-tTA
UseAAVPromoterCMVAvailable SinceDec. 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
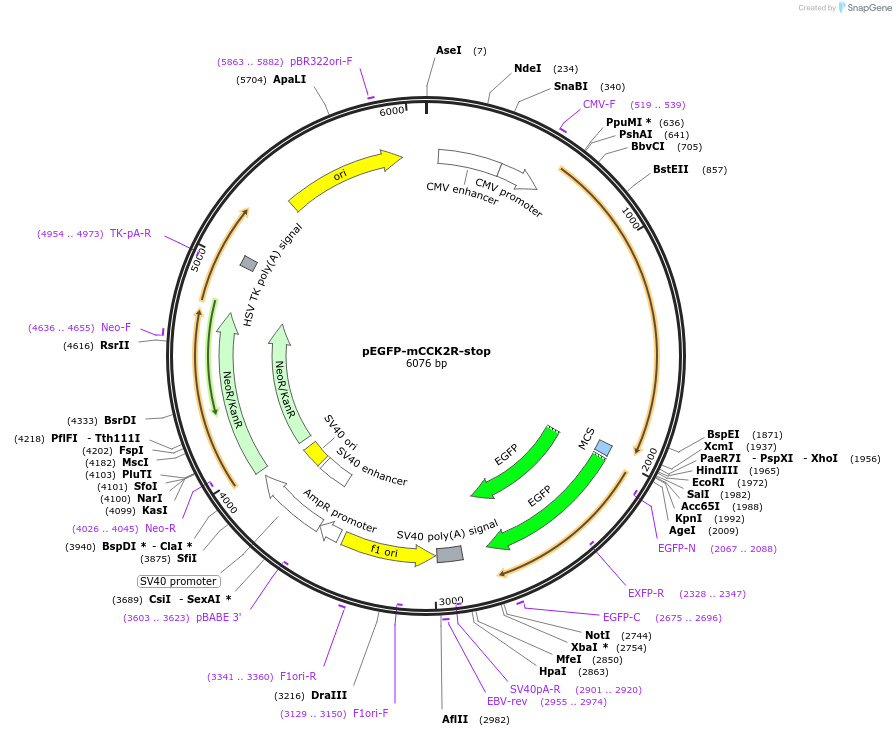
pEGFP-mCCK2R-stop
Plasmid#179321PurposeExpress mouse cholecystokinin 2 receptor (untagged) in mammalian cellsDepositorInsertmouse cholecystokinin2 receptor
ExpressionMammalianMutationstop codon after ORFPromoterCMVAvailable SinceNov. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
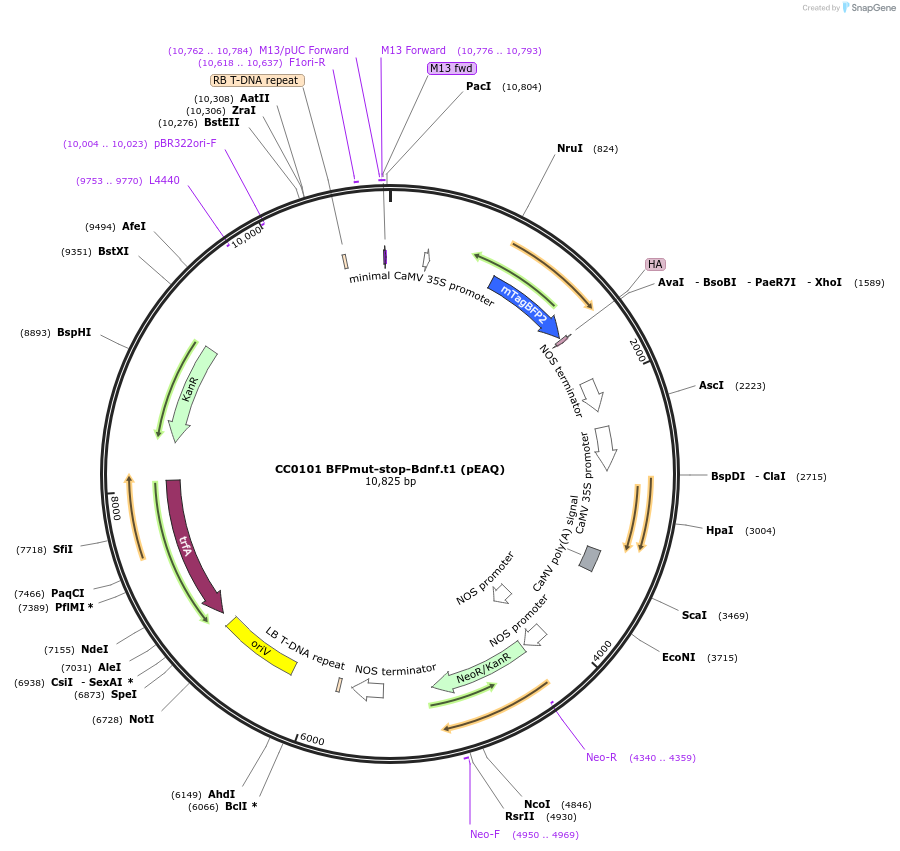
CC0101 BFPmut-stop-Bdnf.t1 (pEAQ)
Plasmid#191173PurposeAn orthogonal trigger sequence used in plant experiments, derived from the 3' UTR of murine BdnfDepositorInsertBdnf.t1 (plant)
ExpressionPlantMutationWTAvailable SinceOct. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
