We narrowed to 30,195 results for: REP
-
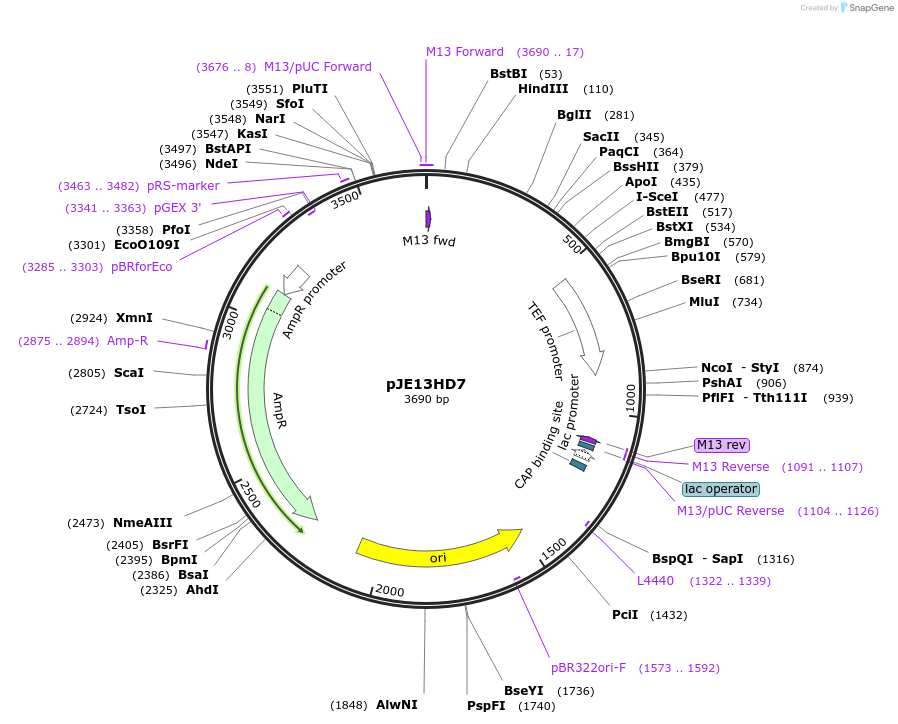
Plasmid#218281PurposeUse together with pITEdv1, or pToxEXP1/pToxAmp plasmids to insert heterologous gene(s) at ho locus.DepositorInsertHO(-253, -1)-Repeat1-ISceI-pAgTEF1>Hph(partial)
ExpressionYeastAvailable SinceMay 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
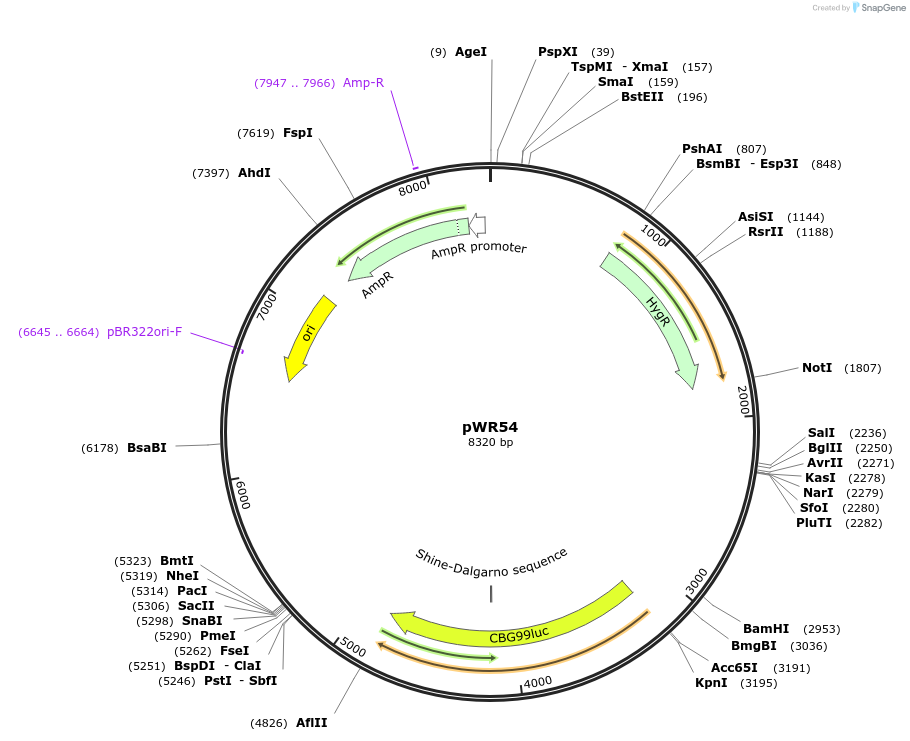
pWR54
Plasmid#203330Purposeepisomal bioluminescent reporter plasmid for S. stipitisDepositorInsertsTEF1 promoter (S. stipitis) (TEF1 S. stipitis (yeast))
coHPH
ACT1 terminator (S. stipitis)
CCW12 promoter (S. stipitis)
coCBG
ADH2 terminator (S. stipitis)
ARS from S. stipitis chromosome 1
ExpressionYeastAvailable SinceDec. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
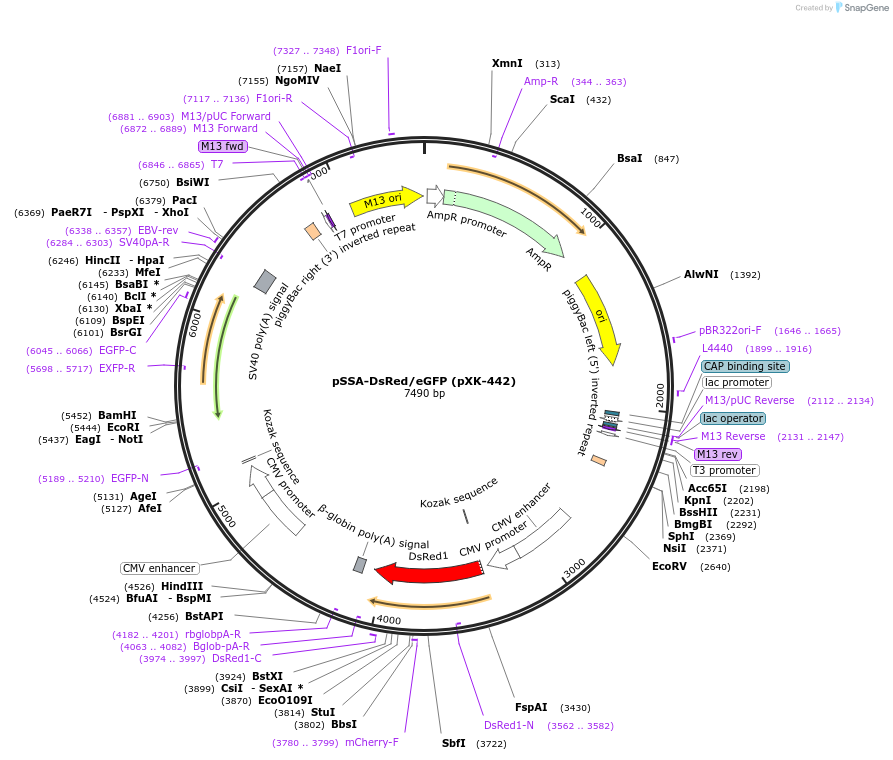
pSSA-DsRed/eGFP (pXK-442)
Plasmid#208827PurposeThe SSA-based surrogate reporter for validating the activity of different gene editing tools as well as for helping to screen gene-editing positive cells. Key Construct: CMV-DsRed-CMV-eGFP*(SSA).DepositorTypeEmpty backboneExpressionMammalianPromoterCMVAvailable SinceDec. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
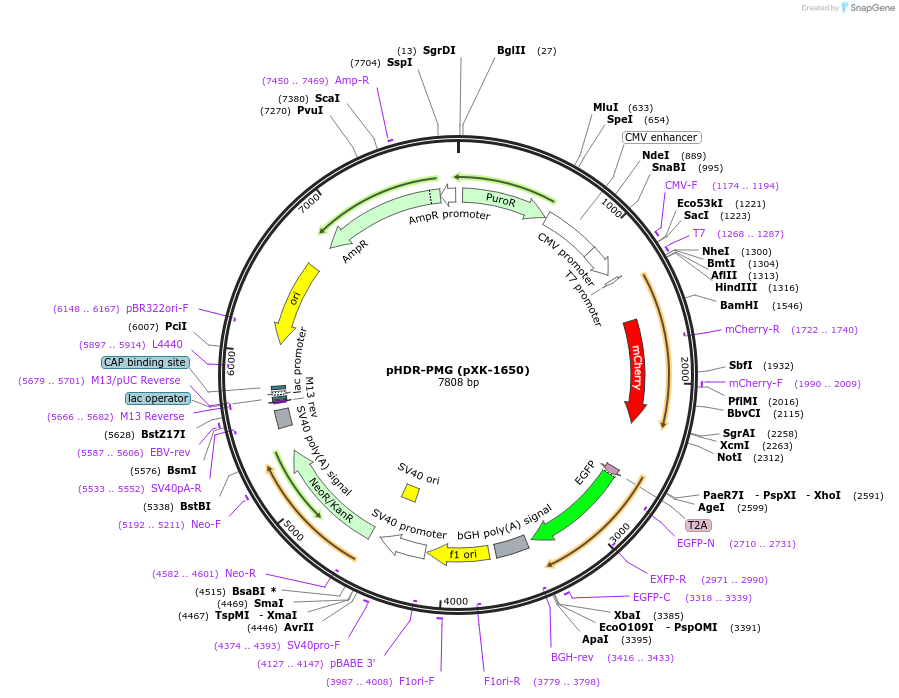
pHDR-PMG (pXK-1650)
