We narrowed to 11,014 results for: phen
-
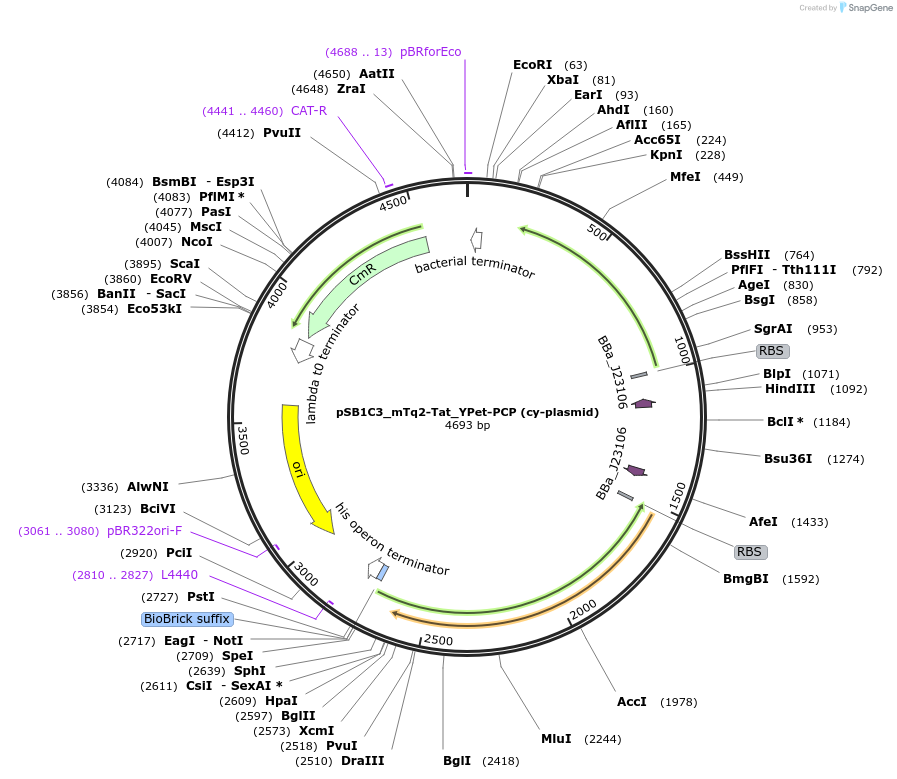
Plasmid#127636PurposeEncodes two fluorecent proteins with RNA binding peptides: mTurquoise2-Tat and YPet-PCP. Expression with constitutive E. coli RNAP promoter (J23106), ribozymes RiboJ10 (for YPet) and PlmJ (for mTq2)DepositorInsertmTurquoiuse2-Tat, YPet-PCP
UseSynthetic BiologyTagsRNA binding peptide: PCP and RNA binding peptide:…ExpressionBacterialAvailable SinceJuly 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
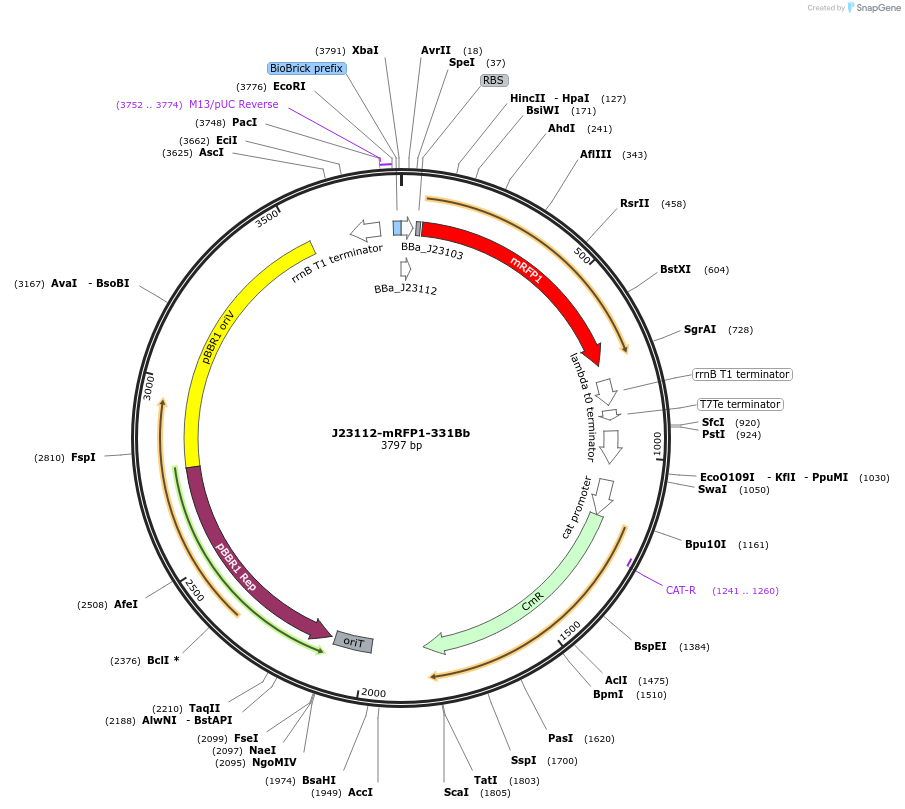
J23112-mRFP1-331Bb
Plasmid#78278PurposemRFP1 behind constitutive promoter J23112 in pSEVA331Bb backboneDepositorInsertJ23112-mRFP1
Available SinceNov. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
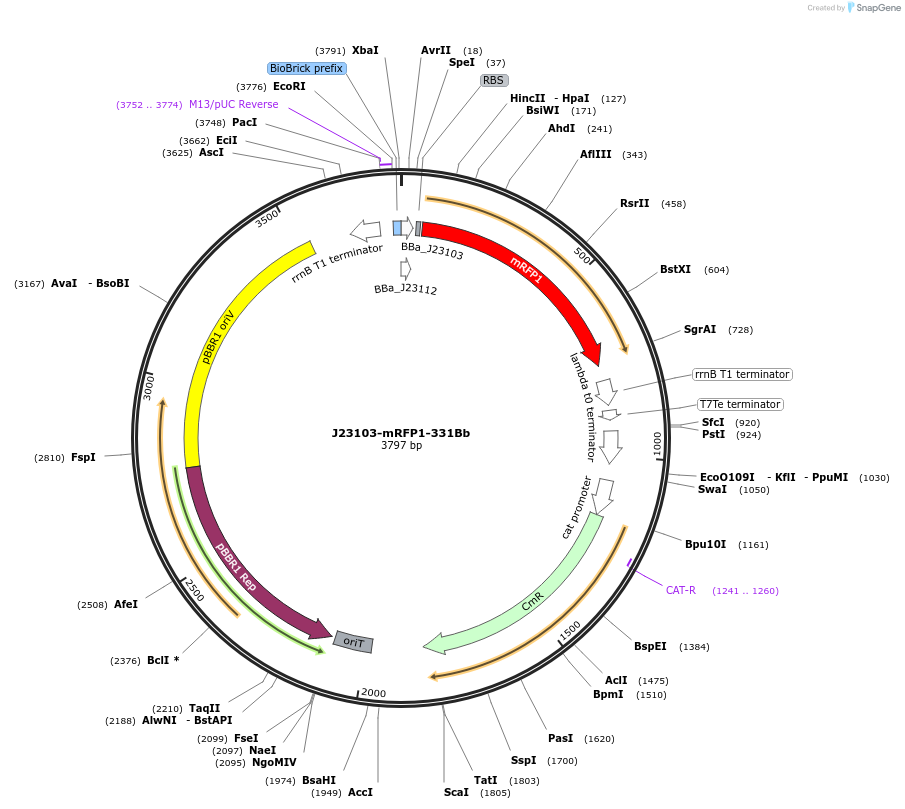
J23103-mRFP1-331Bb
Plasmid#78273PurposemRFP1 behind constitutive promoter J23103 in pSEVA331Bb backboneDepositorInsertJ23103-mRFP1
Available SinceNov. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
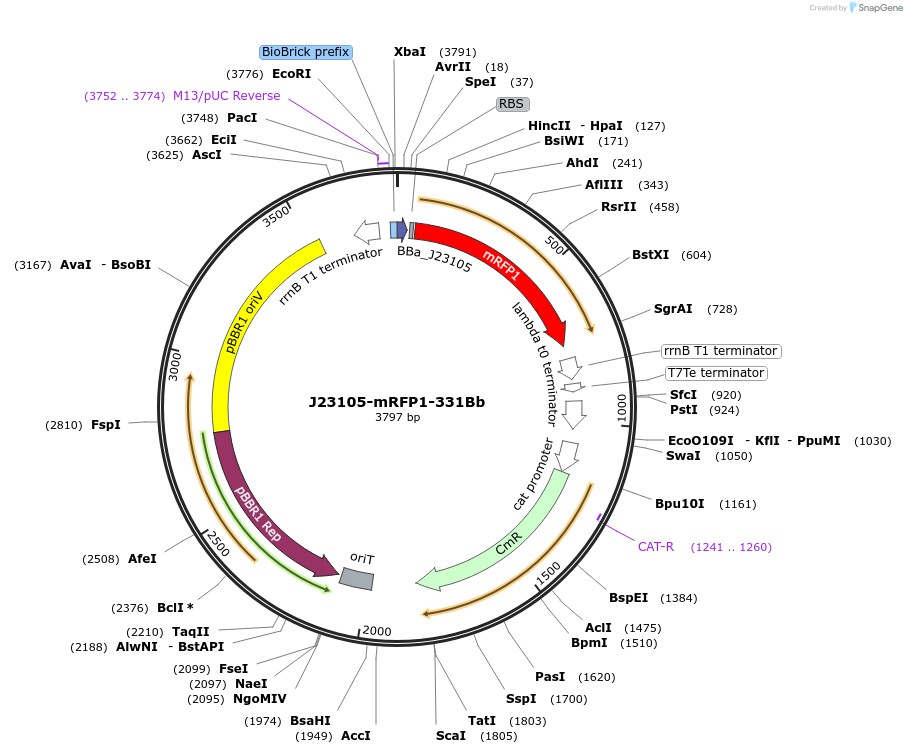
J23105-mRFP1-331Bb
Plasmid#78275PurposemRFP1 behind constitutive promoter J23105 in pSEVA331Bb backboneDepositorInsertJ23105-mRFP1
