We narrowed to 13,839 results for: CRISPR-Cas9
-
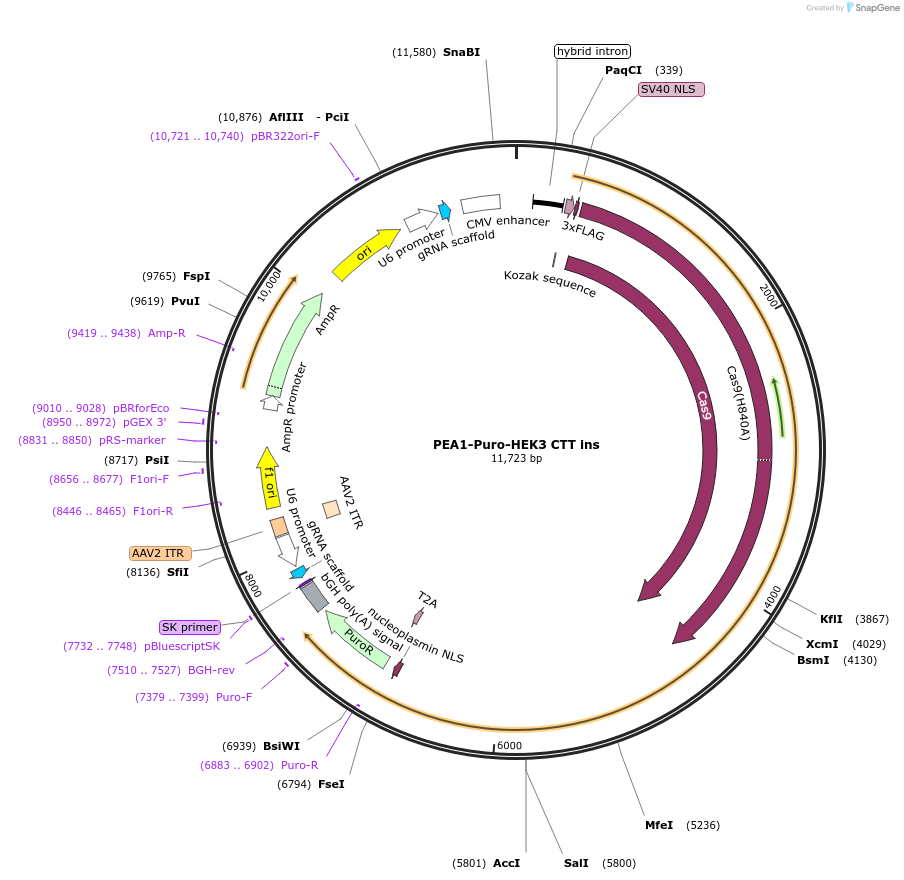
Plasmid#171996PurposeDelivers all prime editing (nickase) components targeting the HEK3 site for a CTT inserrtion, in a single, puromycin selectable plasmidDepositorInsertCbH-Cas9(H840A)-RT-T2A-Puro, hU6-pegRNA HEK3 CTT ins, hU6-sgRNA HEK3 +90
ExpressionMammalianPromoterCMV for Cas9, U6 for gRNAsAvailable SinceAug. 4, 2021AvailabilityAcademic Institutions and Nonprofits only -
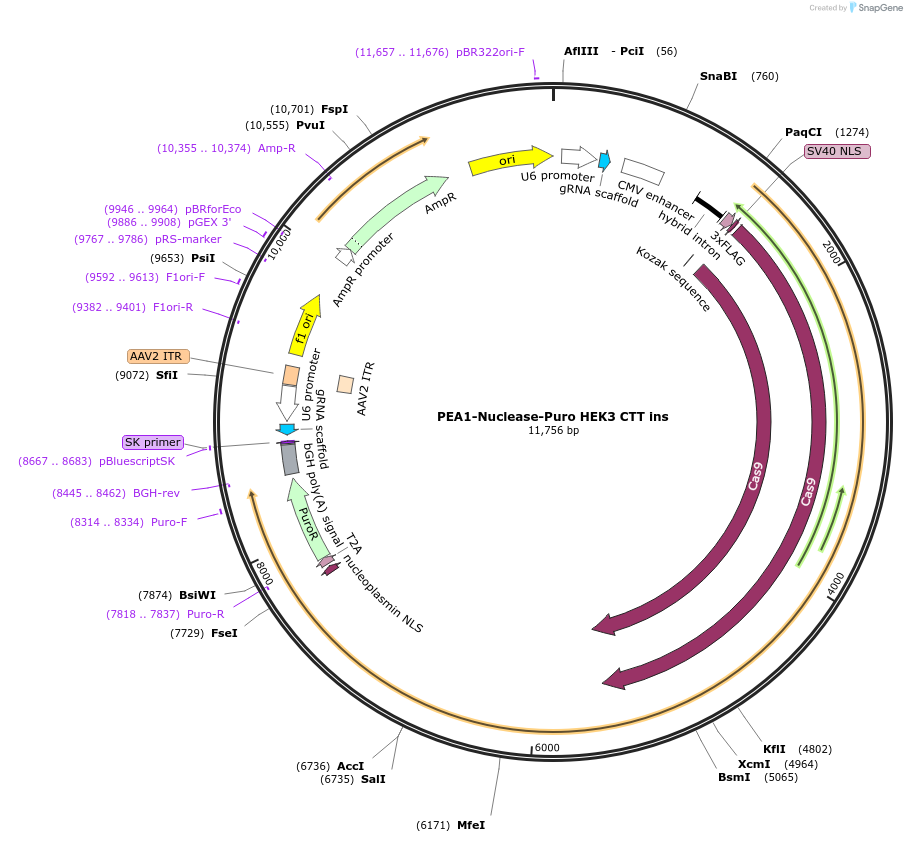
PEA1-Nuclease-Puro HEK3 CTT ins
Plasmid#171995PurposeDelivers all prime editing nuclease components targeting the HEK3 gene for a CTT inserrtion, in a single, puromycin selectable plasmidDepositorInsertHEK3 CTT insertion pegRNA and CbH-Cas9-RT-T2A-Puro, hU6-pegRNA HEK3 CTT ins, hU6-sgRNA sham
