We narrowed to 23,118 results for: Sis
-
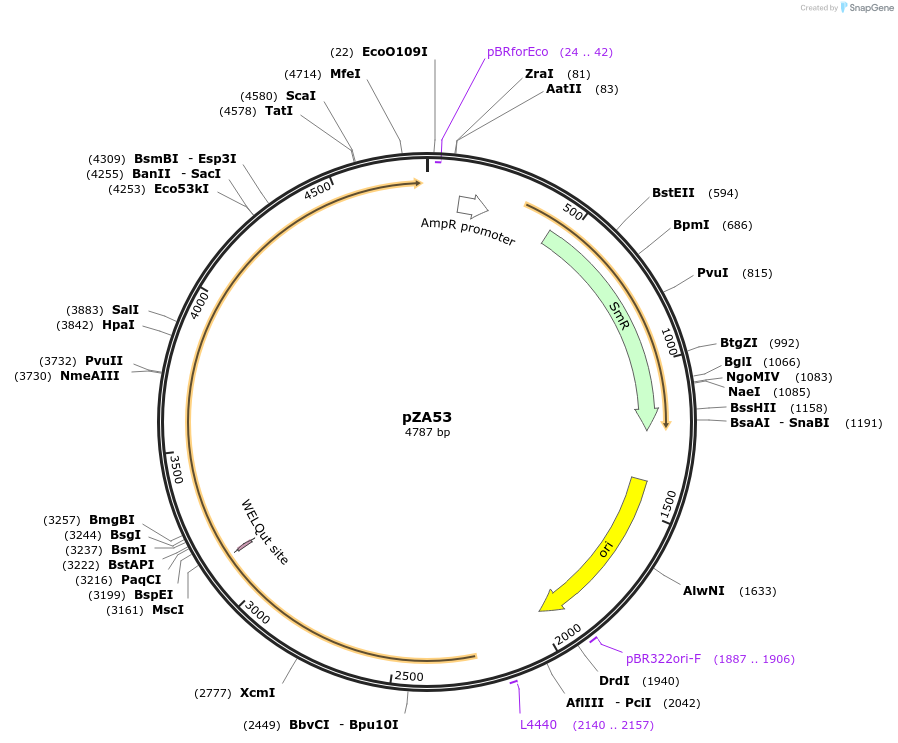
Plasmid#174766PurposeEntry clone for Golden Gate assemblyDepositorInsertGG_NbZAR1syn without STOP codon
UseUnspecifiedMutationWTAvailable SinceOct. 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
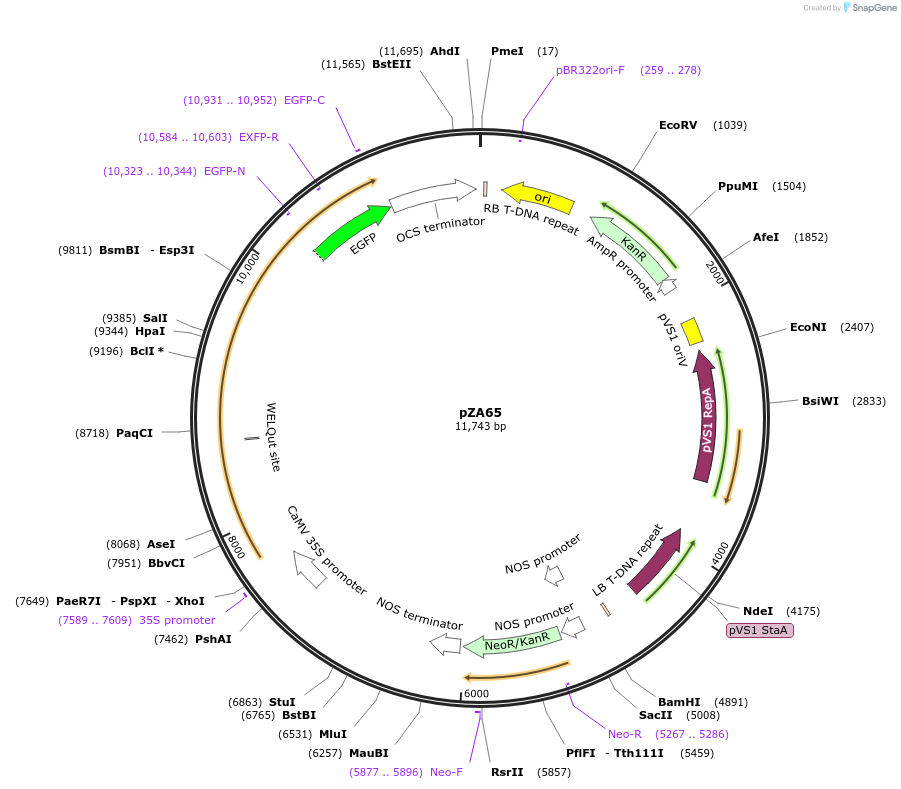
pZA65
Plasmid#174778PurposeIn planta gene expressionDepositorInsertGG_NbZAR1syn-eGFP, Kmr plant selection marker
TagseGFPExpressionPlantMutationWTAvailable SinceOct. 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