Plasmid#208858PurposeHDR-base selectable traffic light reporter (TLR) for helping to screen HDR-editing positive cells. Key Construct: iPuroR*-CMV-PuroR.L-mCherry-PuroR.R-GFP.DepositorTypeEmpty backboneExpressionMammalianPromoterCMVAvailable SinceDec. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
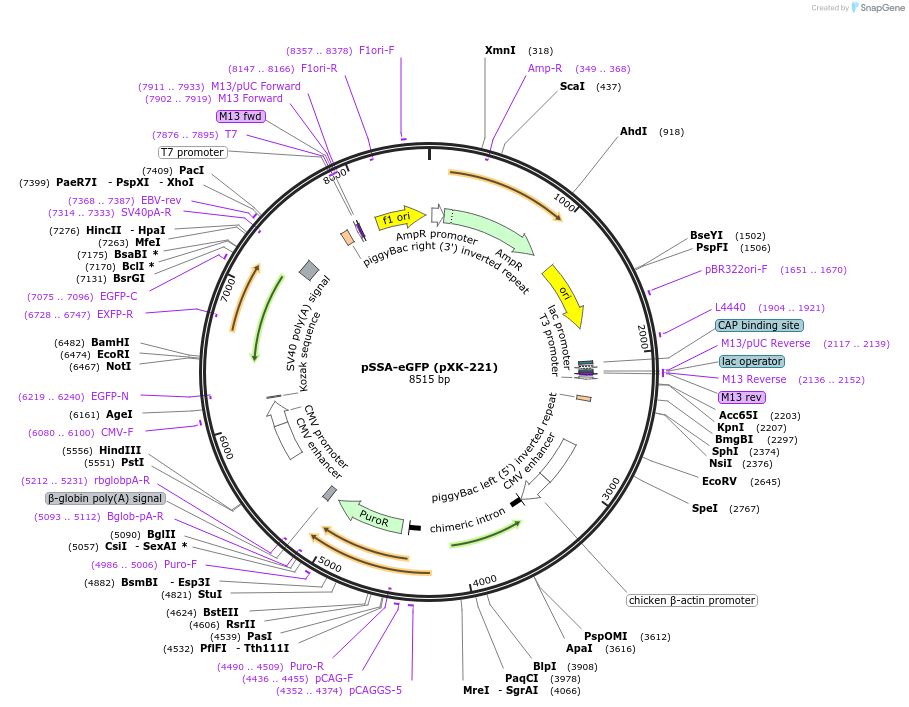
pSSA-eGFP (pXK-221)
Plasmid#208826PurposeThe SSA-based surrogate reporter for validating the activity of gene editing tools as well as for helping to screen gene-editing positive cells. Key Construct: CAG-PuroR-CMV-eGFP*(SSA).DepositorTypeEmpty backboneExpressionMammalianPromoterCMVAvailable SinceDec. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
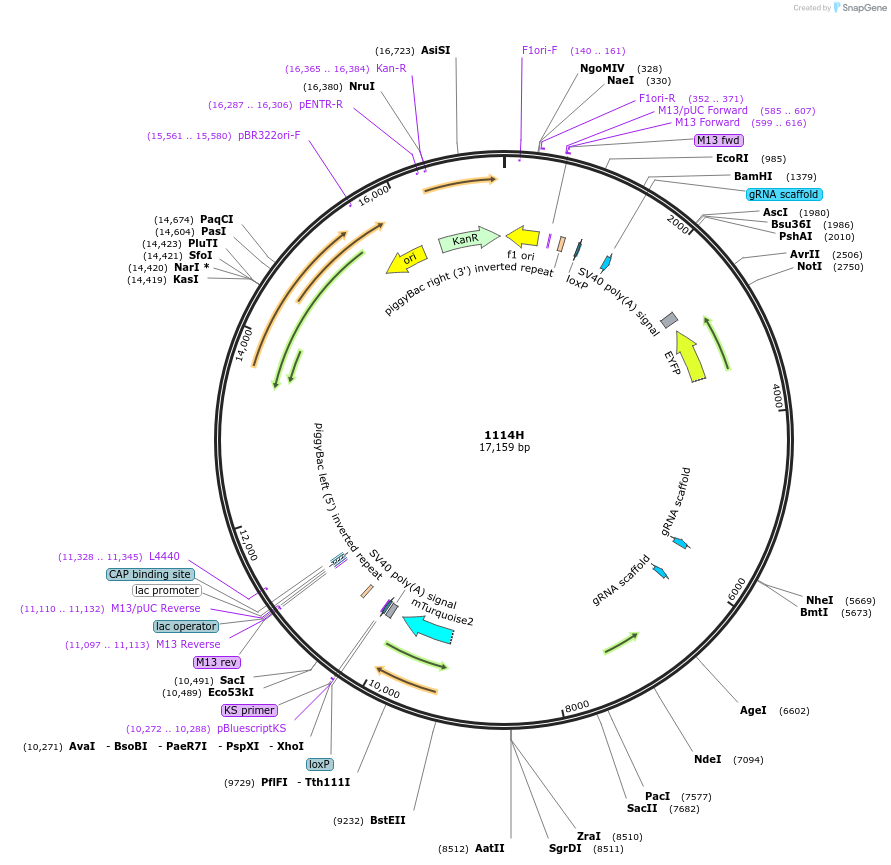
1114H
Plasmid#200640PurposeAn. gambiae transgenesis plasmid. Expresses gRNAs targeting B2-tubulin, ZPG, and DSXF. Crossing to Cas9 generates sterile malesDepositorInsertgRNA targeting DSXF, ZPG, and B2-tubulin. Actin5c-CFP and Vasa2-EYFP reporters.
UseCRISPRExpressionInsectAvailable SinceDec. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
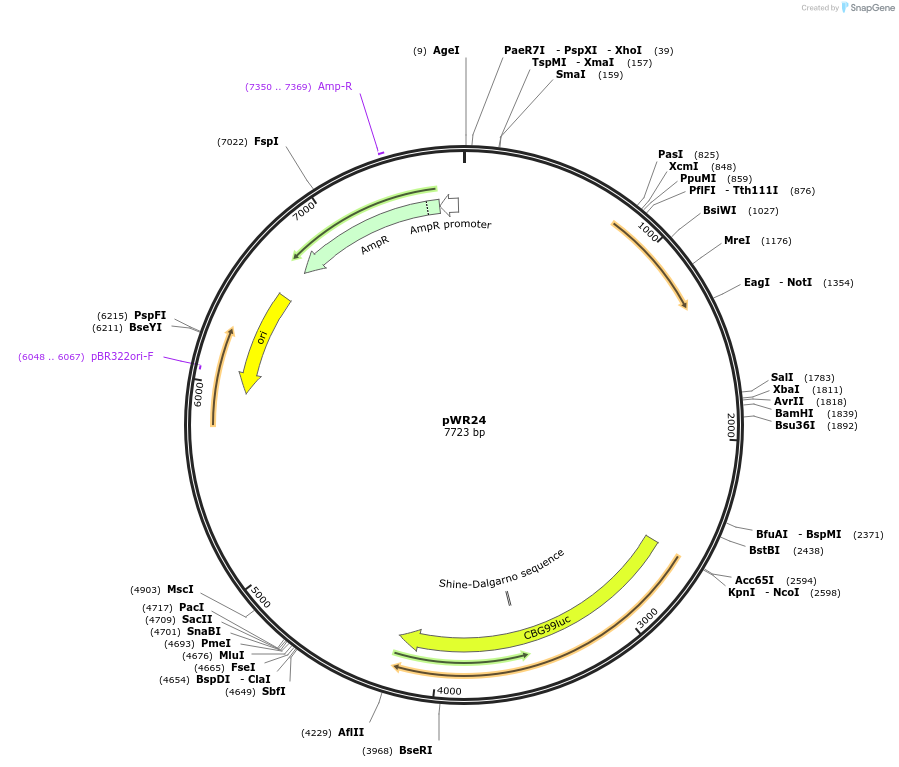
pWR24
Plasmid#202771Purposeepisomal bioluminescent reporter plasmid for S. stipitisDepositorInsertsTEF1 promoter (S. stipitis) (TEF1 S. stipitis (yeast))