Available SinceNov. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
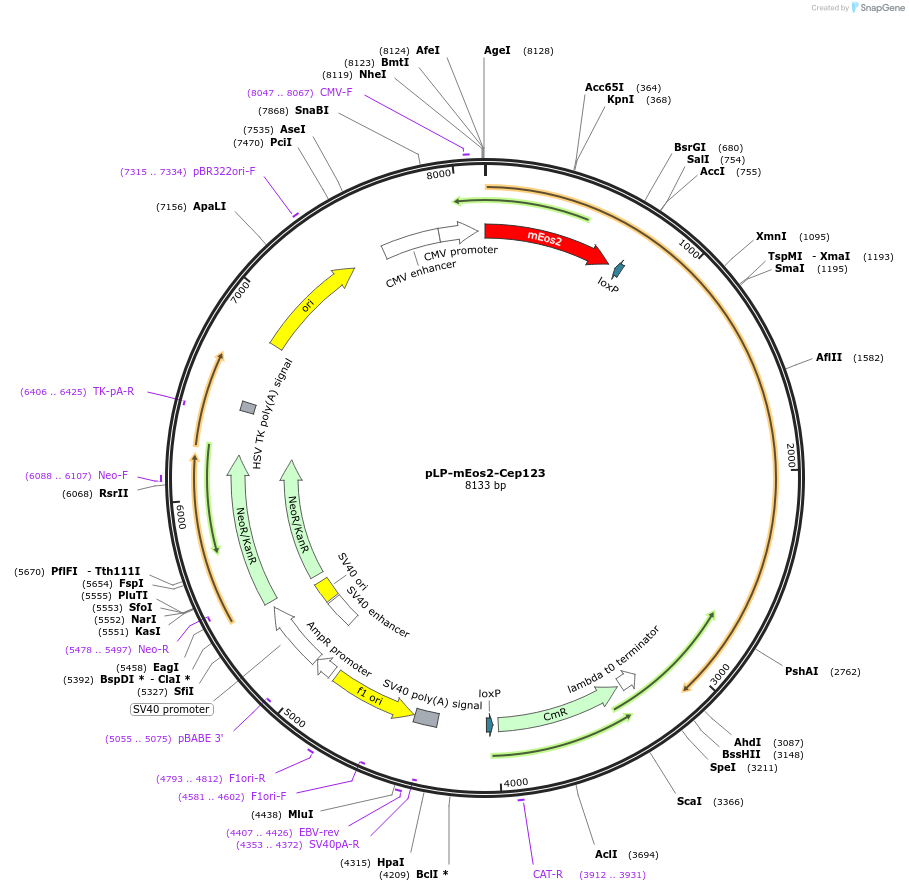
pLP-mEos2-Cep123
Plasmid#72589PurposemEos2 photoconvertible fluorescent protein fused to the N-terminus of isoform 1 of Cep123 (Cep89/CCDC123)DepositorAvailable SinceApril 4, 2016AvailabilityAcademic Institutions and Nonprofits only -
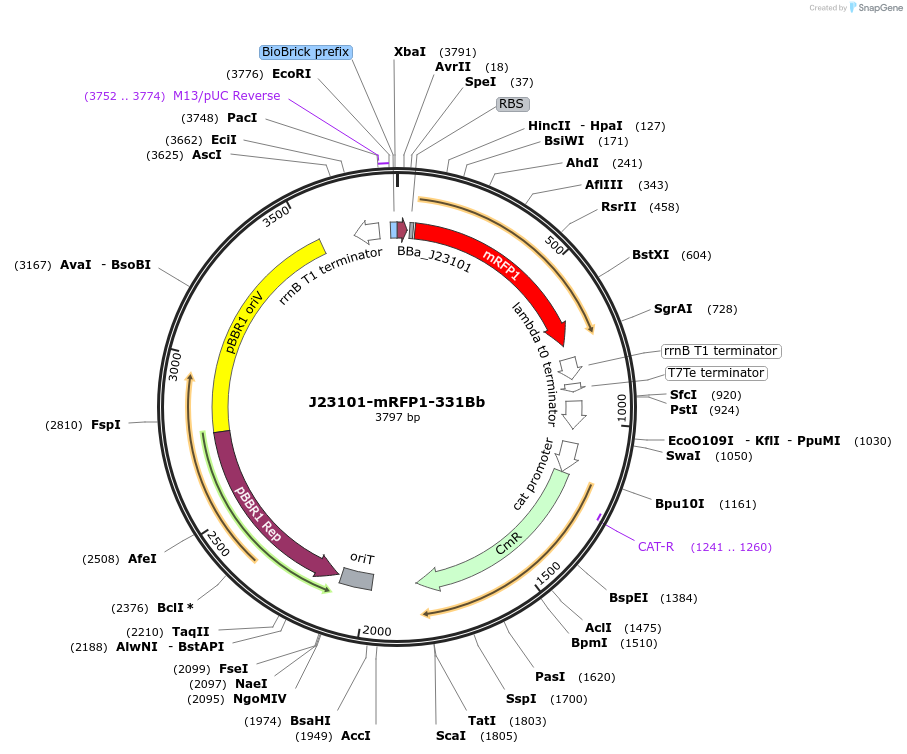
J23101-mRFP1-331Bb
Plasmid#78272PurposemRFP1 behind constitutive promoter J23101 in pSEVA331Bb backboneDepositorInsertJ23101-mRFP1
Available SinceNov. 10, 2016AvailabilityAcademic Institutions and Nonprofits only -
pCINeo-hFX(I154F)-HA
Plasmid#14977DepositorInserthuman frataxin cDNA (FXN Human)
TagsAU1 and HAExpressionMammalianMutationchanged isoleucine 154 to phenylalanineAvailable SinceAug. 17, 2007AvailabilityAcademic Institutions and Nonprofits only -
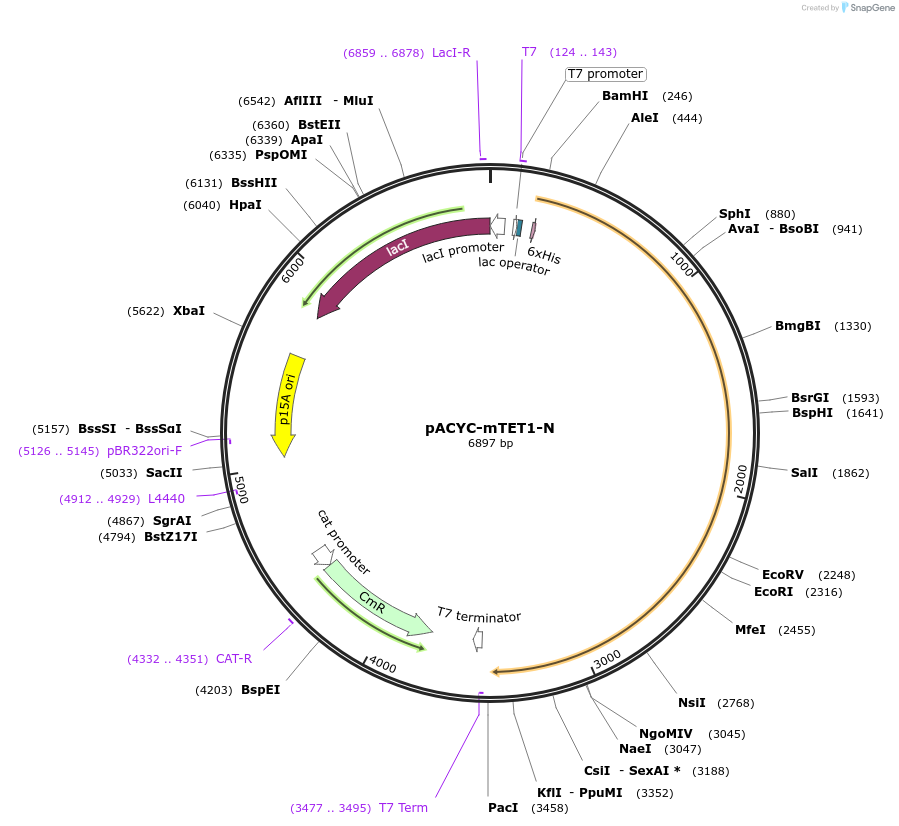
pACYC-mTET1-N
Plasmid#81056Purposebacterial expression of N-terminus of murine TET1DepositorInsertTET1 (Tet1 Mouse)
Tags6HISExpressionBacterialMutationN-terminus, amino acids 301-1366PromoterT7Available SinceAug. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