ExpressionMammalianPromoterCMV for Cas9, U6 for gRNAsAvailable SinceSept. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
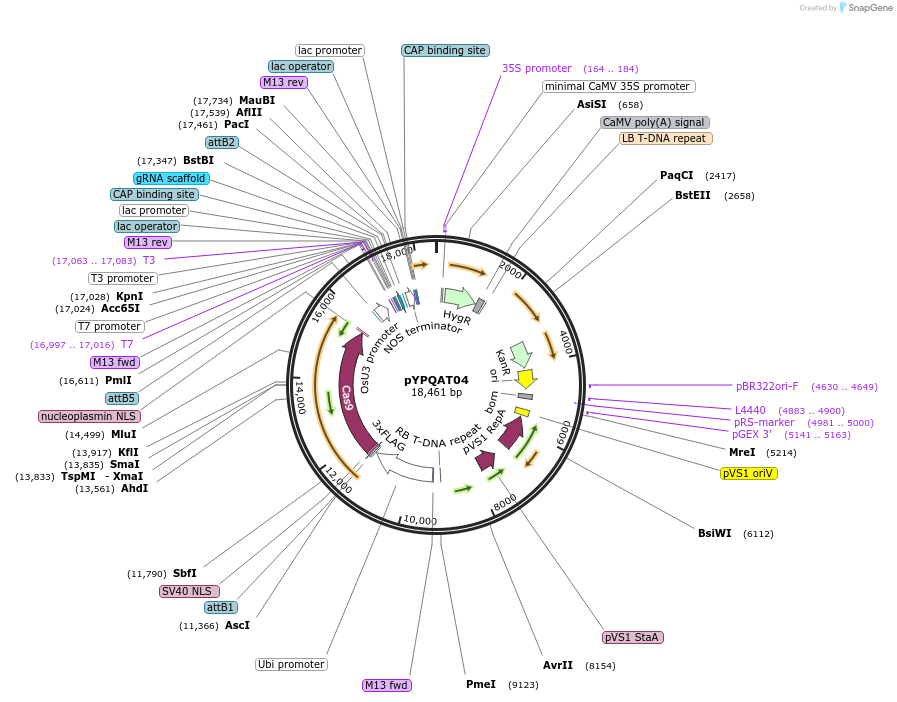
pYPQAT04
Plasmid#223376PurposeT-DNA vector for SpCas9 mediated mutagenesis for monocot plants; NGG PAM; Cas9 was driven by ZmUbi1 and the sgRNA was driven by OsU3 promoter; Hygromycin for plants selection.DepositorInsertZmUbi-SpCas9-OsU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
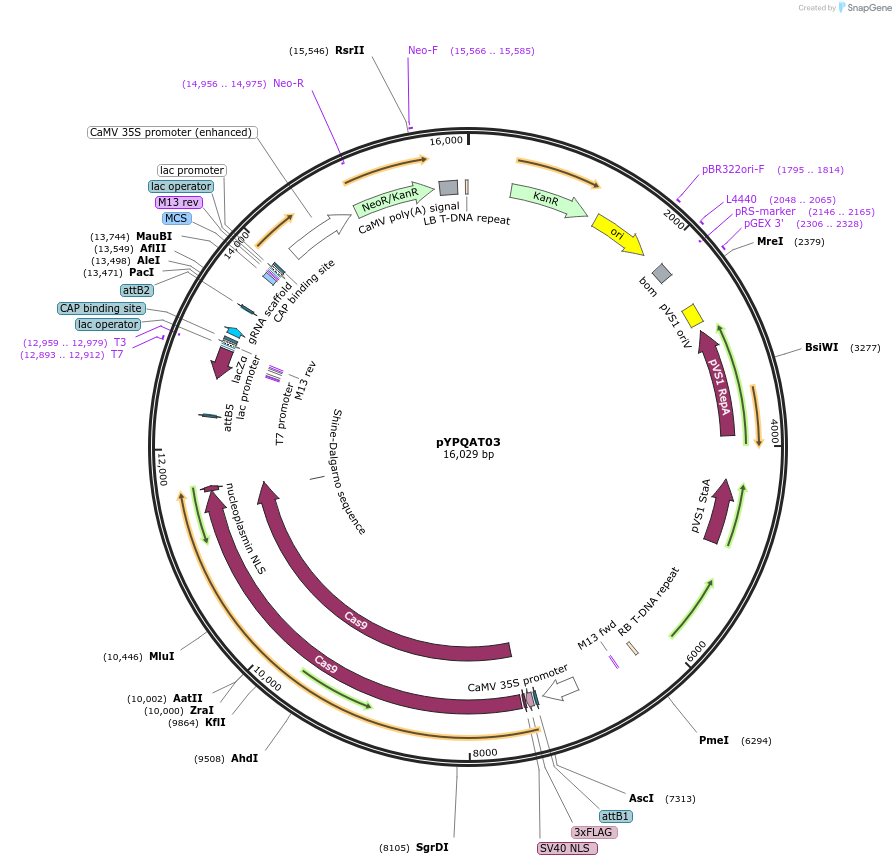
pYPQAT03
Plasmid#223375PurposeT-DNA vector for SpCas9 mediated mutagenesis for dicot plants; NGG PAM; Cas9 was driven by 2x35s and the sgRNA was driven by AtU3 promoter; Kanamycin for plants selection.DepositorInsert2x35s-SpCas9-AtU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
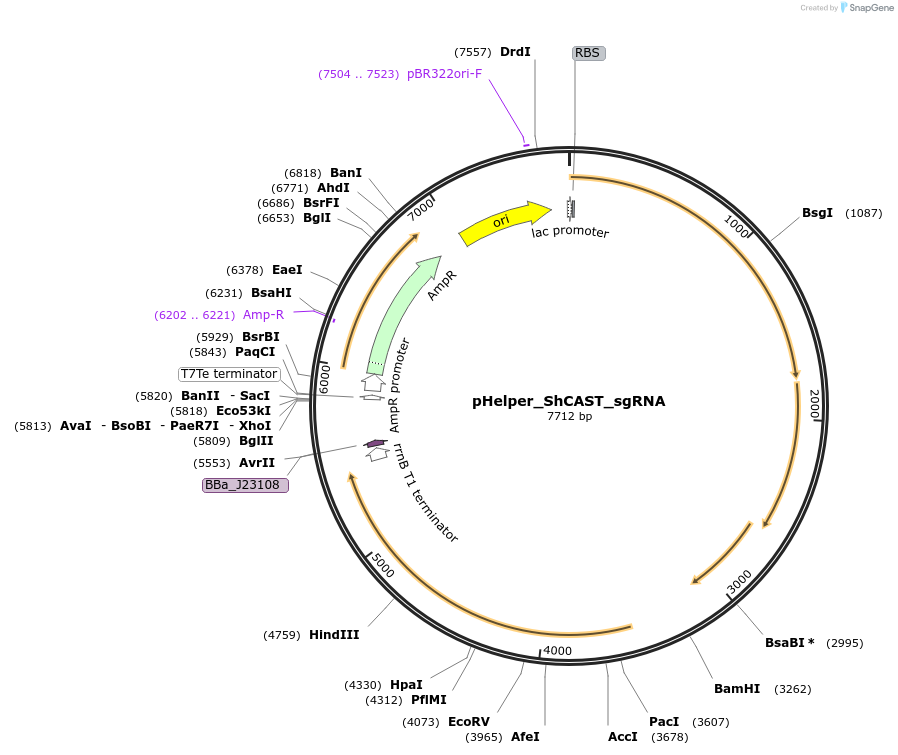
pHelper_ShCAST_sgRNA
