We narrowed to 23,150 results for: Sele
-
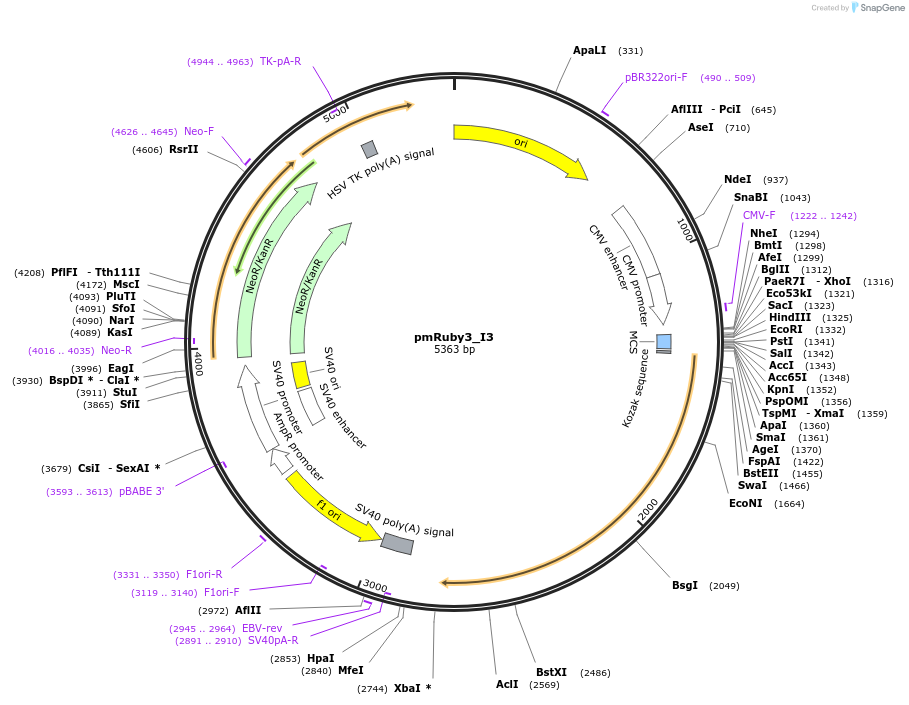
Plasmid#239344PurposeExpresses the I3-01 nanocage subunit N-terminally tagged with mRuby3 in mammalian cells. Assembles into nanocages tagged with 60 FPs.DepositorInsertI3-01
TagsmRuby3ExpressionMammalianMutationK129APromoterCMVAvailable SinceJune 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
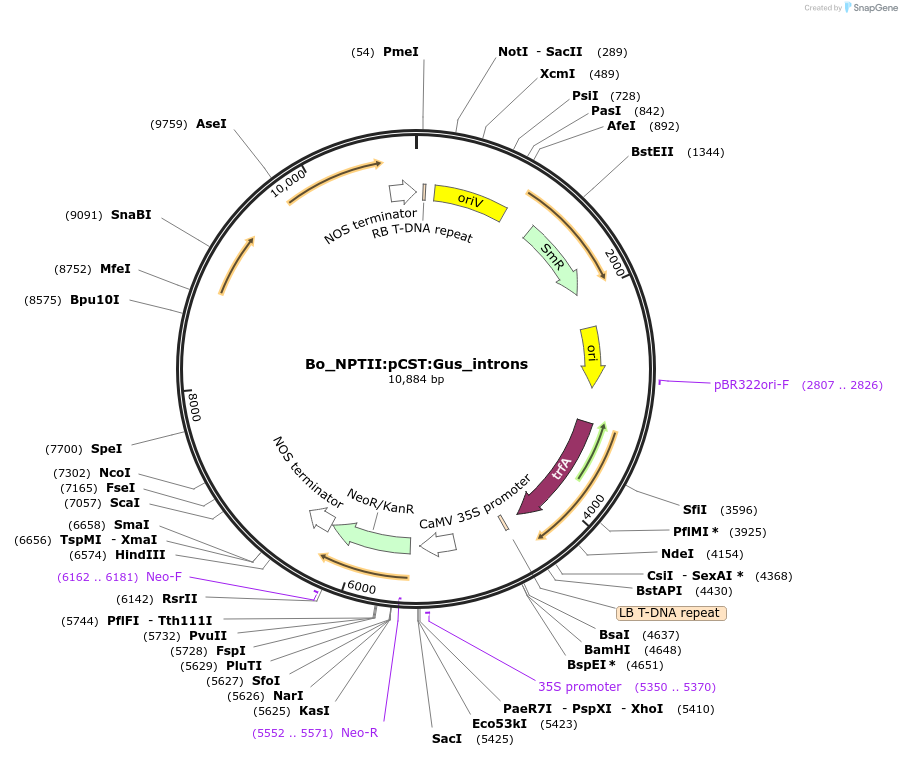
Bo_NPTII:pCST:Gus_introns
Plasmid#215178PurposeEvaluating guard cell specific promotors in BrassicaDepositorInsertClosed stomata1
UseSynthetic BiologyPromoterpCST1Available SinceMay 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
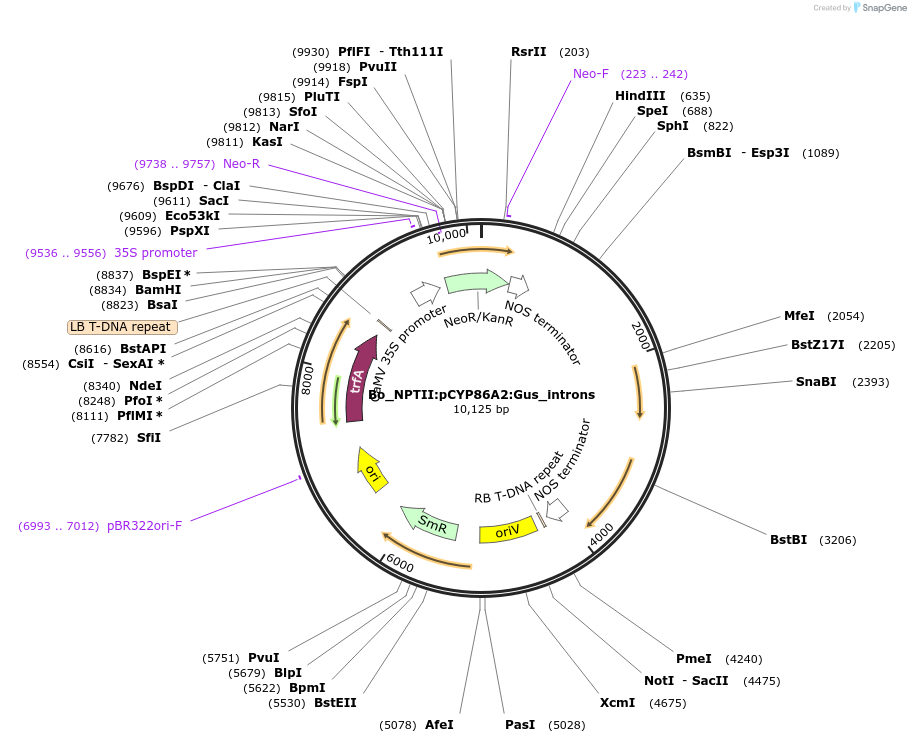
Bo_NPTII:pCYP86A2:Gus_introns
