We narrowed to 14,493 results for: nts
-
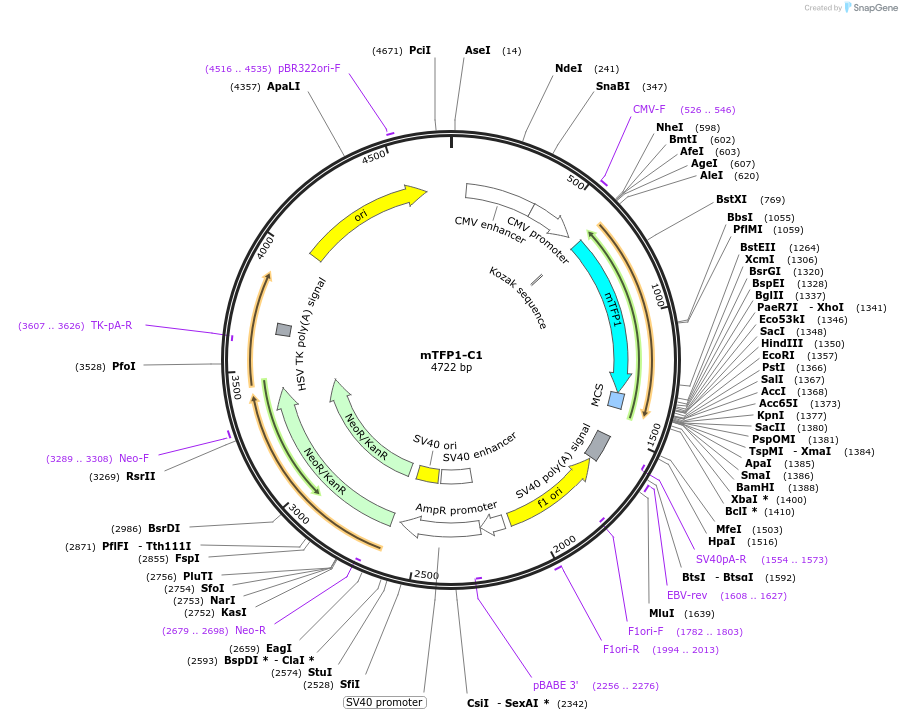
Plasmid#54613PurposeLocalization: C1 Cloning Vector, Excitation: 462, Emission: 492DepositorTypeEmpty backboneTagsmTFP1ExpressionMammalianAvailable SinceJune 20, 2014AvailabilityAcademic Institutions and Nonprofits only
-
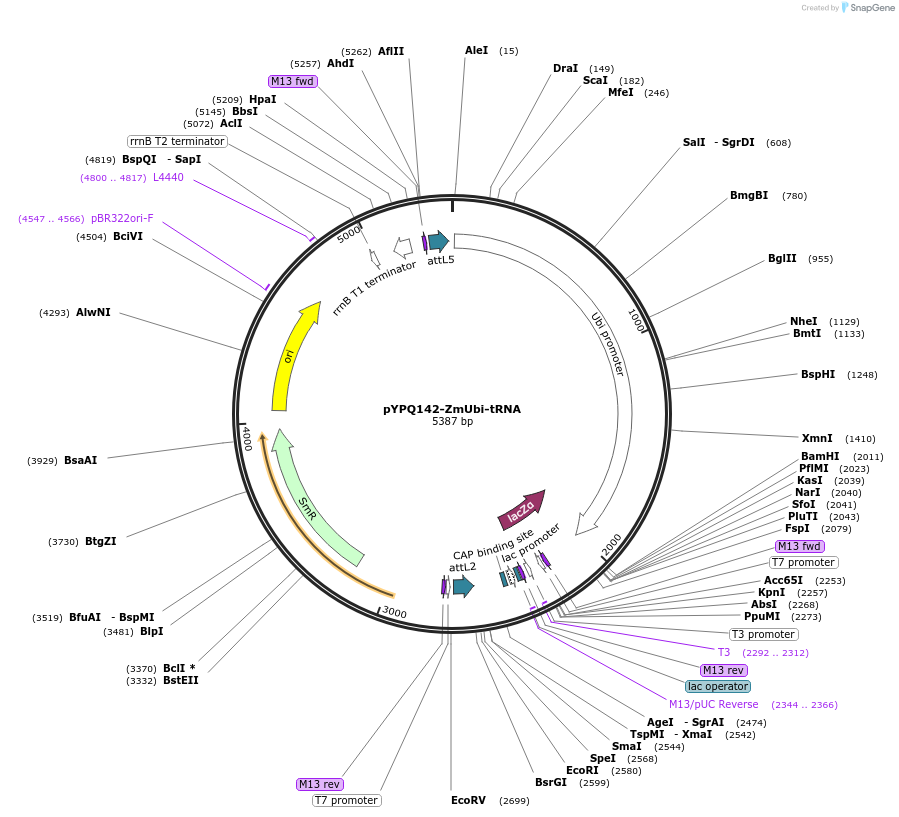
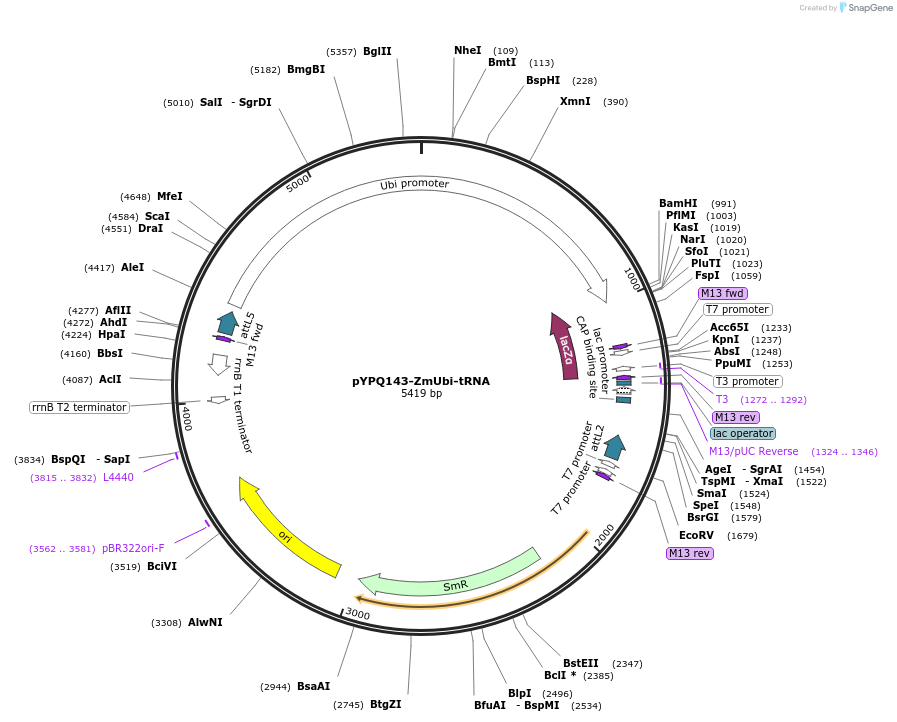
pYPQ142-ZmUbi-tRNA
Plasmid#158578PurposeGateway entry clone and Golden Gate recipient for pYPQ131-tRNA2.0 and pYPQ132-tRNA2.0; assembly of 2 gRNAsDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterMaize ubiquitin 1Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
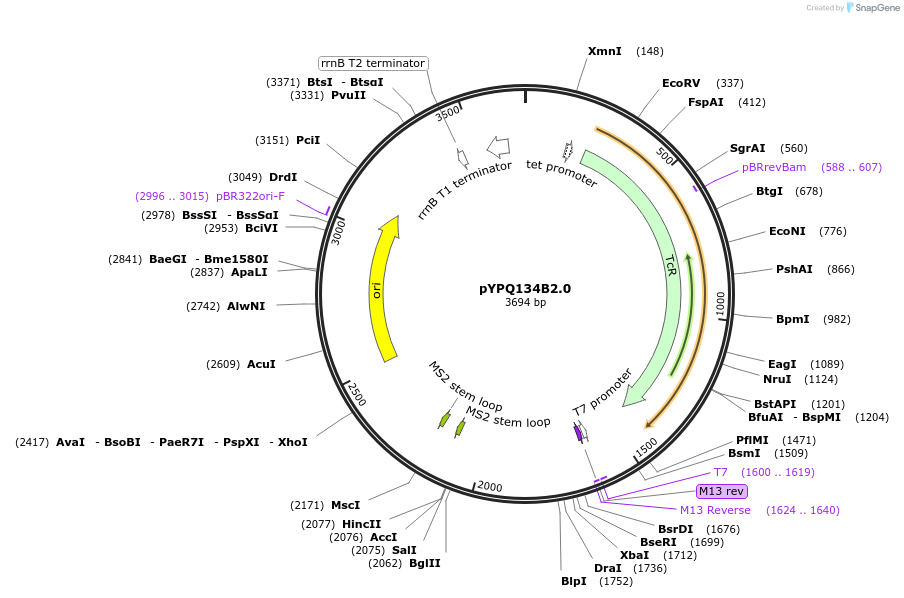
pYPQ134B2.0
Plasmid#167158PurposeGolden Gate entry vector to express the 4th gRNA with gRNA2.0 scaffold (with four MS2 binding sites) under AtU3 promoterDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterAtU3Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
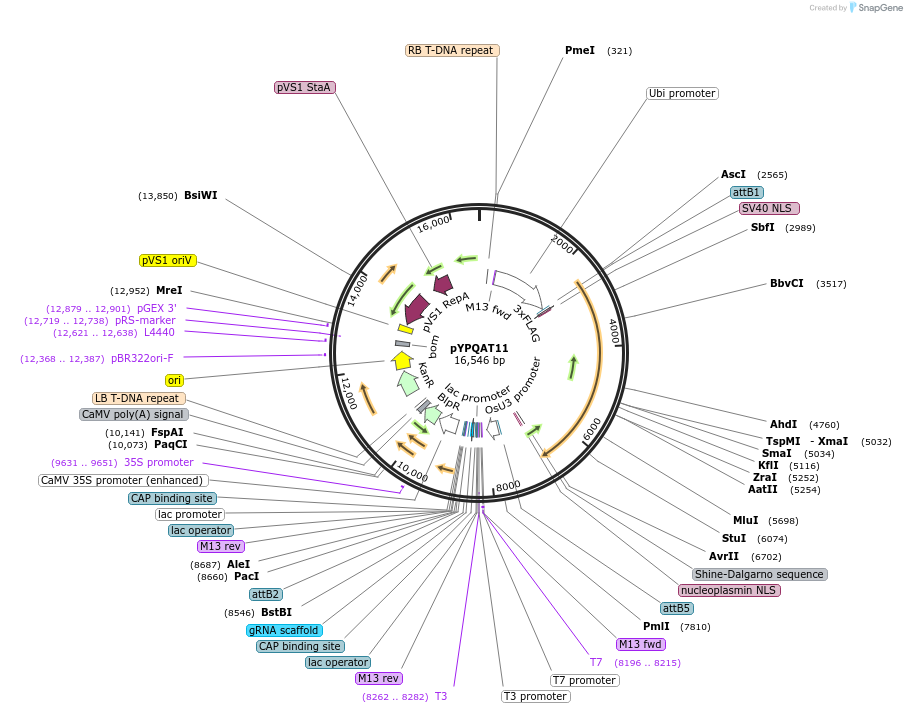
pYPQAT11