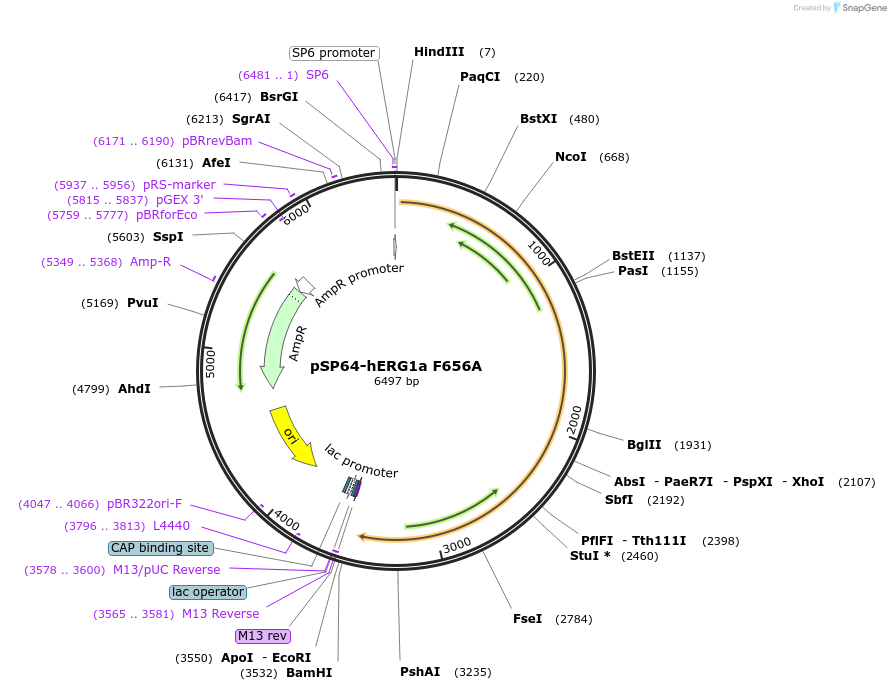
pSP64-hERG1a F656A
Plasmid#53054PurposeExpresses alpha subunit of F656A hERG1a K channel in Xenopus oocytesDepositorInsertPotassium voltage-gated channel subfamily H member 2 (KCNH2 Human)
UseXenopus oocyte expressionMutationchanged Phenylalanine 656 to AlaninePromoterSP6Available SinceJune 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
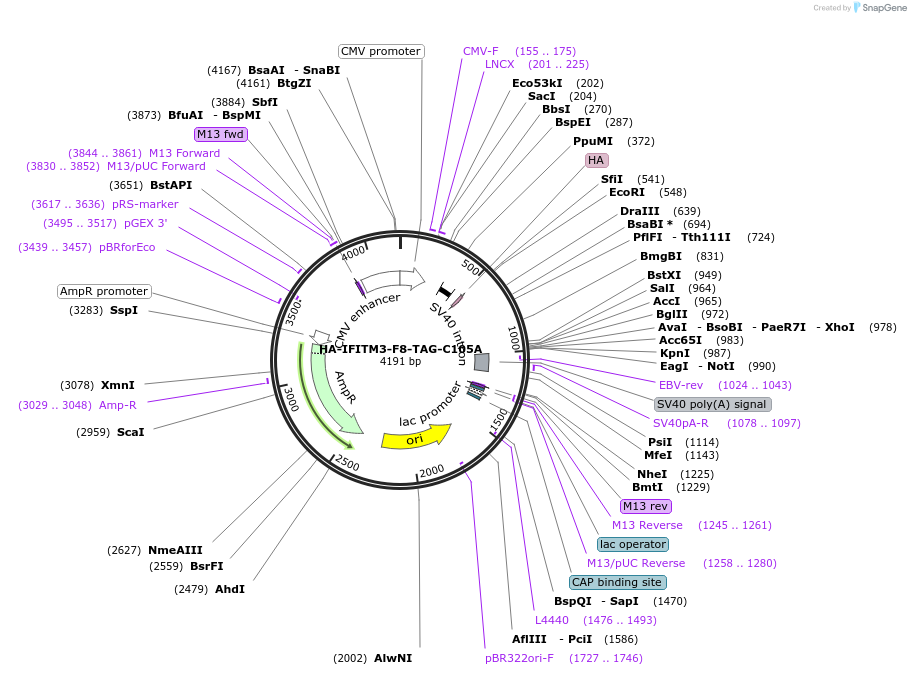
HA-IFITM3-F8-TAG-C105A
Plasmid#122478Purposeexpresses human IFITM3-C105A with unnatural amino acid incorporated site specifically in response to the amber codon TAG in mammalian cells when co-transfected with tRNA synthetase/tRNA pair.DepositorInsertIFITM3 (IFITM3 Human)
TagsHAExpressionMammalianMutationChanged Phenylalanine 8 to amber codon, cysteine …Available SinceMarch 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
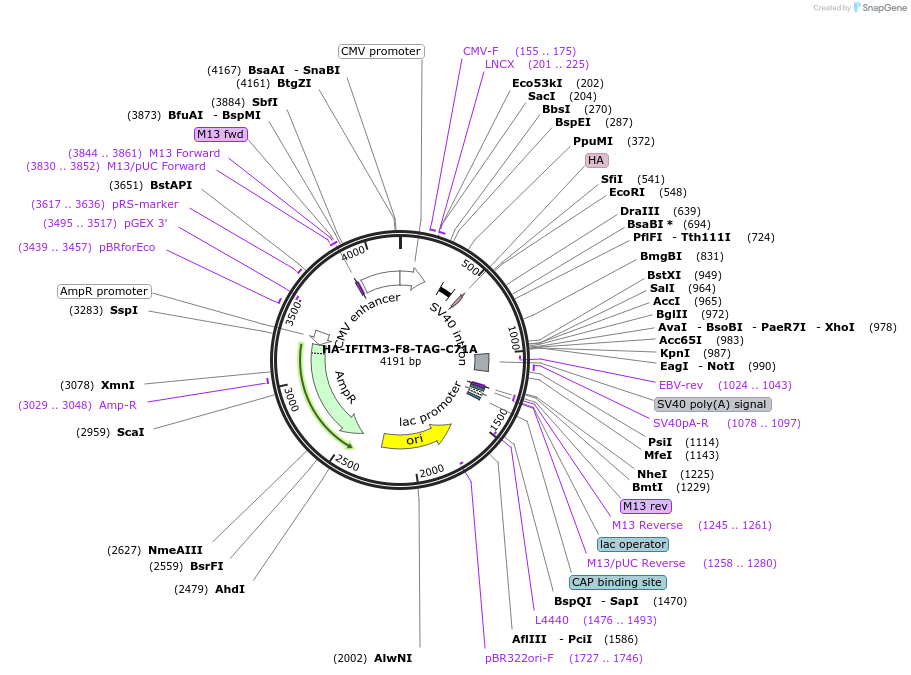
HA-IFITM3-F8-TAG-C71A
Plasmid#122476Purposeexpresses human IFITM3-C71A with unnatural amino acid incorporated site specifically in response to the amber codon TAG in mammalian cells when co-transfected with tRNA synthetase/tRNA pair.DepositorInsertIFITM3 (IFITM3 Human)