Plasmid#215179PurposeEvaluating guard cell specific promotors in BrassicaDepositorInsertCytochrome p450 86a2 monooxygenase
UseSynthetic BiologyPromoterpCYP86A2Available SinceMay 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
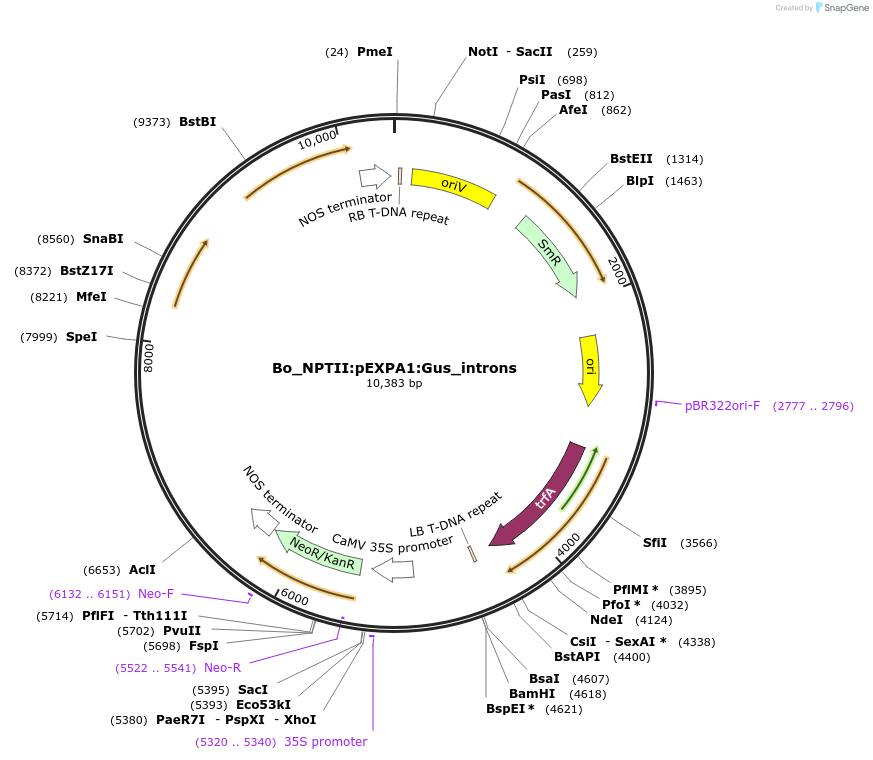
Bo_NPTII:pEXPA1:Gus_introns
Plasmid#215180PurposeEvaluating guard cell specific promotors in BrassicaDepositorInsertExpansin1
UseSynthetic BiologyPromoterpEXPA1Available SinceMay 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
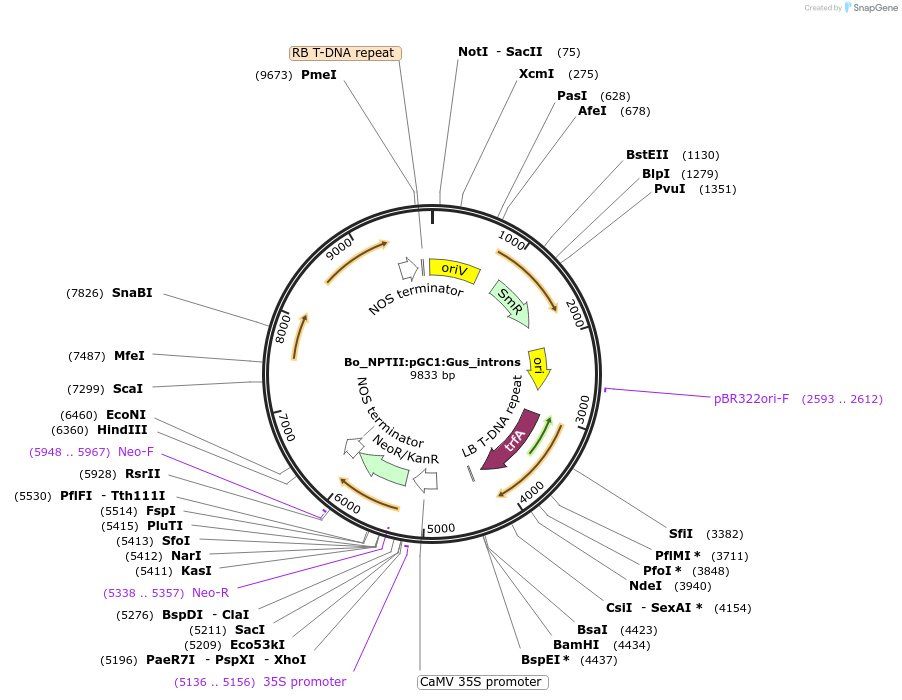
Bo_NPTII:pGC1:Gus_introns
Plasmid#215181PurposeEvaluating guard cell specific promotors in BrassicaDepositorInsertGA-stimulated regulatory protein
