We narrowed to 32,226 results for: Eng
-
Plasmid#52149PurposeEncodes module 3 with amino acids 12N-16Q for recognition of nt 6UDepositorInsertPumilio homology domain amino acids 97-132 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12N in Module 3Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-3SR
Plasmid#52150PurposeEncodes module 3 with amino acids 12S-16R for recognition of nt 6CDepositorInsertPumilio homology domain amino acids 97-132 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12S and 16Q to 16R in Module 3Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4CQ
Plasmid#52151PurposeEncodes module 4 with amino acids 12C-16Q for recognition of nt 5ADepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12C in Module 4Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4SR
Plasmid#52154PurposeEncodes module 4 with amino acids 12S-16R for recognition of nt 5CDepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16R in Module 4Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4CYQ
Plasmid#52155PurposeEncodes module 4 with amino acids 12C-13Y-16Q for recognition of nt 5ADepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12C and 13H to 13Y in Module 4Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4SYE
Plasmid#52156PurposeEncodes module 4 with amino acids 12S-13Y-16E for recognition of nt 5GDepositorInsertD43951 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S, 13H to 13Y, and 16Q to 16E in…Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4NYQ
Plasmid#52157PurposeEncodes module 4 with amino acids 12N-13Y-16Q for recognition of nt 5UDepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 13H to 13Y in Module 4Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-5SE
Plasmid#52160PurposeEncodes module 5 with amino acids 12S-16E for recognition of nt 4GDepositorInsertPumilio homology domain amino acids 169-204 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12S and 16Q to 16E in Module 5Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-5NQ
Plasmid#52161PurposeEncodes module 5 with amino acids 12N-16Q for recognition of nt 4UDepositorInsertPumilio homology domain amino acids 169-204 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12N in Module 5Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-6CQ