Plasmid#127921PurposeExpresses ShCAST and an empty sgRNA scaffold. New targets can be added using Golden Gate assembly (LguI sites).DepositorInsertShTnsB, ShTnsC, ShTniQ, ShCas12k, sgRNA scaffold for guide cloning (LguI sites)
Available SinceJuly 10, 2019AvailabilityAcademic Institutions and Nonprofits only -
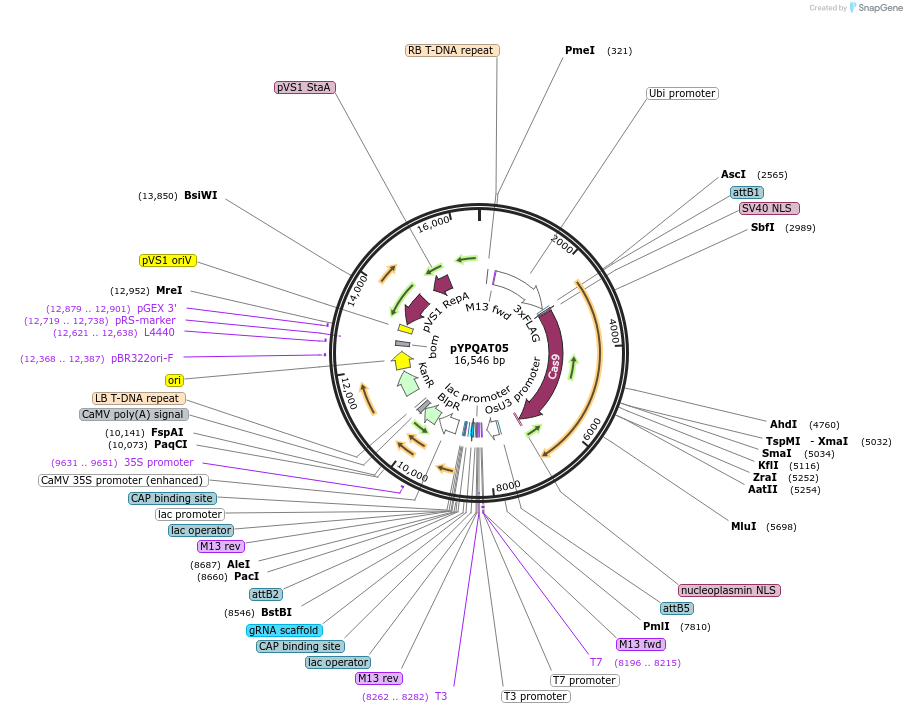
pYPQAT05
Plasmid#223377PurposeT-DNA vector for SpCas9 mediated mutagenesis for monocot plants; NGG PAM; Cas9 was driven by ZmUbi1 and the sgRNA was driven by OsU3 promoter; BASTA for plants selection.DepositorInsertZmUbi-SpCas9-OsU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
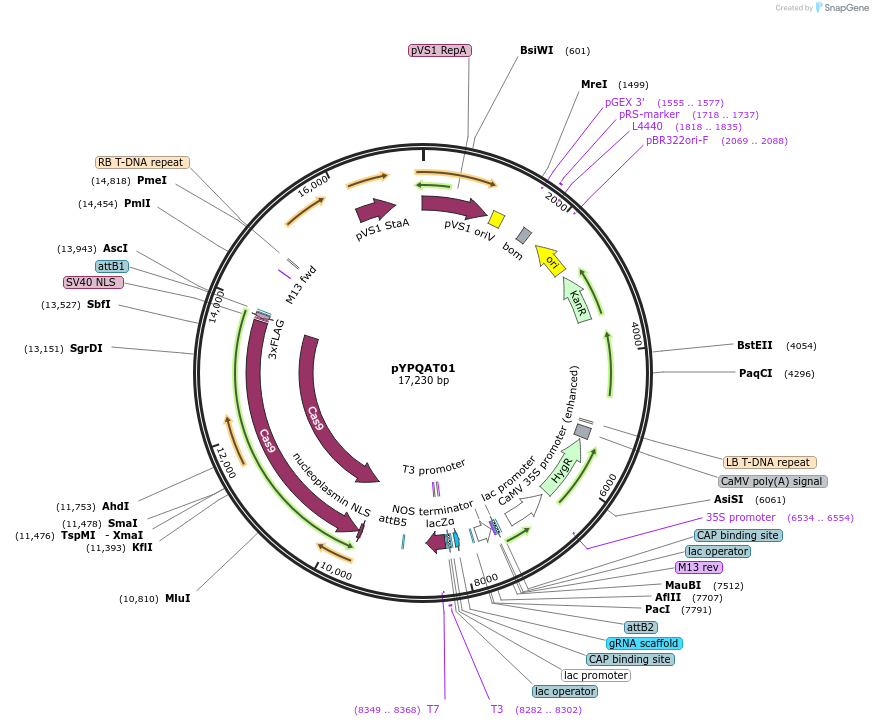
pYPQAT01
Plasmid#223373PurposeT-DNA vector for SpCas9 mediated mutagenesis for dicot plants; NGG PAM; Cas9 was driven by AtUBQ10 and the sgRNA was driven by AtU3 promoter; Hygromycin for plants selection.DepositorInsertAtUBQ10-SpCas9-AtU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
