We narrowed to 4,562 results for: TIL
-
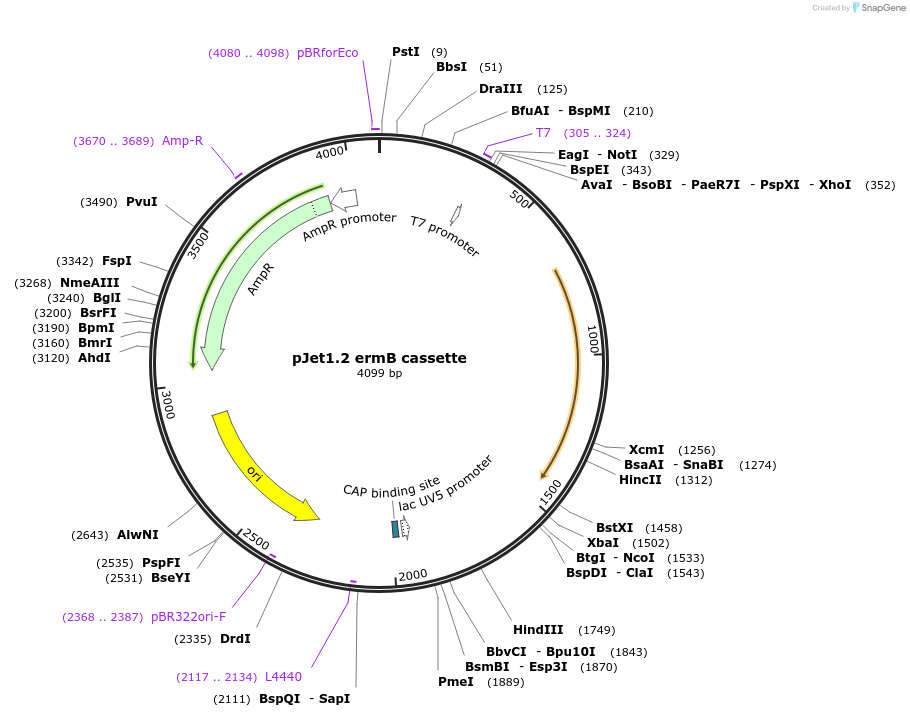
Plasmid#117121PurposeErythromycin resistance cassetteDepositorInsertErythromycin resistance cassette for Bacillus subtilis selection
UseGolden gate donor vector for gene targeting in ba…Available SinceNov. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
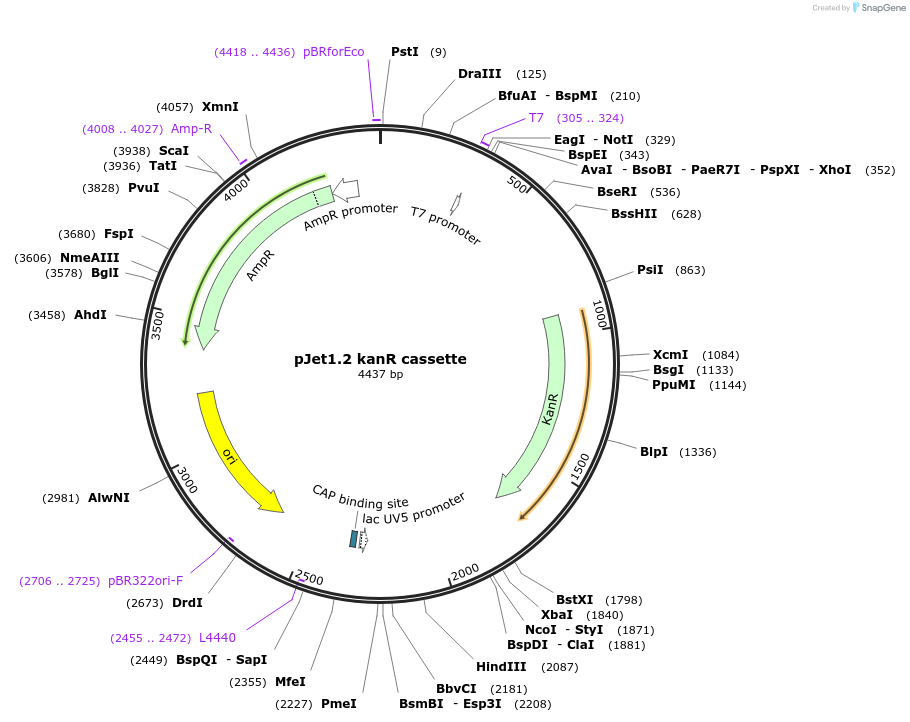
pJet1.2 kanR cassette
Plasmid#117122PurposeKanamycin resistance cassetteDepositorInsertKanamycin resistance cassette for Bacillus subtilis selection
UseGolden gate donor vector for gene targeting in ba…Available SinceDec. 3, 2018AvailabilityAcademic Institutions and Nonprofits only -
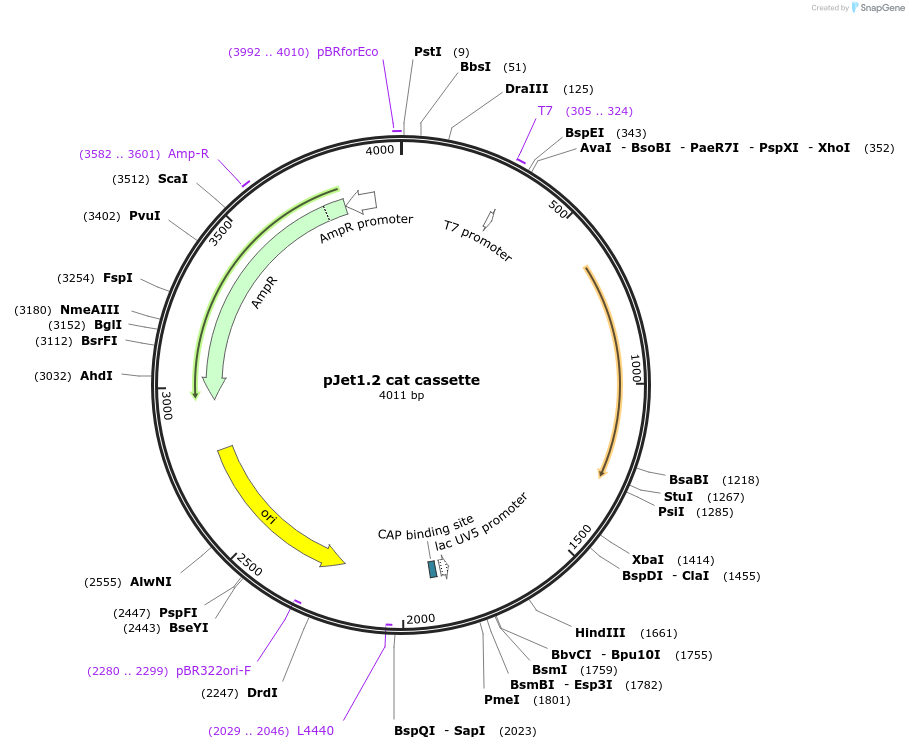
pJet1.2 cat cassette