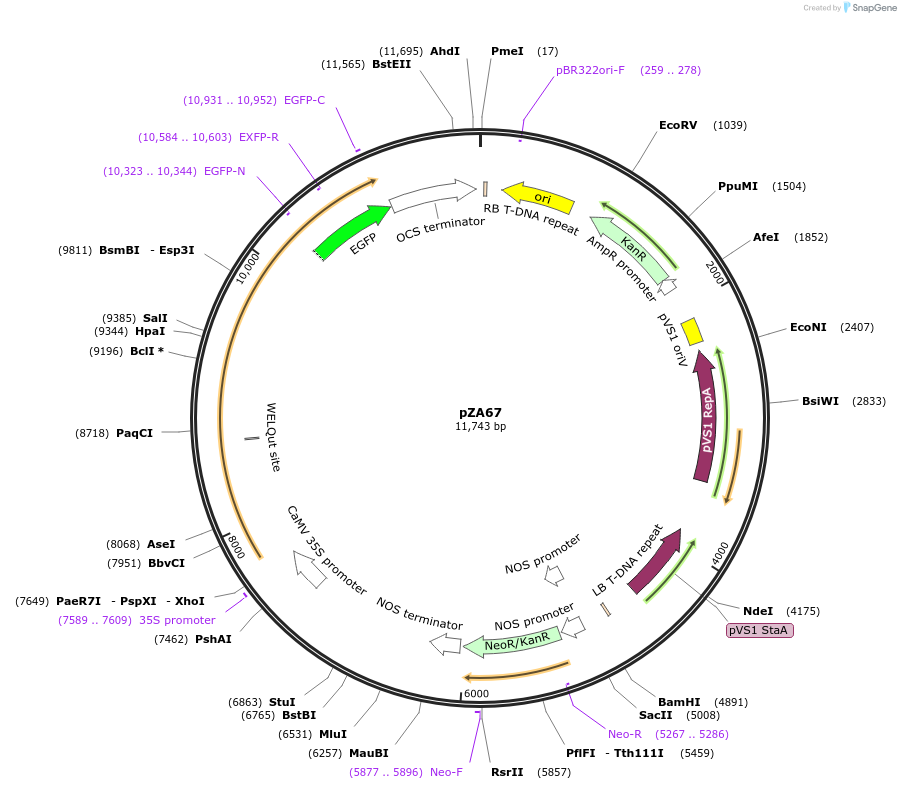
pZA67
Plasmid#174780PurposeIn planta gene expressionDepositorInsertGG_NbZAR1syn-L17E-eGFP, Kmr plant selection marker
TagseGFPExpressionPlantMutationL17EAvailable SinceOct. 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
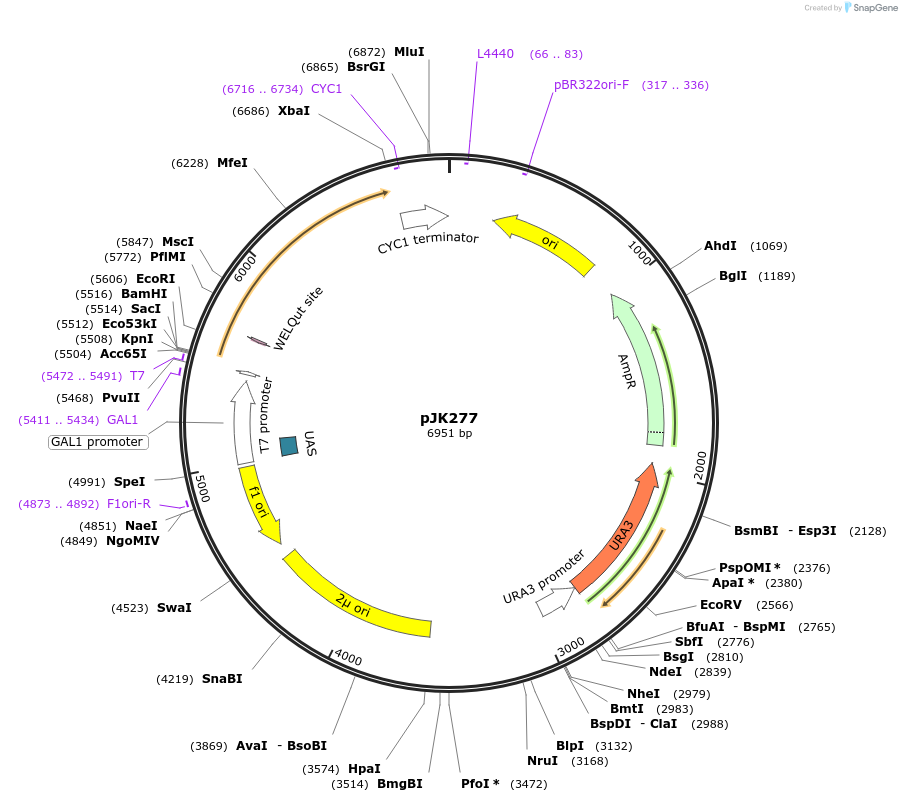
pJK277
Plasmid#71565PurposeProduces Crepis alpina delta15 fatty acid desaturase (FAD3) in yeastDepositorInsertdelta15 fatty acid desaturase
ExpressionBacterial and YeastPromoterGAL1Available SinceJan. 14, 2016AvailabilityAcademic Institutions and Nonprofits only -