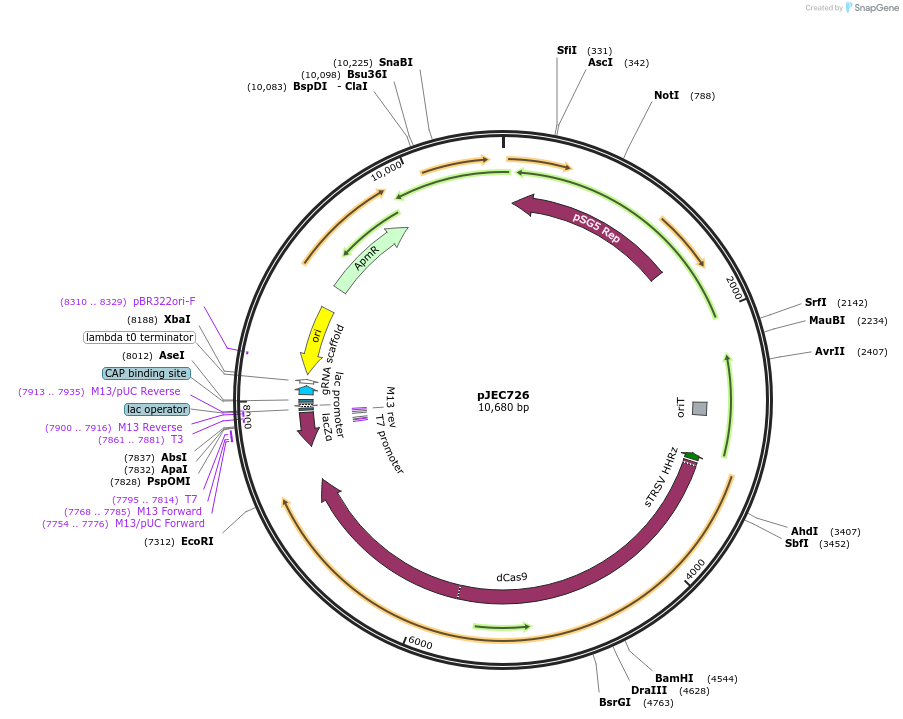
pJEC726
Plasmid#190814PurposeCRISPRi backbone plasmid harboring BbsI sites for easy cloning of sgRNA targeting regions via Golden gate.DepositorInsertdCas9
UseCRISPR and Synthetic BiologyMutationS. pyogenes dCas9 codon optimized for Streptomyce…PromoterSP30Available SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
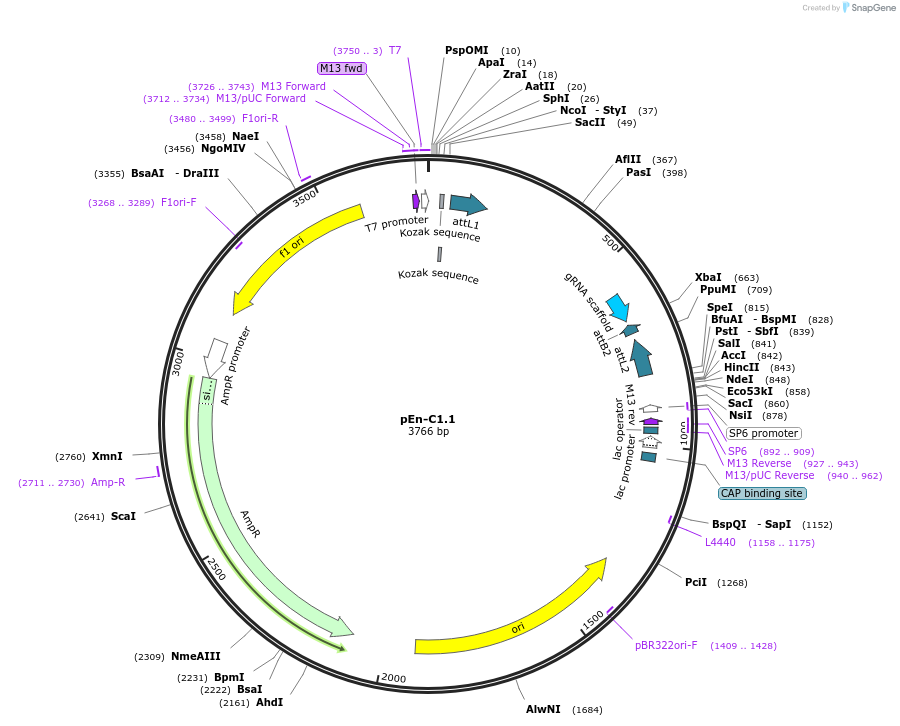
pEn-C1.1
Plasmid#61479PurposeEncodes CRISPR sgRNA for Gateway or conventional cloning of 1-3 sgRNAs into pDe-CAS9 or pCAS9-TPCDepositorInsertAtU6-26:sgRNA
UseCRISPRExpressionPlantPromoterU6-26Available SinceSept. 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
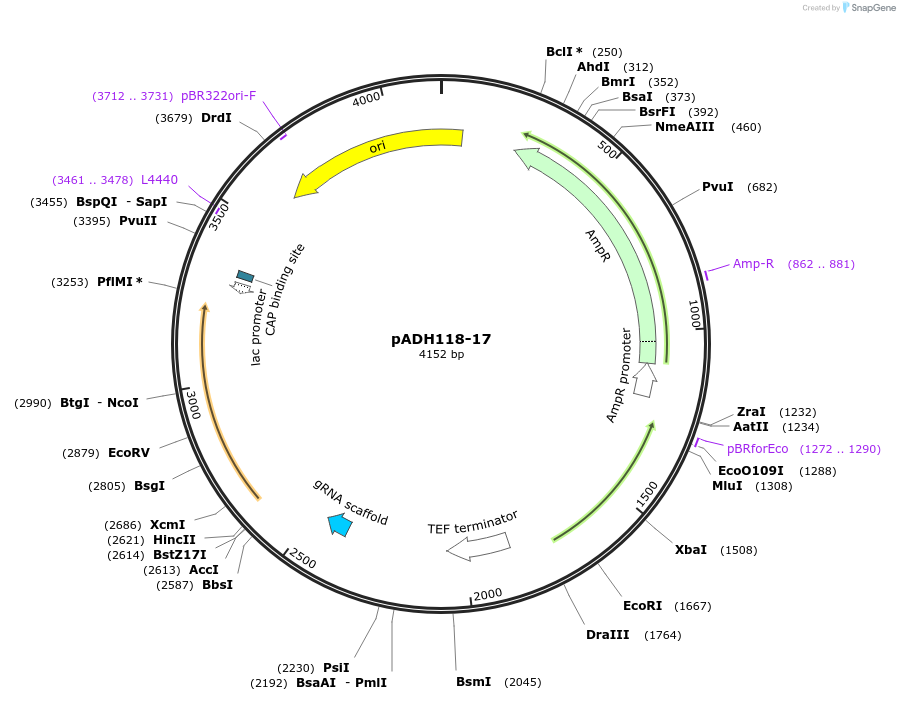
pADH118-17