Plasmid#223383PurposeT-DNA vector for SpRY mediated mutagenesis for monocot plants; NG or NA PAM preference; SpRY was driven by ZmUbi1 and the sgRNA was driven by OsU3 promoter; BASTA for plants selection.DepositorInsertZmUbi-SpRY-OsU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceJuly 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
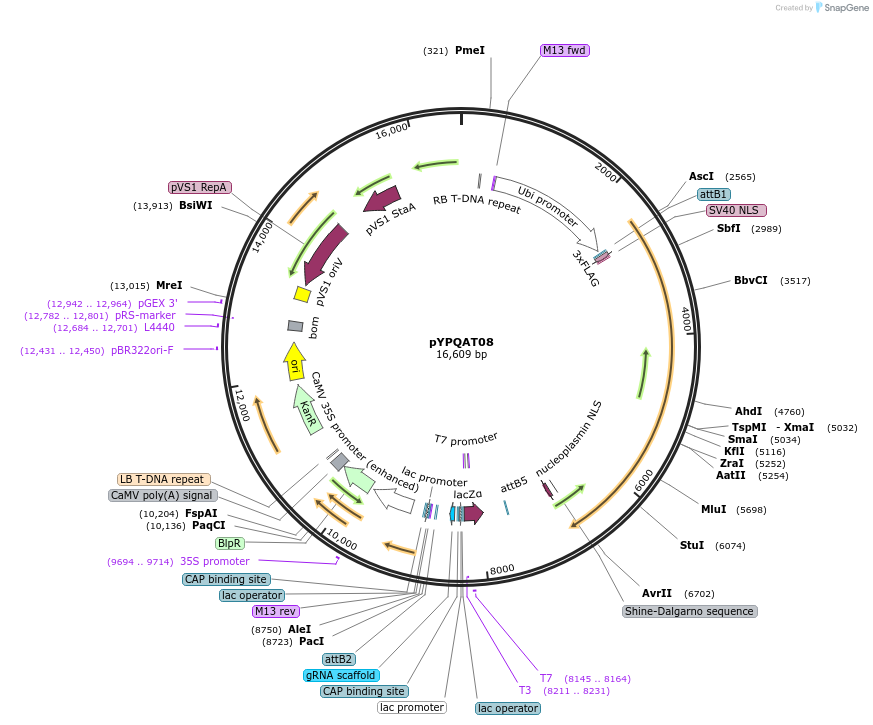
pYPQAT08
Plasmid#223380PurposeT-DNA vector for SpRY mediated mutagenesis for dicot plants; NG or NA PAM preference; SpRY was driven by ZmUbi1 and the sgRNA was driven by AtU3 promoter; BASTA for plants selection.DepositorInsertZmUbi-SpRY-AtU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
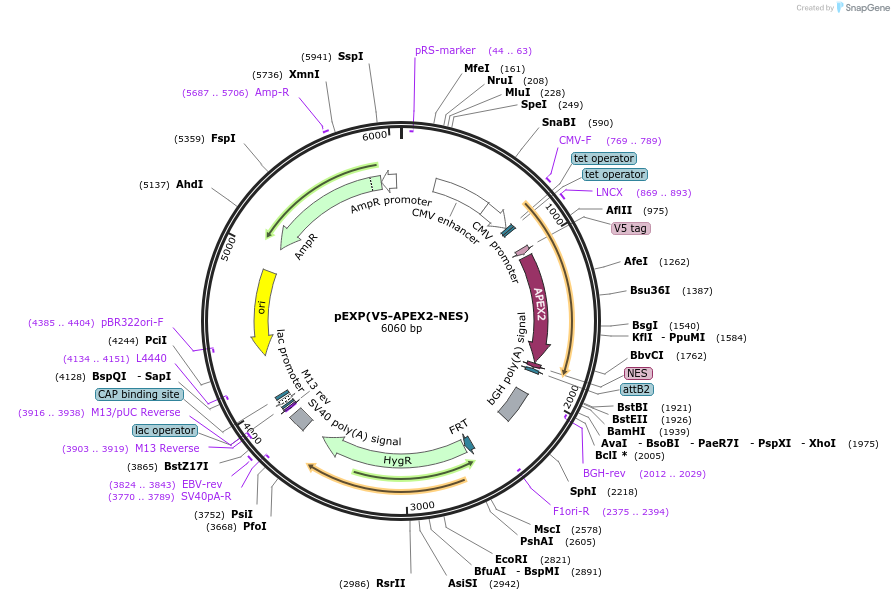
pEXP(V5-APEX2-NES)
Plasmid#107596PurposeExpresses V5-APEX2-NES in mammalian cellsDepositorInsertAPEX2-NES
UseFrt site to insert into hek293 t-rex genomeTagsNES and V5ExpressionMammalianPromoterCMV Dox inducibleAvailable SinceMay 9, 2018AvailabilityAcademic Institutions and Nonprofits only -
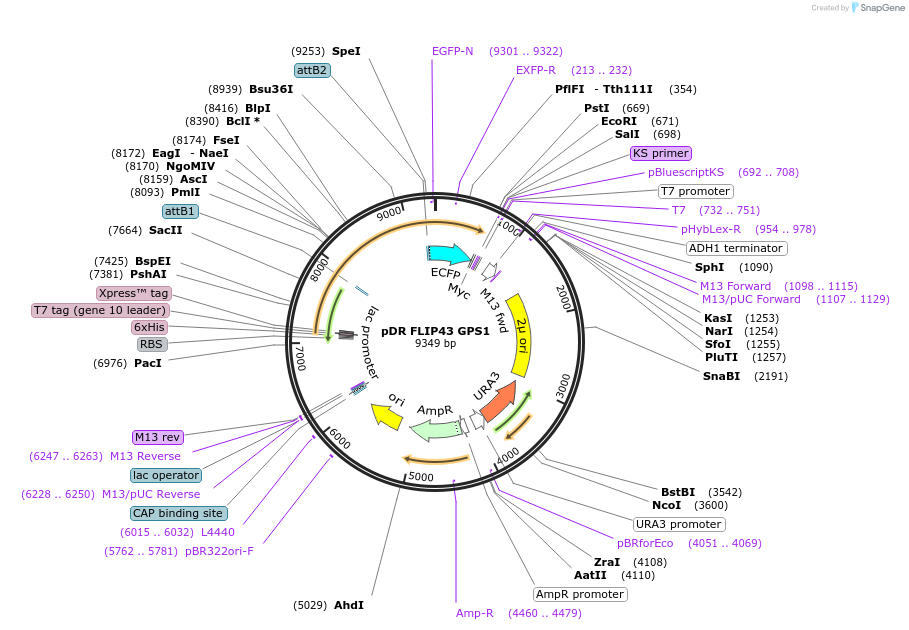