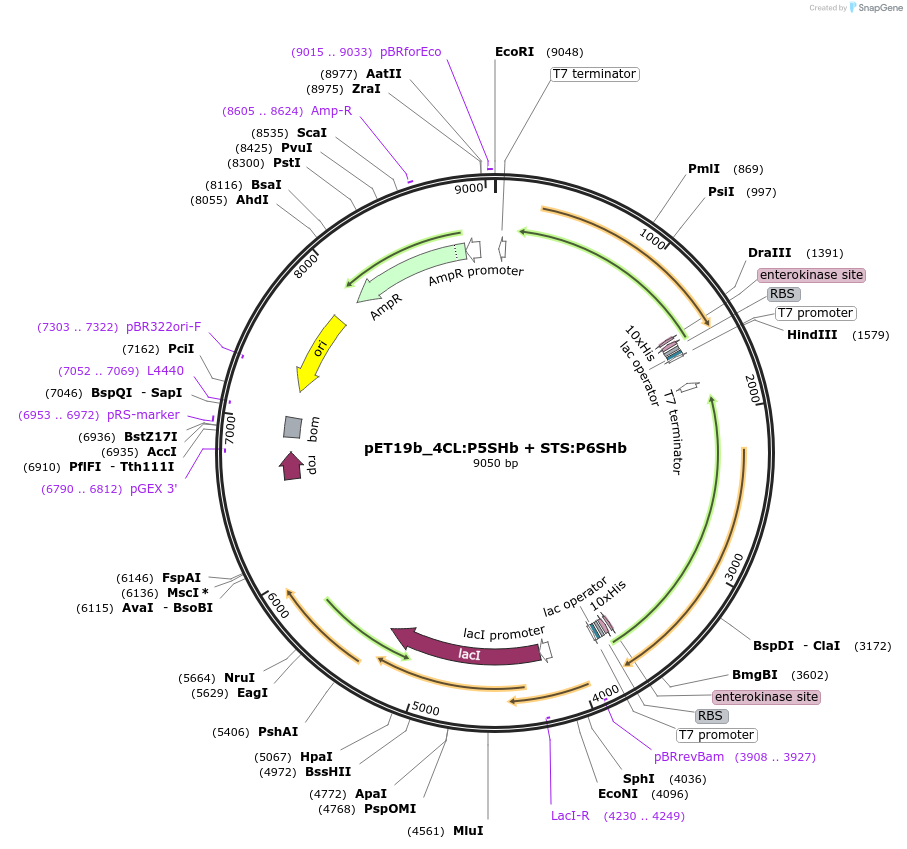
pET19b_4CL:P5SHb + STS:P6SHb
Plasmid#139792Purposeexpression of biosynthetic enzymes 4CL and STS in fusion with CCDepositorInserts4CL:P5SHb
STS:P6SHb
ExpressionBacterialAvailable SinceJune 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
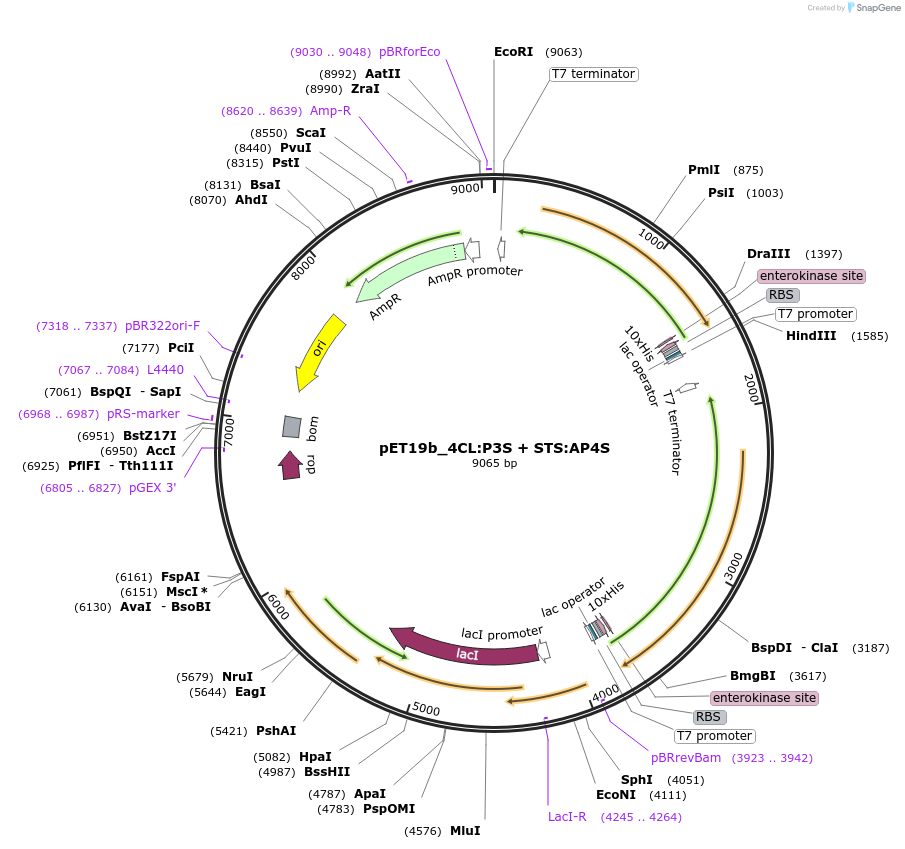
pET19b_4CL:P3S + STS:AP4S
Plasmid#139795Purposeexpression of biosynthetic enzymes 4CL and STS in fusion with CCDepositorInserts4CL:P3S
STS:AP4S
ExpressionBacterialAvailable SinceMay 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