Plasmid#117119PurposeChloramphenicol resistance cassetteDepositorInsertChloramphenicol resistance cassette for Bacillus subtilis selection
UseGolden gate donor vector for gene targeting in ba…Available SinceNov. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
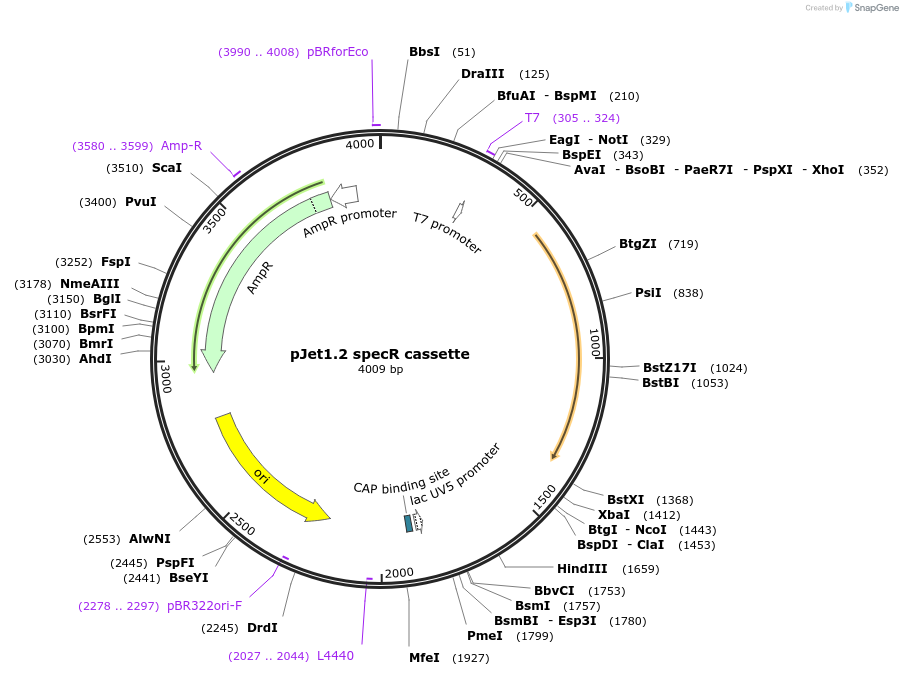
pJet1.2 specR cassette
Plasmid#117120PurposeSpectinomycin resistance cassetteDepositorInsertSpectinomycin resistance cassette for Bacillus subtilis selection
UseGolden gate donor vector for gene targeting in ba…Available SinceNov. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
BS.YxiN.Twin1.a1-479.E152Q
Plasmid#17518DepositorInsertB. subtilis DEAD-box helicase YxiN
Tagsintein-chitin binding domainExpressionBacterialMutationE152Q mutationAvailable SinceMarch 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
