We narrowed to 9,432 results for: tre promoter
-
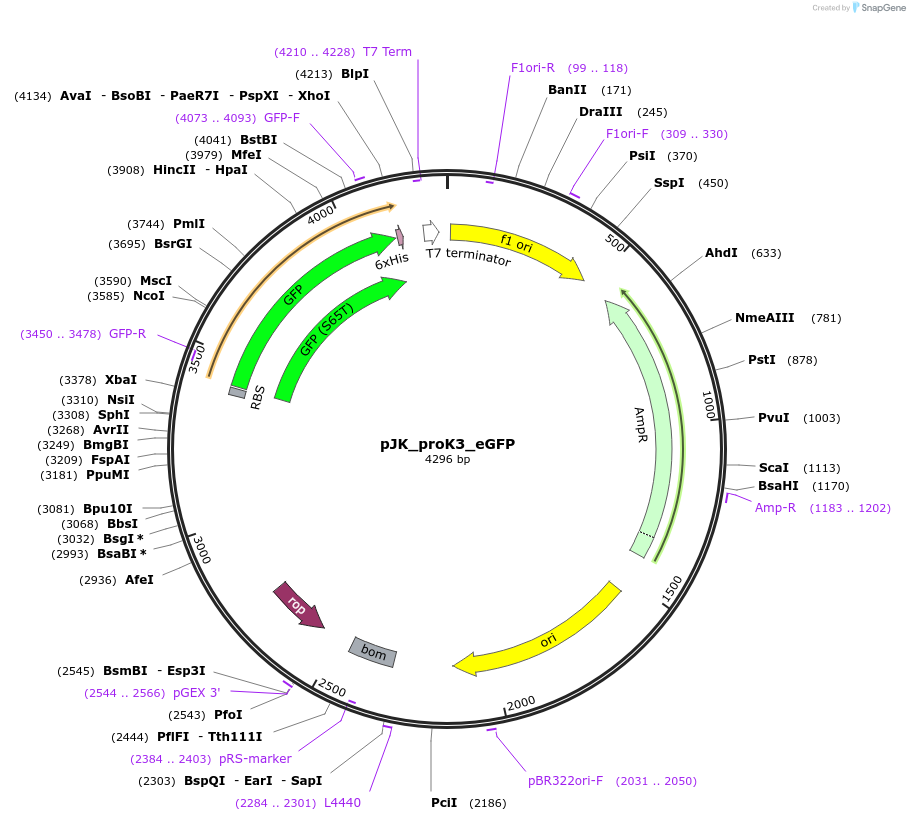
Plasmid#59852PurposeConstitutively expresses eGFP with proK3 promoterDepositorInsertEnhanced Green Fluorescent Protein
Tags6x-HisExpressionBacterialMutationS2R,S65G,S72APromoterproK3Available SinceOct. 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
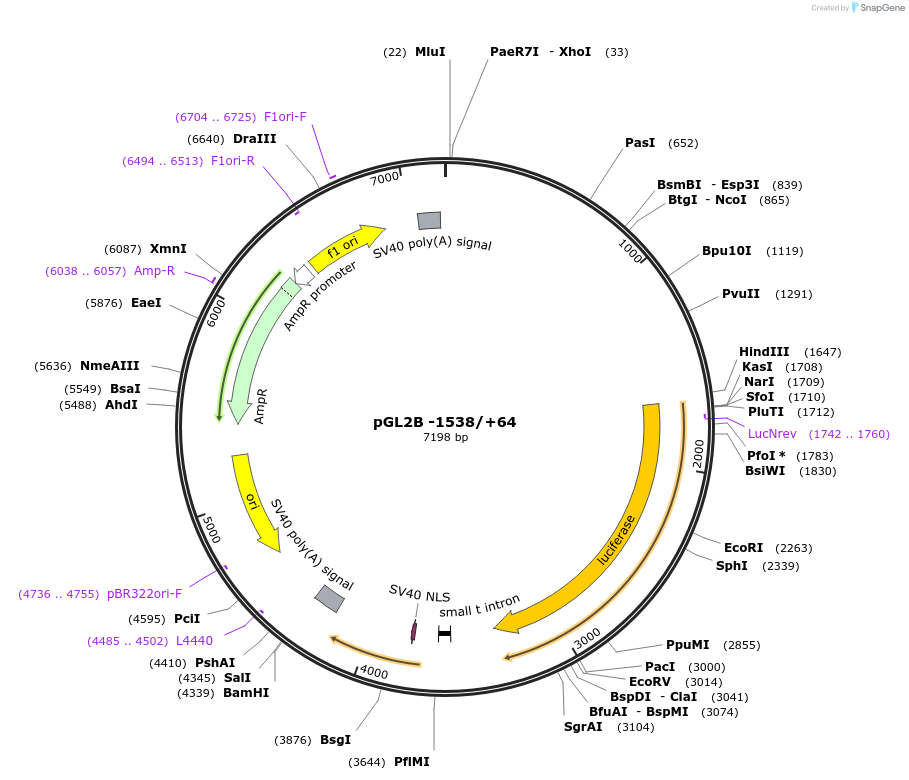
pGL2B -1538/+64
Plasmid#24942DepositorAvailable SinceAug. 18, 2010AvailabilityAcademic Institutions and Nonprofits only -
pGL2B -938/+64
Plasmid#24943DepositorAvailable SinceAug. 18, 2010AvailabilityAcademic Institutions and Nonprofits only -