Plasmid#52163PurposeEncodes module 6 with amino acids 12C-16Q for recognition of nt 3ADepositorInsertPumilio homology domain amino acids 205-240 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12C in Module 6Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-6SR
Plasmid#52166PurposeEncodes module 6 with amino acids 12S-16R for recognition of nt 3CDepositorInsertPumilio homology domain amino acids 205-240 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16R in Module 6Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
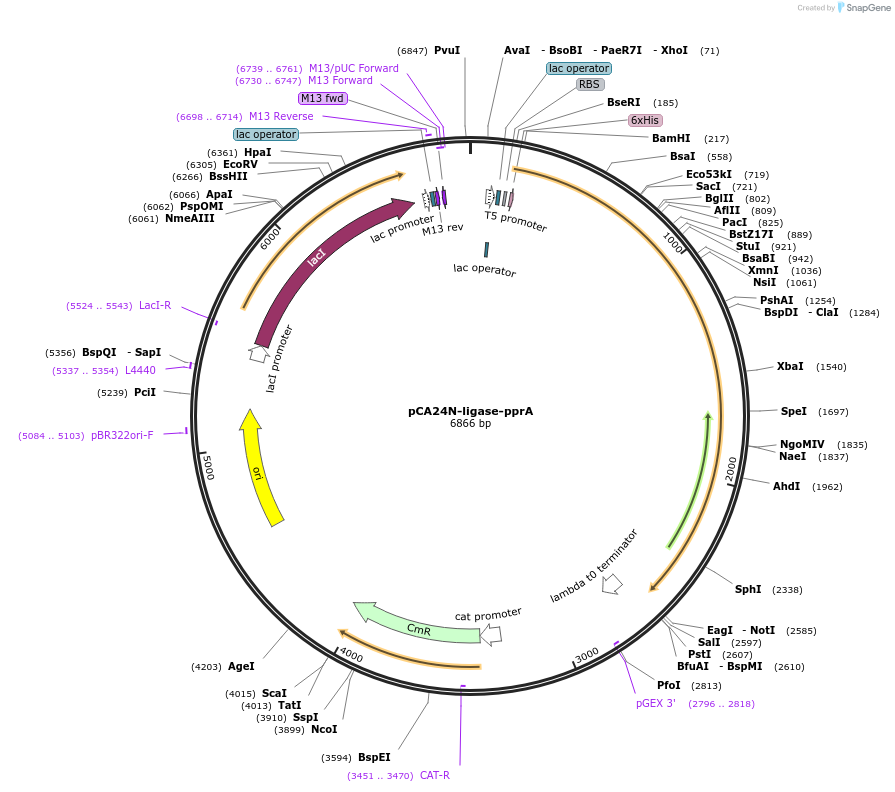
pCA24N-ligase-pprA
Plasmid#87744PurposeIPTG-inducible expression of ligase-pprA fusion for protein purificationDepositorInsertT4 DNA ligase (30 )
Tags6xHis and codon optimized pprA (from Deinococcus …ExpressionBacterialPromoterT5-lacAvailable SinceMarch 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
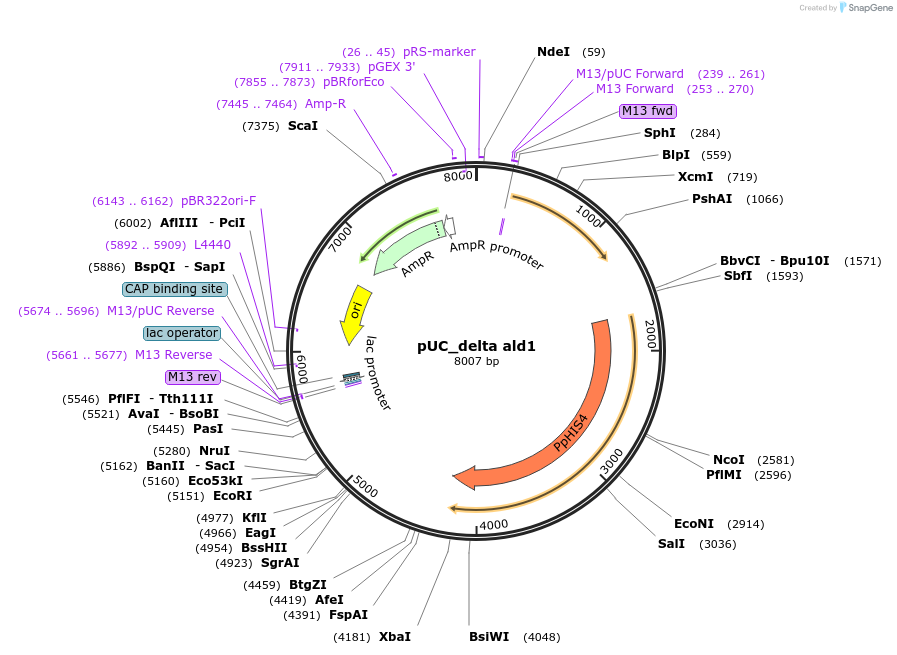
pUC_delta ald1
Plasmid#126708Purposevector for knocking out ald1 in Komagataella phaffiiDepositorInsertHIS4
ExpressionYeastPromoterHisAvailable SinceJune 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
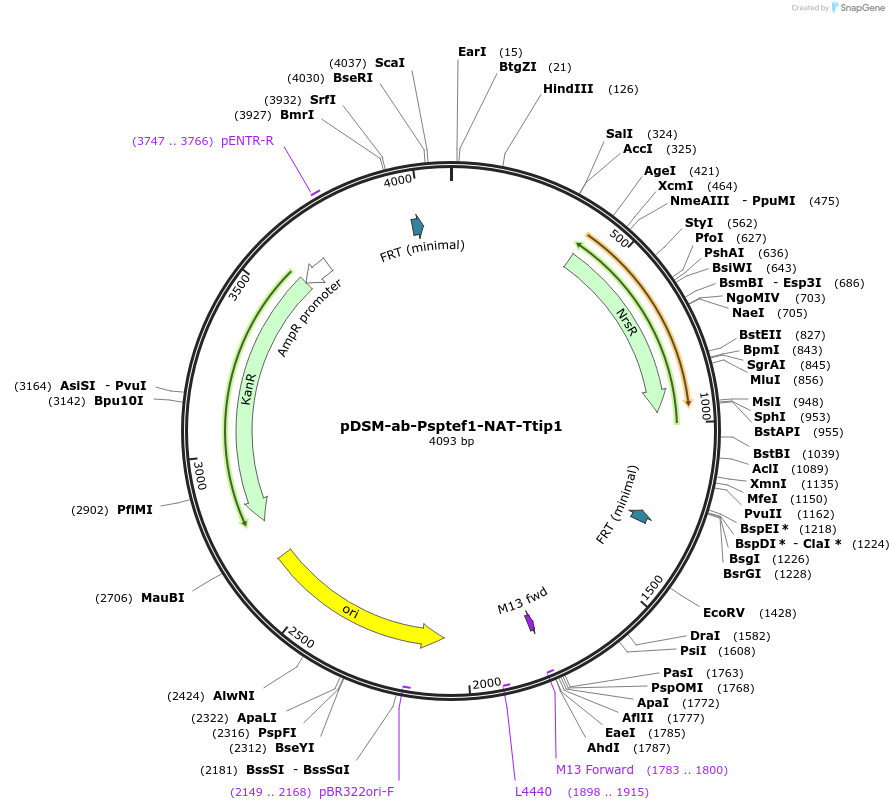