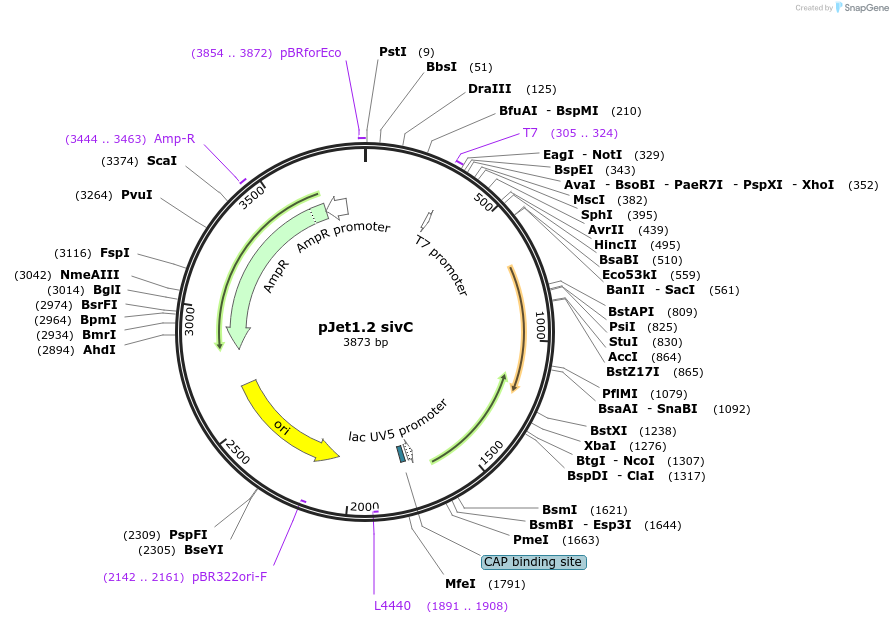
pJet1.2 sivC
Plasmid#117131Purposesmc downstream homology regionDepositorInsertsivC
UseGolden gate donor vector for gene targeting in ba…Available SinceNov. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
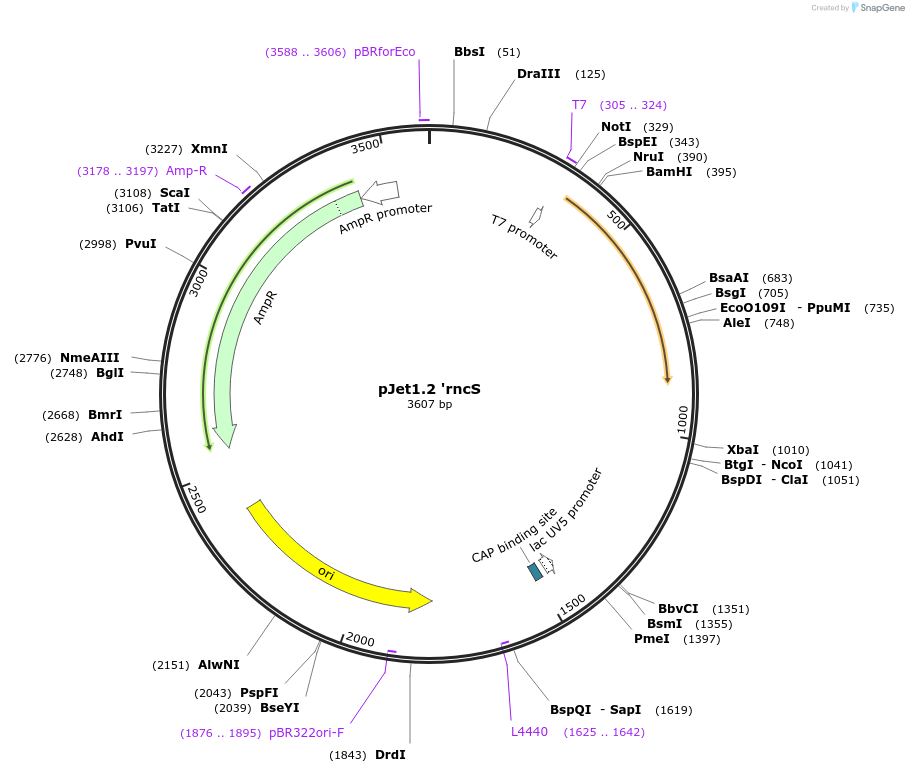
pJet1.2 'rncS
Plasmid#117129Purposesmc gene upstream homology regionDepositorInsert'rncS
UseGolden gate donor vector for gene targeting in ba…Available SinceNov. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
BS.YxiN.Twin1.a207-479.wt
Plasmid#17516DepositorInsertB. subtilis DEAD-box helicase YxiN second domain
Tagsintein-chitin binding domainExpressionBacterialMutationdeleted amino acid residues 1-206Available SinceMarch 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