pGL2B -158/+64
Plasmid#24946DepositorAvailable SinceJune 4, 2010AvailabilityAcademic Institutions and Nonprofits only -
pGL2B -376/+64
Plasmid#24945DepositorAvailable SinceAug. 18, 2010AvailabilityAcademic Institutions and Nonprofits only -
pGL2B -688/+64
Plasmid#24944DepositorAvailable SinceAug. 18, 2010AvailabilityAcademic Institutions and Nonprofits only -
pGL2B -78/+64
Plasmid#24947DepositorAvailable SinceAug. 18, 2010AvailabilityAcademic Institutions and Nonprofits only -
pGL2B -55/+64
Plasmid#24948DepositorAvailable SinceJune 4, 2010AvailabilityAcademic Institutions and Nonprofits only -
-
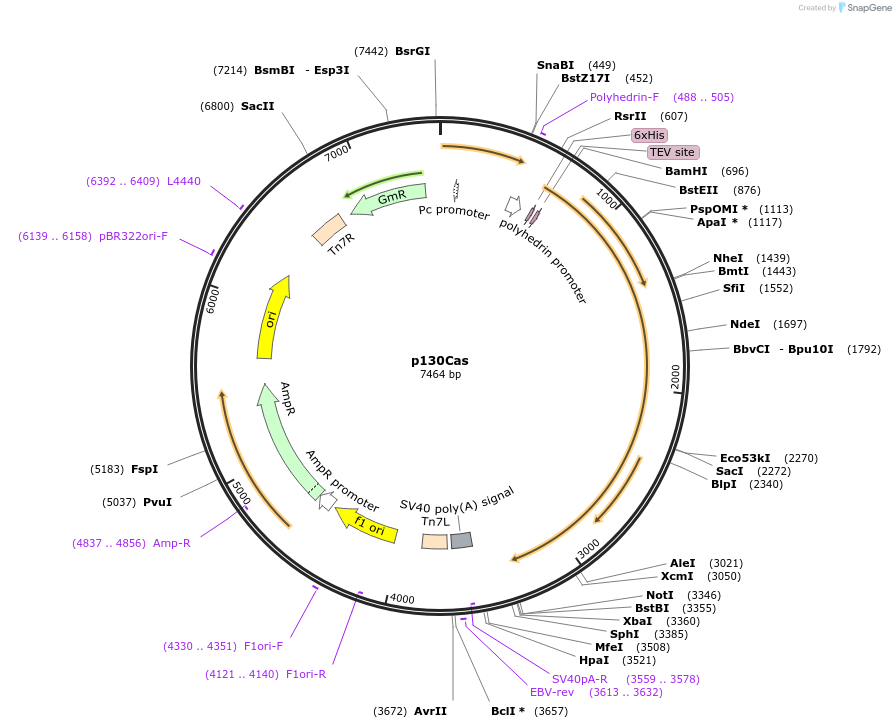
p130Cas
Plasmid#186131PurposeFor expression of recombinant p130Cas in SF9 cells.DepositorAvailable SinceJune 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
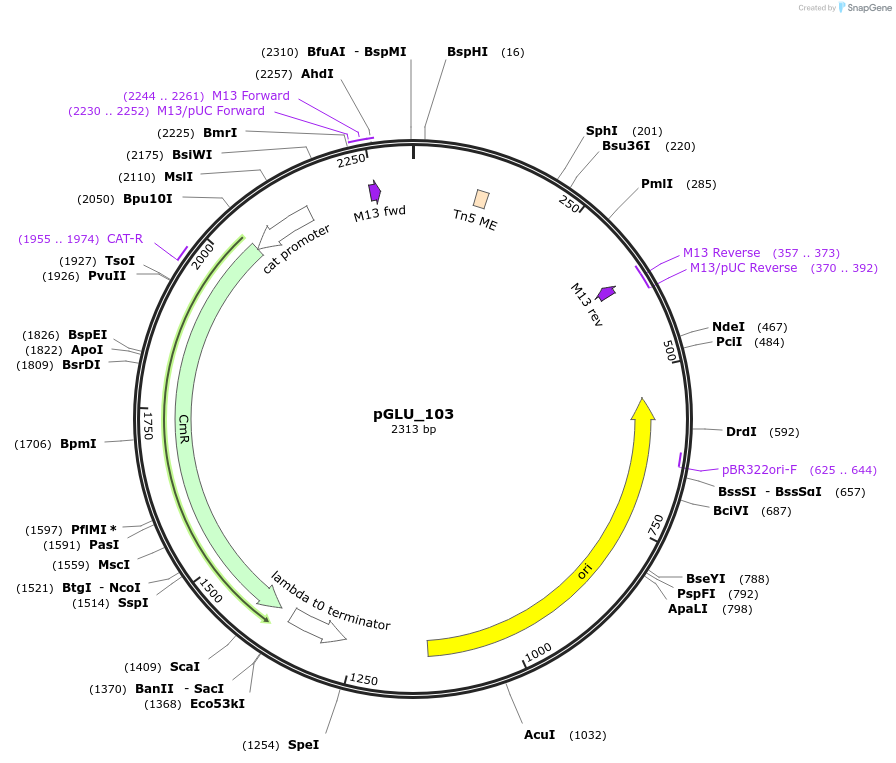
pGLU_103
Plasmid#225785PurposeThis is the promoter region taken from upstream of Tn5 transposase in pBAM1. It can be considered the "default" promoter to use.DepositorInsertPtn5_[Tn5]
UseSynthetic BiologyMutationN/AAvailable SinceSept. 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