UseSynthetic BiologyPromoterAtGC1Available SinceMay 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
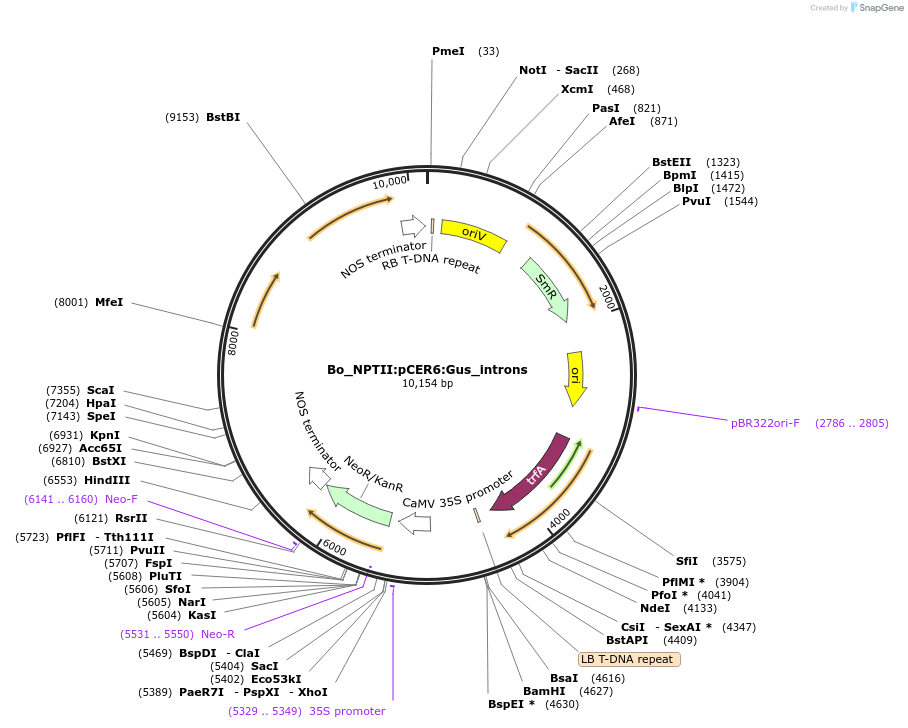
Bo_NPTII:pCER6:Gus_introns
Plasmid#215182PurposeEvaluating guard cell specific promotors in BrassicaDepositorInsert3-ketoacyl-coa synthase6
UseSynthetic BiologyPromoterAtCER6Available SinceMay 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
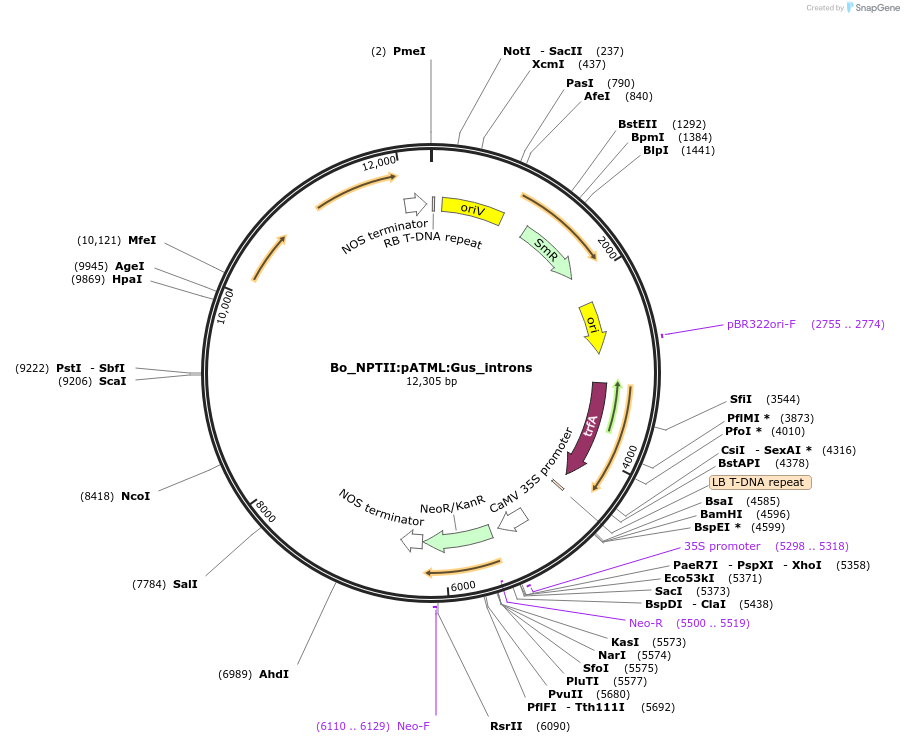
Bo_NPTII:pATML:Gus_introns
Plasmid#215183PurposeEvaluating guard cell specific promotors in BrassicaDepositorInsertMeristem layer1
UseSynthetic BiologyPromoterAtML1Available SinceMay 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
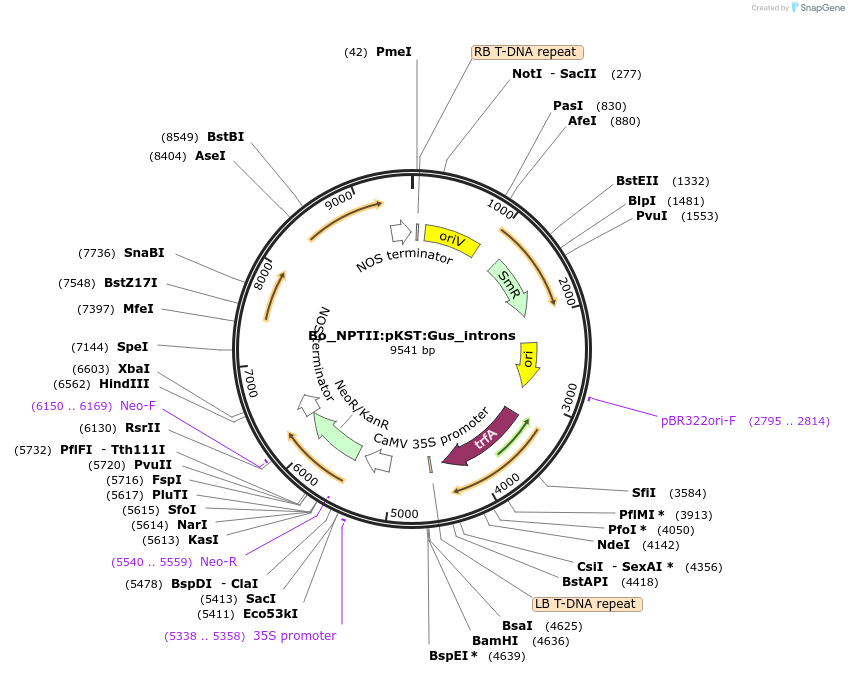
Bo_NPTII:pKST:Gus_introns