pDR FLIP43 GPS1
Plasmid#101084PurposeGA biosensor expression plasmidDepositorInsertGPS1
Tags6HIS and cMycExpressionYeastPromoterpPMA1Available SinceOct. 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
pYPQ143-ZmUbi-tRNA
Plasmid#158400PurposeGateway entry clone and Golden Gate recipient for pYPQ131-tRNA2.0 to pYPQ133-tRNA2.0; assembly of 3 gRNAsDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterMaize ubiquitin 1Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
pYPQ135B2.0
Plasmid#167159PurposeGolden Gate entry vector to express the 5th gRNA with gRNA2.0 scaffold (with four MS2 binding sites) under AtU3 promoterDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterAtU3Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
pRPS5A-PTP-3Xflag-16S rRNA Left_G1397C-ABE
Plasmid#189642Purpose16S rRNA Left_G1397C-ABEDepositorInsertTALE and DddAtox and TadA 8e
UseTALENExpressionPlantPromoterRPS5AAvailable SinceDec. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
pYPQ144-ZmUbi-tRNA
Plasmid#158402PurposeGateway entry clone and Golden Gate recipient for pYPQ131-tRNA2.0 to pYPQ134-tRNA2.0; assembly of 4 gRNAsDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterMaize ubiquitin 1Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
pEYFP-p23
Plasmid#108225PurposeMammalian expression vector for EYFP fusion to human p23DepositorAvailable SinceApril 5, 2018AvailabilityAcademic Institutions and Nonprofits only -
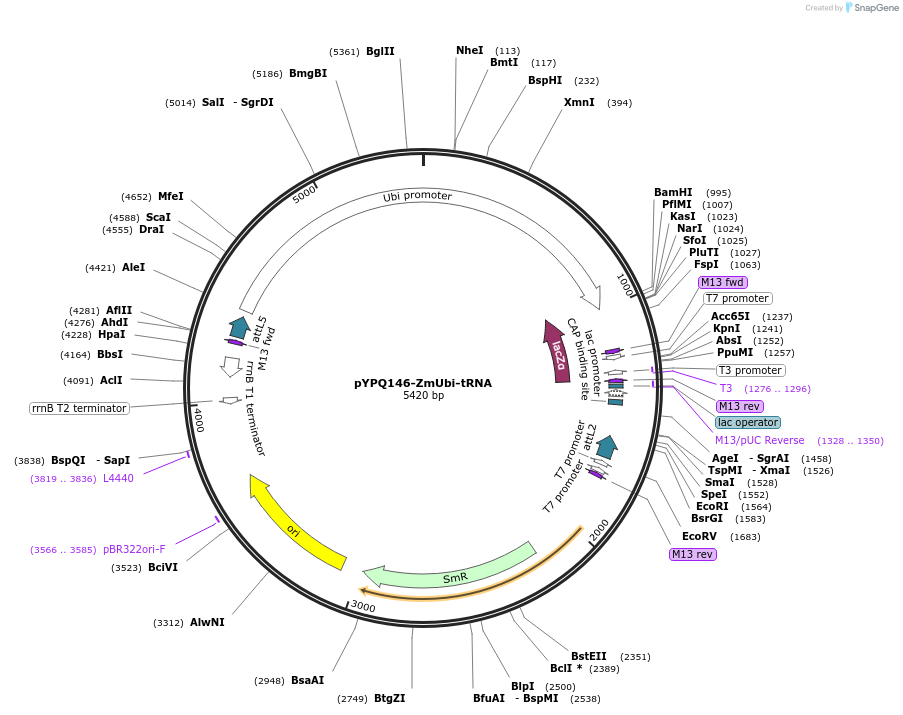
pYPQ146-ZmUbi-tRNA
Plasmid#158404PurposeGateway entry clone and Golden Gate recipient for pYPQ131-tRNA2.0 to pYPQ136-tRNA2.0; assembly of 6 gRNAsDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterMaize ubiquitin 1Available SinceJuly 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
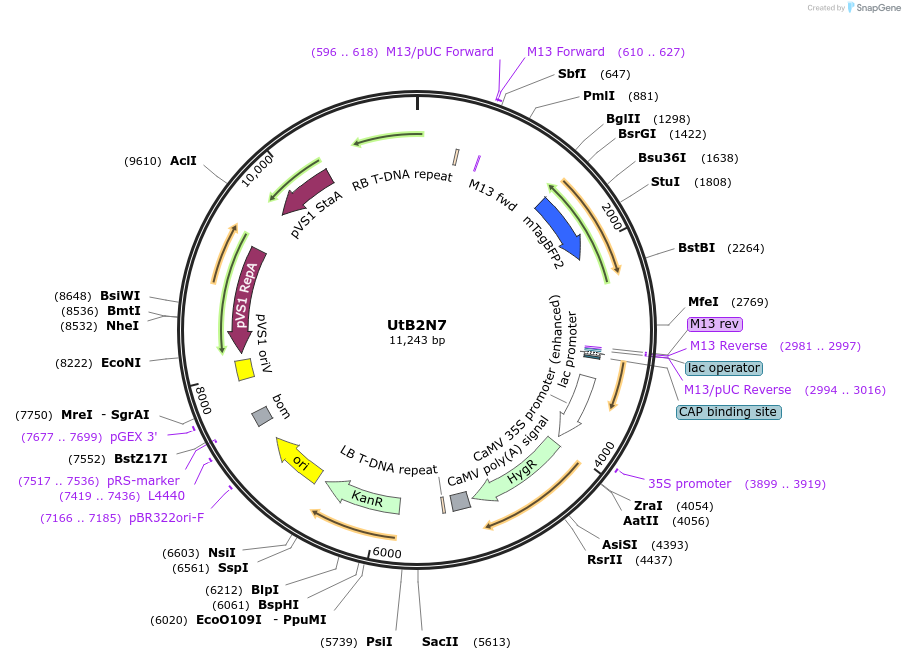
UtB2N7
Plasmid#161015Purposeubiquitously expressed nuclear localized tagBFP2DepositorInserttagBFP2
ExpressionPlantAvailable SinceDec. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
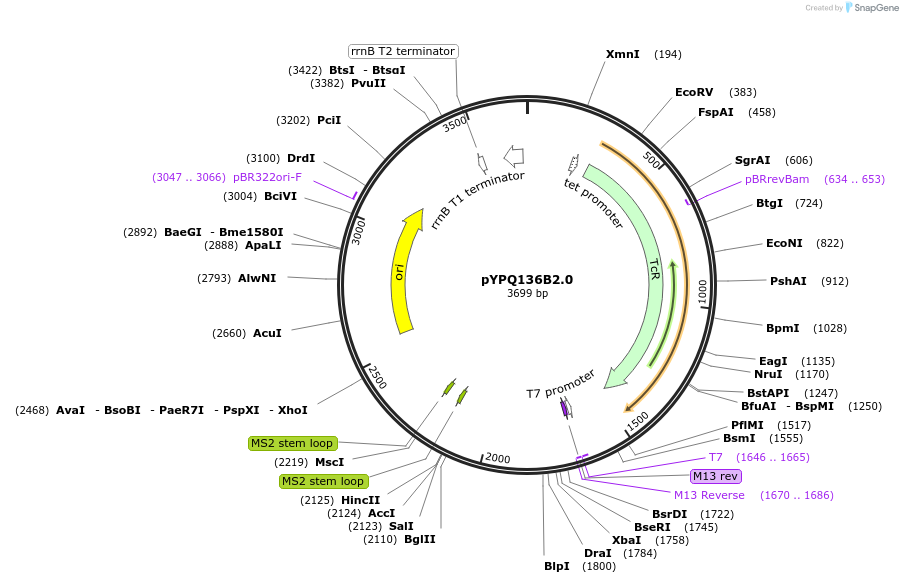
pYPQ136B2.0