coNAT
ACT1 terminator (S. stipitis)
TDH3 promoter (S. stipitis)
coCBG
ADH2 terminator (S. stipitis)
ARS from S. stipitis chromosome 1
ExpressionYeastAvailable SinceOct. 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
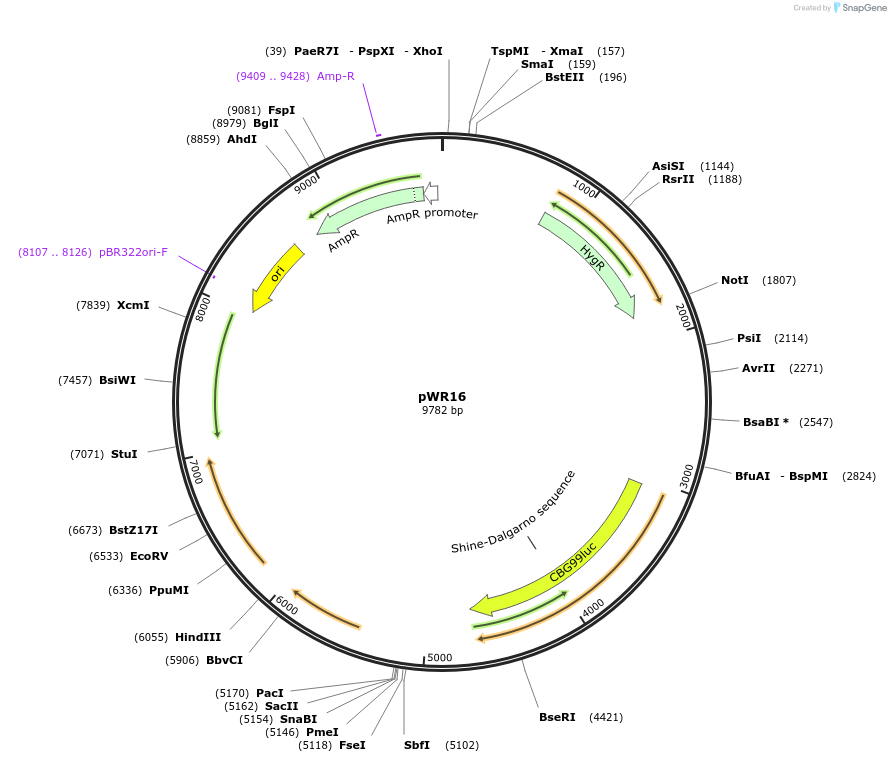
pWR16
Plasmid#202711Purposeintegrating bioluminescent reporter plasmid for S. stipitisDepositorInsertsTEF1 promoter (S. stipitis) (TEF1 S. stipitis (yeast))
coHPH
ACT1 terminator (S. stipitis)
TDH3 promoter (S. stipitis)
coCBG
ADH2 terminator (S. stipitis)
URA3 and flanking sequence
ExpressionYeastAvailable SinceOct. 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
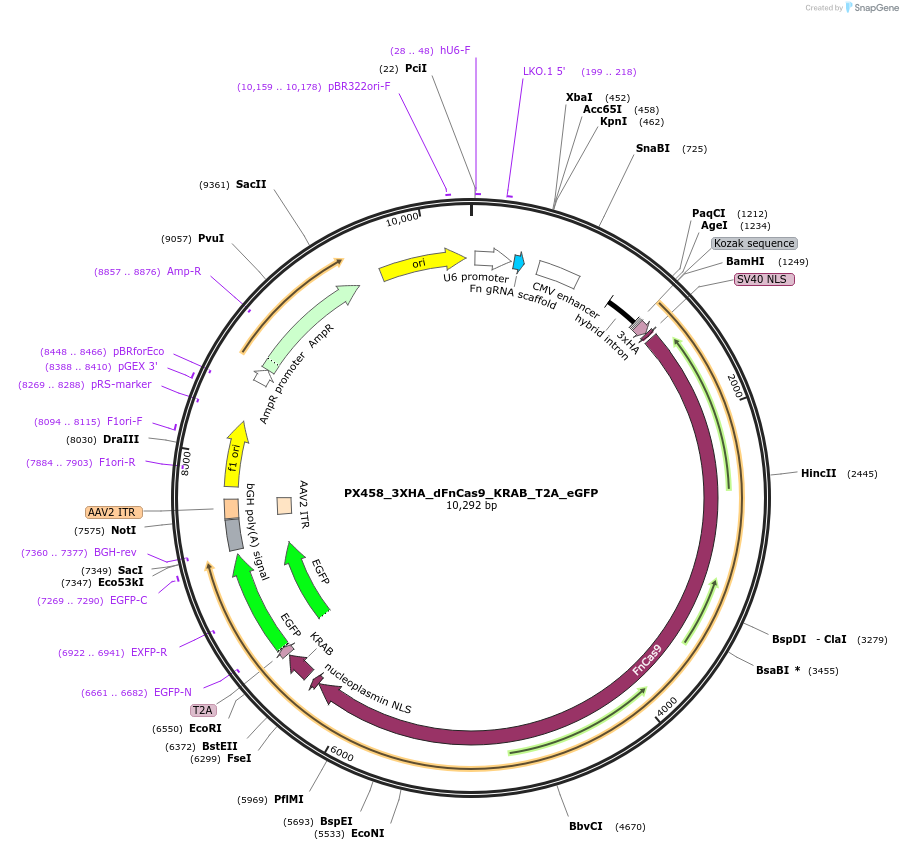
PX458_3XHA_dFnCas9_KRAB_T2A_eGFP
Plasmid#200389Purpose3XHA tagged dead Cas9 from F. novicida with KRAB repressor domain, T2A self-cleaving peptide and enhanced GFP biomarker for mammalian expressionDepositorInsertPX458_3XHA_dFnCas9_KRAB_eGFP
UseCRISPRTags3xHA-NLS and SV40NLS-KRAB-T2A-EGFPExpressionMammalianPromoterCBHAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
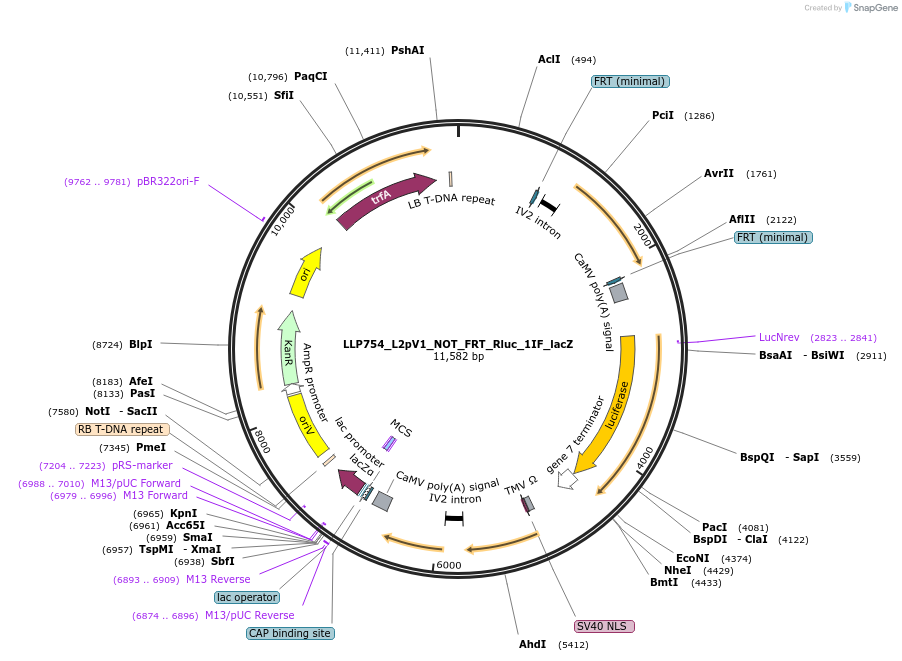
LLP754_L2pV1_NOT_FRT_Rluc_1IF_lacZ