TagsHAExpressionMammalianMutationChanged Phenylalanine 8 to amber codon, cysteine …Available SinceMarch 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
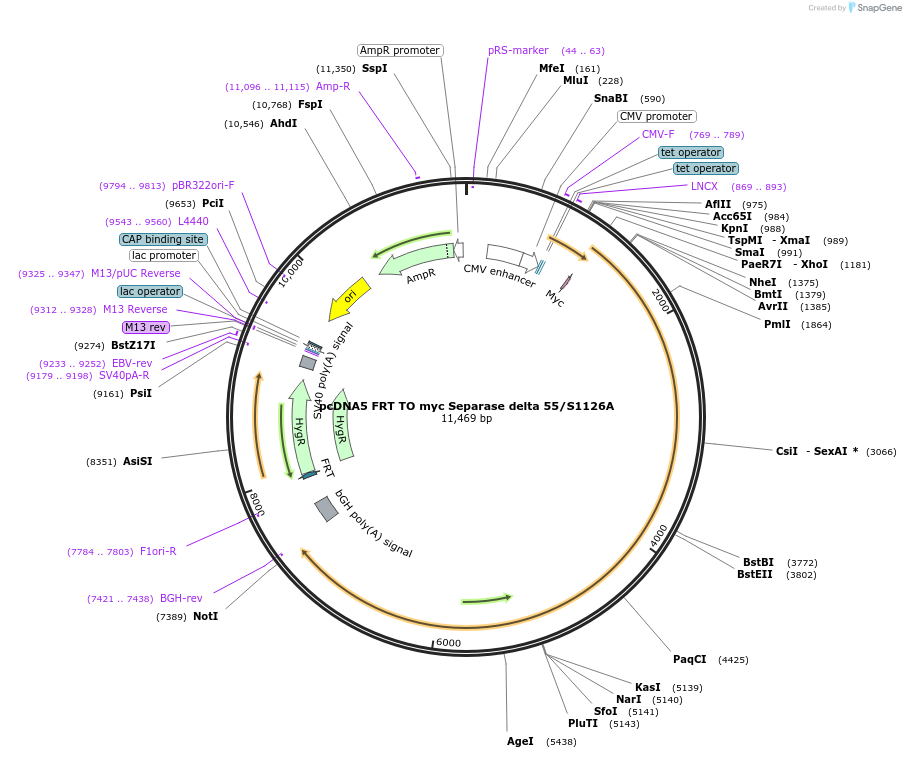
pcDNA5 FRT TO myc Separase delta 55/S1126A
Plasmid#59825PurposeAllows the integration of myc Separase delta 55/S1126A in the genome and Tet-inducible expression.DepositorInsertSeparase (ESPL1 Human)
TagsMycExpressionMammalianMutationdelta 55/S1126APromoterhybrid human cytomegalovirus (CMV)/TetO2 promoterAvailable SinceMay 12, 2015AvailabilityAcademic Institutions and Nonprofits only -
-
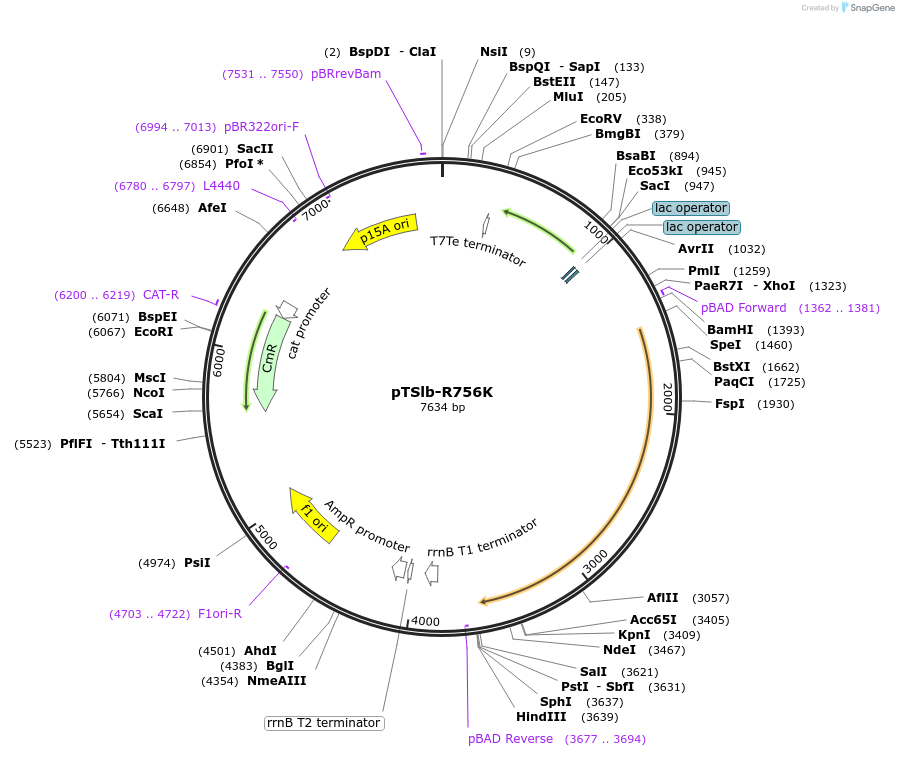
pTSlb-R756K
Plasmid#60731PurposeExpresses both fragments of T7 RNAP split at position 179. The N-terminal fragment is driven by PLac while the C-terminal fragment is driven by PBAD. C-terminal fragment has the point mutations R756SDepositorInsertsResidues 1-179 of split T7 RNAP
Residues 180-880 of split T7 RNAP
UseSynthetic BiologyExpressionBacterialMutationResidues 1-180 of split T7 RNAP and Residues 180-…Available SinceNov. 4, 2014AvailabilityAcademic Institutions and Nonprofits only -