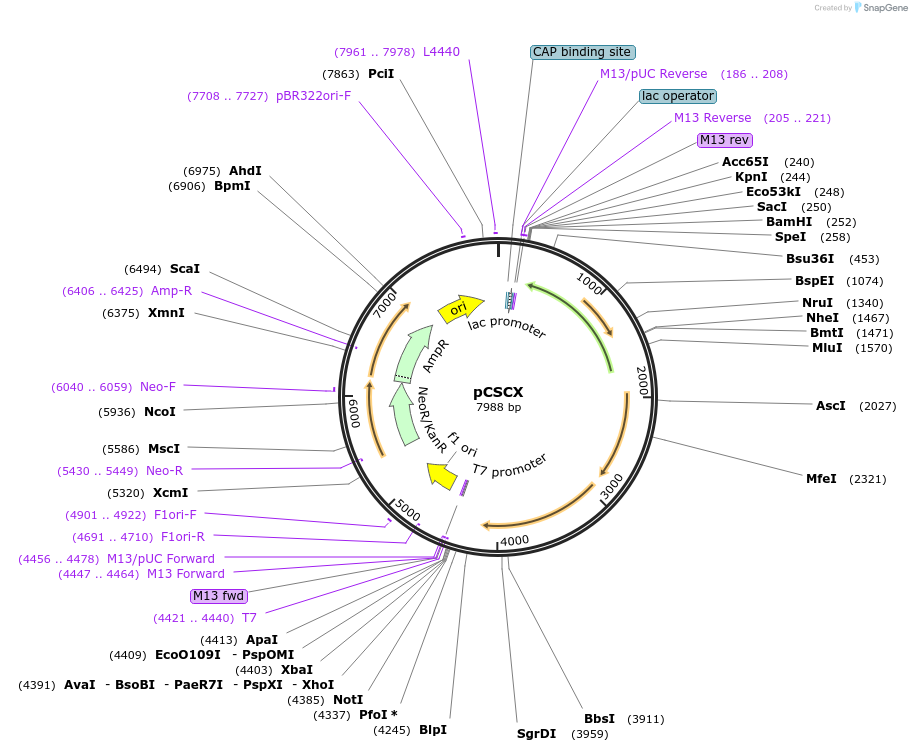
pCSCX
Plasmid#63918PurposeExpression of the sucrose utilization phenotypeDepositorInsertcscAKB
UseBacterial cloning vectorAvailable SinceMay 19, 2015AvailabilityAcademic Institutions and Nonprofits only -
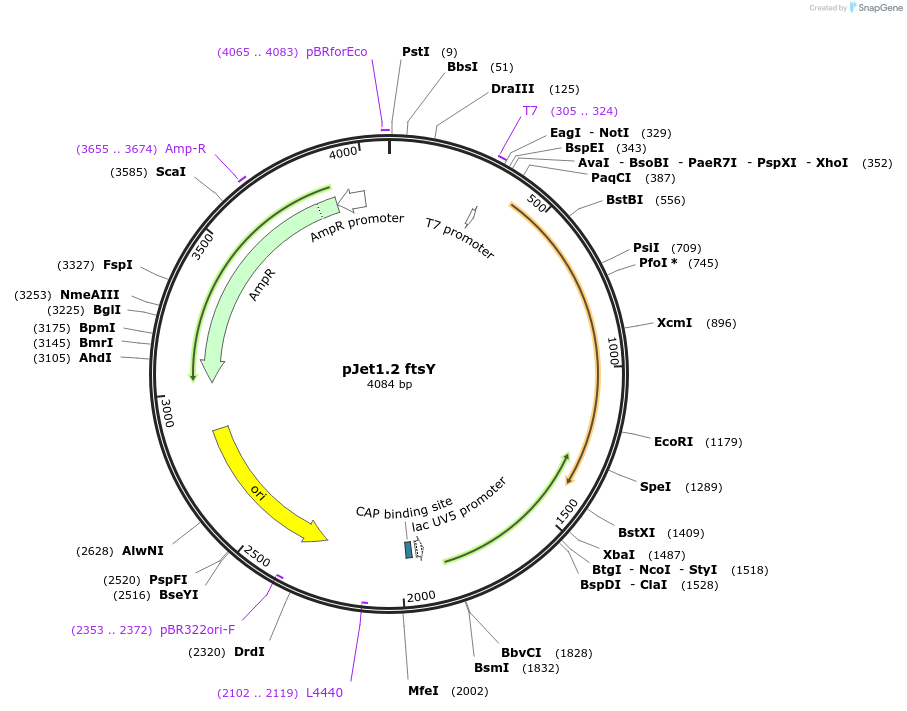
pJet1.2 ftsY
Plasmid#117130PurposeFtsY gene cassetteDepositorInsertftsY
UseGolden gate donor vector for gene targeting in ba…Available SinceNov. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
BS.YxiN.Twin1.a1-368.E152Q
Plasmid#17517DepositorInsertB. subtilis DEAD-box helicase YxiN first two domains
Tagsintein-chitin binding domainExpressionBacterialMutationdeleted amino acid residues 369-479; E152Q mutat…Available SinceMarch 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