Plasmid#192399PurposeOff state for the first generation of NOT gates, which target the Rluc CDS to repress expression via its FRT sites.DepositorInsertAct2::FRT-Rluc-FRT
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
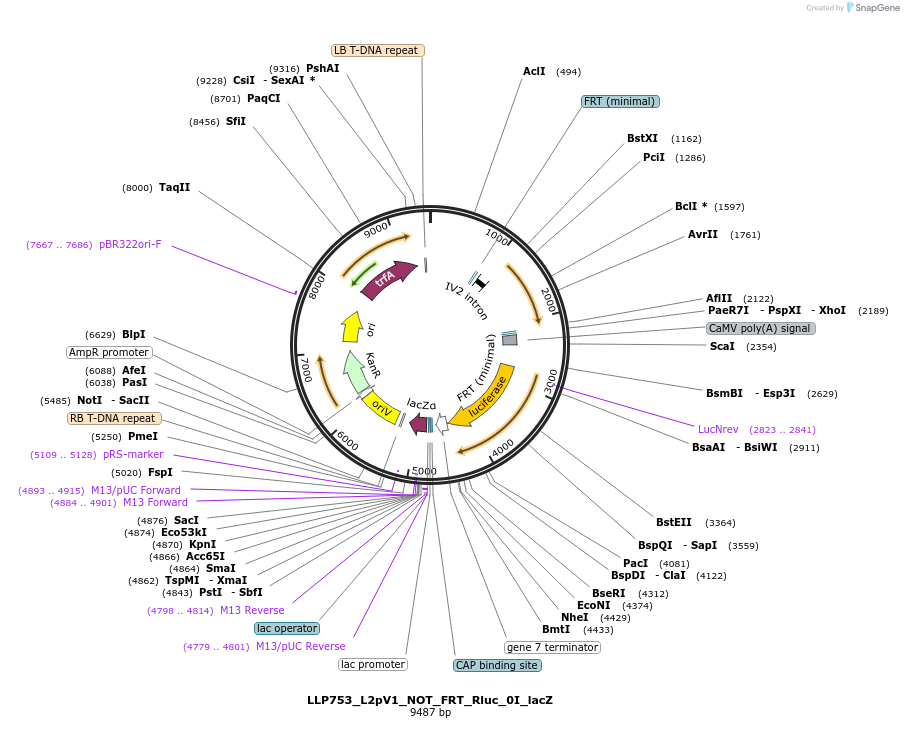
LLP753_L2pV1_NOT_FRT_Rluc_0I_lacZ
Plasmid#192398PurposeOn state for the first generation of NOT gates, which target the Rluc CDS to repress expression via its FRT sites.DepositorInsertAct2::FRT-Rluc-FRT
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
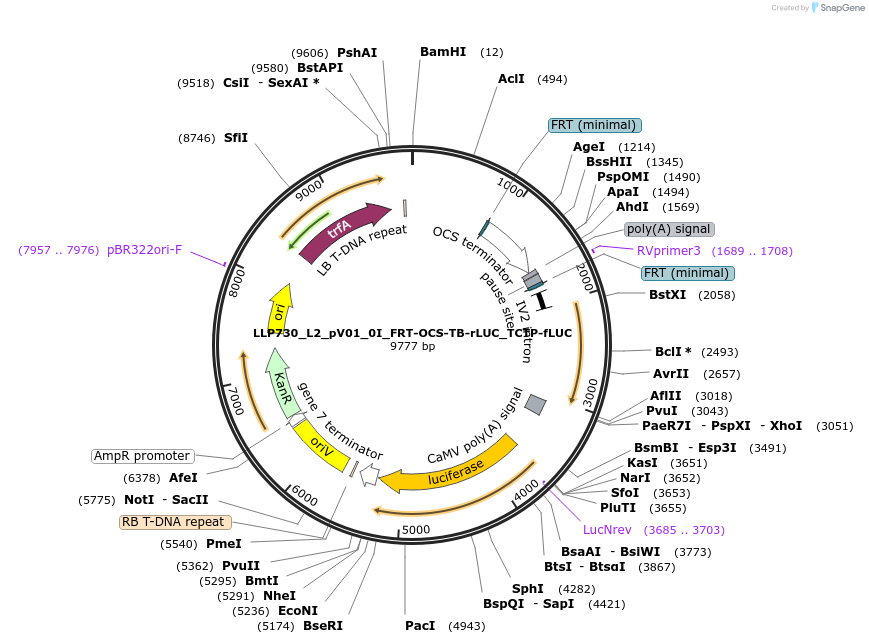
LLP730_L2_pV01_0I_FRT-OCS-TB-rLUC_TCTP-fLUC
Plasmid#192373PurposeTo test for the repressive potential of the OCS and Transcription Block (TB) fusion with the FRT sites around it.DepositorInsertAct2::FRT-OCS-TB-FRT::Rluc
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
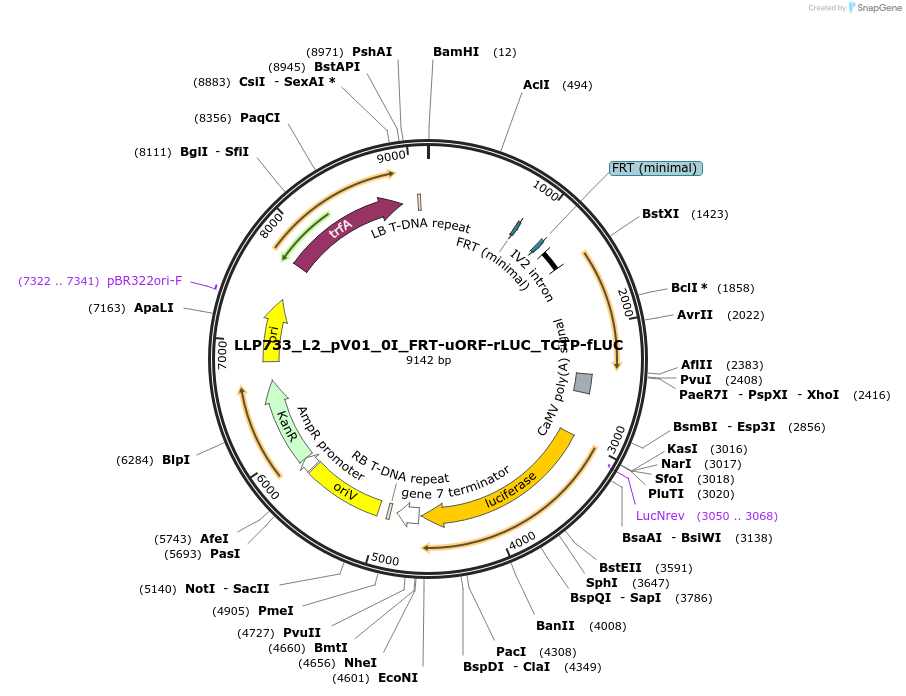
LLP733_L2_pV01_0I_FRT-uORF-rLUC_TCTP-fLUC
Plasmid#192376PurposeTo test for the repressive potential of the conserved peptide uORF from AT1G36730 with the FRT sites around it.DepositorInsertAct2::FRT-uORF-FRT::Rluc
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
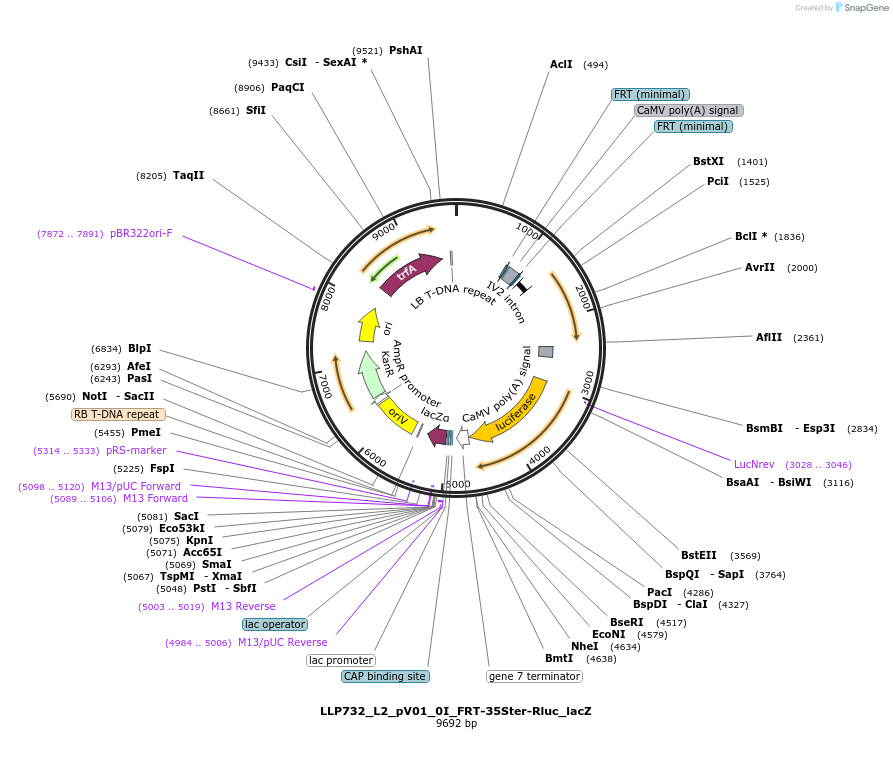
LLP732_L2_pV01_0I_FRT-35Ster-Rluc_lacZ