Plasmid#215175PurposeEvaluating guard cell specific promotors in BrassicaDepositorInsertInwardly rectifying K+ channel1
UseSynthetic BiologyPromoterpKST1Available SinceMay 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
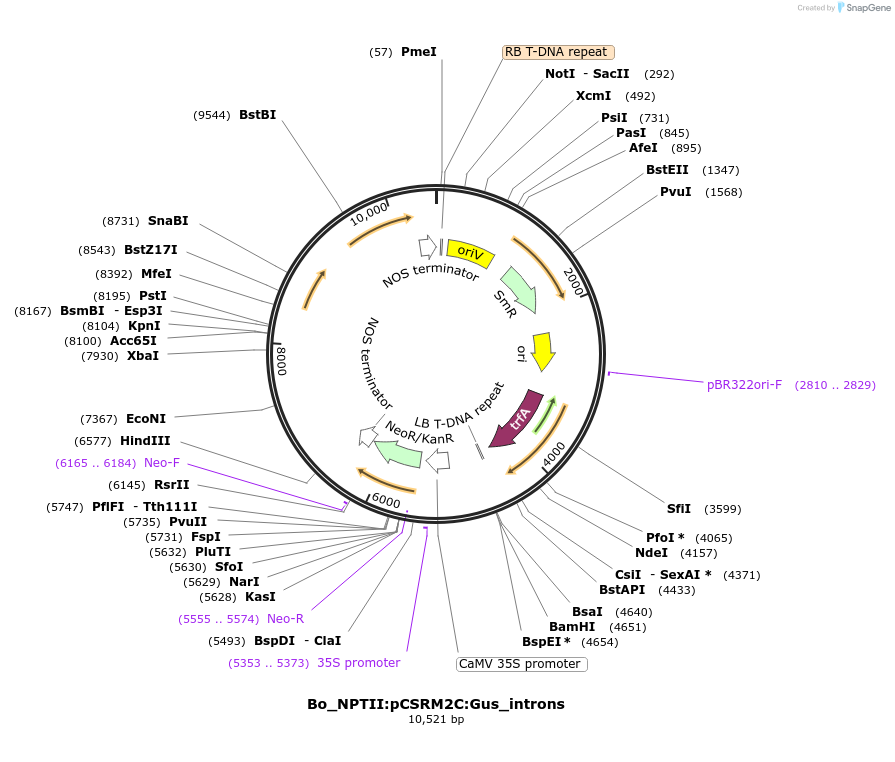
Bo_NPTII:pCSRM2C:Gus_introns
Plasmid#215177PurposeEvaluating guard cell specific promotors in BrassicaDepositorInsertScream2
UseSynthetic BiologyPromoterpSCRM2Available SinceMay 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
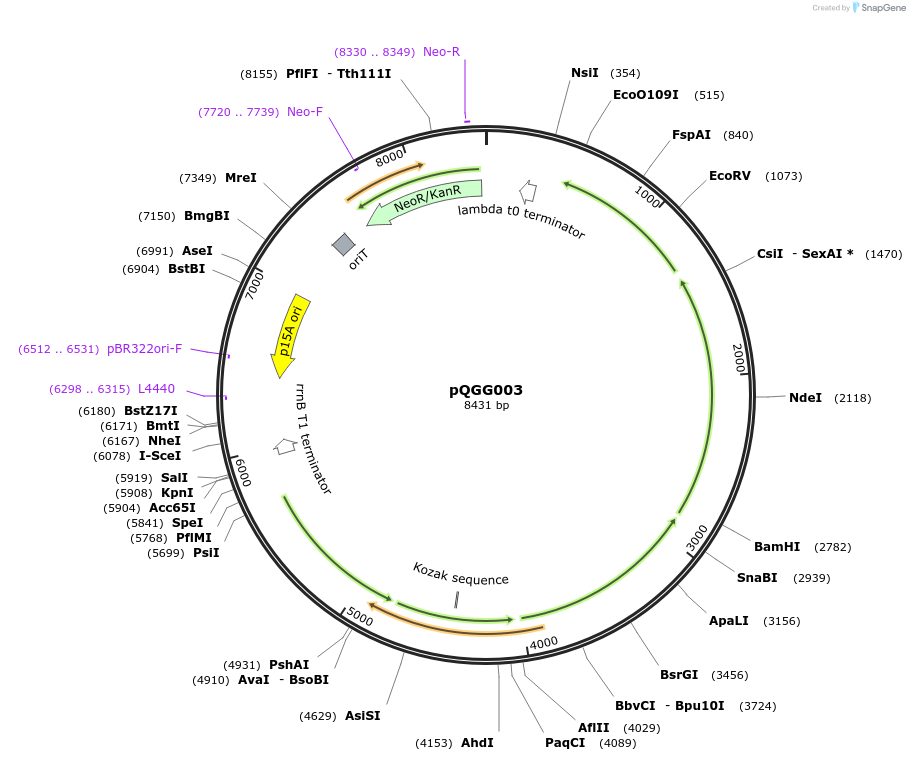
pQGG003
Plasmid#231330PurposeBEVA Golden Gate cloning vector; BEVA2.0 Narrow host range plasmid with I-SceI counterselectable site for BpiI cloning; KmR/NmR.DepositorTypeEmpty backboneExpressionBacterialAvailable SinceMay 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