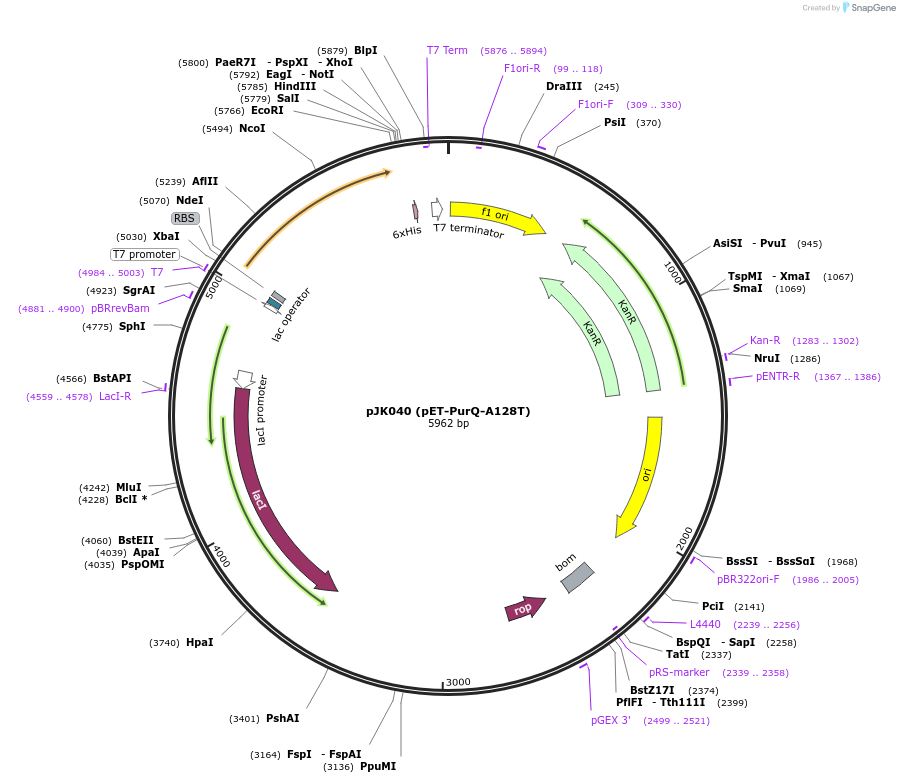
pJK040 (pET-PurQ-A128T)
Plasmid#73840PurposeProduces Bacillus subtilis formylglycinamidine ribonucleotide synthetase glutaminase subunit, A128T mutant (PurQ-A128T)DepositorInsertformylglycinamidine ribonucleotide synthetase, glutaminase subunit
ExpressionBacterialMutationA128T mutantPromoterT7Available SinceApril 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
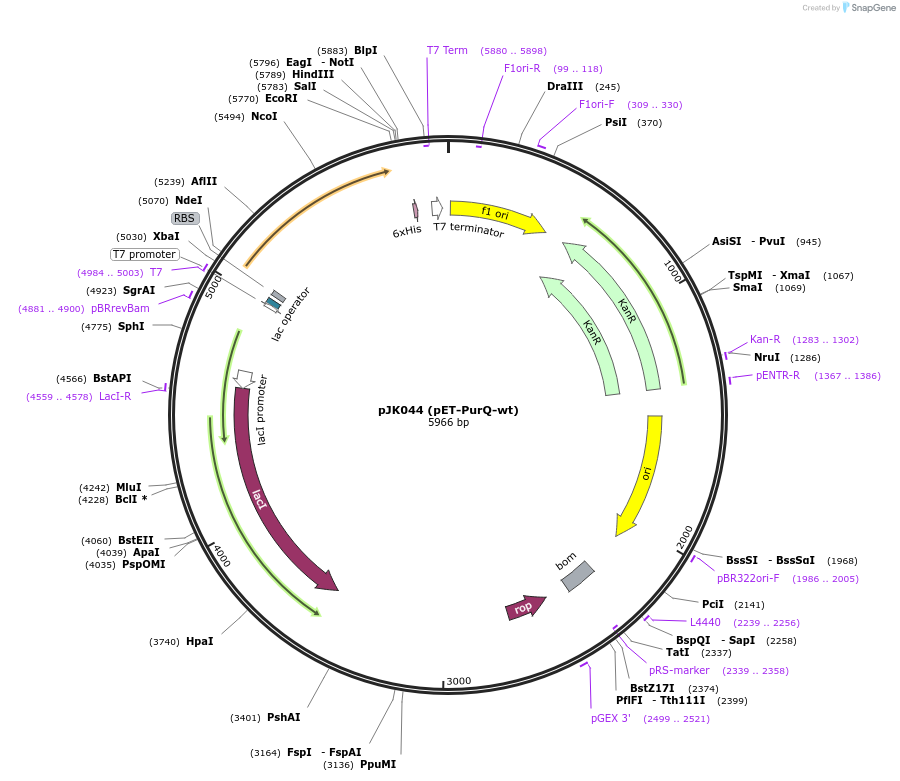
pJK044 (pET-PurQ-wt)
Plasmid#73839PurposeProduces Bacillus subtilis formylglycinamidine ribonucleotide synthetase glutaminase subunit, (PurQ)DepositorInsertformylglycinamidine ribonucleotide synthetase, glutaminase subunit