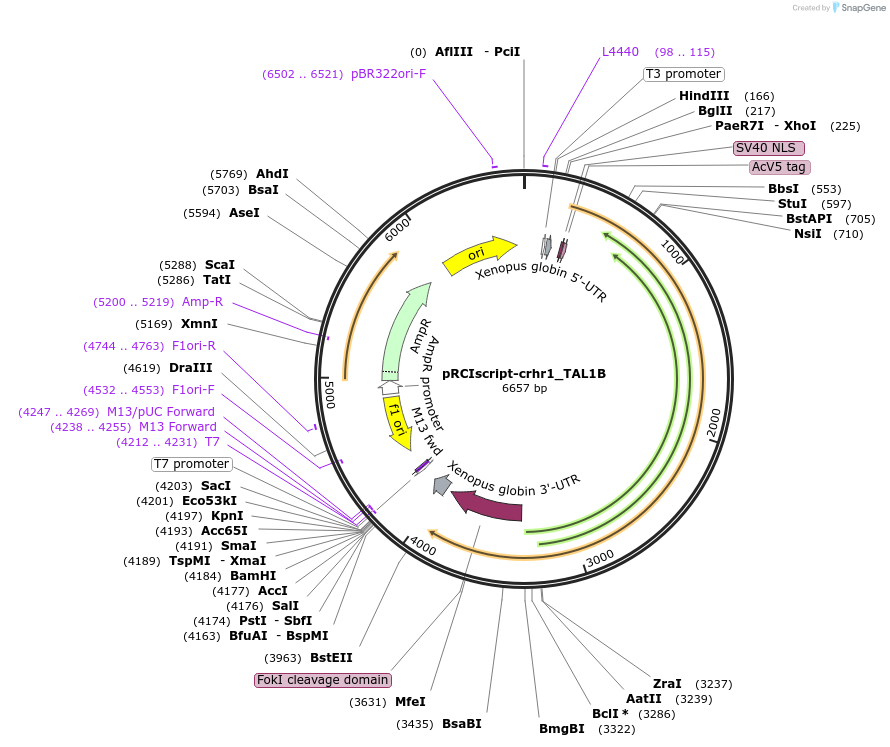
pRCIscript-crhr1_TAL1B
Plasmid#44235DepositorAvailable SinceApril 10, 2013AvailabilityAcademic Institutions and Nonprofits only -
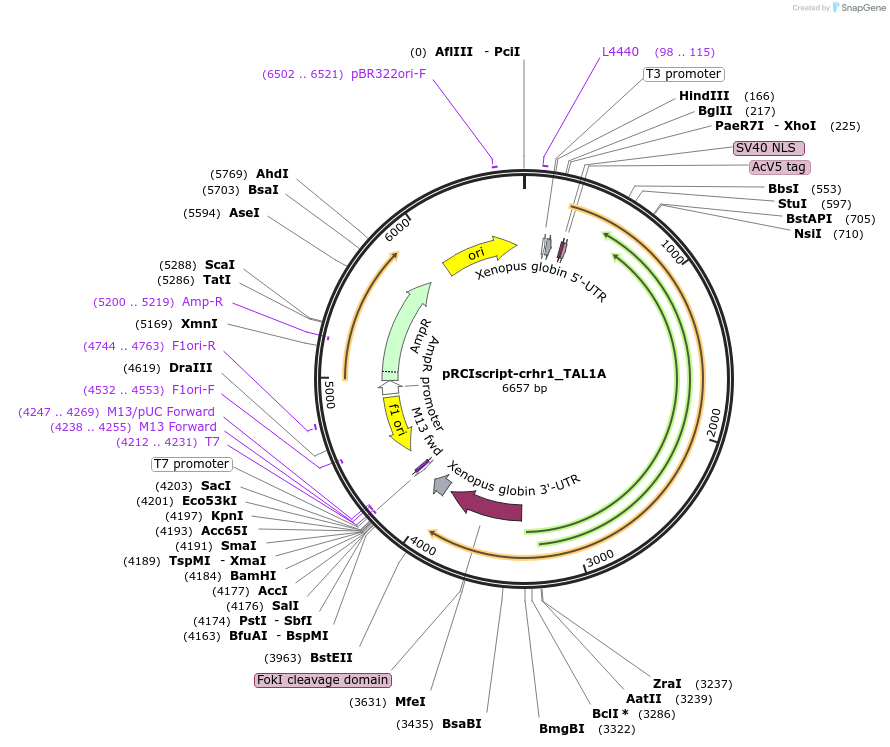
pRCIscript-crhr1_TAL1A
Plasmid#44234DepositorAvailable SinceApril 10, 2013AvailabilityAcademic Institutions and Nonprofits only -
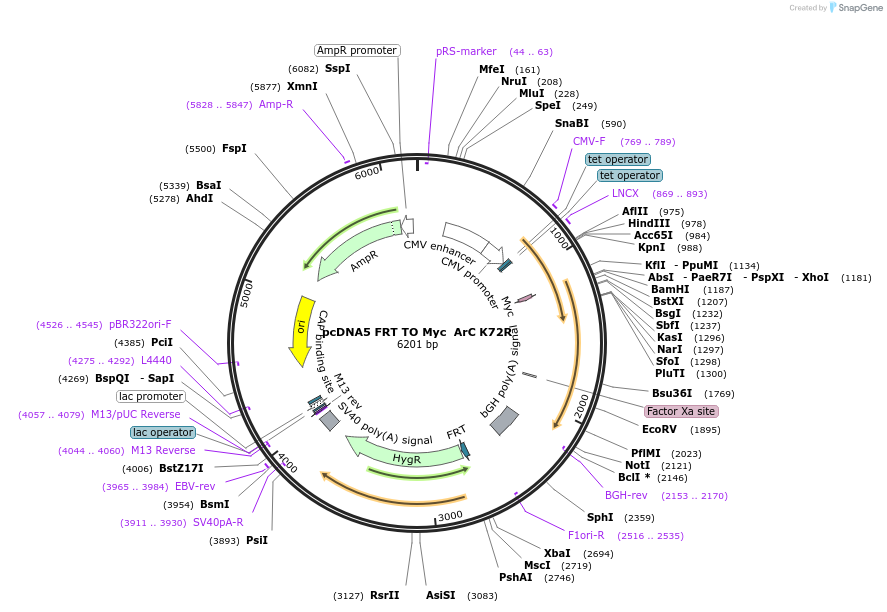
pcDNA5 FRT TO Myc ArC K72R
Plasmid#59811PurposeAllows the integration of Myc ArC K72R in the genome and Tet-inducible expression.DepositorInsertAurora C (AURKC Human)
TagsMycExpressionMammalianMutationK72R, kinase mutantPromoterhybrid human cytomegalovirus (CMV)/TetO2 promoterAvailable SinceFeb. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
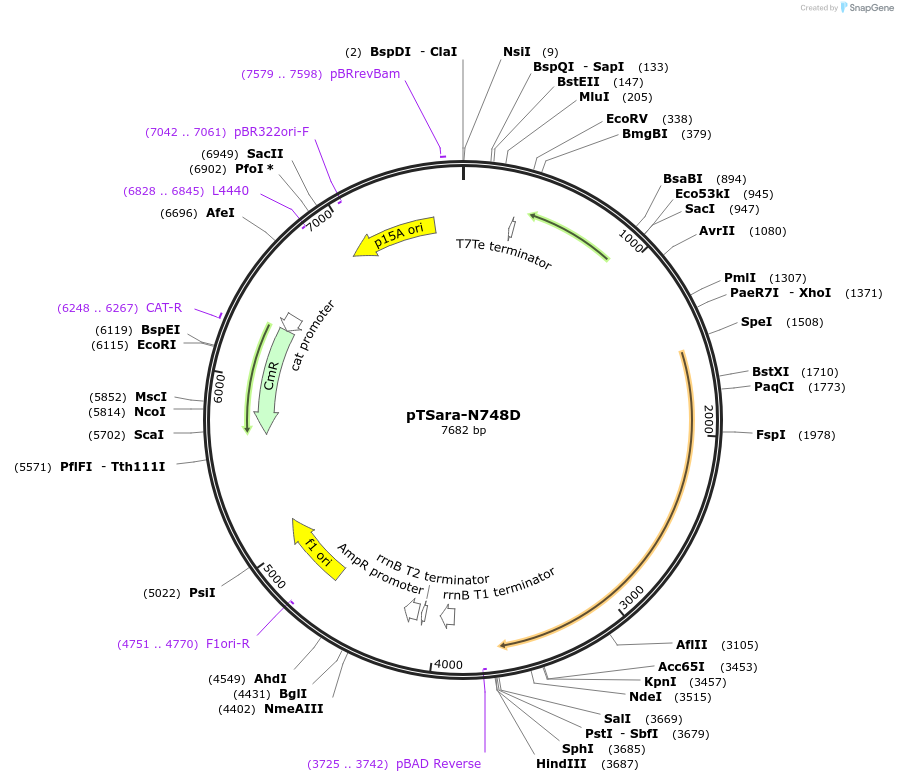
pTSara-N748D
Plasmid#60726PurposeExpresses both fragments of T7 RNAP split at position 179. Both fragments are driven by PBAD. The C-terminal fragment has the point mutation N748D. Contains a constitutive araC ORFDepositorInsertsResidues 1-179 of split T7 RNAP
Residues 180-880 of split T7 RNAP
