We narrowed to 9,265 results for: yeast expression
-
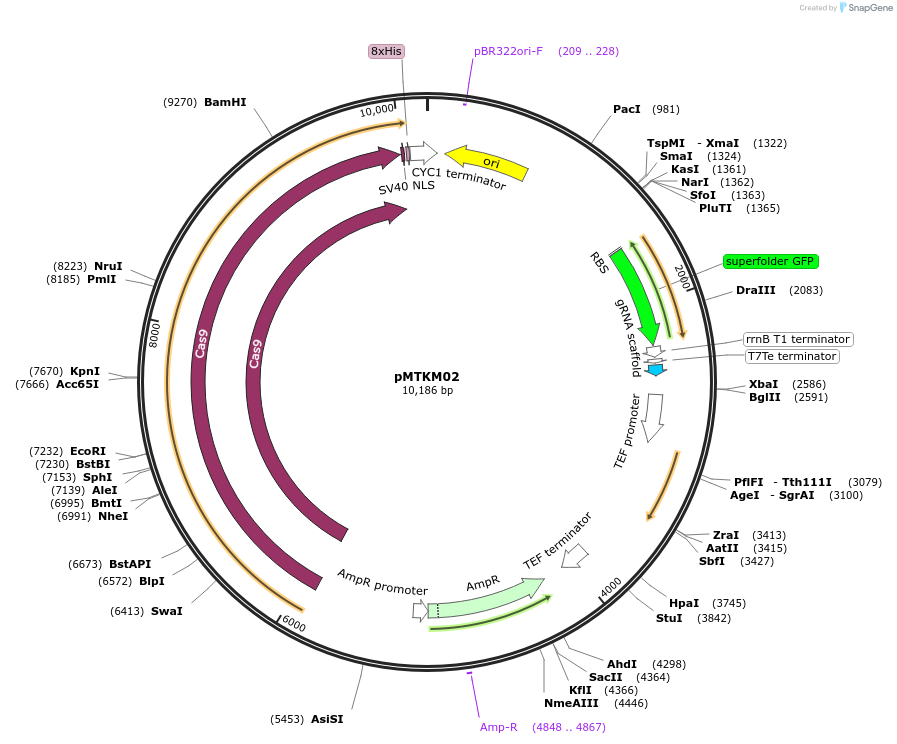
Plasmid#233474PurposeSpCAS9-Tyr-[gRNA: BsaI GFP dropout] with pan(OPT)ARS replication origin and split-Nat selection.DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyTagsExpressionYeastMutationPromoterAvailable SinceJuly 2, 2025AvailabilityAcademic Institutions and Nonprofits only
-
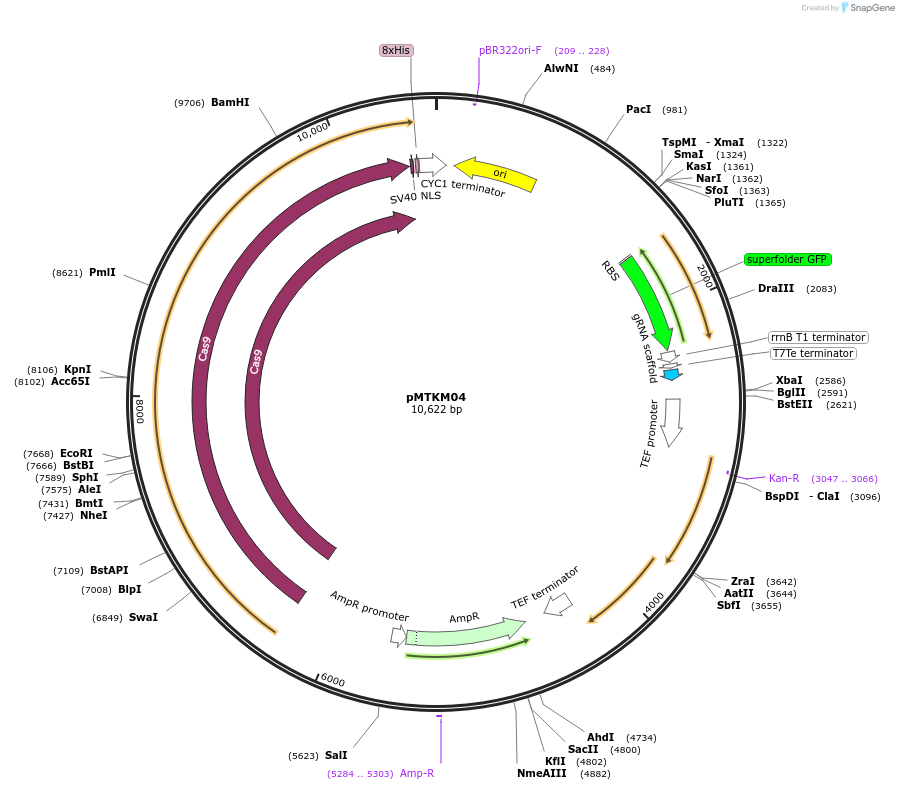
pMTKM04
Plasmid#233476PurposeSpCAS9-Tyr-[gRNA: BsaI GFP dropout] with pan(OPT)ARS replication origin and split-G418 selection.DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyTagsExpressionYeastMutationPromoterAvailable SinceJuly 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
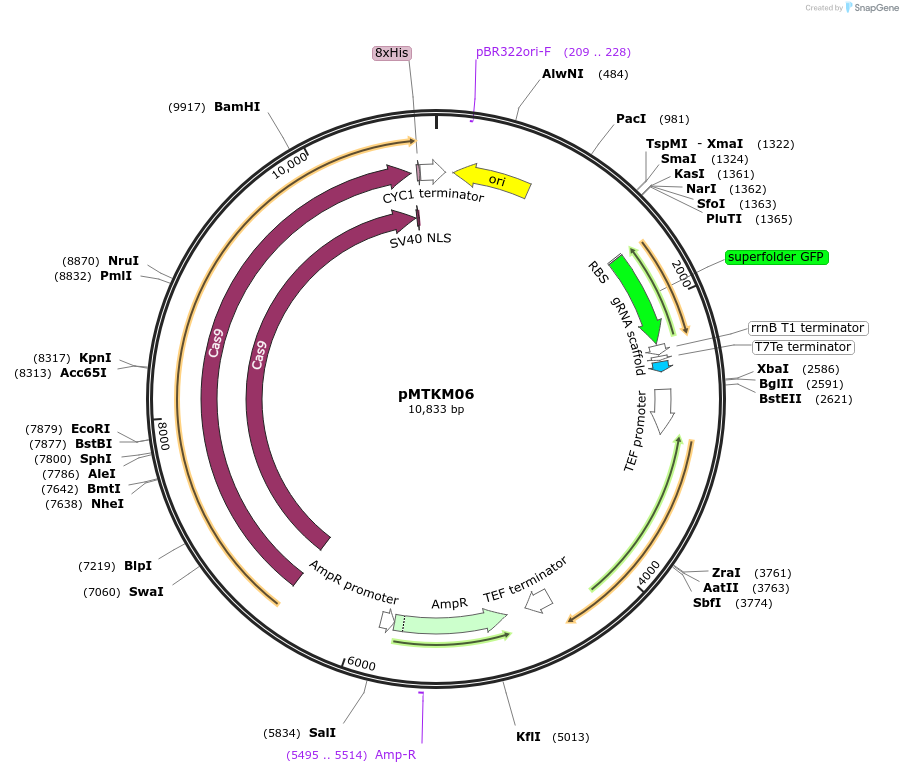
pMTKM06
Plasmid#233478PurposeSpCAS9-Tyr-[gRNA: BsaI GFP dropout] with pan(OPT)ARS replication origin and split-Hyg selection.DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyTagsExpressionYeastMutationPromoterAvailable SinceJuly 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
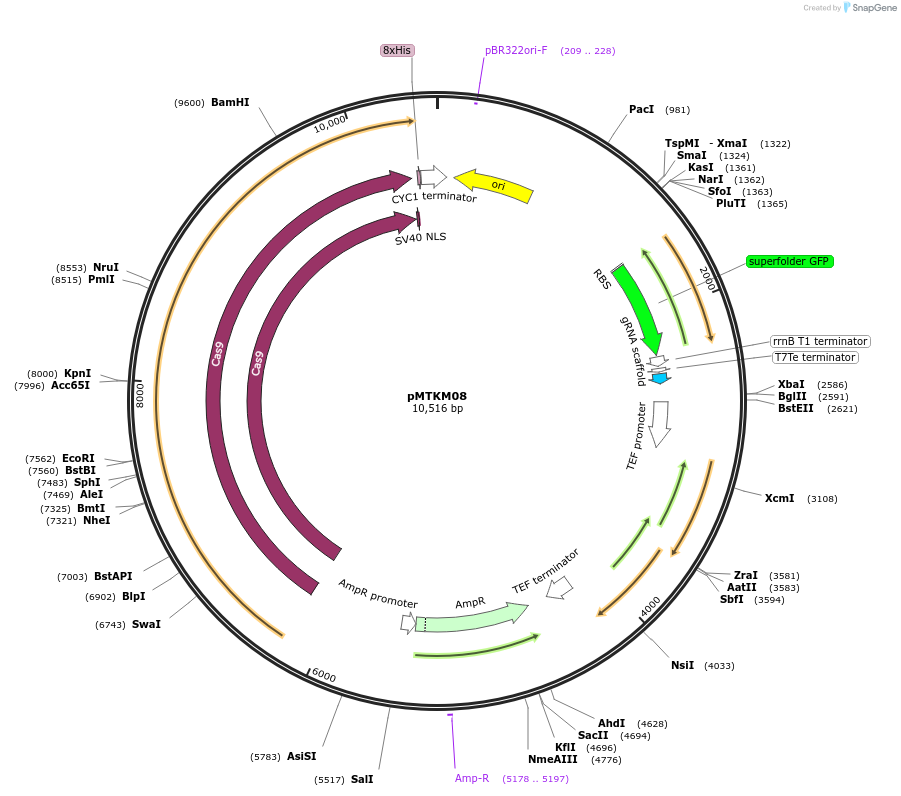
pMTKM08
Plasmid#233480PurposeSpCAS9-Tyr-[gRNA: BsaI GFP dropout] with pan(OPT)ARS replication origin and split-URA selection.DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyTagsExpressionYeastMutationPromoterAvailable SinceJuly 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
185_pETcon_SARS2_BQ1-1