Plasmid#167160PurposeGolden Gate entry vector to express the 6th gRNA with gRNA2.0 scaffold (with four MS2 binding sites) under AtU3 promoterDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterAtU3Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
AL13Rb-35S
Plasmid#161008PurposePP7-tagged reporter driven by the 35S promoterDepositorInsertPP7 and H2B-mScarlet
ExpressionPlantAvailable SinceDec. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
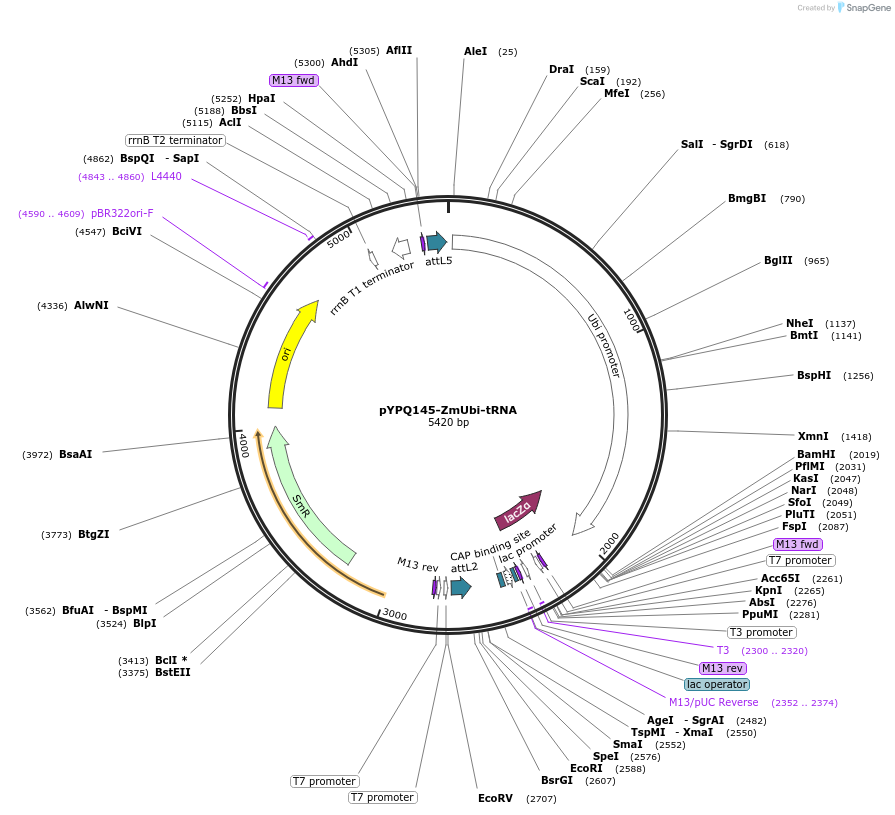
pYPQ145-ZmUbi-tRNA
Plasmid#158403PurposeGateway entry clone and Golden Gate recipient for pYPQ131-tRNA2.0 to pYPQ135-tRNA2.0; assembly of 5 gRNAsDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterMaize ubiquitin 1Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
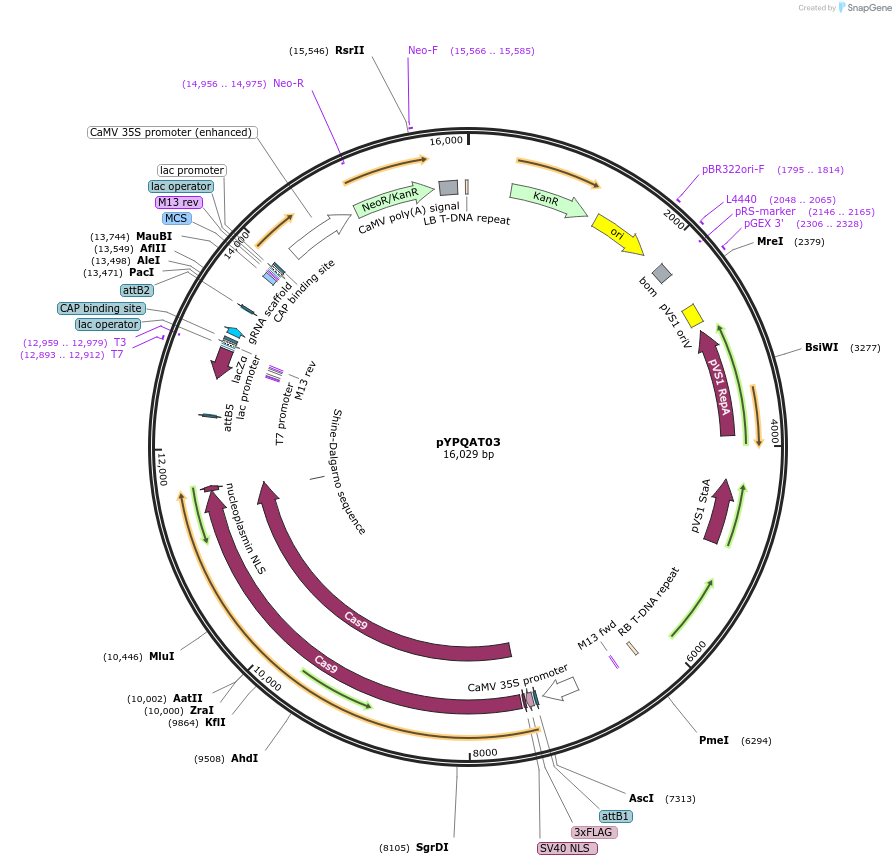
pYPQAT03
Plasmid#223375PurposeT-DNA vector for SpCas9 mediated mutagenesis for dicot plants; NGG PAM; Cas9 was driven by 2x35s and the sgRNA was driven by AtU3 promoter; Kanamycin for plants selection.DepositorInsert2x35s-SpCas9-AtU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