UseSynthetic BiologyExpressionBacterialMutationResidues 1-180 of split T7 RNAP and Residues 180-…Available SinceOct. 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
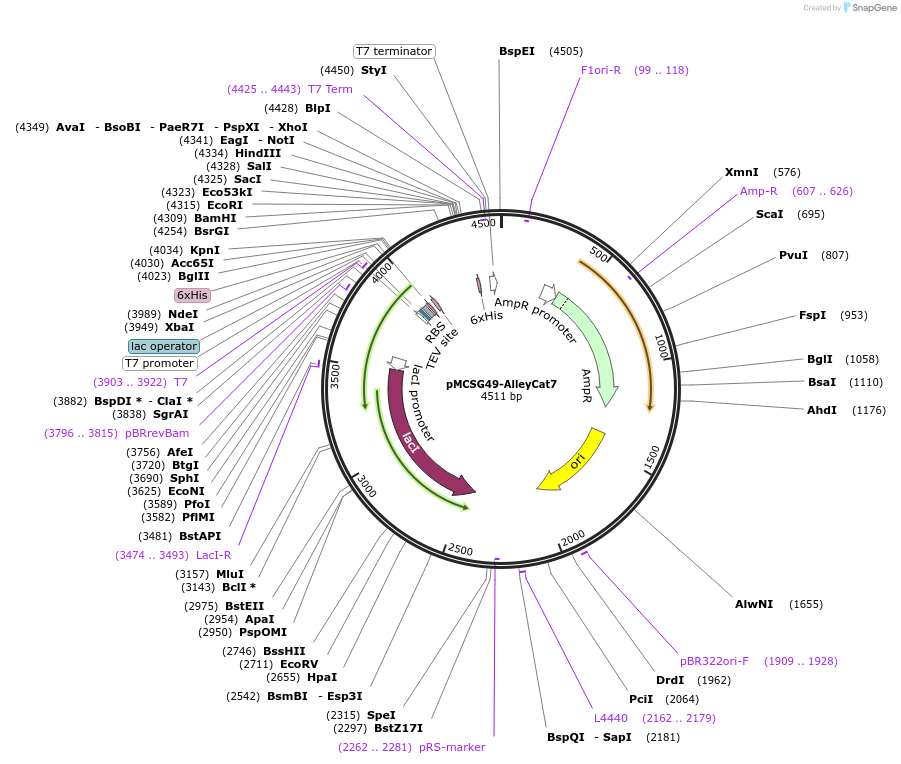
pMCSG49-AlleyCat7
Plasmid#42591DepositorInsertmutated C-terminal portion of calmodulin gene (M76-K148)
TagsHis and TEVExpressionBacterialMutationchanged Phenylalanine 92 to Glutamine acid, Isole…PromoterT7Available SinceMarch 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
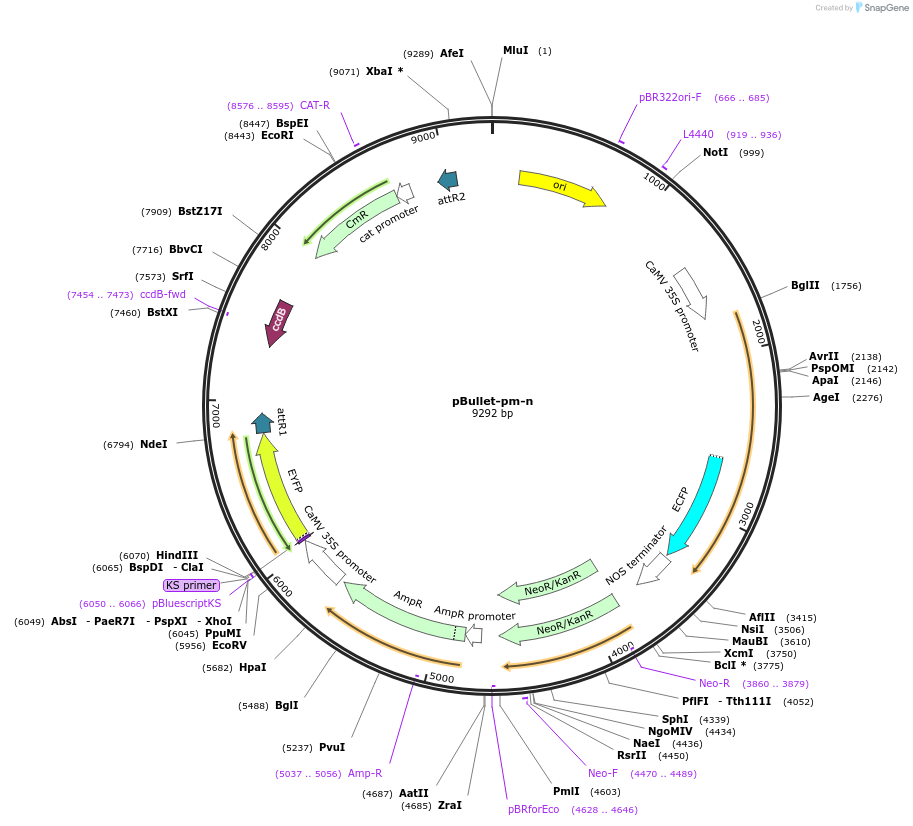
pBullet-pm-n
Plasmid#53077Purposedestination vector with PIP2a-ECFP (plasma membrane marker) for tagged fluorescent protein (N-ter-EYFP) of plant geneDepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceJuly 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
-
-
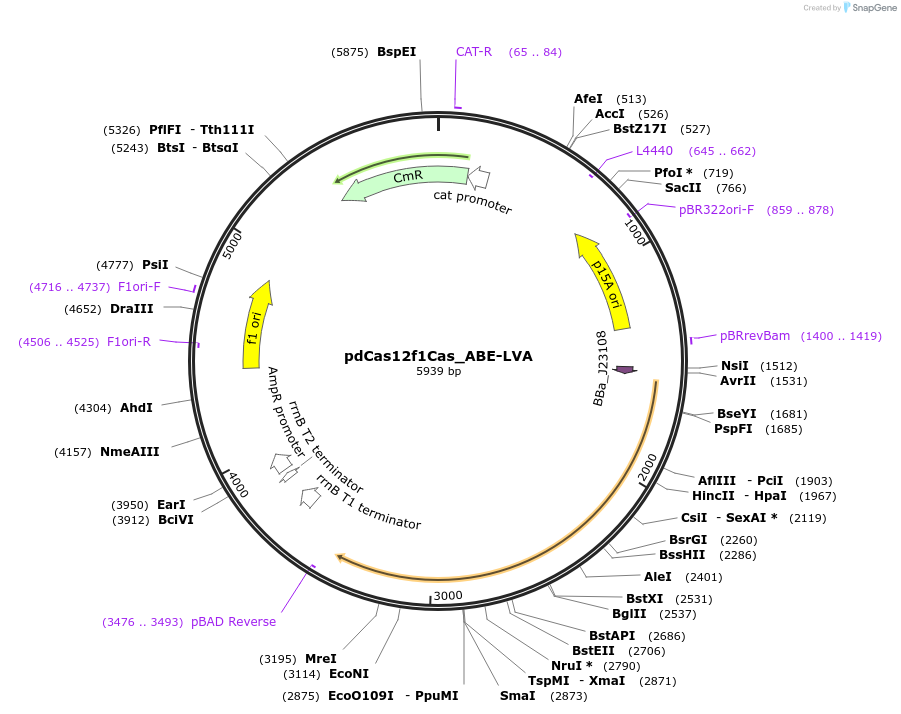
pdCas12f1Cas_ABE-LVA
Plasmid#220995PurposeProkaryotic and constitutive expression of AsCas12f1/TadA8e-LVA fusionDepositorInsertAcidibacillus sulfuroxidans Cas12f1-ABE
UseCRISPRTagsLVAExpressionBacterialMutationD225AAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
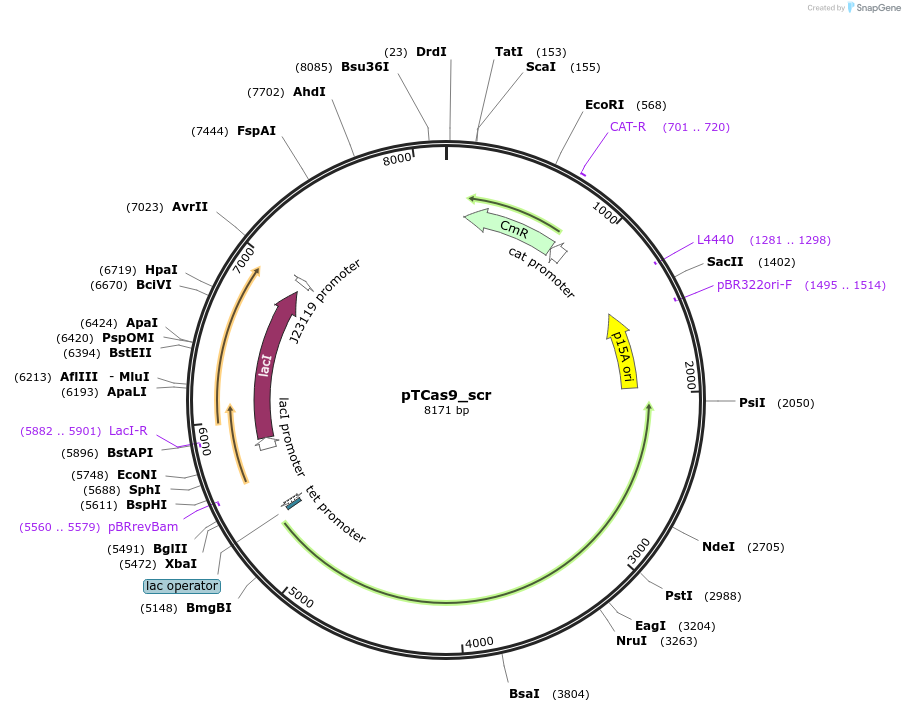
pTCas9_scr