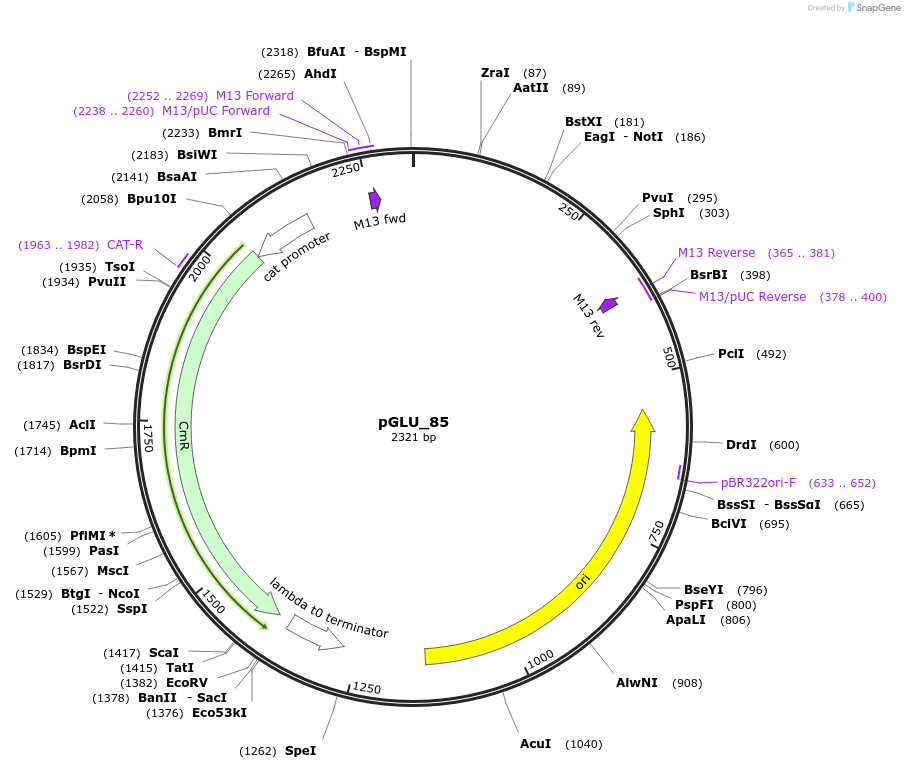
pGLU_85
Plasmid#225800PurposeThis is the promoter region taken from upstream of Himar transposase in pMarC9-Kan. It can be considered the "default" promoter to use.DepositorInsertPmarC9_[Himar]
UseSynthetic BiologyMutationN/AAvailable SinceSept. 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
-
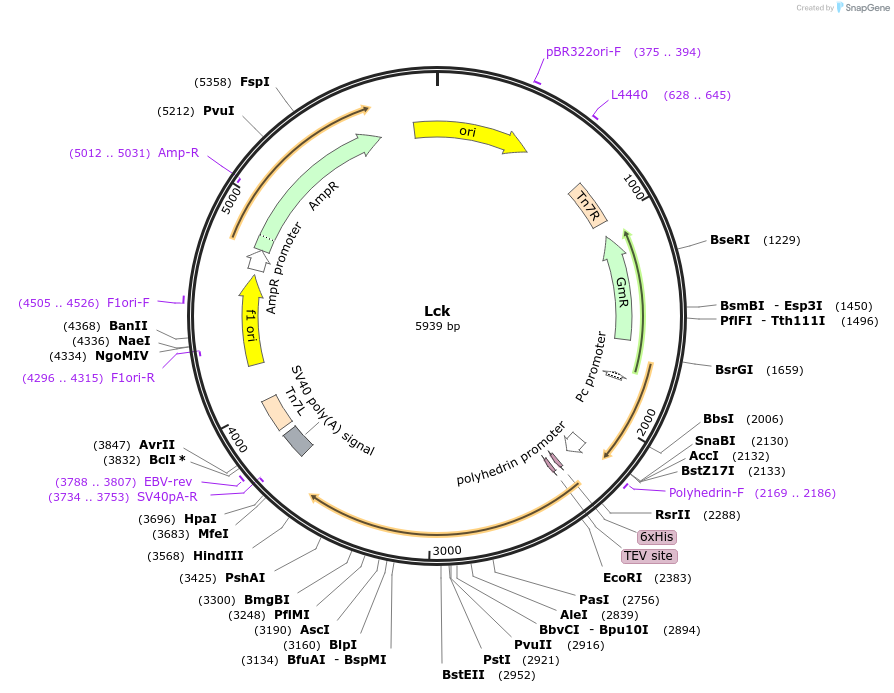
Lck
Plasmid#186130PurposeFor expression of recombinant Lck in SF9 cells.DepositorAvailable SinceJune 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