Plasmid#192375PurposeTo test for the repressive potential of the 35S terminator from CaMV with the FRT sites around it.DepositorInsertAct2::FRT-35Ster-FRT::Rluc
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
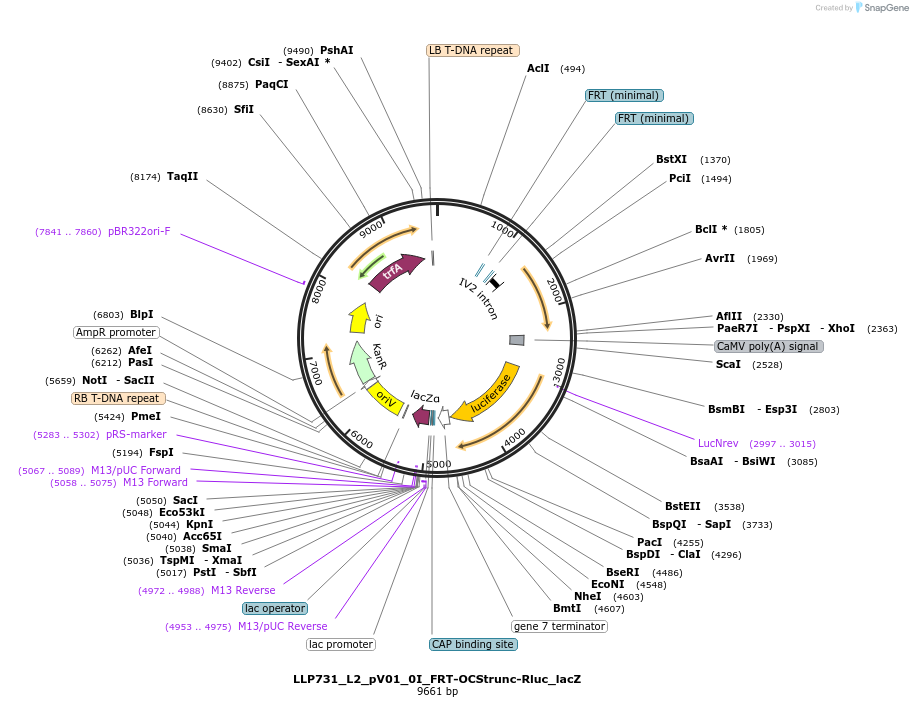
LLP731_L2_pV01_0I_FRT-OCStrunc-Rluc_lacZ
Plasmid#192374PurposeTo test for the repressive potential of the OCS variant (truncation) with the FRT sites around it.DepositorInsertAct2::FRT-OCStrunc-FRT::Rluc
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
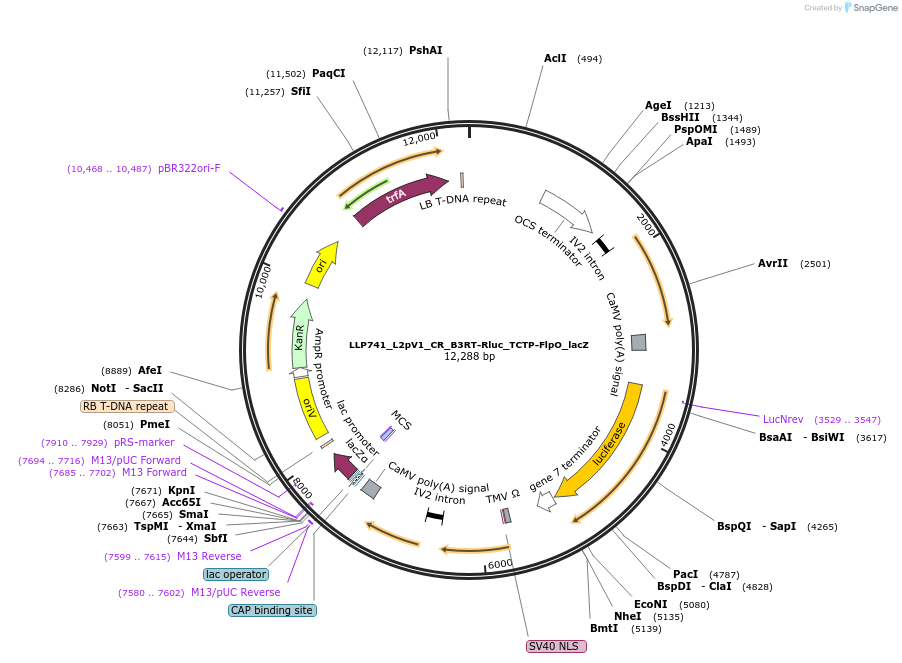
LLP741_L2pV1_CR_B3RT-Rluc_TCTP-FlpO_lacZ
Plasmid#192385PurposeTo test for if Flp is able to de-repress a circuit with B3RT, to rule out cross-reactivity between these recombinases.DepositorInsertAct2::B3RT-OCS-B3RT::Rluc
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
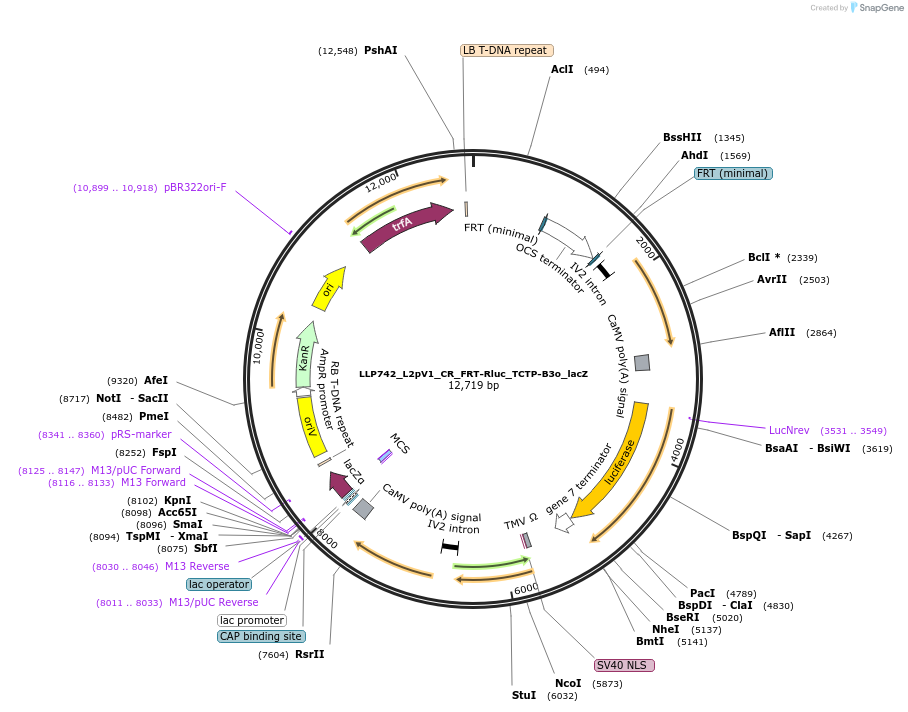
LLP742_L2pV1_CR_FRT-Rluc_TCTP-B3o_lacZ
Plasmid#192386PurposeTo test for if B3 is able to de-repress a circuit with FRT, to rule out cross-reactivity between these recombinases.DepositorInsertAct2::FRT-OCS-FRT::Rluc
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
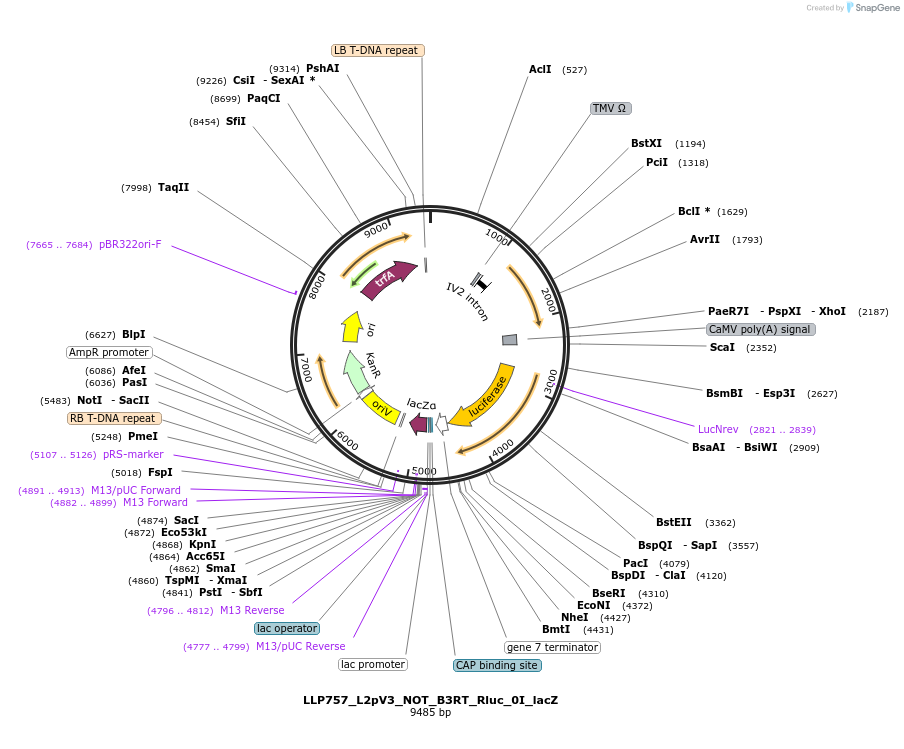
LLP757_L2pV3_NOT_B3RT_Rluc_0I_lacZ