Plasmid#208085Purposeyeast surface display of the SARS-CoV-2 BQ.1.1 variant RBDDepositorInsertSARS-CoV-2 BQ.1.1 RBD (S SARS-CoV-2 virus, Budding Yeast)
UseTagsHA and c-MycExpressionYeastMutationPromoterAvailable SinceOct. 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
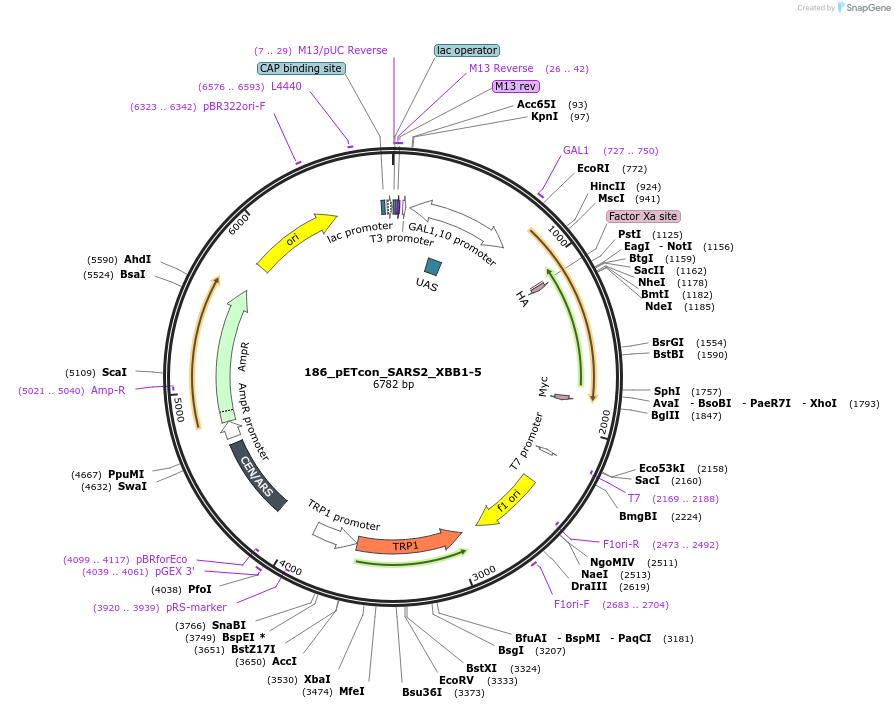
186_pETcon_SARS2_XBB1-5
Plasmid#208086Purposeyeast surface display of the SARS-CoV-2 XBB.1.5 variant RBDDepositorInsertSARS-CoV-2 XBB.1.5 RBD (S SARS-CoV-2 virus, Budding Yeast)
UseTagsHA and c-MycExpressionYeastMutationPromoterAvailable SinceOct. 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
SEC7-2EGFP-ter-NAT
Plasmid#184764PurposeMarker for yeast late Golgi/early endosome . Integration of 2xGFP tag at SEC7 C terminus. Uses antibiotic resistance marker natMX6.DepositorAvailable SinceFeb. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
CIT1-2EGFP-ter-URA3
Plasmid#184767PurposeMarker for yeast mitochondria. Integration of 2xGFP tag at CIT1 C terminus. Uses auxotrophic marker URA3(Candida albicans).DepositorAvailable SinceFeb. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
EMC1-2EGFP-ter-NAT
Plasmid#184763PurposeMarker for yeast endoplasmic reticulum (ER). Integration of 2xGFP tag at EMC1 C terminus. Uses antibiotic resistance marker natMX6.DepositorAvailable SinceFeb. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
ACH1-2EGFP-ter-URA3