Plasmid#207566PurposeThermoCas9-mediated cleavage of genomic DNA in E. coliDepositorInsertPlac/tet_ThermoCas9_PlacI_lacI_PJ23119_sgRNA(scrambled non-targeting spacer)
ExpressionBacterialMutationWild-typeAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
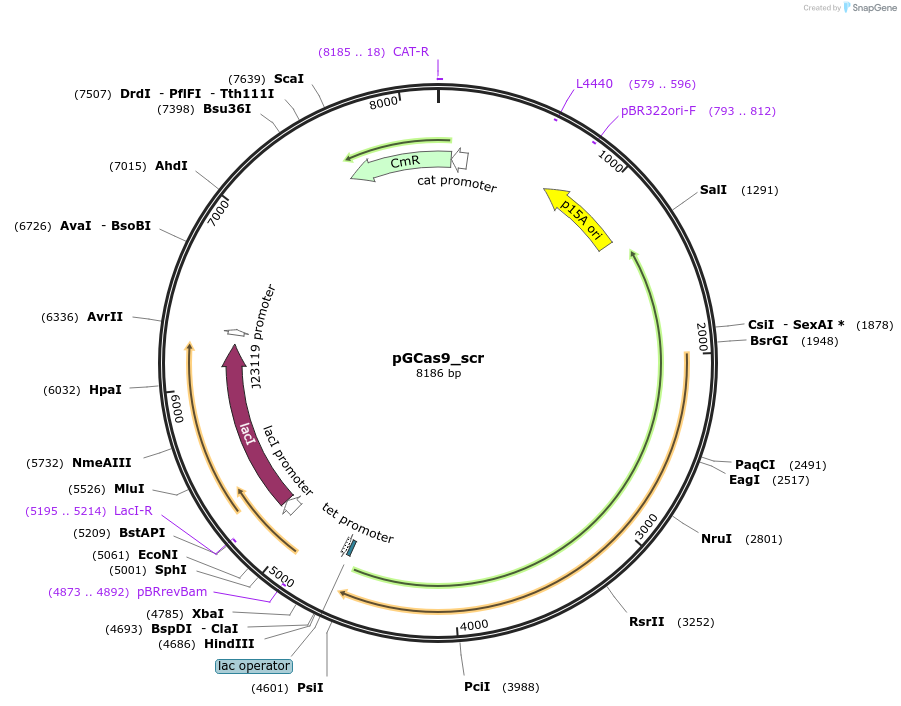
pGCas9_scr
Plasmid#207567PurposeGeoCas9-mediated cleavage of genomic DNA in E. coliDepositorInsertPlac/tet_GeoCas9_PlacI_lacI_PJ23119_sgRNA(scrambled non-targeting spacer)
ExpressionBacterialMutationWild-typeAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
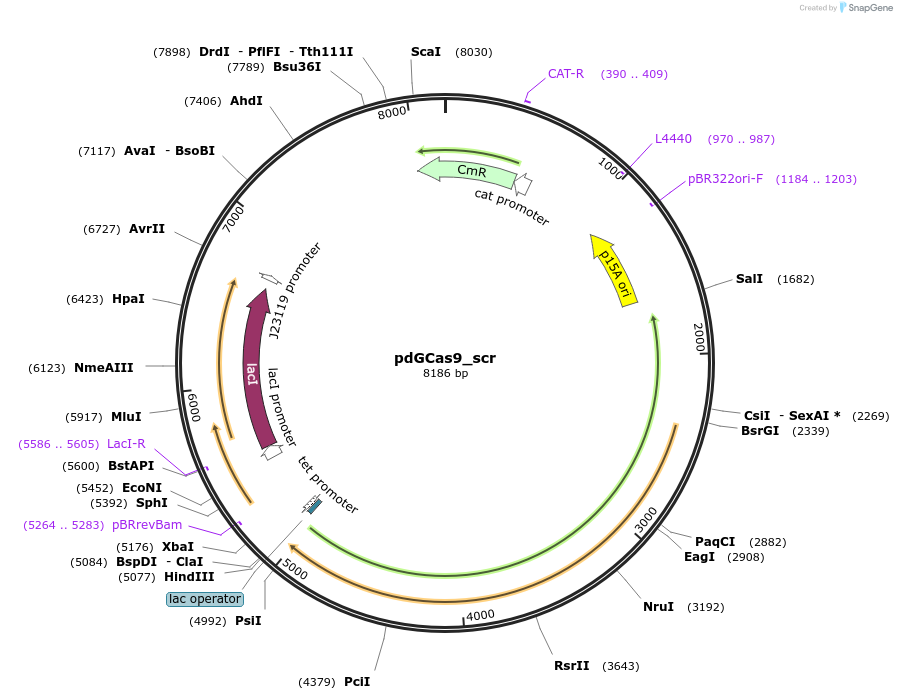
pdTCas9_scr
Plasmid#207568PurposedThermoCas9-mediated silencing of genomic gene in E. coliDepositorInsertPlac/tet_dThermoCas9_PlacI_lacI_PJ23119_sgRNA(scrambled non-targeting spacer)
ExpressionBacterialMutationThermoCas9(D8A,H582A)Available SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
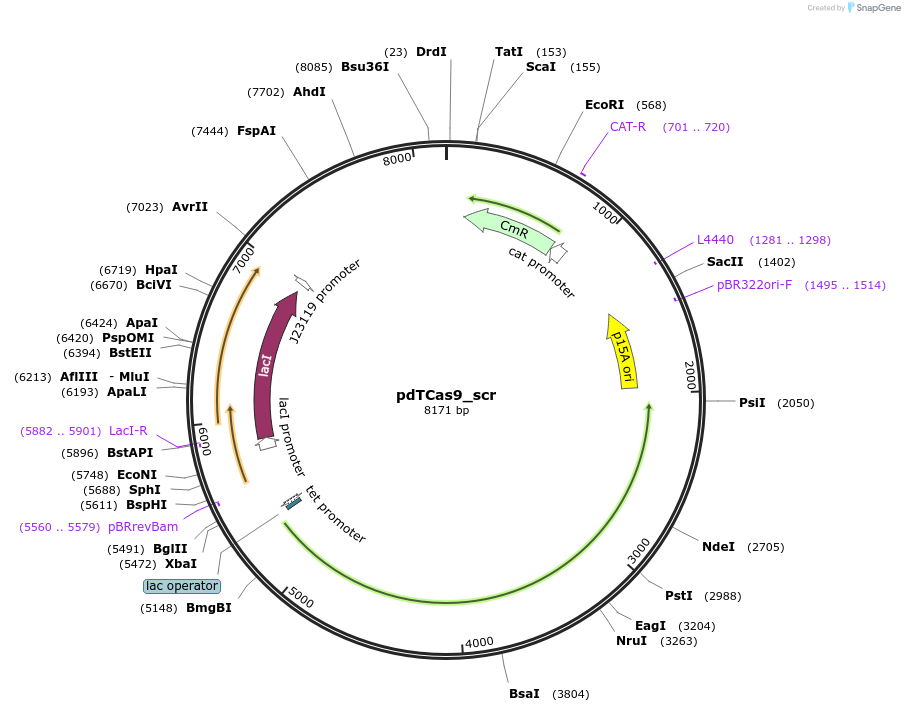
pdGCas9_scr
Plasmid#207569PurposedGeoCas9-mediated silencing of genomic gene in E. coliDepositorInsertPlac/tet_dGeoCas9_PlacI_lacI_PJ23119_sgRNA(scrambled non-targeting spacer)
ExpressionBacterialMutationGeoCas9(D8A,H582A)Available SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