Plasmid#192404PurposeOn state for the second generation of NOT gates, which target the Rluc promoter to repress expression via its B3RT sites.DepositorInsertB3RT-Act2-B3RT::Rluc
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
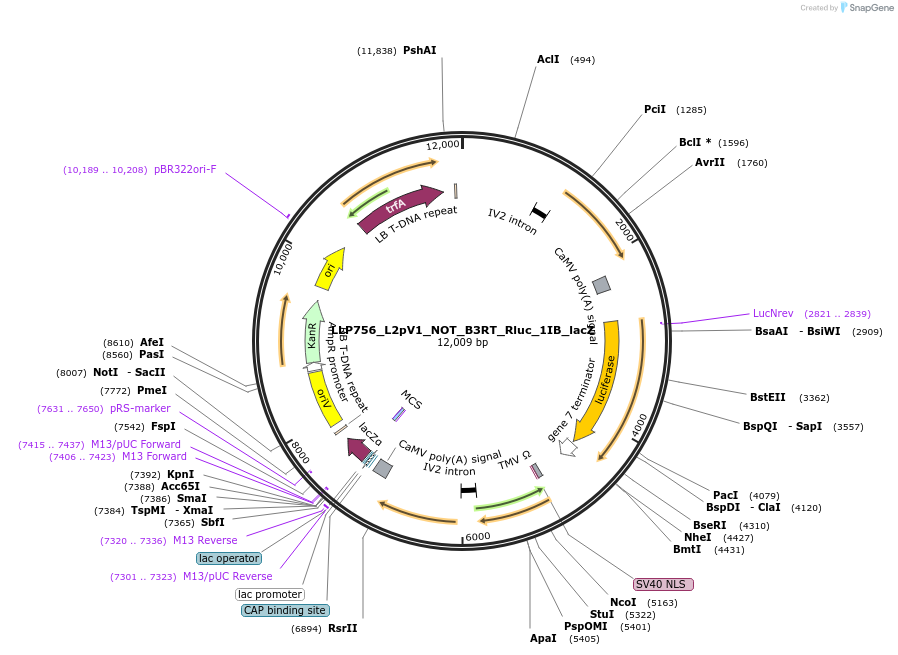
LLP756_L2pV1_NOT_B3RT_Rluc_1IB_lacZ
Plasmid#192401PurposeOff state for the first generation of NOT gates, which target the Rluc CDS to repress expression via its B3RT sites.DepositorInsertAct2::B3RT-Rluc-B3RT
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
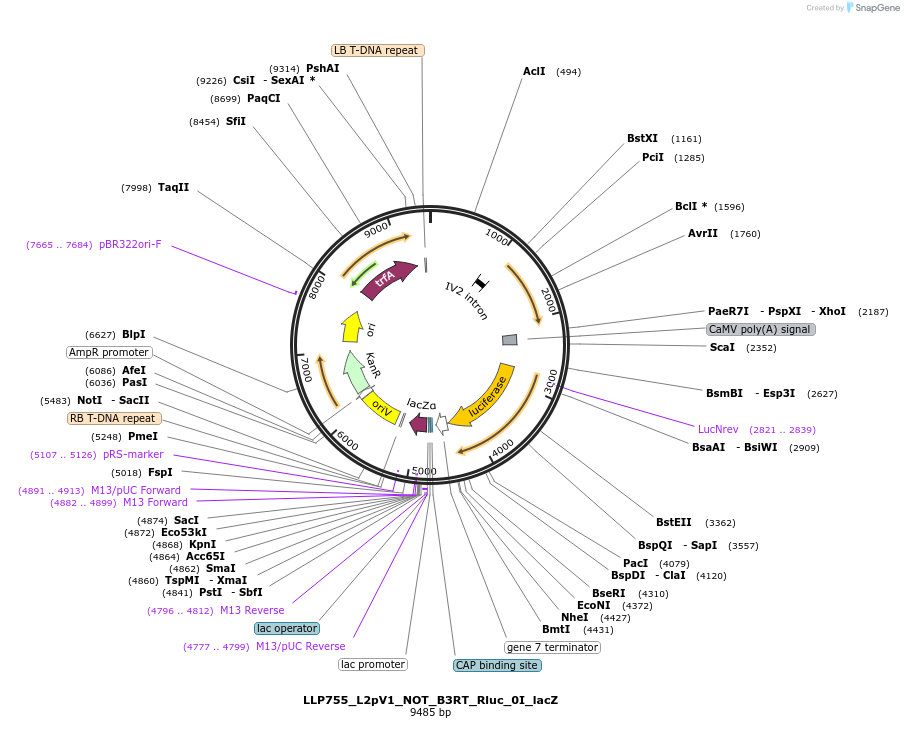
LLP755_L2pV1_NOT_B3RT_Rluc_0I_lacZ
Plasmid#192400PurposeOn state for the first generation of NOT gates, which target the Rluc CDS to repress expression via its B3RT sites.DepositorInsertAct2::B3RT-Rluc-B3RT
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
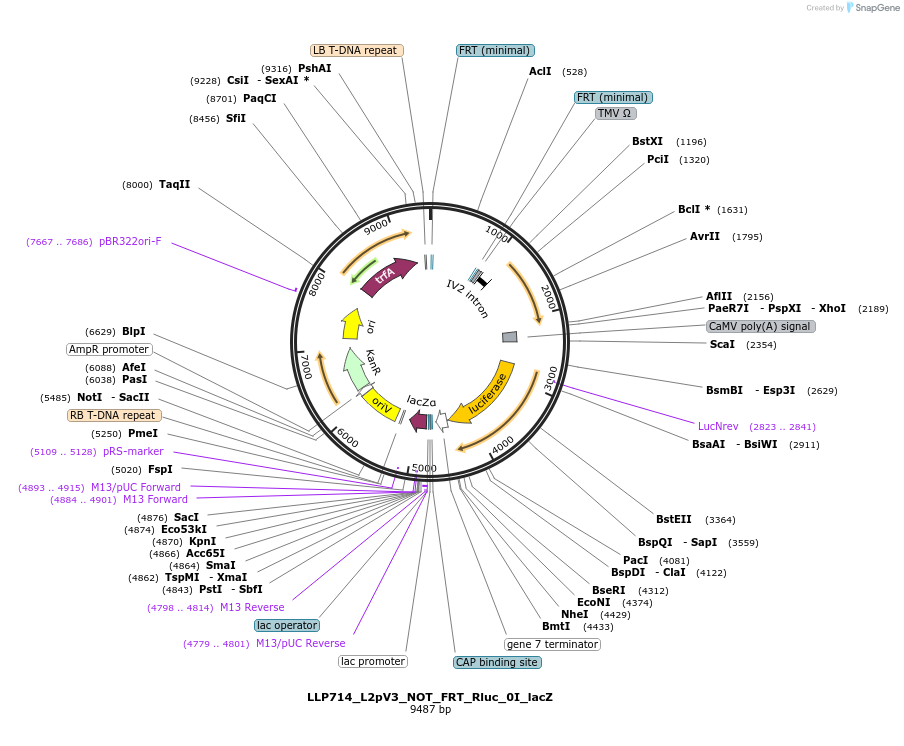
LLP714_L2pV3_NOT_FRT_Rluc_0I_lacZ
Plasmid#192402PurposeOn state for the second generation of NOT gates, which target the Rluc promoter to repress expression via its FRT sites.DepositorInsertFRT-Act2-FRT::Rluc
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
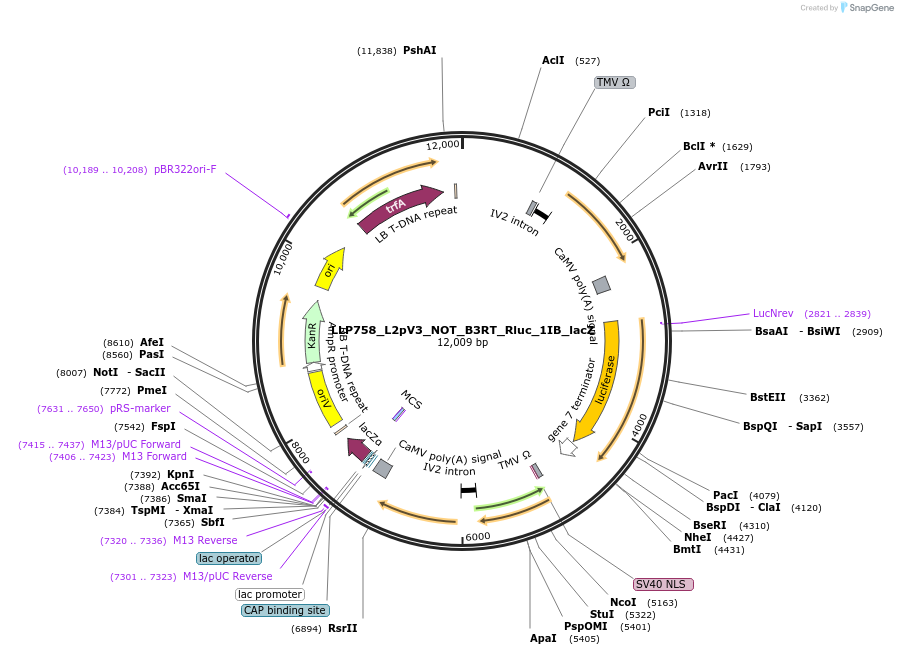
LLP758_L2pV3_NOT_B3RT_Rluc_1IB_lacZ
Plasmid#192405PurposeOff state for the second generation of NOT gates, which target the Rluc promoter to repress expression via its B3RT sites.DepositorInsertB3RT-Act2-B3RT::Rluc