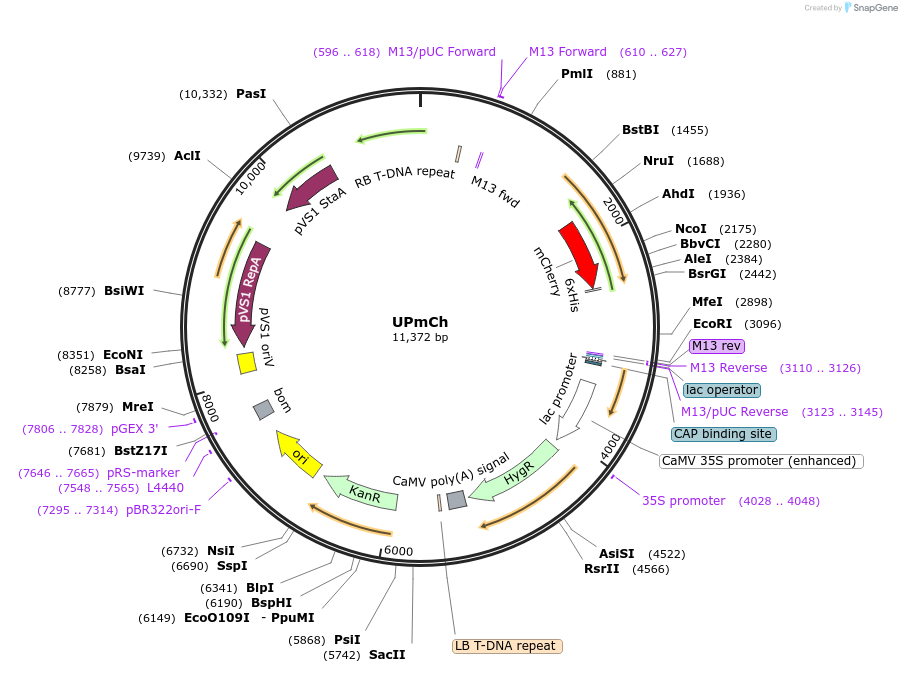
UPmCh
Plasmid#161004Purposeubiquitous expression of PCP-mCherryDepositorInsertPCP-mCherry
ExpressionPlantAvailable SinceDec. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
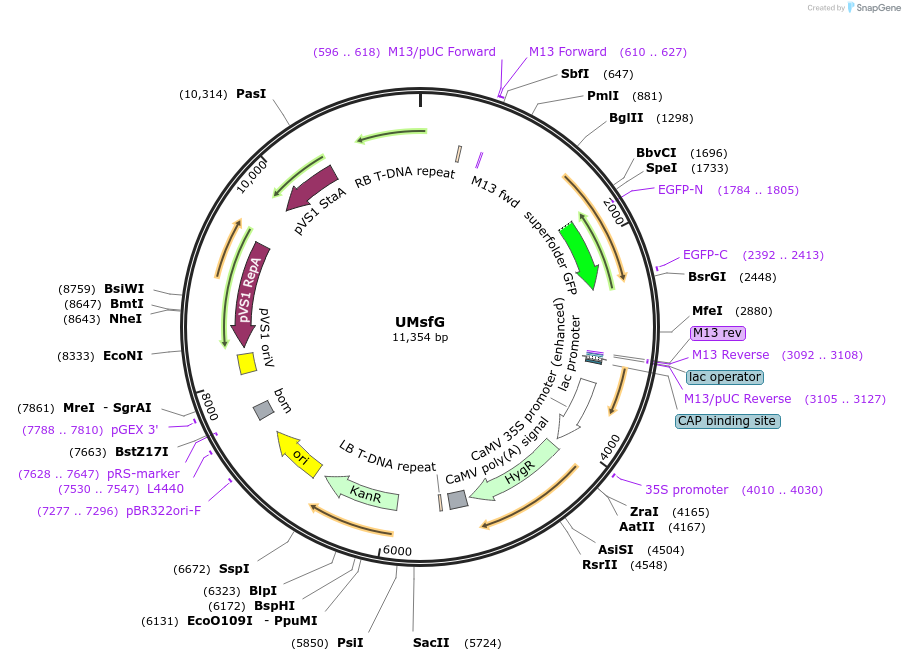
UMsfG
Plasmid#161005Purposeubiquitous expression of MCP-sfGFPDepositorInsertMCP-sfGFP
ExpressionPlantAvailable SinceDec. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
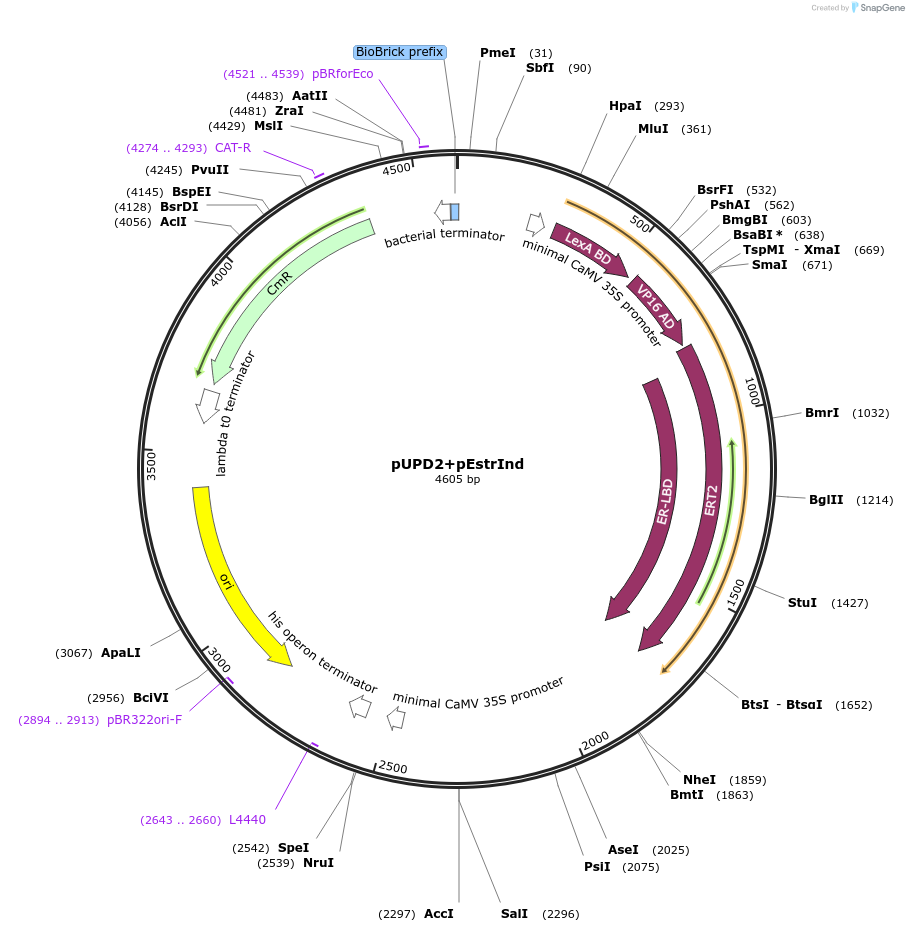
pUPD2+pEstrInd
Plasmid#170880PurposePhytobrick - Estradiol Inducible PromoterDepositorInsertpEstradiol_Inducible
ExpressionPlantMutationc.837C>T and c.1353G>A to remove BsaI and E…Available SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
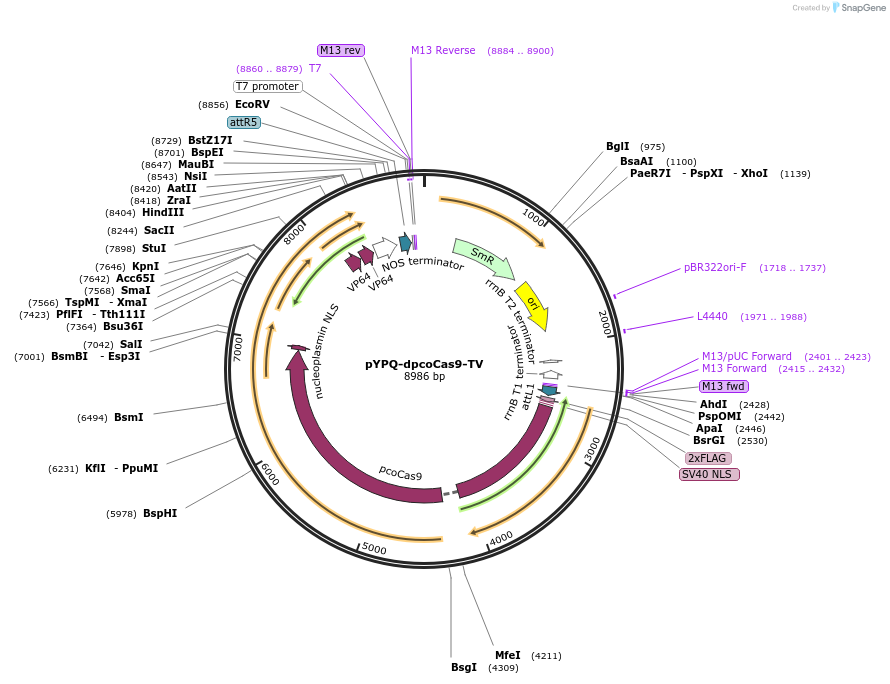
pYPQ-dpcoCas9-TV
Plasmid#158409PurposeCRISPR-dpcoCas9 fused with TV activator for transcriptional activationDepositorInsertdpcoCas9-TV(6xTAL-2xVP64)
UseCRISPRExpressionPlantAvailable SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