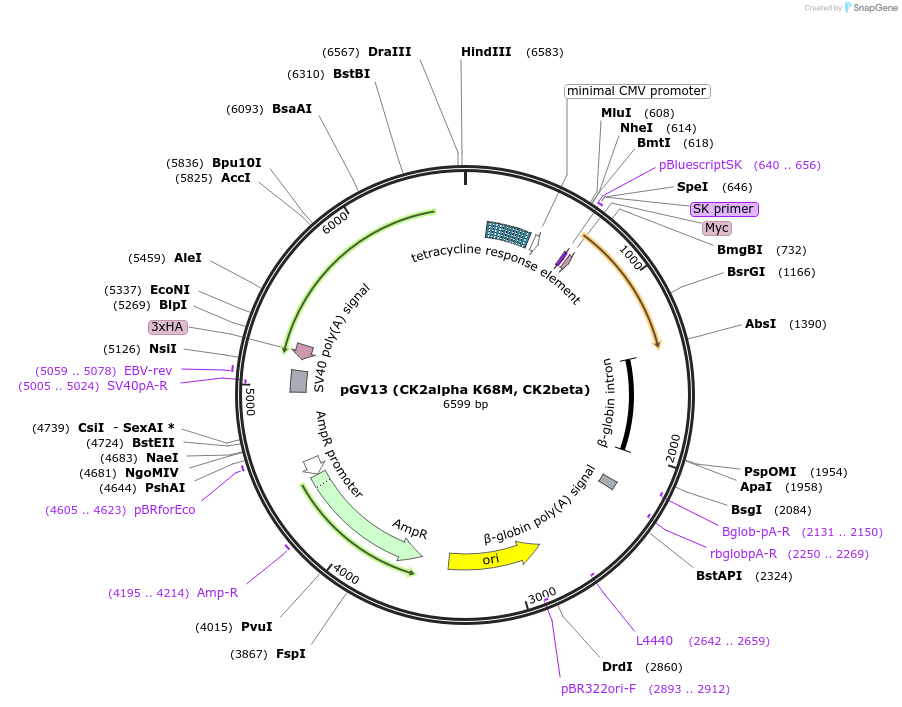
pGV13 (CK2alpha K68M, CK2beta)
Plasmid#27094DepositorUseTetracycline-regulated expressionTagsHA on CK2alpha and Myc on CK2betaExpressionMammalianMutationK68M on CK2alphaPromoterbidirectional tet-responsive promoter and bidirec…Available SinceOct. 14, 2011AvailabilityAcademic Institutions and Nonprofits only -
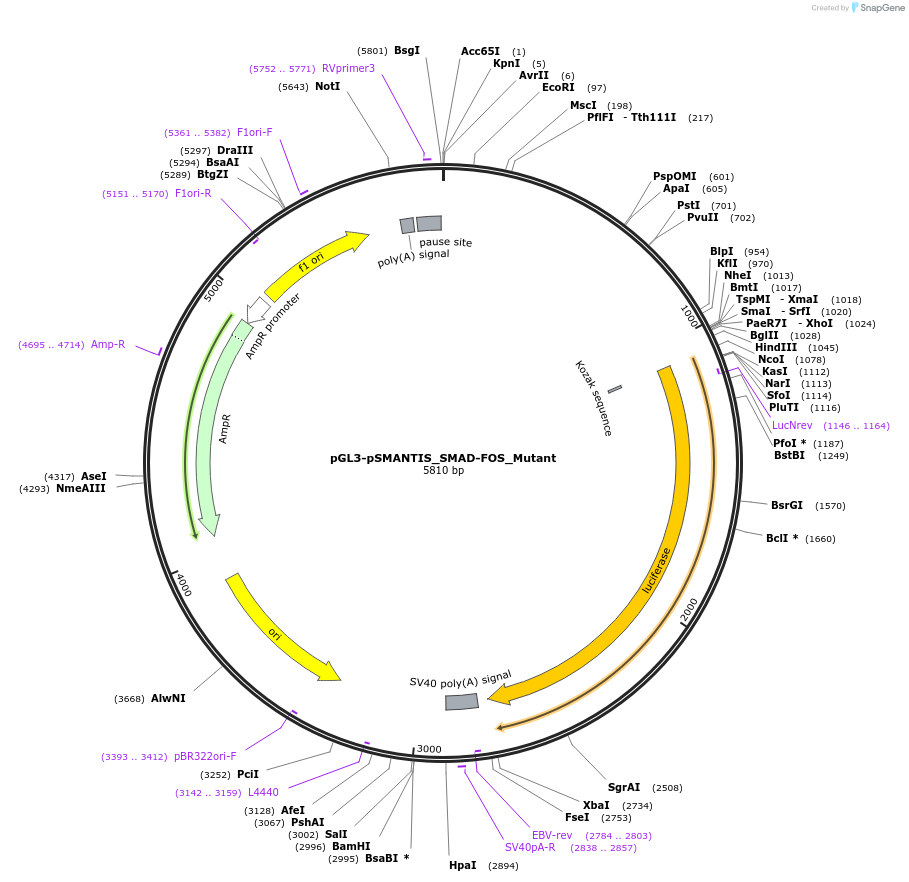
pGL3-pSMANTIS_SMAD-FOS_Mutant
Plasmid#194188PurposeIncludes the promoter (1kb) of SMTS with mutated SMAD/FOS binding siteDepositorInsertSMANTIS promoter (1kb) (SMANTIS Human)