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
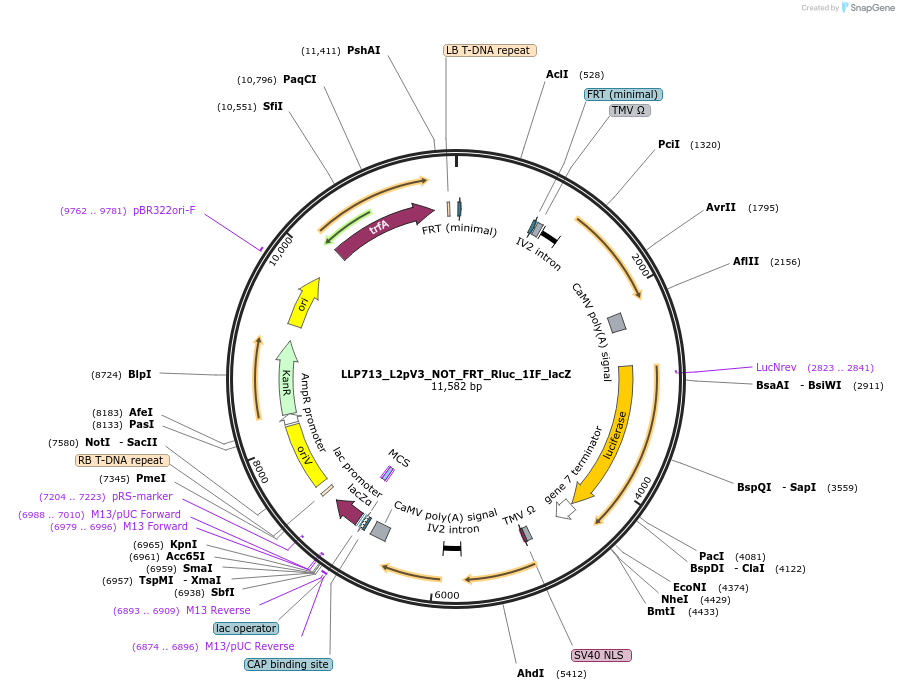
LLP713_L2pV3_NOT_FRT_Rluc_1IF_lacZ
Plasmid#192403PurposeOff state for the second generation of NOT gates, which target the Rluc promoter to repress expression via its FRT sites.DepositorInsertFRT-Act2-FRT::Rluc
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceNov. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
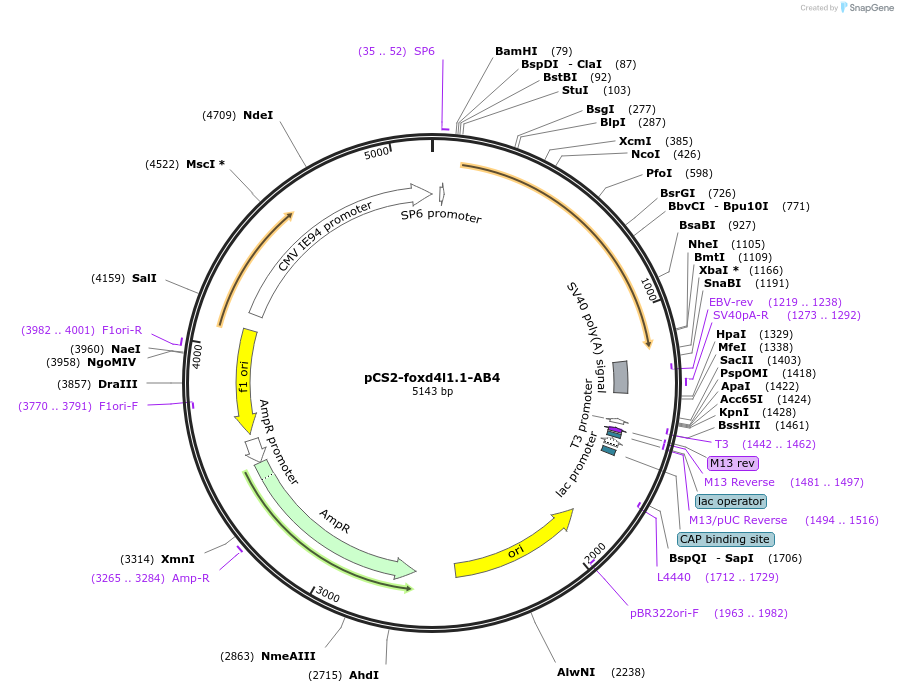
pCS2-foxd4l1.1-AB4
Plasmid#185513PurposeExpression of Xenopus laevis foxd4DepositorInsertfoxd4l1.1
UseIn vitro transcriptionMutationIDILGE replaced with AAAAAAAvailable SinceJuly 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
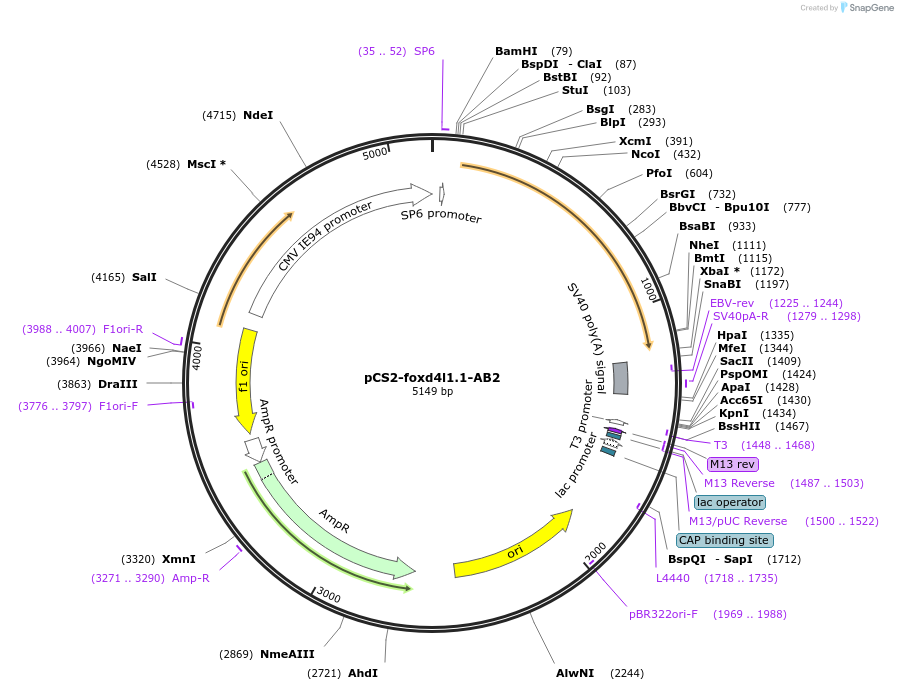
pCS2-foxd4l1.1-AB2
Plasmid#185512PurposeExpression of Xenopus laevis foxd4DepositorInsertfoxd4l1.1
UseIn vitro transcriptionMutationIDIL replaced with AAAAAAAvailable SinceJuly 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
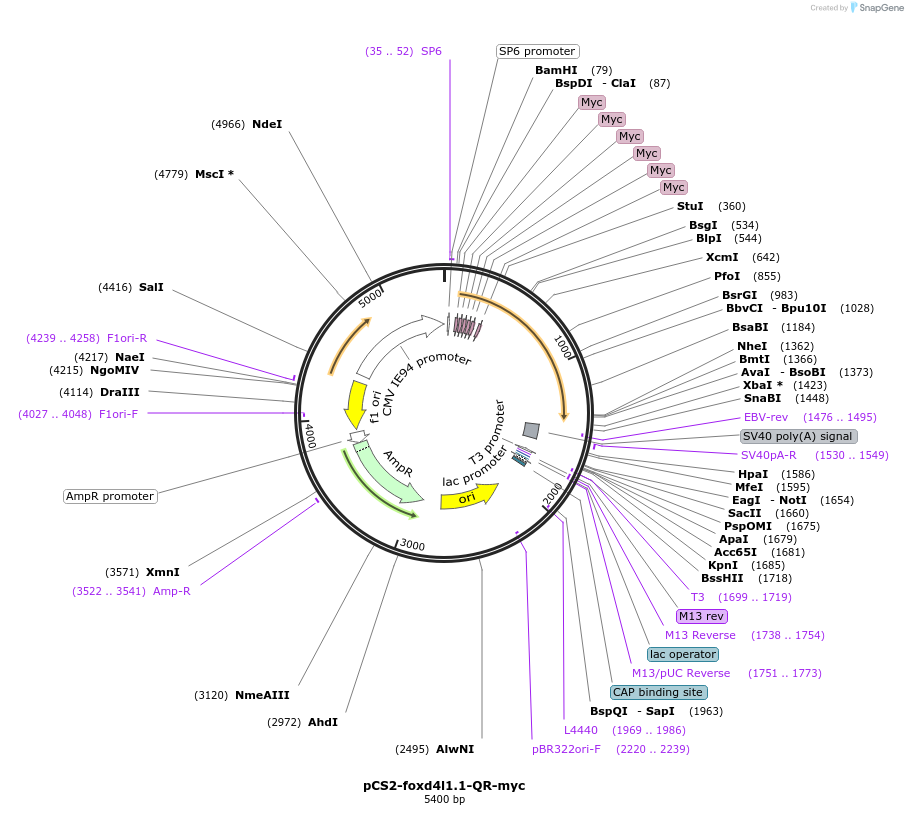
pCS2-foxd4l1.1-QR-myc
Plasmid#185521PurposeExpression of Xenopus laevis foxd4DepositorInsertfoxd4l1.1