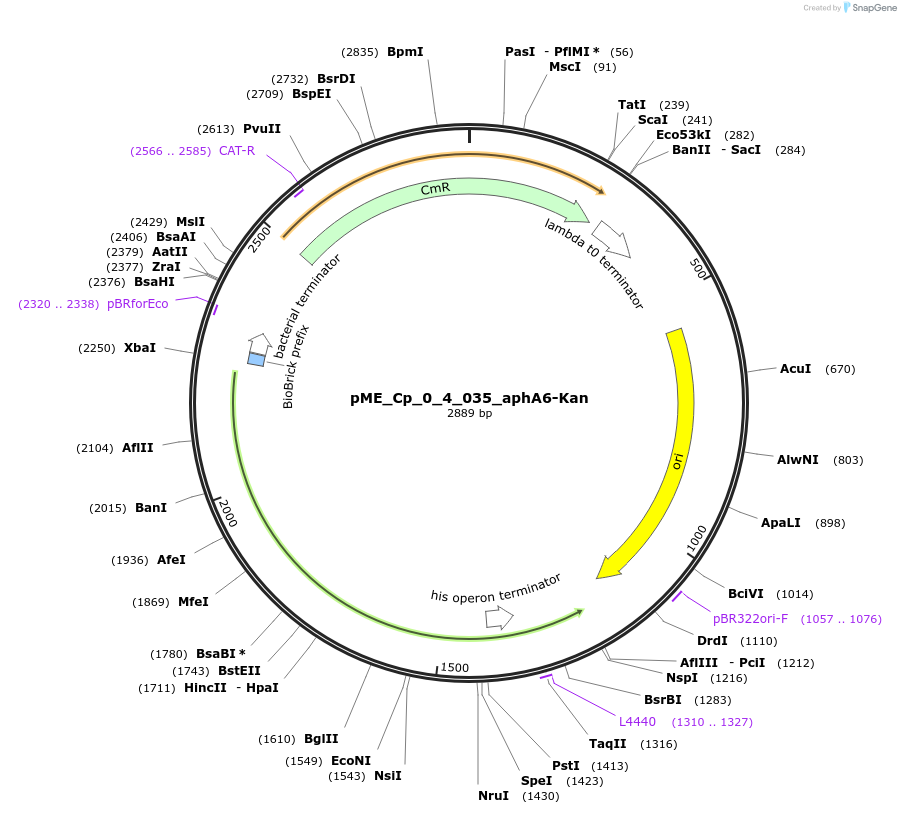
pME_Cp_0_4_035_aphA6-Kan
Plasmid#235904PurposeAminoglycoside-3'-phosphotransferase selection gene, codon optimized for Chlamydomonas reinhardtiiDepositorInsertaphA6-Kann
UseSynthetic BiologyAvailable SinceMay 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
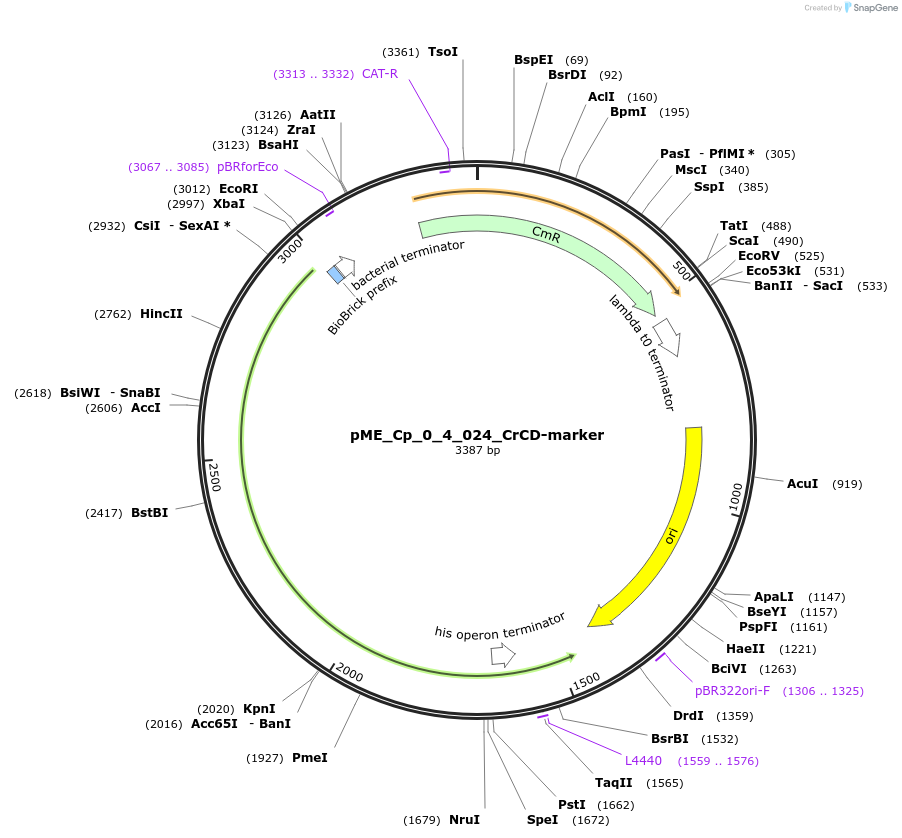
pME_Cp_0_4_024_CrCD-marker
Plasmid#235900PurposeCytosine deaminase selection gene, codon optimized for Chlamydomonas reinhardtiiDepositorInsertCrCD-marker
UseSynthetic BiologyAvailable SinceMay 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
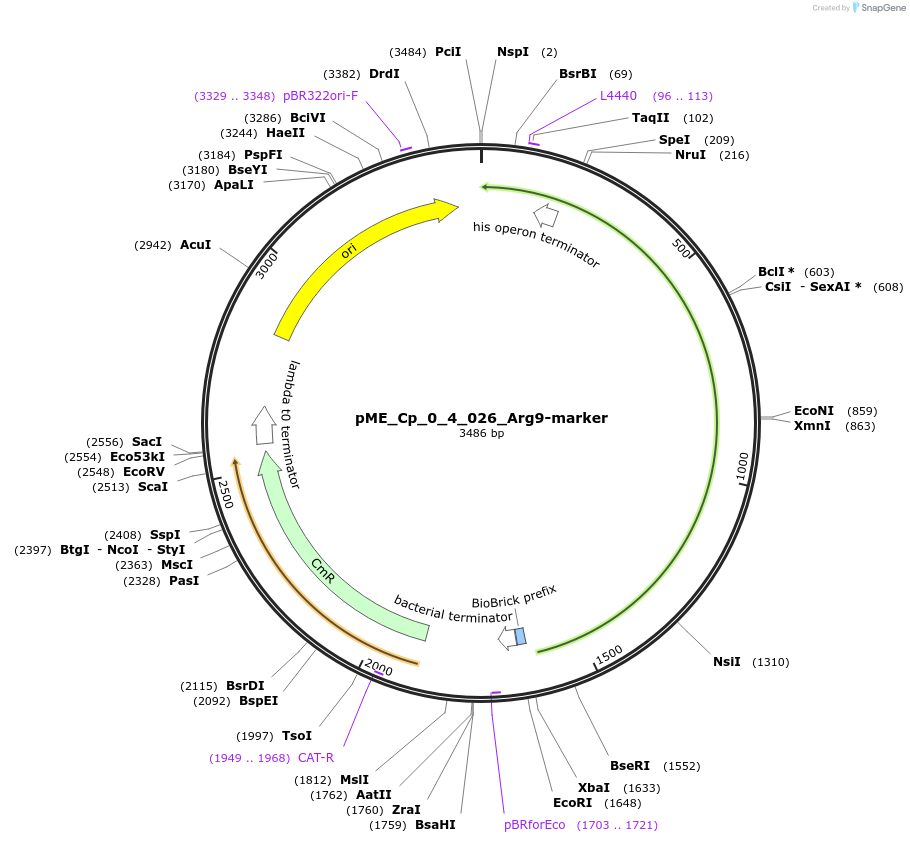
pME_Cp_0_4_026_Arg9-marker