ExpressionBacterialPromoterT7Available SinceApril 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
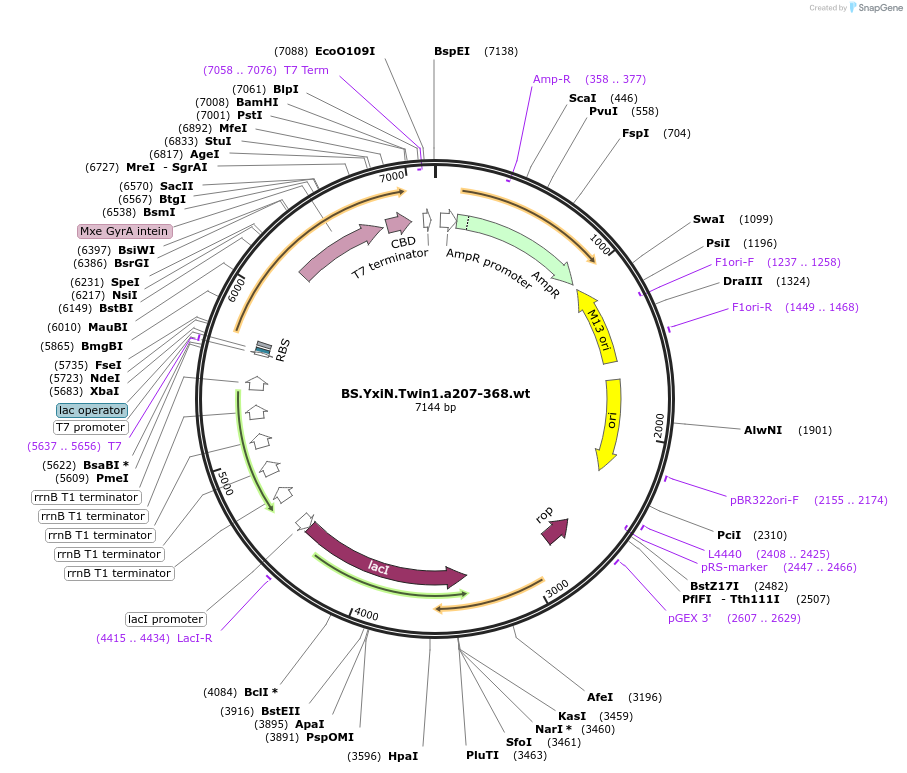
BS.YxiN.Twin1.a207-368.wt
Plasmid#13531DepositorInsertB. subtilis DEAD-box helicase YxiN second domain
Tagsintein-chitin binding domainExpressionBacterialMutationdeleted amino acid residues 1-206 and 369-479Available SinceJan. 4, 2007AvailabilityAcademic Institutions and Nonprofits only -
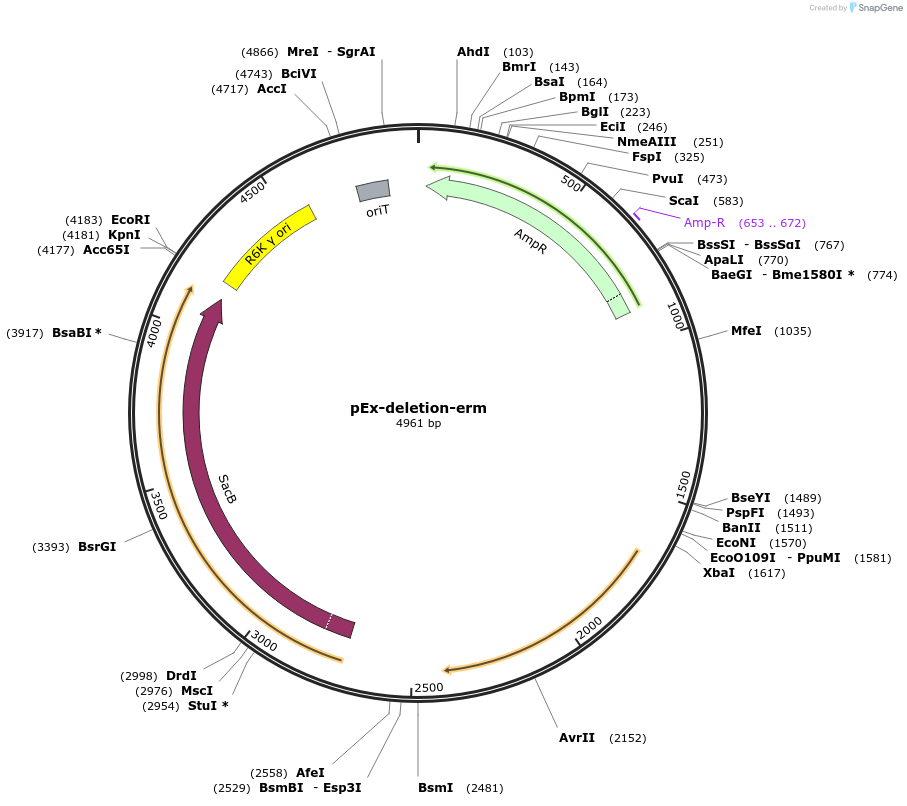
pEx-deletion-erm
Plasmid#172213PurposeFor allelic exchange in erythromycin-sensitive Prevotella copri strainsDepositorTypeEmpty backboneUseAllelic exchange in prevotella copriAvailable SinceMay 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
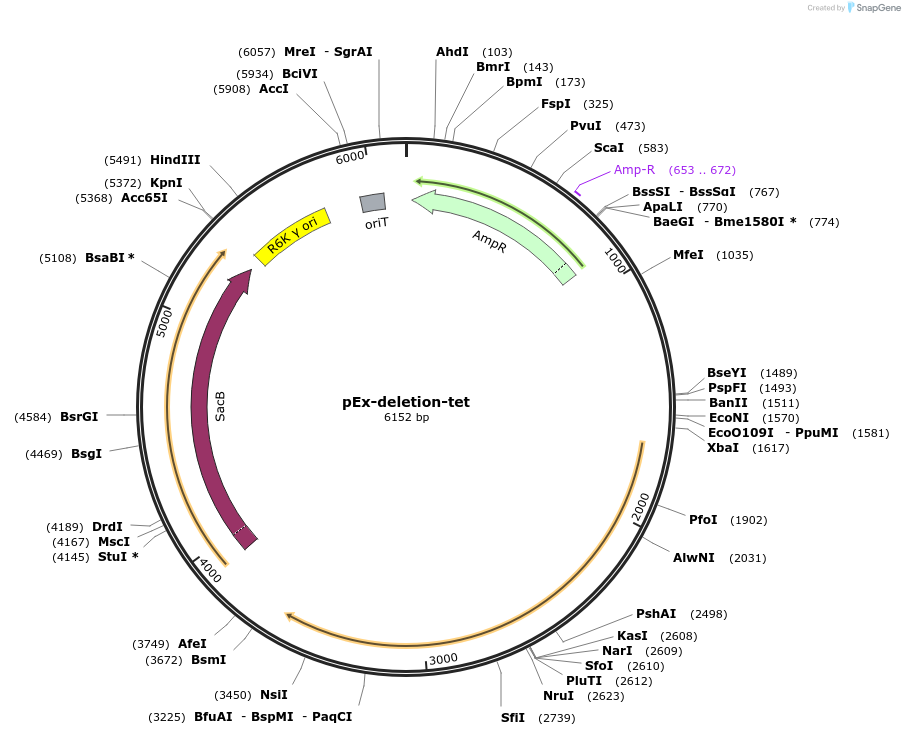