Plasmid#184769PurposeMarker for yeast mitochondria. Integration of 2xGFP tag at ACH1 C terminus. Uses auxotrophic marker URA3(Candida albicans).DepositorAvailable SinceFeb. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
AFG3-2EGFP-ter-URA3
Plasmid#184770PurposeMarker for yeast mitochondria. Integration of 2xGFP tag at AFG3 C terminus. Uses auxotrophic marker URA3(Candida albicans).DepositorAvailable SinceFeb. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
SYS1-2EGFP-ter-URA3
Plasmid#184771PurposeMarker for yeast early Golgi. Integration of 2xGFP tag at SYS1 C terminus. Uses auxotrophic marker URA3(Candida albicans).DepositorAvailable SinceFeb. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
SEC7-2Katushka-ter-Hygromycin
Plasmid#184772PurposeRed marker for yeast late Golgi/early endosome. Integration of 2xKatushka tag at SEC7 C terminus. Uses antibiotic resistance marker hphMX6.DepositorAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
pSCL.040
Plasmid#184974PurposeTest effect of extending a1/a2 on ADE2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 27 v2
UseTagsExpressionYeastMutationADE2 donor P272stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
pSCL.109
Plasmid#184978PurposeTest effect of extending a1/a2 on LYP1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 27 v1