Plasmid#235901PurposeN-acetyl ornithine aminotransferase selection gene, codon optimized for Chlamydomonas reinhardtiiDepositorInsertArg9-marker
UseSynthetic BiologyAvailable SinceMay 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
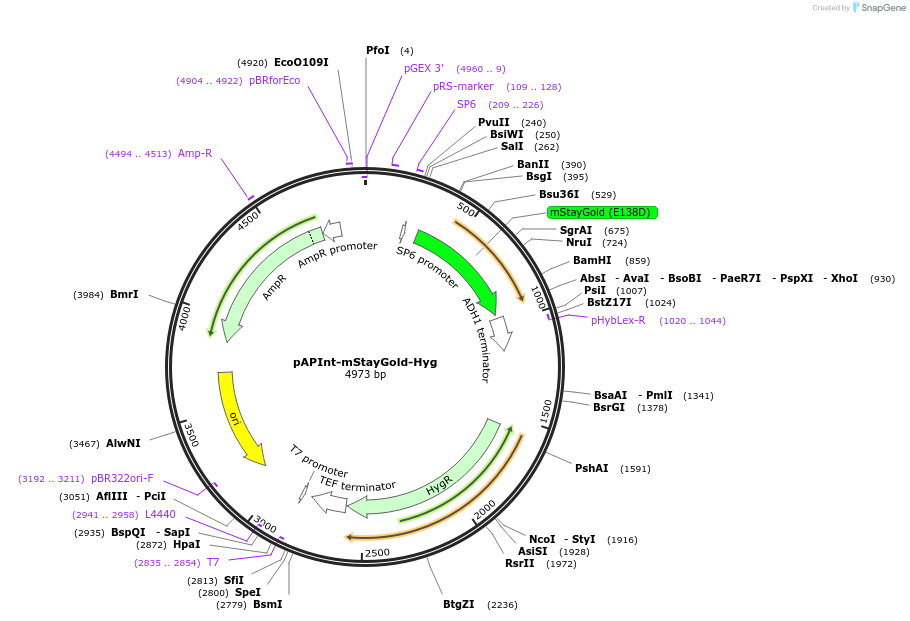
pAPInt-mStayGold-Hyg
Plasmid#236486PurposePlasmid for deleting or C-terminally tagging endogenous genes with mStayGold and selection on Hyg.DepositorInsertmStayGold
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
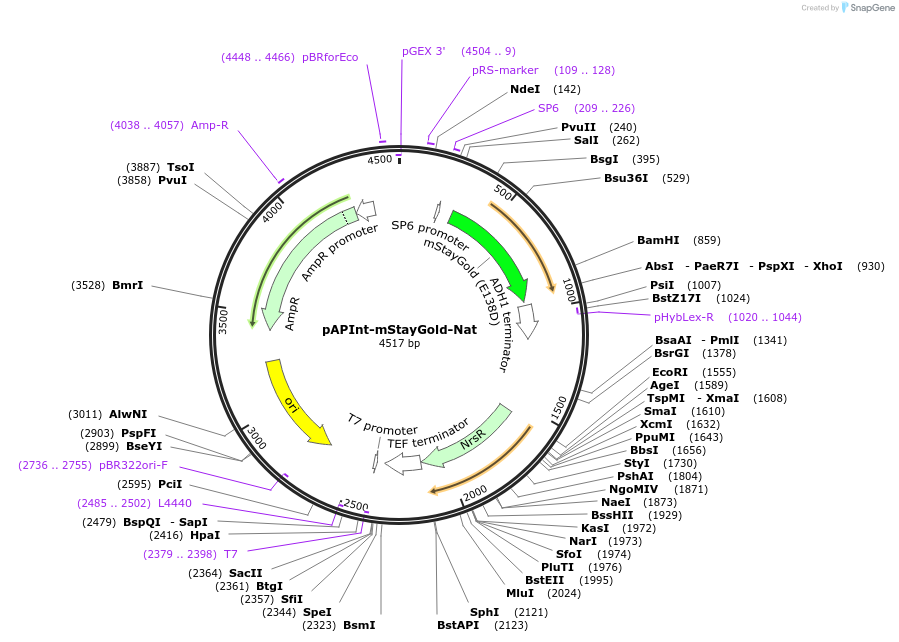
pAPInt-mStayGold-Nat
Plasmid#236485PurposePlasmid for deleting or C-terminally tagging endogenous genes with mStayGold and selection on Nat.DepositorInsertmStayGold
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