Plasmid#91727PurposeNAT-marked C. albicans URA3-specific gRNA expression construct; part 2 of 2 of C.alb LEUpOUT CRISPR system. Use with pADH137 CAS9 expression construct.DepositorInsertNAT 2of2, pSNR52, C. albicans URA3-specific gRNA, LEU2 2of2
Available SinceJune 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
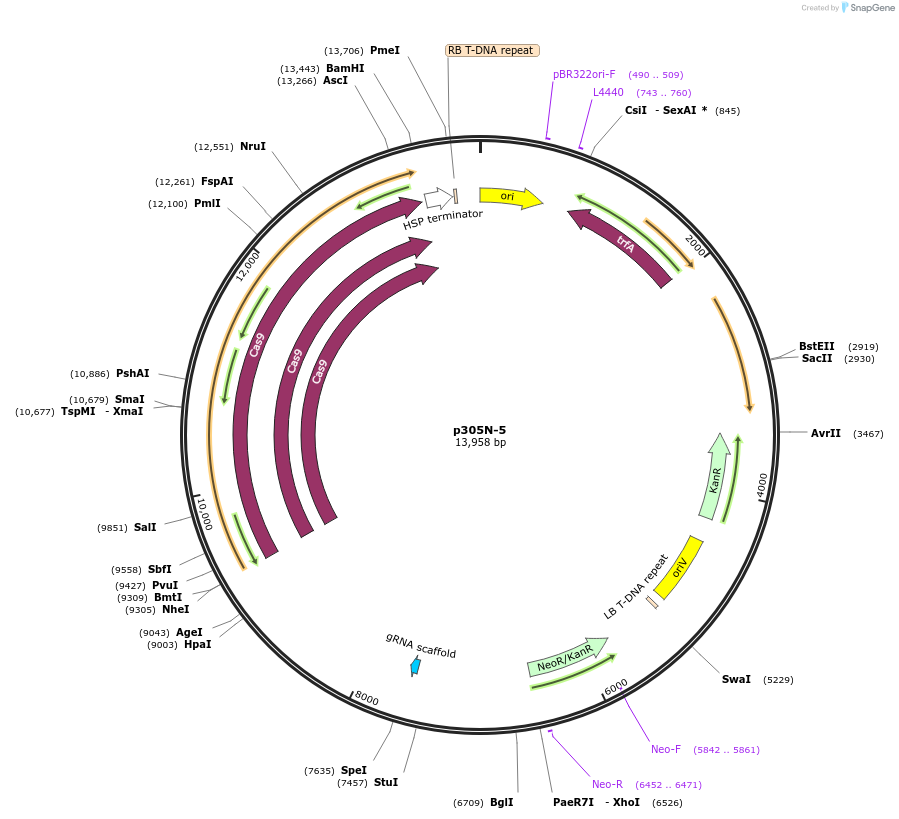
p305N-5
Plasmid#246288PurposeEvaluation of AtU6.1m2 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.1m2 promoter
UseCRISPRExpressionPlantMutationA-to-C (-63 from TSS) and G-to-T (-57) within USEPromoterAtU6.1m2 promoterAvailable SinceDec. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
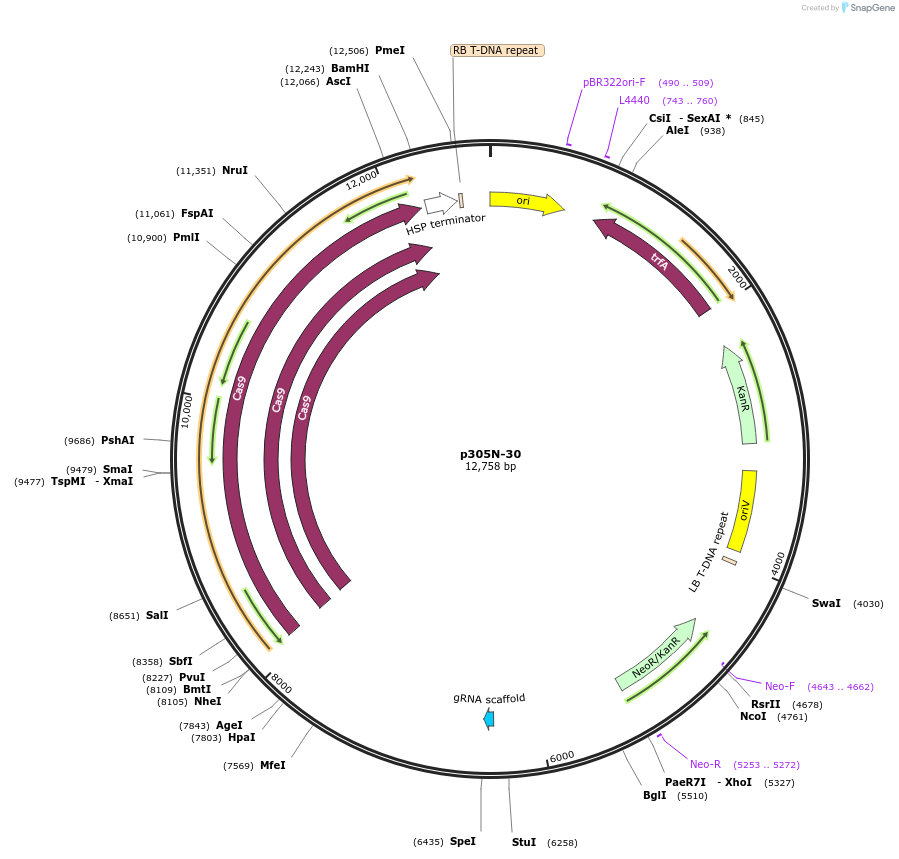
p305N-30
Plasmid#246313PurposeEvaluation of PtU6.2c3 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c3 promoter
UseCRISPRExpressionPlantMutationdeletion: -T (-29 from TSS) within TATAPromoterPtU6.2c3 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