pDSM-ab-Psptef1-NAT-Ttip1
Plasmid#127747PurposeYeast pathway position 2. NatMX transcription unit with the S. paradoxus TEF1 promoter and TIP1 terminator. Nourseothricin resistance.DepositorInsertnourseothricin N-acetyltransferase
UseSynthetic BiologyExpressionYeastPromoterPsptef1Available SinceNov. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
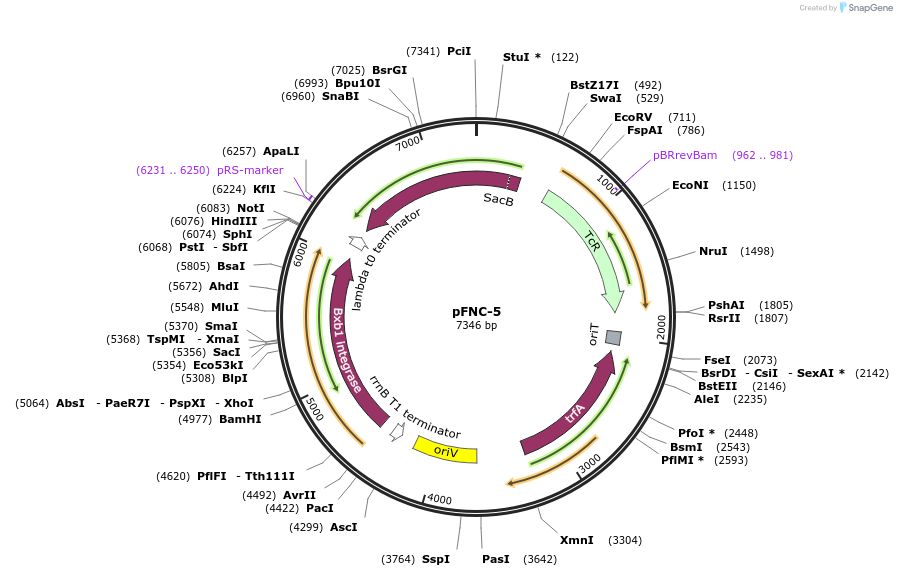
pFNC-5
Plasmid#233297PurposePlasmid encoding the BxbI phage integrase. It can be used to remove the antibiotic resistance cassette integrated with pCIFR. TcR, easily curable via sucrose counterselection.DepositorInsertsBxbI phage integrase
sacB
ExpressionBacterialPromoterPEM7 and unknownAvailable SinceApril 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
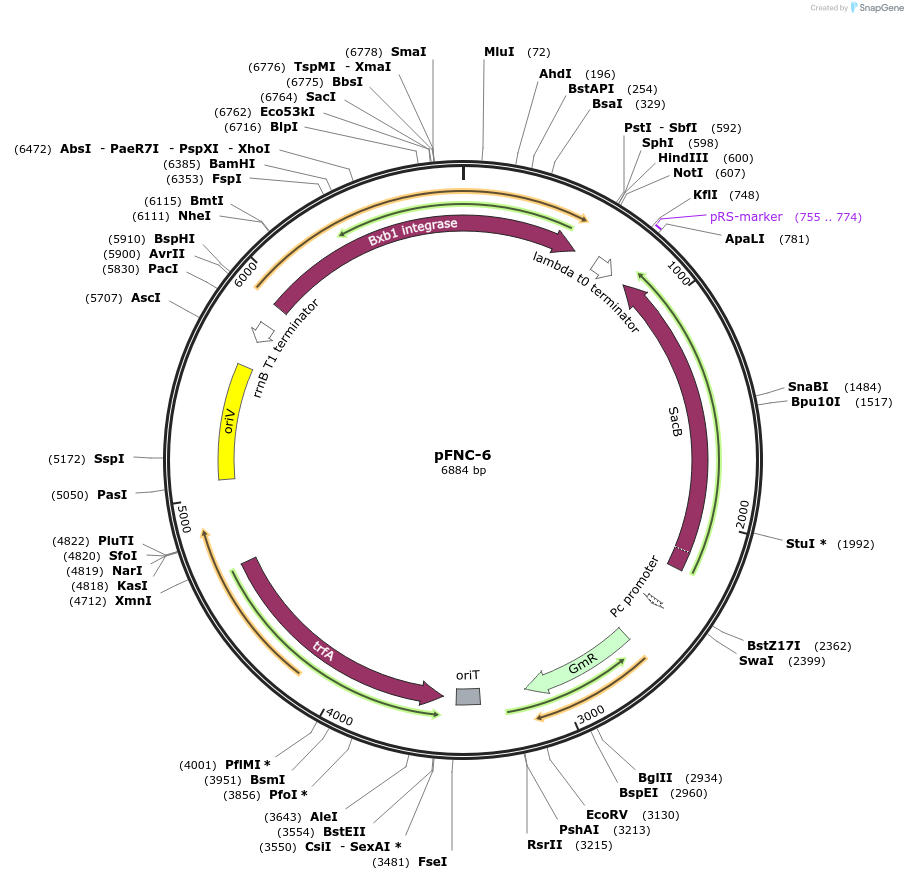
pFNC-6
Plasmid#227669PurposePlasmid encoding the BxbI phage integrase. It can be used to remove the antibiotic resistance cassette integrated with pCIFR. GmR, easily curable via sucrose counterselection.DepositorInsertsBxbI phage integrase
sacB
ExpressionBacterialPromoterPEM7Available SinceJan. 22, 2025AvailabilityAcademic Institutions and Nonprofits only -