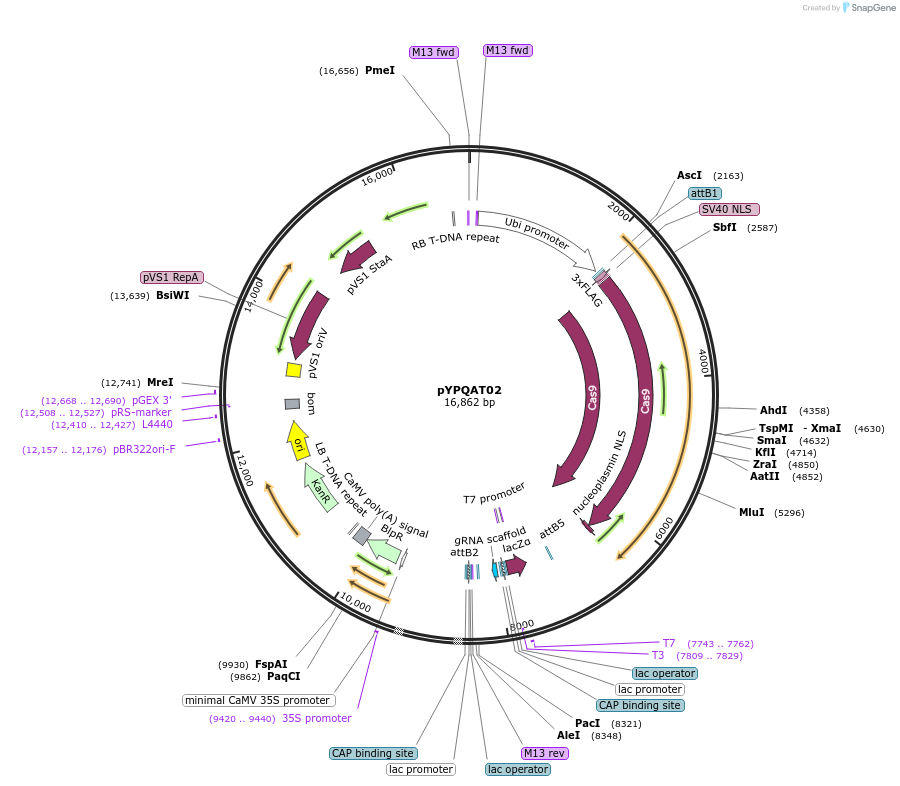
pYPQAT02
Plasmid#223374PurposeT-DNA vector for SpCas9 mediated mutagenesis for plants; NGG PAM; Cas9 was driven by ZmUbi1 and the sgRNA was driven by AtU3 promoter; BASTA for plants selection.DepositorInsertZmUbi-SpCas9-AtU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
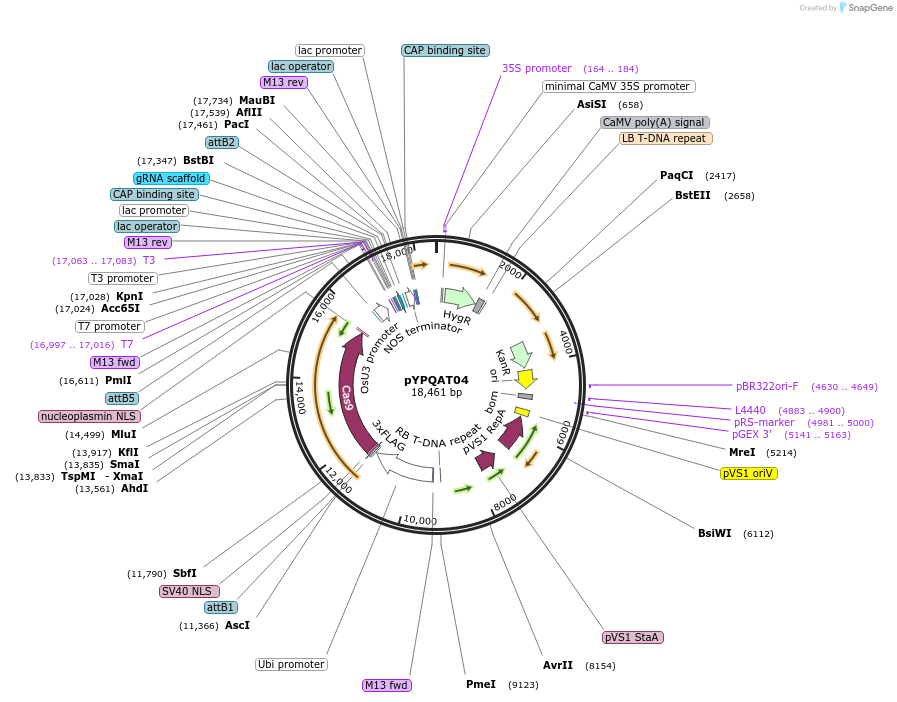
pYPQAT04
Plasmid#223376PurposeT-DNA vector for SpCas9 mediated mutagenesis for monocot plants; NGG PAM; Cas9 was driven by ZmUbi1 and the sgRNA was driven by OsU3 promoter; Hygromycin for plants selection.DepositorInsertZmUbi-SpCas9-OsU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
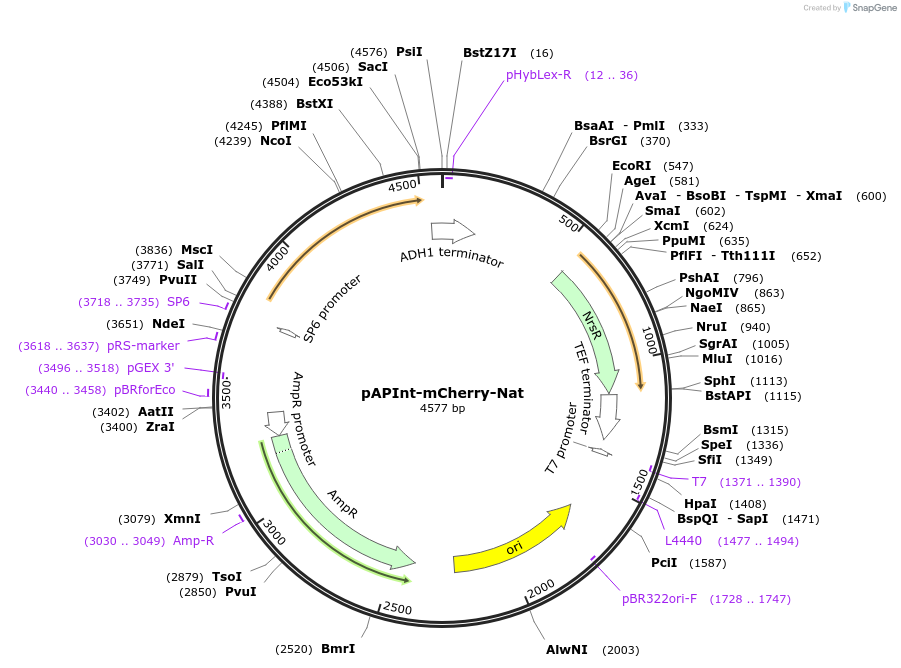
pAPInt-mCherry-Nat