UPG
Plasmid#161003Purposeubiquitous expression of PCP-GFPDepositorInsertPCP-GFP
ExpressionPlantAvailable SinceDec. 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
mTFP1-Lifeact-7
Plasmid#54749PurposeLocalization: Actin, Excitation: 462, Emission: 492DepositorInsertLifeact-mTFP1
ExpressionMammalianAvailable SinceJune 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
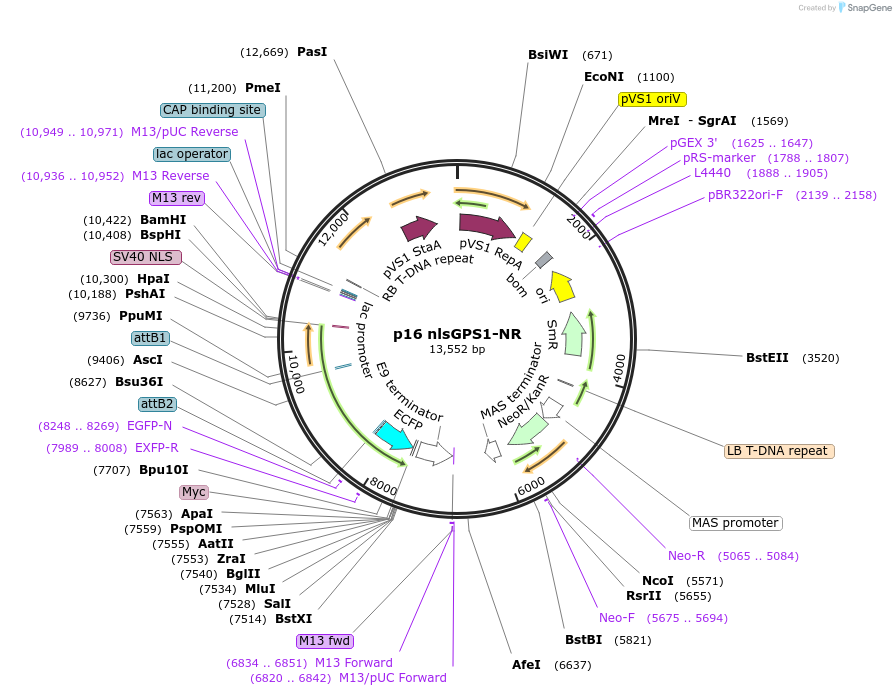
p16 nlsGPS1-NR
Plasmid#101082PurposeGA biosensor expression plasmidDepositorInsertnlsGPS1-NR
TagscMycExpressionPlantMutationAtGAI E51K E54R, AtGID1C S114A F115APromoterp16 (AT3G60245)Available SinceOct. 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
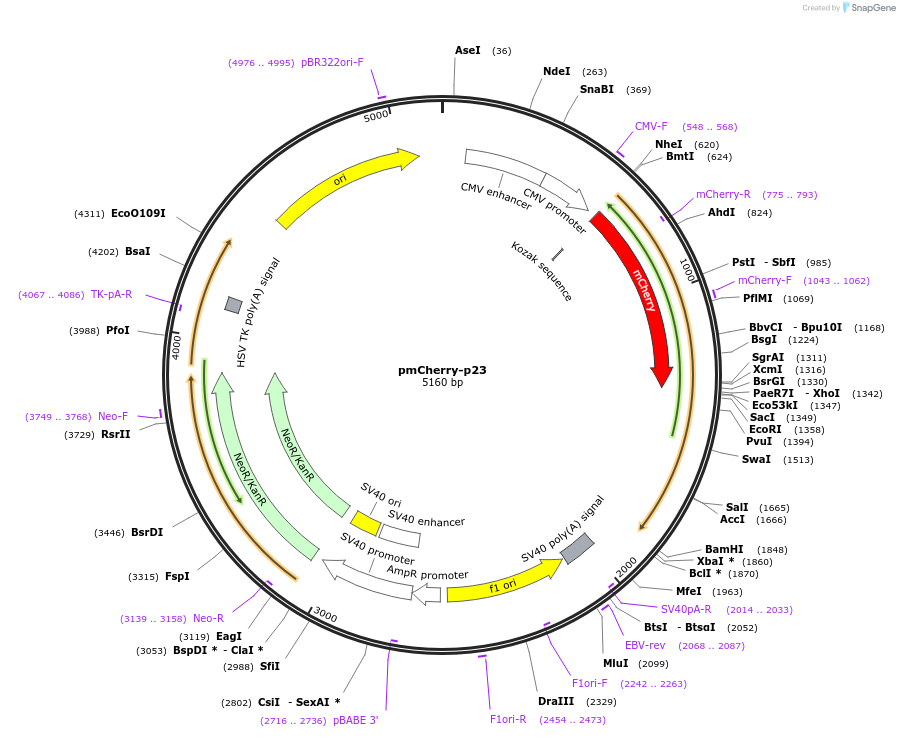
pmCherry-p23
Plasmid#108224PurposeMammalian expression vector for mCherry fusion to human p23DepositorAvailable SinceApril 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
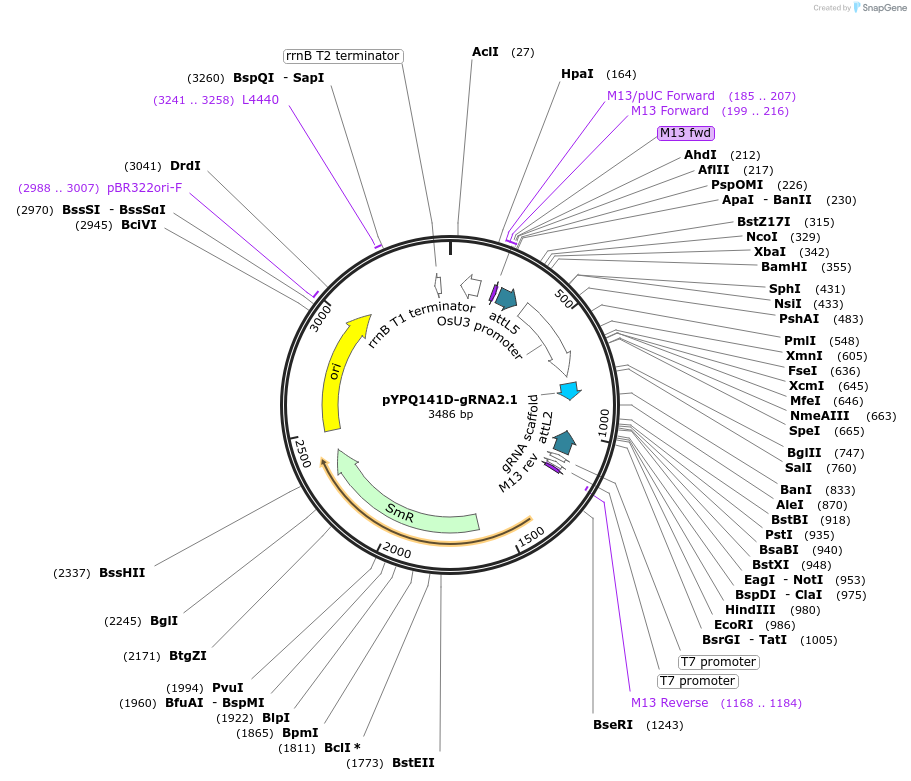
pYPQ141D-gRNA2.1