UseLuciferaseMutationmutated SMAD/FOS Site, Region 3-15nt of 1000ntPromoterSMANTIS promoter (1kb), mutatedAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
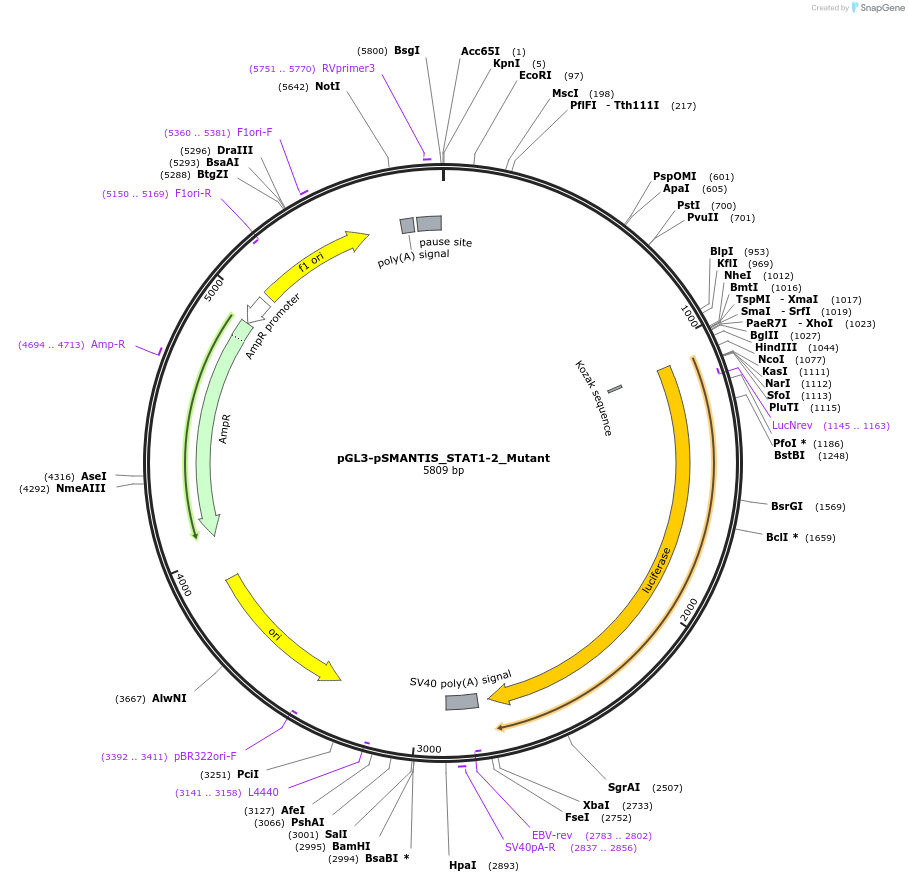
pGL3-pSMANTIS_STAT1-2_Mutant
Plasmid#194189PurposeIncludes the promoter (1kb) of SMTS with mutated STAT1/2 binding siteDepositorInsertSMANTIS promoter (1kb) (SMANTIS Human)
UseLuciferaseMutationmutated STAT1/2 site, Region 653-673nt of 1000ntPromoterSMANTIS promoter (1kb), mutatedAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
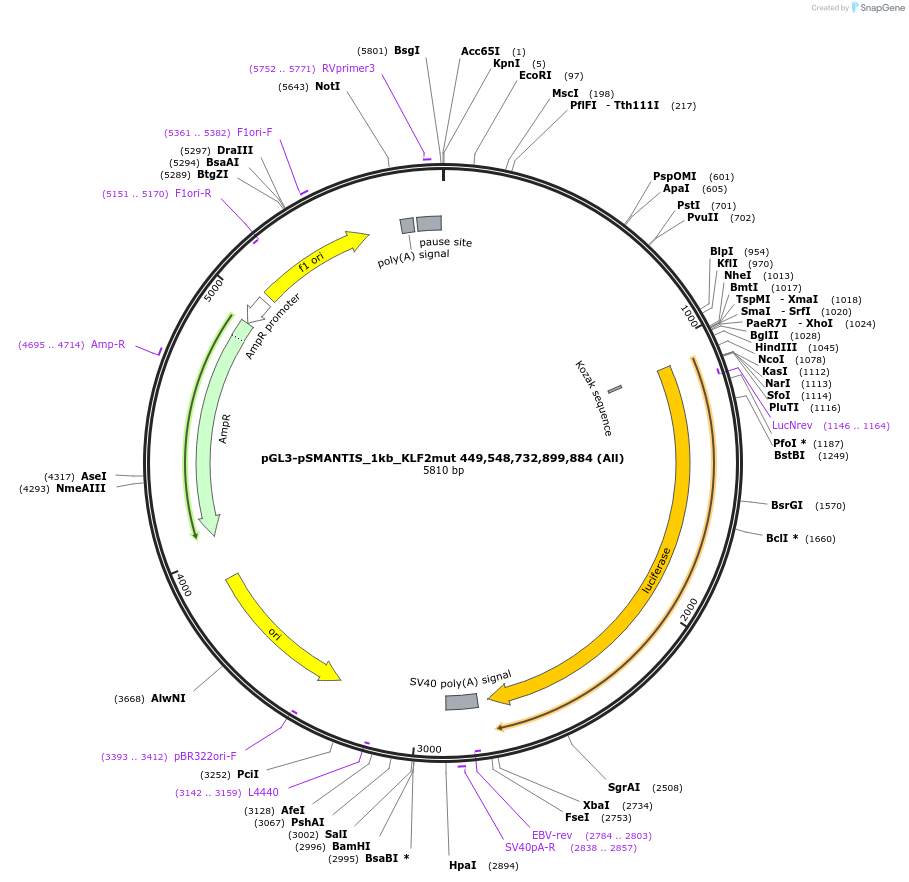