Plasmid#236488PurposePlasmid for deleting or C-terminally tagging endogenous genes with mCherry and selection on Nat.DepositorInsertmCherry
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
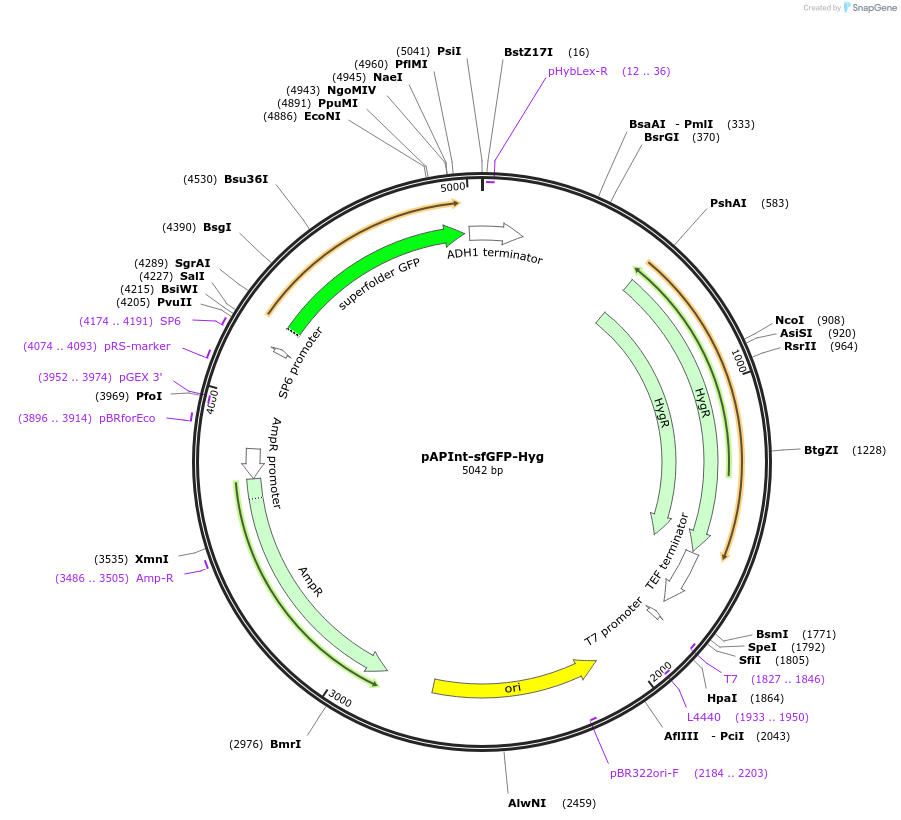
pAPInt-sfGFP-Hyg
Plasmid#236480PurposePlasmid for deleting or C-terminally tagging endogenous genes with sfGFP and selection on Hyg.DepositorInsertsfGFP
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
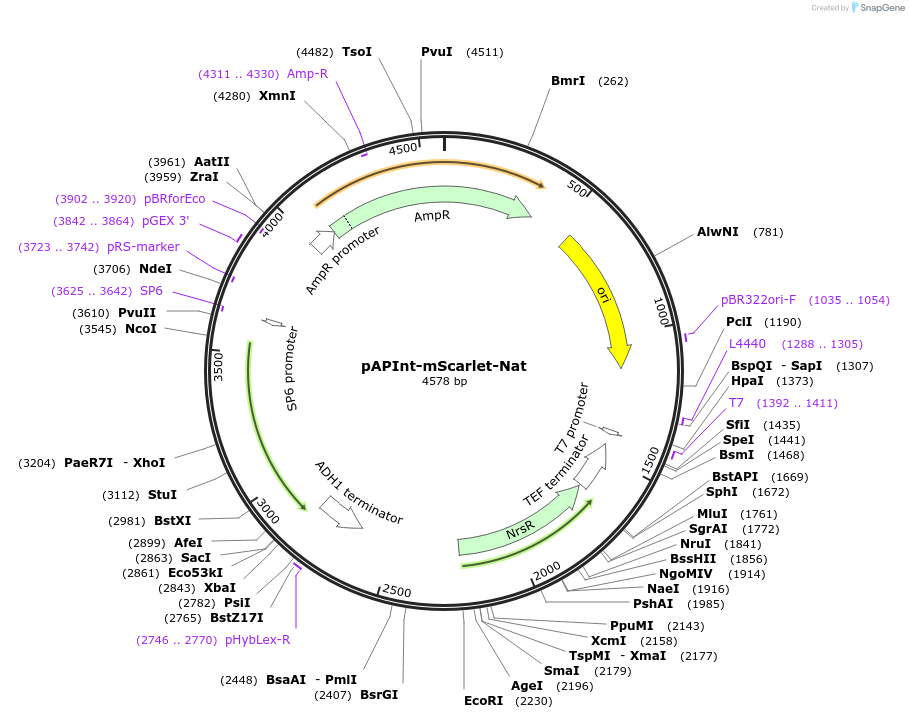
pAPInt-mScarlet-Nat
Plasmid#236491PurposePlasmid for deleting or C-terminally tagging endogenous genes with mScarlet and selection on Nat.DepositorInsertmScarlet
ExpressionYeastMutationCodon optimized for A. pullulansAvailable SinceApril 9, 2025AvailabilityAcademic Institutions and Nonprofits only