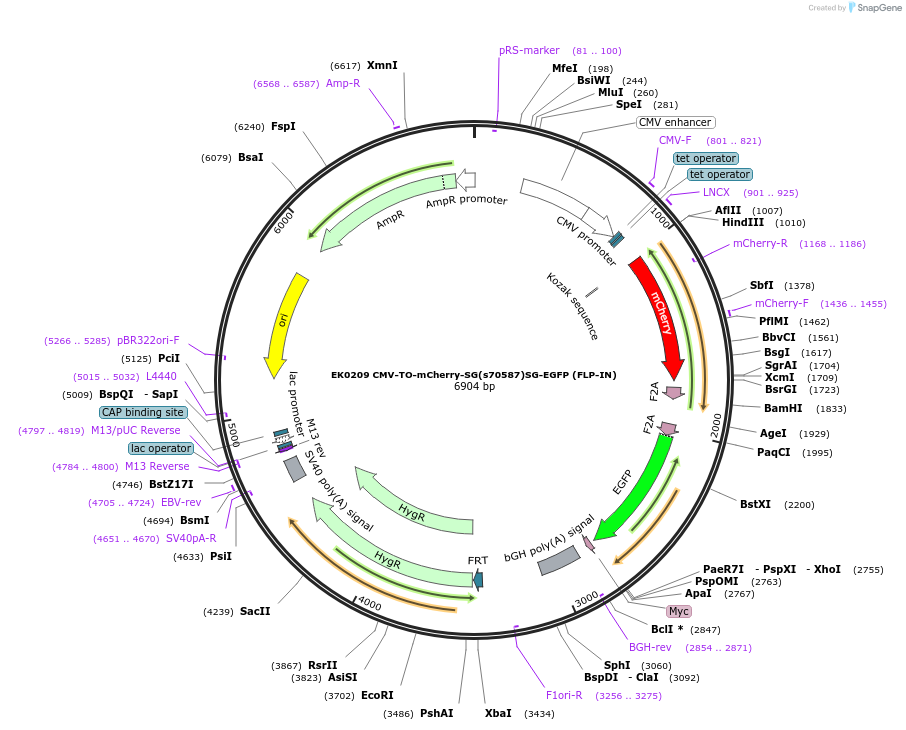
EK0209 CMV-TO-mCherry-SG(s70587)SG-EGFP (FLP-IN)
Plasmid#191174PurposeMammalian expression of EK0285 with CMV-tetO promoter instead of SFFV.DepositorInsertCMV-TO:mCherry:s70587:EGFP
ExpressionMammalianMutationWTPromoterCMV-TOAvailable SinceOct. 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
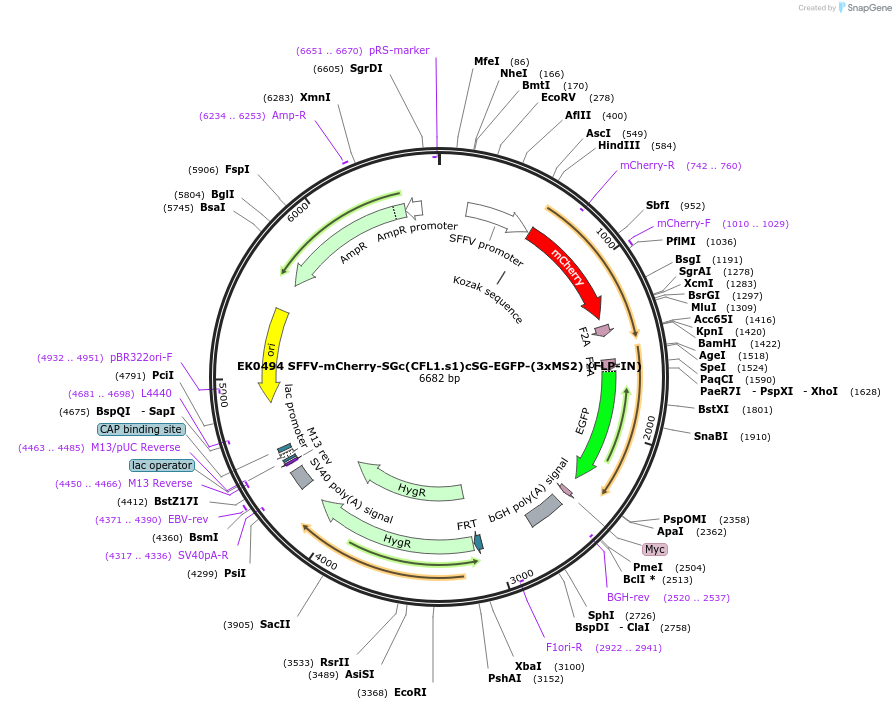
EK0494 SFFV-mCherry-SGc(CFL1.s1)cSG-EGFP-(3xMS2) (FLP-IN)
Plasmid#191181PurposeExpression of CFL1 sensor in mammalian cellsDepositorInsertmCherry:CFL1.s1:EGFP-3xMS2
ExpressionMammalianMutationWTPromoterSFFVAvailable SinceOct. 26, 2022AvailabilityAcademic Institutions and Nonprofits only