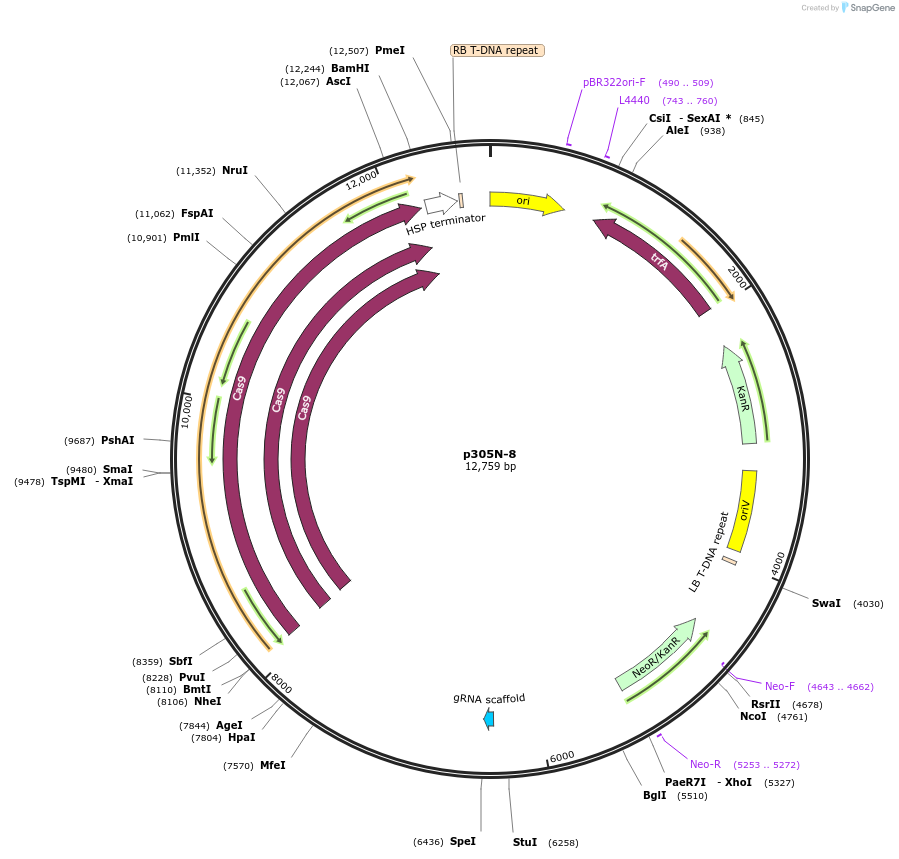
p305N-8
Plasmid#246291PurposeEvaluation of AtU6.29c14 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.29c14 promoter
UseCRISPRExpressionPlantMutationT-to-C (-1 from TSS)PromoterAtU6.29c14 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
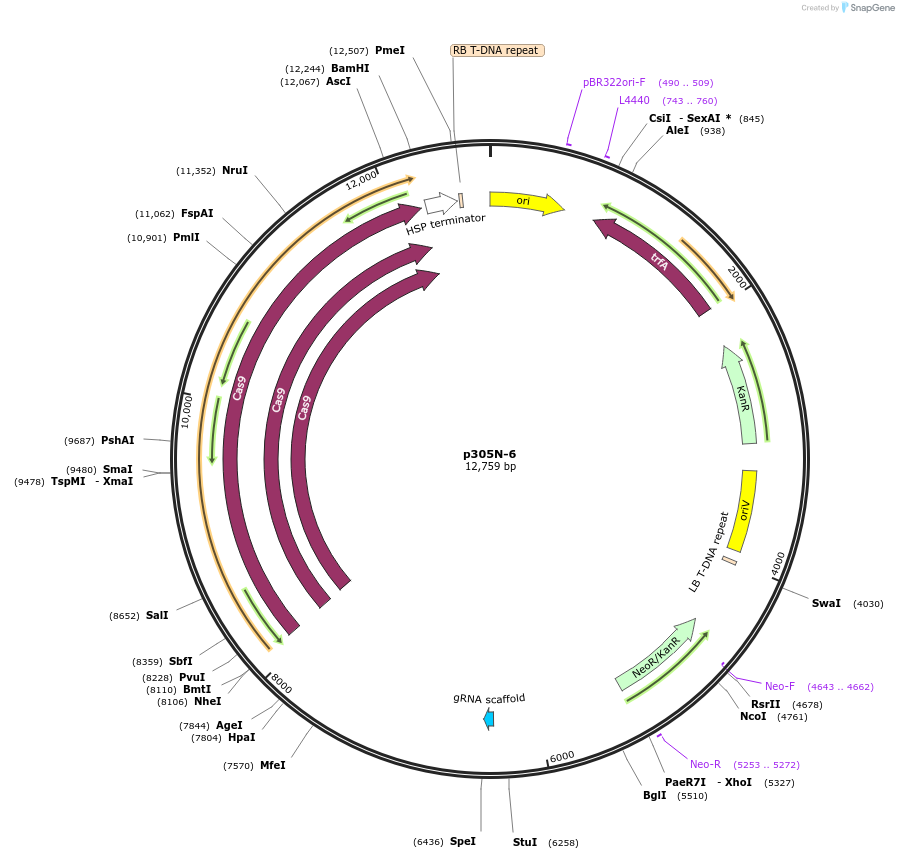
p305N-6
Plasmid#246289PurposeEvaluation of AtU6.1m3 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.1m3 promoter
UseCRISPRExpressionPlantMutationG-to-T (-57 from TSS) within USEPromoterAtU6.1m3 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
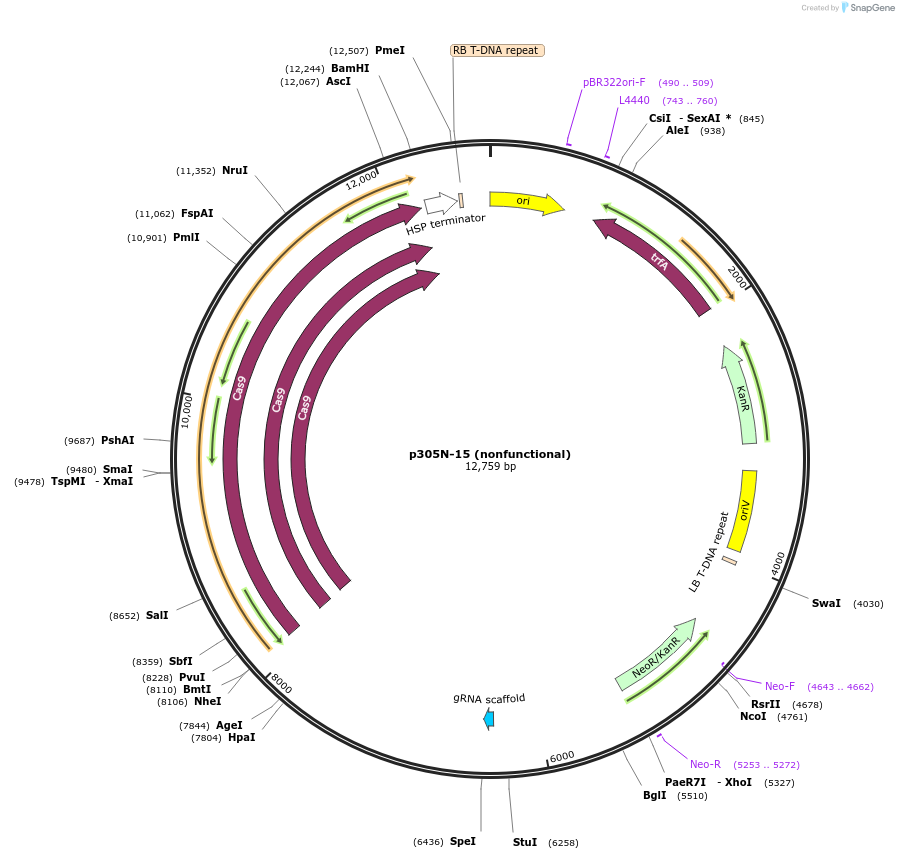
p305N-15 (nonfunctional)
Plasmid#246298PurposeEvaluation of HbU6.2 promoter (nonfunctional) (Pol III promoter) for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertHbU6.2 promoter (nonfunctional)