pEx-deletion-tet
Plasmid#172214PurposeFor allelic exchange in tetracyclin-sensitive Prevotella copri strainsDepositorTypeEmpty backboneUseAllelic exchange in prevotella copriAvailable SinceApril 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
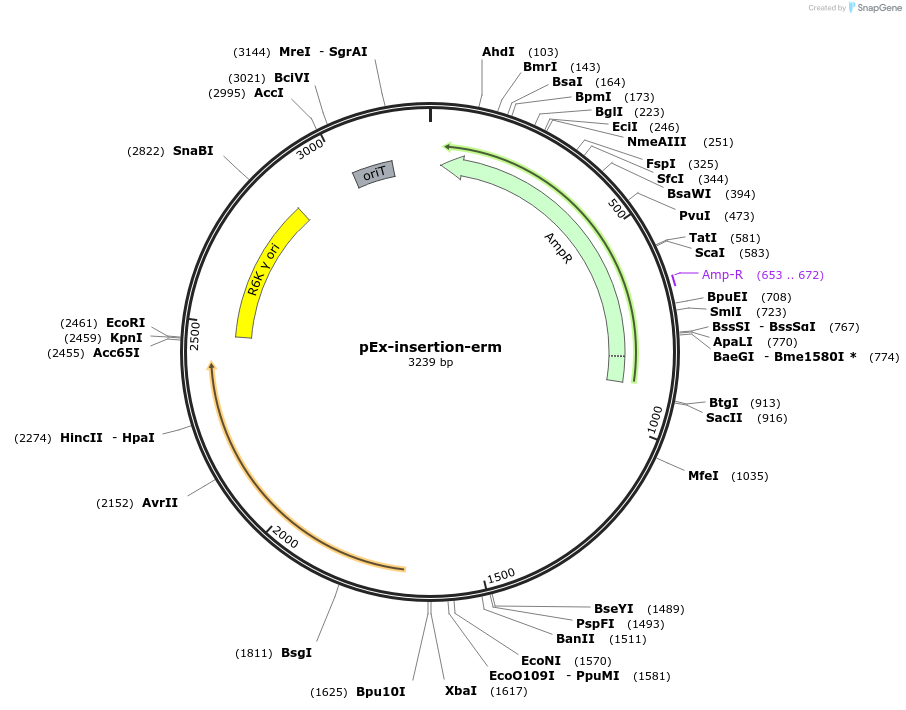
pEx-insertion-erm
Plasmid#172210PurposeFor genetic insertion in erythromycin-sensitive Prevotella copri strainsDepositorTypeEmpty backboneUseGenetic insertion in prevotella copriAvailable SinceApril 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
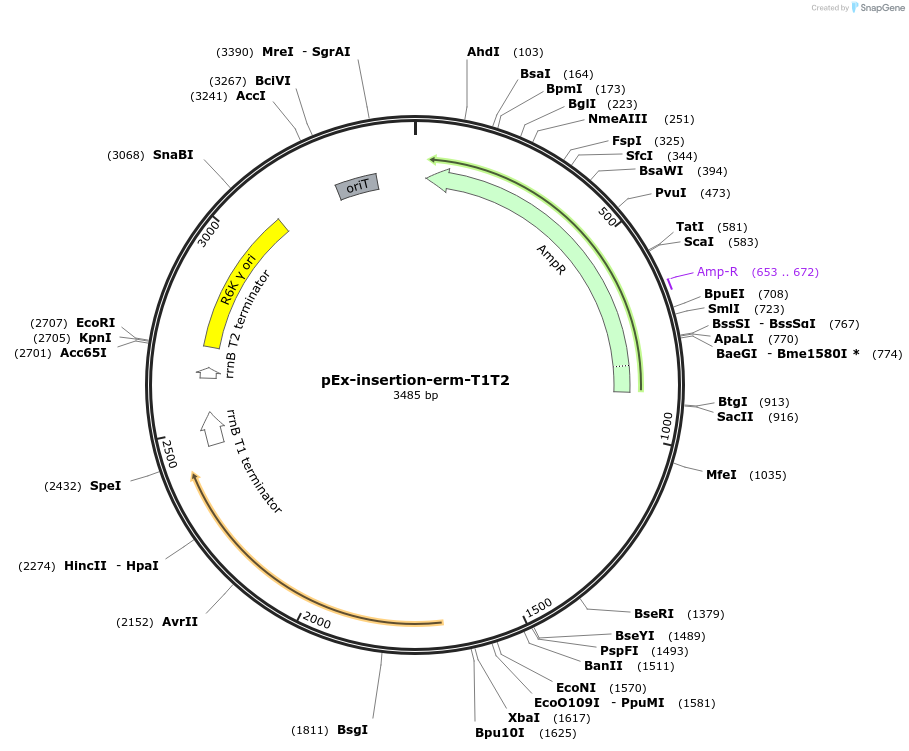
pEx-insertion-erm-T1T2
Plasmid#172212PurposeFor genetic insertion in erythromycin-sensitive Prevotella copri strains, with T1-T2 terminatorDepositorTypeEmpty