UseIn vitro transcriptionTagsmycMutationreplaces Q>R upstream of GARQYLNIAvailable SinceJuly 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
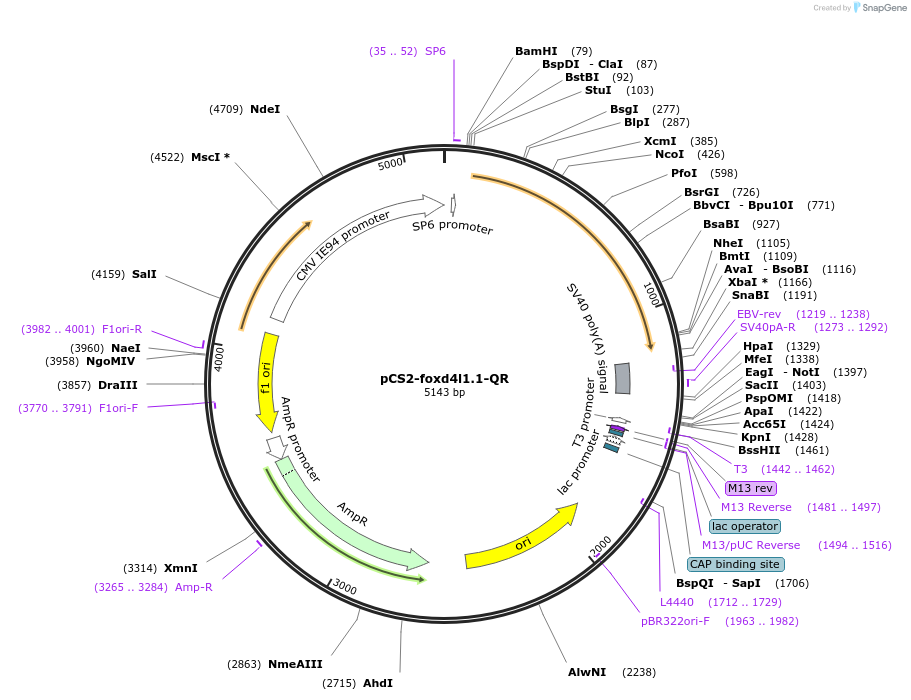
pCS2-foxd4l1.1-QR
Plasmid#185520PurposeExpression of Xenopus laevis foxd4DepositorInsertfoxd4l1.1
UseIn vitro transcriptionMutationreplaces Q>R upstream of GARQYLNIAvailable SinceJuly 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
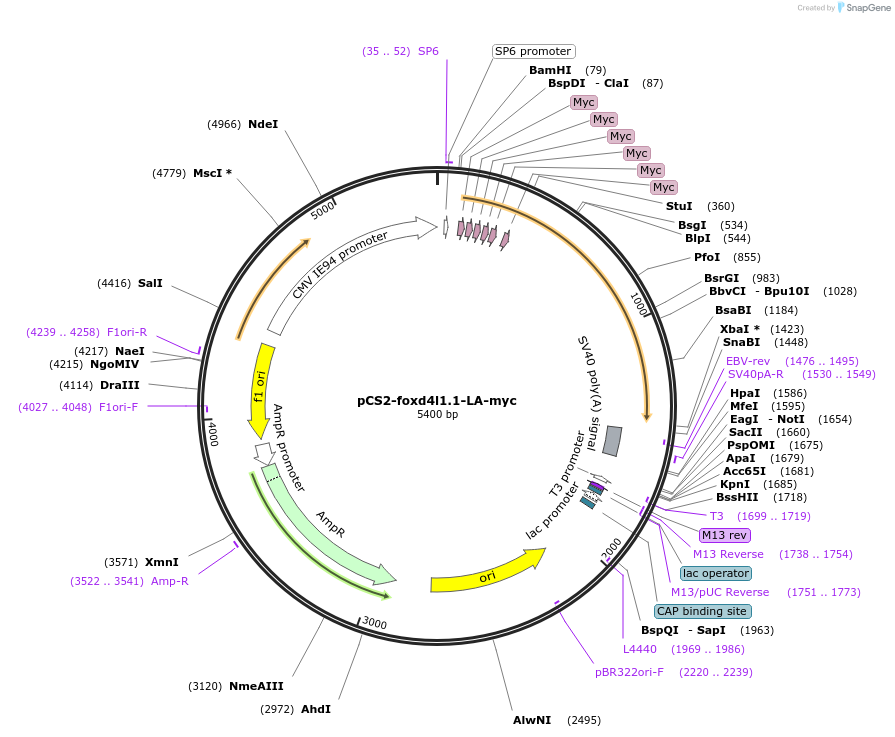
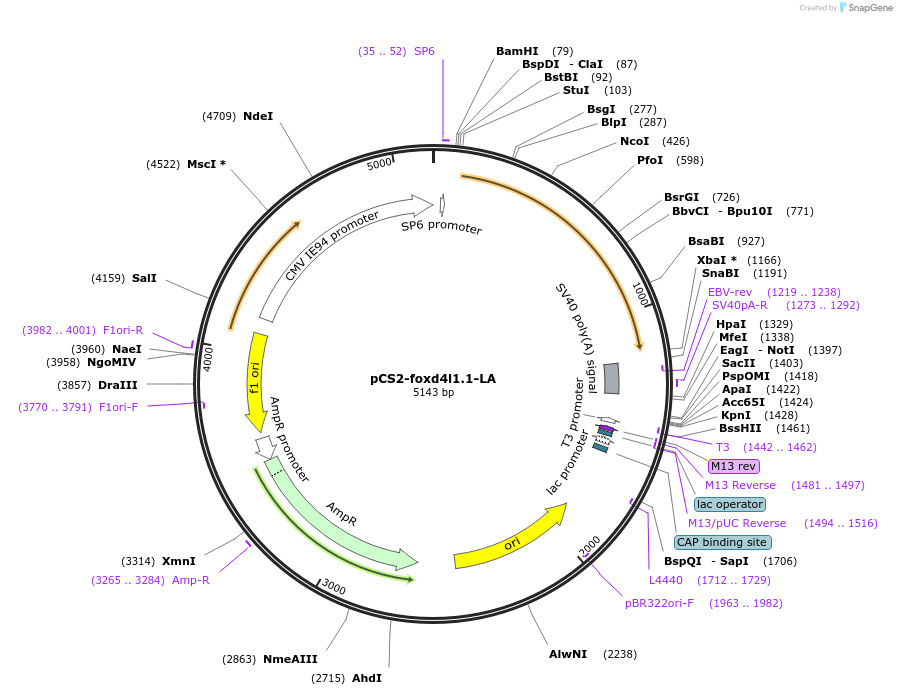
pCS2-foxd4l1.1-LA-myc
Plasmid#185519PurposeExpression of Xenopus laevis foxd4DepositorInsertfoxd4l1.1
UseIn vitro transcriptionTagsmycMutationreplaces L>A upstream of GARQYLNAvailable SinceJuly 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
pCS2-foxd4l1.1-LA
Plasmid#185518PurposeExpression of Xenopus laevis foxd4DepositorInsertfoxd4l1.1
UseIn vitro transcriptionMutationreplaces L>A upstream of GARQYLNIAvailable SinceJuly 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
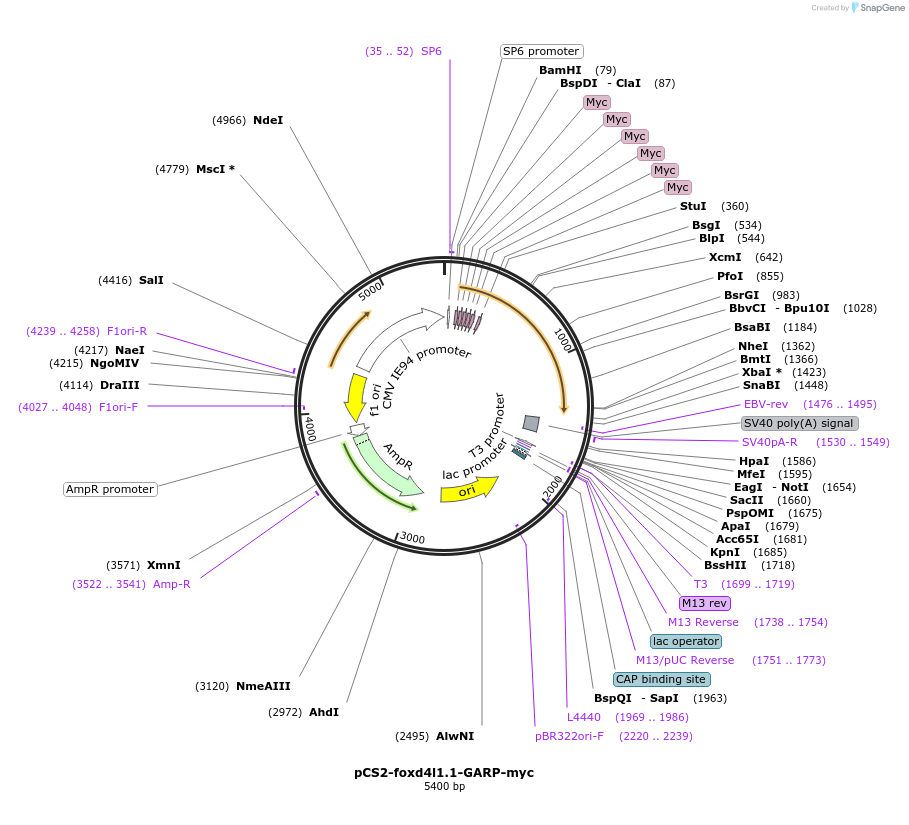
pCS2-foxd4l1.1-GARP-myc
Plasmid#185517PurposeExpression of Xenopus laevis foxd4DepositorInsertfoxd4l1.1
UseIn vitro transcriptionTagsmycMutationReplaces Q to P in GARQYNLIAvailable SinceJuly 12, 2022AvailabilityAcademic Institutions and Nonprofits only