Plasmid#167161PurposeExpress single gRNA with gRNA2.1 scaffold (with four MS2 binding sites) under OsU3 promoterDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterOsU3Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
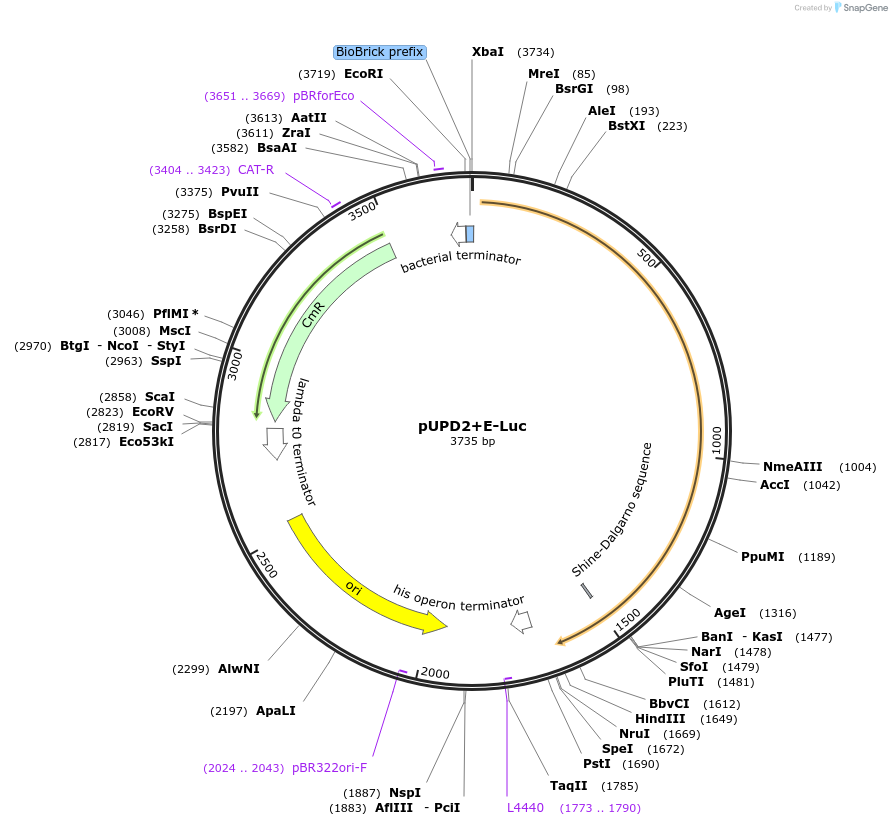
pUPD2+E-Luc
Plasmid#170873PurposePhytobrick - E-Luc Luciferase CDSDepositorInsertE-Luc
UseLuciferase and Synthetic BiologyMutationc.375C>T to remove BbsI siteAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
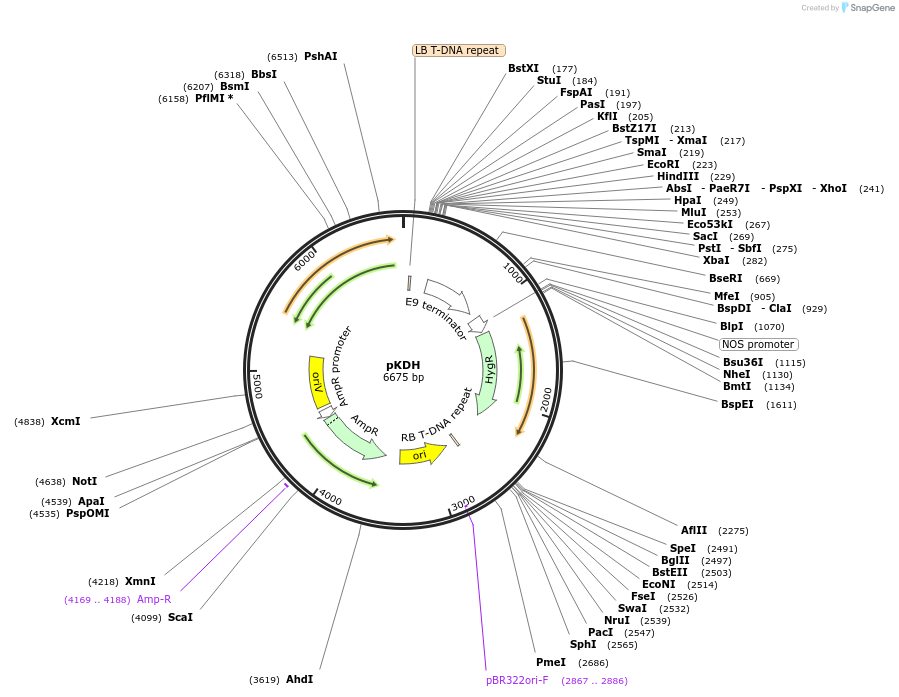
pKDH
Plasmid#67584PurposeGene expression in plants. A promoter and/or reporter can be cloned into this plasmidDepositorTypeEmpty backboneExpressionPlantPromoterNoneAvailable SinceJuly 29, 2015AvailabilityAcademic Institutions and Nonprofits only -
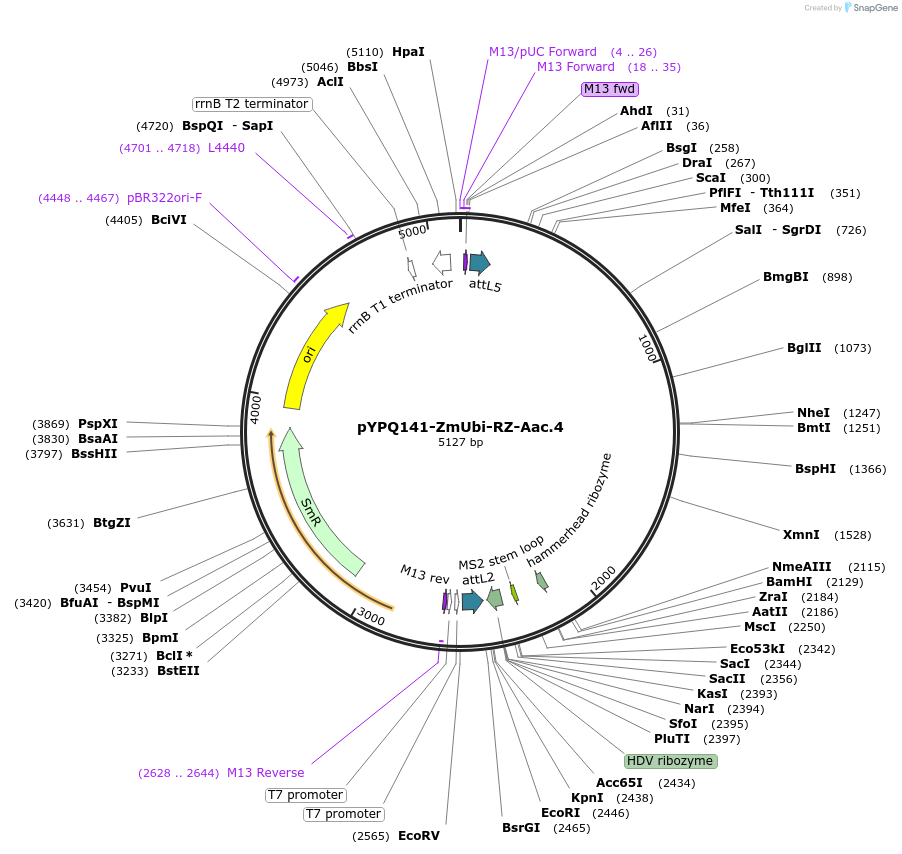
pYPQ141-ZmUbi-RZ-Aac.4
Plasmid#158406PurposeExpress single gRNA with Aac.4 scaffold (with four MS2 binding sites) under ZmUbi promoterDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterMaize ubiquitin 1Available SinceJune 25, 2021AvailabilityAcademic Institutions and Nonprofits only