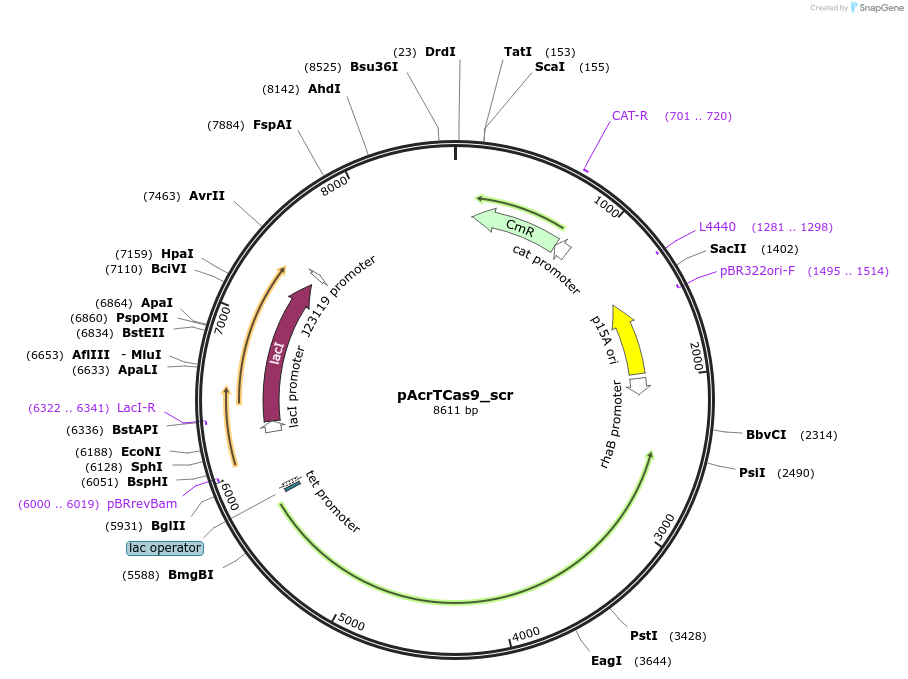
pAcrTCas9_scr
Plasmid#207571PurposeAcrIIC1-mediated inhibition of ThermoCas9 cleavage activity in E. coliDepositorInsertPrha_AcrIIC1Nme_Plac/tet_ThermoCas9_PlacI_lacI_PJ23119_sgRNA(scrambled non-targeting spacer)
ExpressionBacterialMutationWild-typeAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
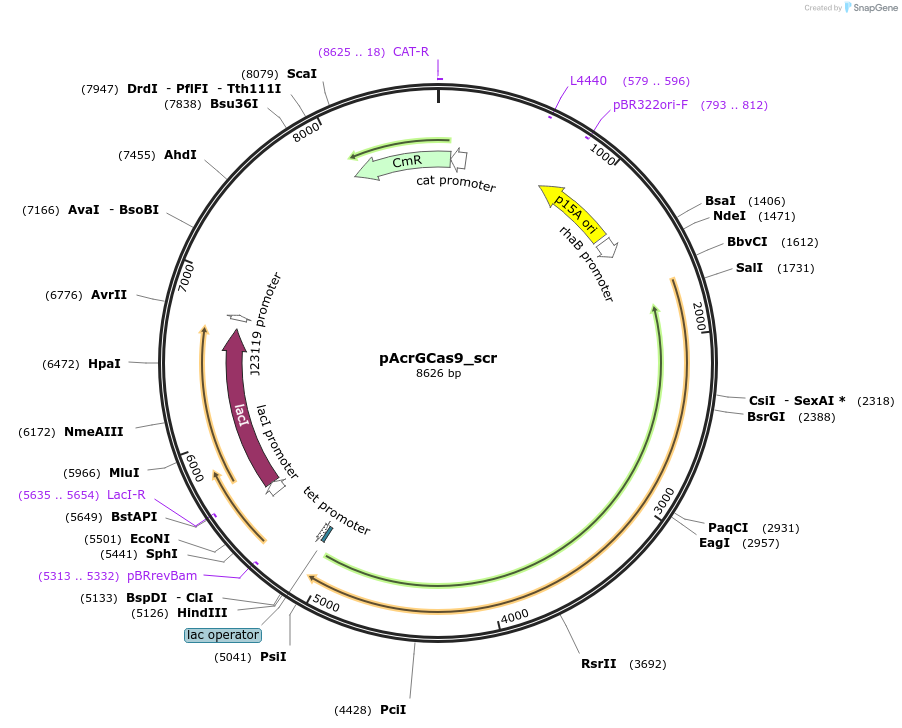
pAcrGCas9_scr
Plasmid#207572PurposeAcrIIC1-mediated inhibition of GeoCas9 cleavage activity in E. coliDepositorInsertPrha_AcrIIC1Nme_Plac/tet_GeoCas9_PlacI_lacI_PJ23119_sgRNA(scrambled non-targeting spacer)
ExpressionBacterialMutationWild-typeAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
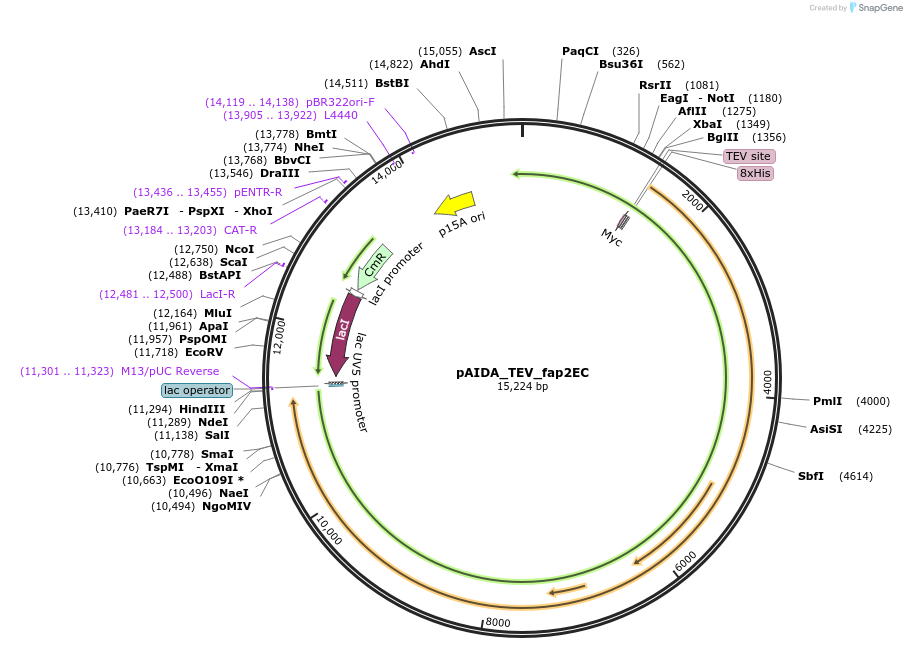
pAIDA_TEV_fap2EC
Plasmid#246866PurposeExpression of Fusobacterium nucleatum ATCC23726 Fap2 extracellular domain (42-3271) with histag and TEV site in fusion with E. coli autotransporter AIDADepositorInsertF. nucleatum ATCC23726 Fap2 (extracellular domain, res. 42–3271)
Tags8x histidine, AIDA autotransporter, AIDA signal s…ExpressionBacterialAvailable SinceNov. 18, 2025AvailabilityAcademic Institutions and Nonprofits only