UseTagsExpressionYeastMutationLYP1 donor E27stop, a1/a2 length extended to 27 bpPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
pSCL.107
Plasmid#184976PurposeTest effect of extending a1/a2 on CAN1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, CAN1 G444X, a1/a2 length: 27 v1
UseTagsExpressionYeastMutationCAN1 donor G444stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
pSCL.111
Plasmid#184980PurposeTest effect of extending a1/a2 on TRP2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, TRP2 E64X, a1/a2 length: 27 v1
UseTagsExpressionYeastMutationTRP2 donor E64stop, a1/a2 length extended to 27 bpPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
pSCL.113
Plasmid#184982PurposeTest effect of extending a1/a2 on FAA1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, FAA1 P233X, a1/a2 length: 27 v1
UseTagsExpressionYeastMutationFAA1 donor P233stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
pSCL.039
Plasmid#184973PurposeTest effect of extending a1/a2 on ADE2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 27 v1
UseTagsExpressionYeastMutationADE2 donor P272stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
pGAD-Atg17(L313D I314K)
Plasmid#121390PurposeFor Yeast-two-hybrid analysisDepositorInsertATG17
UseTagsExpressionYeastMutationLeucine 313 to Aspartic acid; Isoleucine 314 to L…PromoterADH1 promoterAvailable SinceMarch 8, 2019AvailabilityAcademic Institutions and Nonprofits only