UseCRISPRExpressionPlantPromoterHbU6.2 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
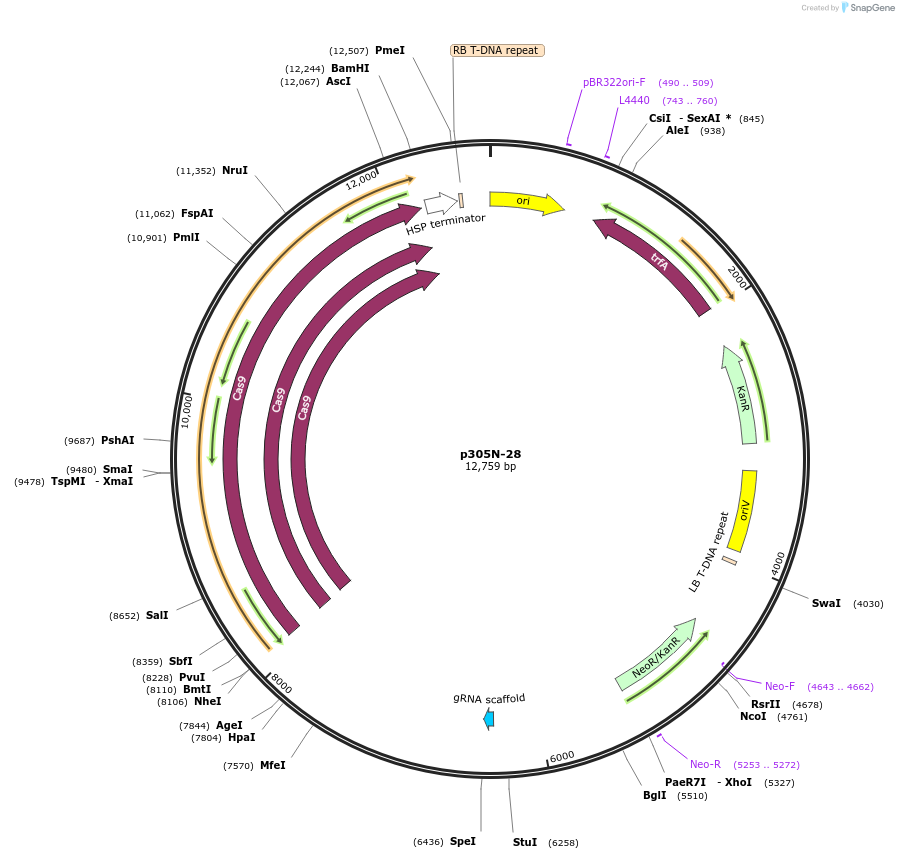
p305N-28
Plasmid#246311PurposeEvaluation of PtU6.1c1 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.1c1 promoter
UseCRISPRExpressionPlantMutationC-to-A (-42 from TSS), A-to-C (-41)PromoterPtU6.1c1 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
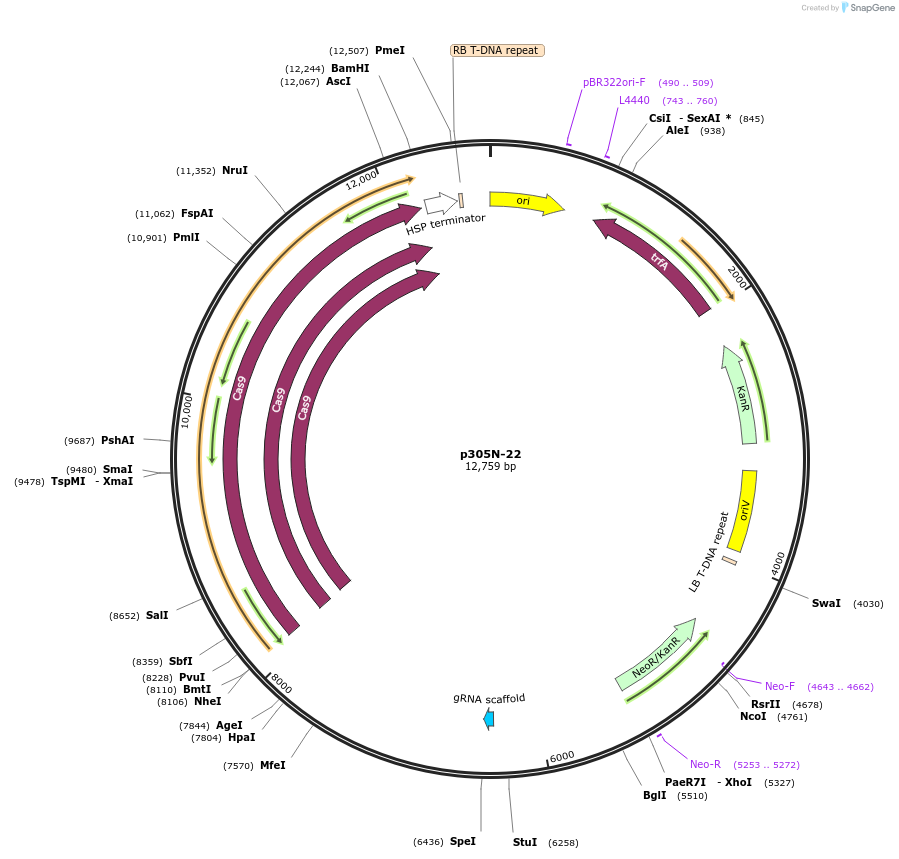
p305N-22
Plasmid#246305PurposeEvaluation of MtU6.6-70 promoter (Pol III promoter) deletion (70 bp) for CRISPR-Cas9 editing of mEGFP in Nicotiana benthamiana and poplar reporter linesDepositorInsertMtU6.6-70 promoter
UseCRISPRExpressionPlantPromoterMtU6.6-70 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
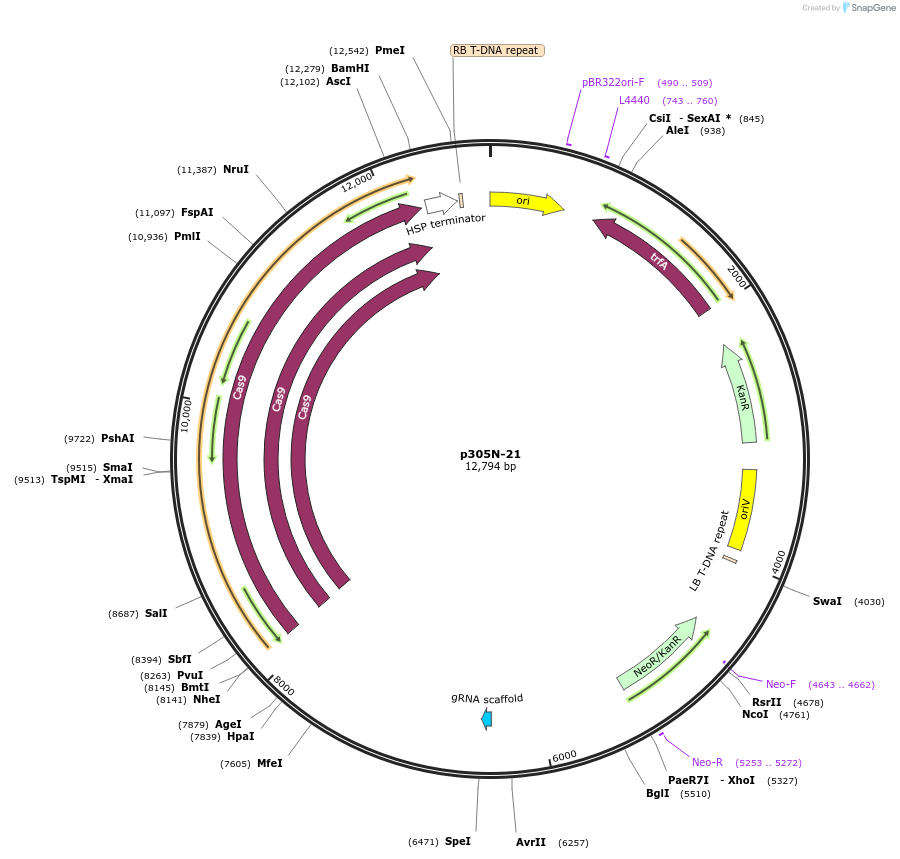
p305N-21