pGL3-pSMANTIS_1kb_KLF2mut 449,548,732,899,884 (All)
Plasmid#194190PurposeIncludes the promoter (1kb) of SMTS with 5 mutated KLF binding sites (positions 449,548,732,899,884)DepositorInsertSMANTIS promoter (1kb) (SMANTIS Human)
UseLuciferaseMutation5 mutated KLF binding sites (positions 449,548,73…PromoterSMANTIS promoter (1kb), mutatedAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
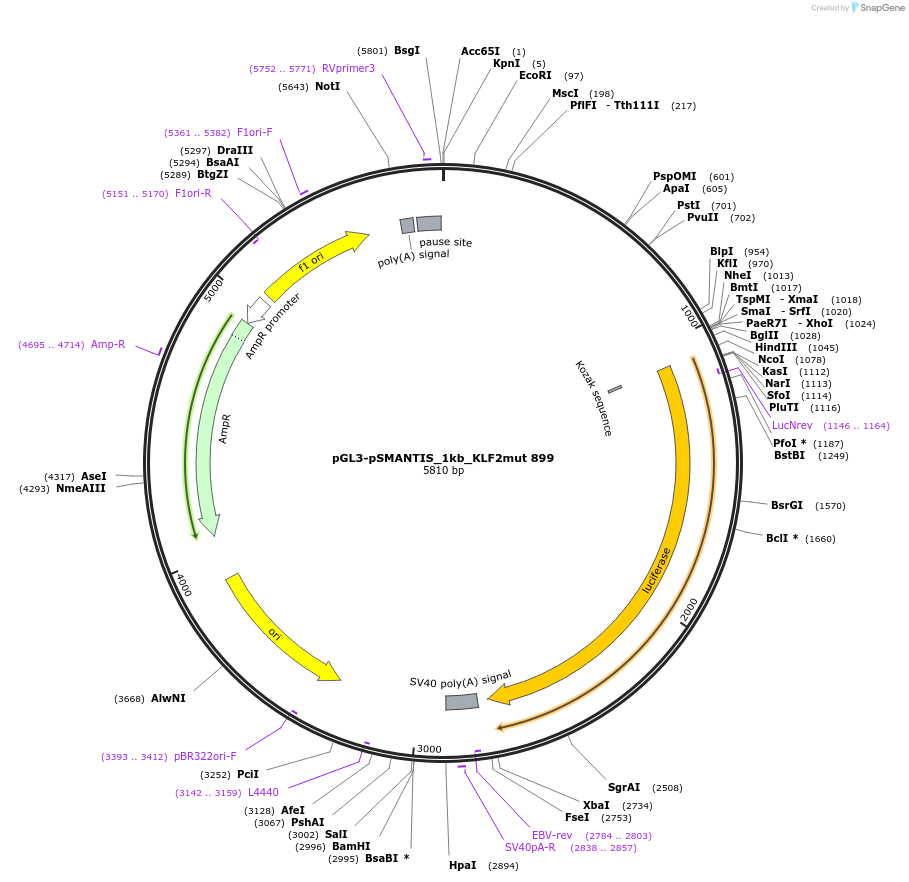
pGL3-pSMANTIS_1kb_KLF2mut 899
Plasmid#194193PurposeIncludes the promoter (1kb) of SMTS with mutated KLF binding sites (position 899)DepositorInsertSMANTIS promoter (1kb) (SMANTIS Human)