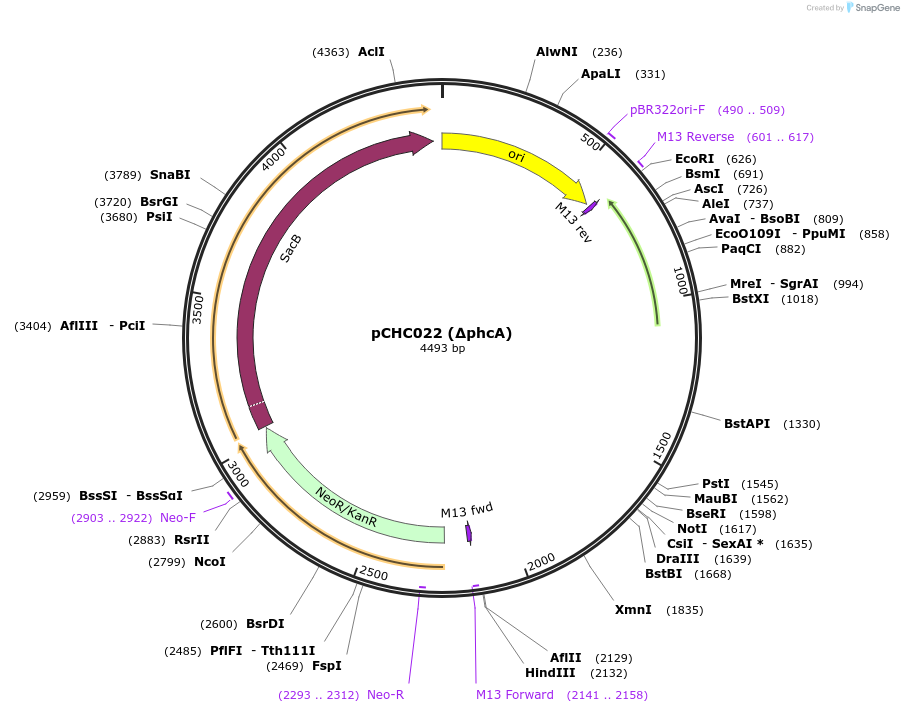
pCHC022 (∆phcA)
Plasmid#226200PurposeElectroporation knockout plasmid (pK18sB) to delete the C. necator LysR-type transcriptional regulator PhcA.DepositorInsertHomology arms for PhcA
UseSynthetic BiologyAvailable SinceNov. 18, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
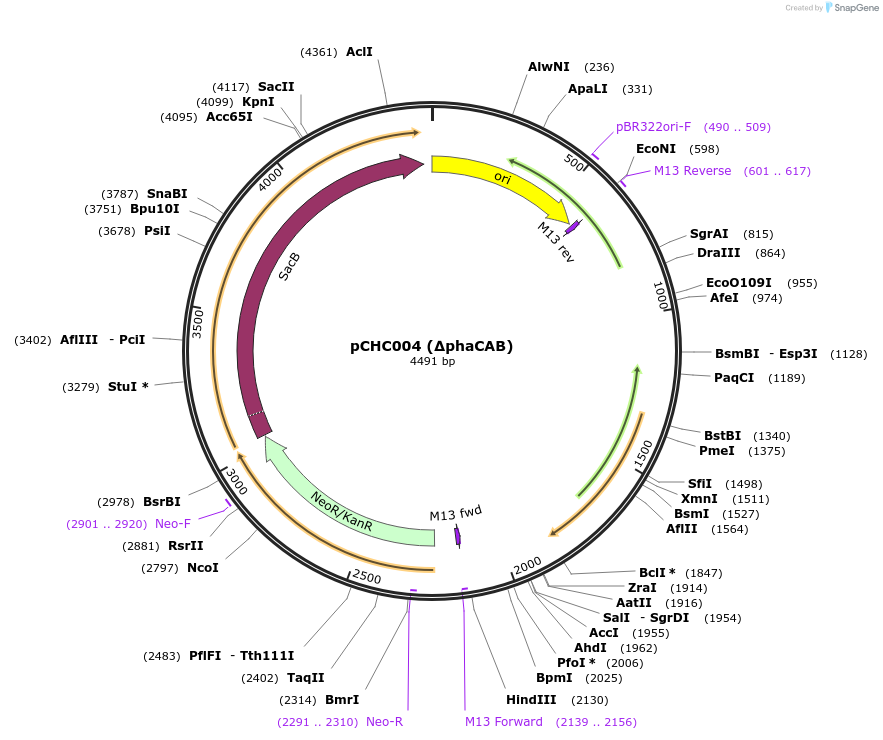
pCHC004 (∆phaCAB)
Plasmid#226198PurposeElectroporation knockout plasmid (pK18sB) to delete the C. necator phaCAB operon, to prevent accumulation of PHB.DepositorInsertHomology arms for phaCAB
UseSynthetic BiologyAvailable SinceNov. 15, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
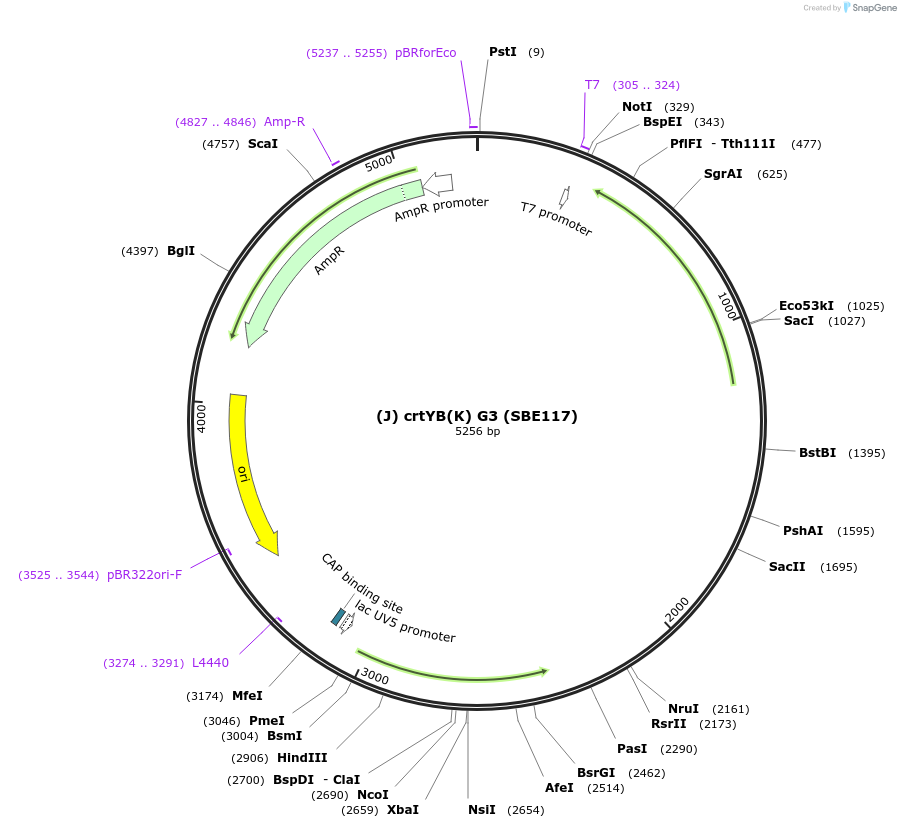
(J) crtYB(K) G3 (SBE117)
Plasmid#194996PurposeRhodotorula toruloides gene phytoene synthase/lycopene cyclase (crtYB, RHTO_04605) for G3DepositorInsert(J) crtYB(K)
ExpressionBacterialAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
(G) crtI (H) G2 (SBE116)
Plasmid#194995PurposeRhodotorula toruloides gene phytoene dehydrogenase (crtI, RHTO_04602) for G2DepositorInsert(G) crtI (H)
ExpressionBacterialAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only