Plasmid#246304PurposeEvaluation of MtU6.6-104 promoter (Pol III promoter) deletion (104 bp) for CRISPR-Cas9 editing of mEGFP in Nicotiana benthamiana and poplar reporter linesDepositorInsertMtU6.6-104 promoter
UseCRISPRExpressionPlantPromoterMtU6.6-104 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
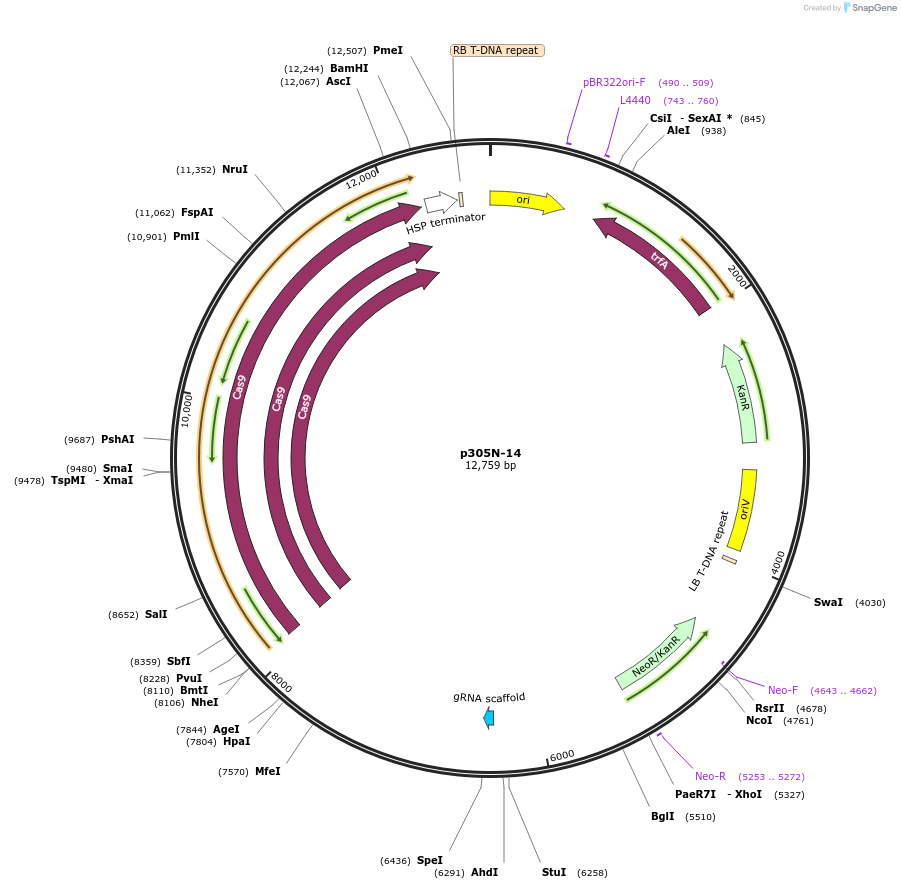
p305N-14
Plasmid#246297PurposeEvaluation of CiU6.6c16 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertCiU6.6c16 promoter
UseCRISPRExpressionPlantMutationT-to-C (-2 from TSS)PromoterCiU6.6c16 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
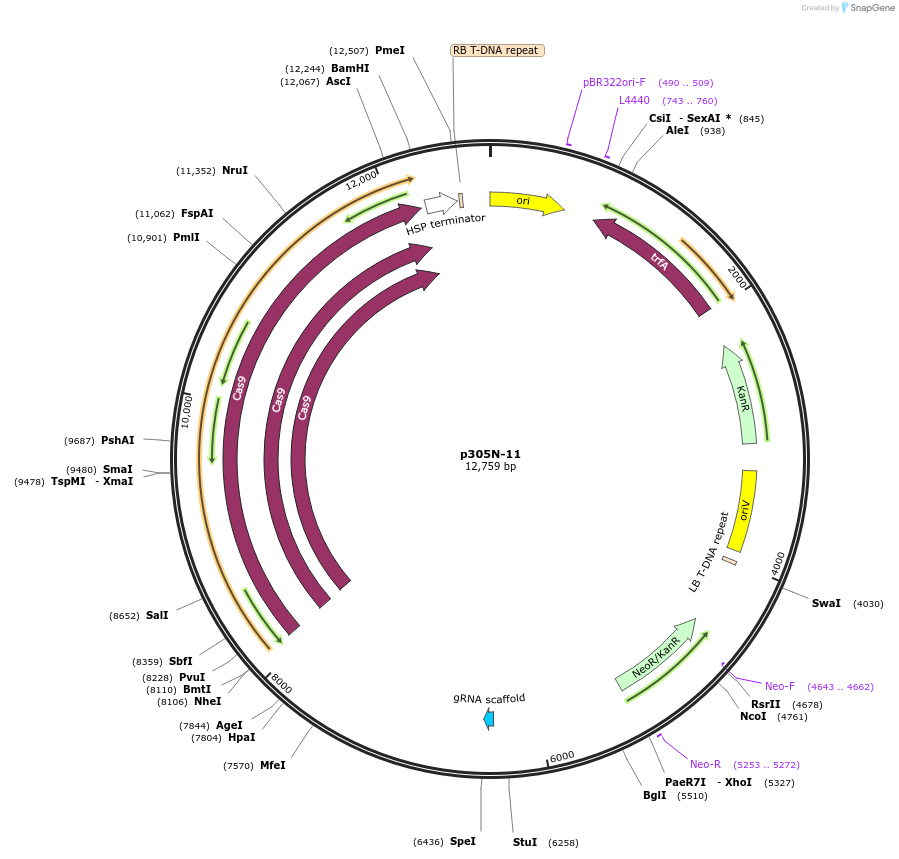
p305N-11
Plasmid#246294PurposeEvaluation of CiU6.3c8 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertCiU6.3c8 promoter
UseCRISPRExpressionPlantMutationA-to-C (-23 from TSS)PromoterCiU6.3c8 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only