backboneUseGenetic insertion in prevotella copriAvailable SinceApril 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
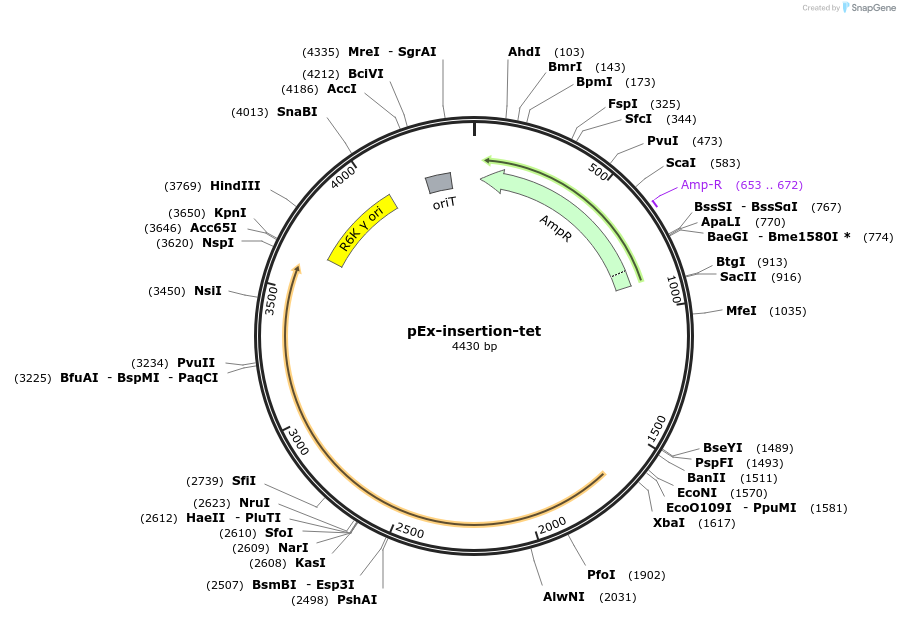
pEx-insertion-tet
Plasmid#172211PurposeFor genetic insertion in tetracyclin-sensitive Prevotella copri strainsDepositorTypeEmpty backboneUseGenetic insertion in prevotella copriAvailable SinceApril 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
p123shy1-mCherry-HA
Plasmid#67875Purposeencodes shy1 with a C-terminal mCherry-HA tag under control of its endogenous promoter.It's suitable for localization studies in Ustilago maydis.DepositorInsertshy1
UseExpression in ustilago maydisTagsHA and mCherryPromotershy1Available SinceAug. 7, 2015AvailabilityAcademic Institutions and Nonprofits only