UseLuciferaseMutationmutated KLF binding sites (position 899)PromoterSMANTIS promoter (1kb), mutatedAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
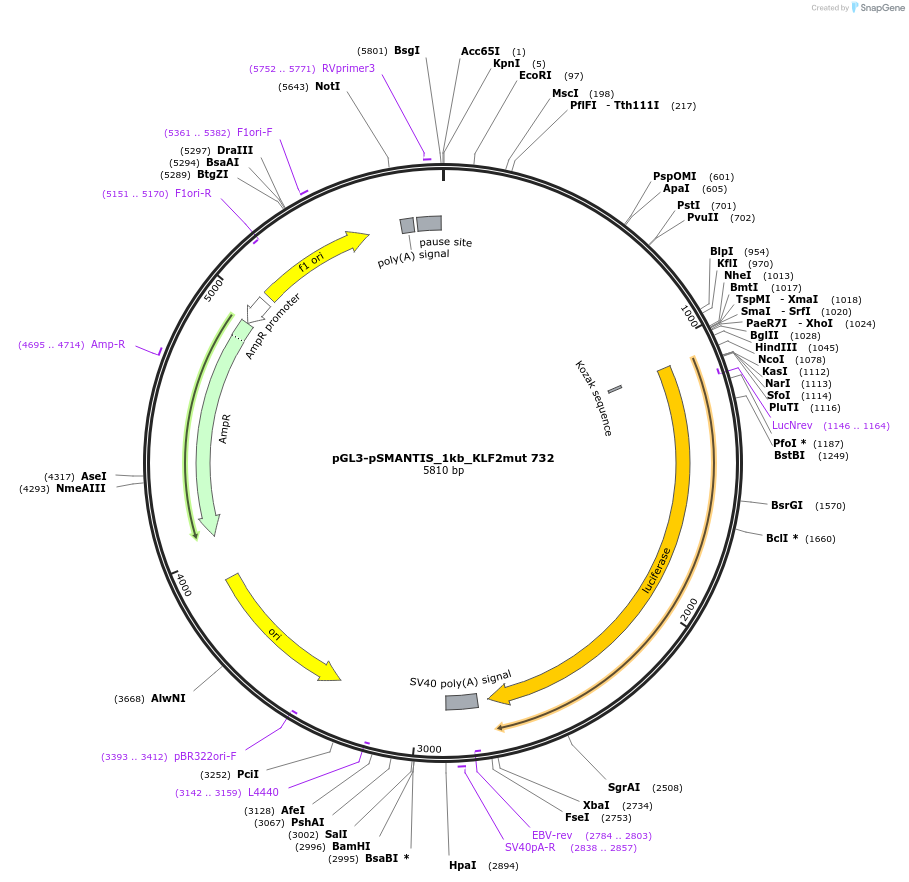
pGL3-pSMANTIS_1kb_KLF2mut 732
Plasmid#194195PurposeIncludes the promoter (1kb) of SMTS with mutated KLF binding sites (position 732)DepositorInsertSMANTIS promoter (1kb) (SMANTIS Human)
UseLuciferaseMutationmutated KLF binding sites (position 732)PromoterSMANTIS promoter (1kb), mutatedAvailable SinceJan. 24, 2023AvailabilityAcademic Institutions and Nonprofits only