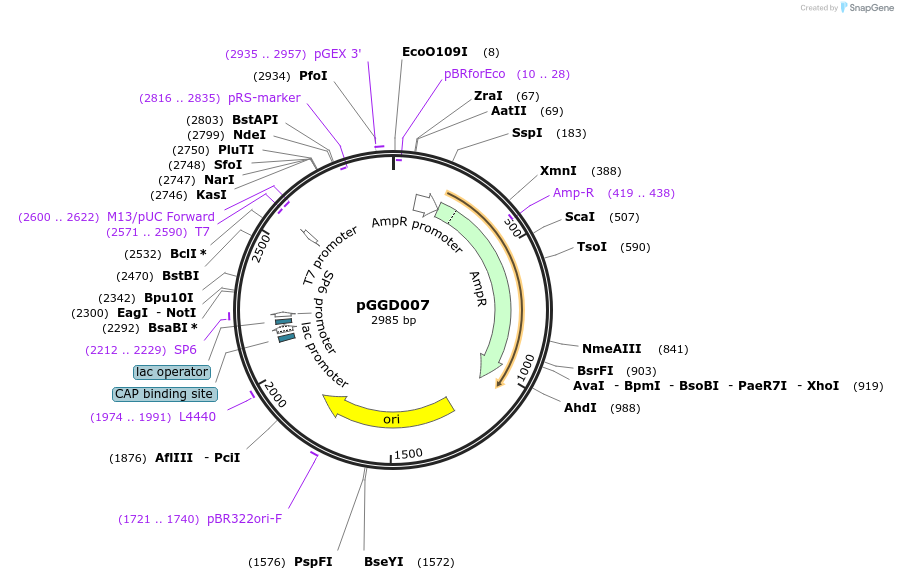
pGGD007
Plasmid#48837PurposeProvides linker:nuclear localization signal as GreenGate C-terminal tag module.DepositorInsertlinker:nuclear localization signal
UseGolden gate compatible cloning vectorAvailable SinceJan. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
pcDNA-FLAG-Nv-NF-kB-Cys
Plasmid#27260DepositorInsertNv-NF-kB
TagsFLAGExpressionMammalianMutation"Cys allele" = Cysteine at aa 67Available SinceFeb. 1, 2011AvailabilityAcademic Institutions and Nonprofits only -
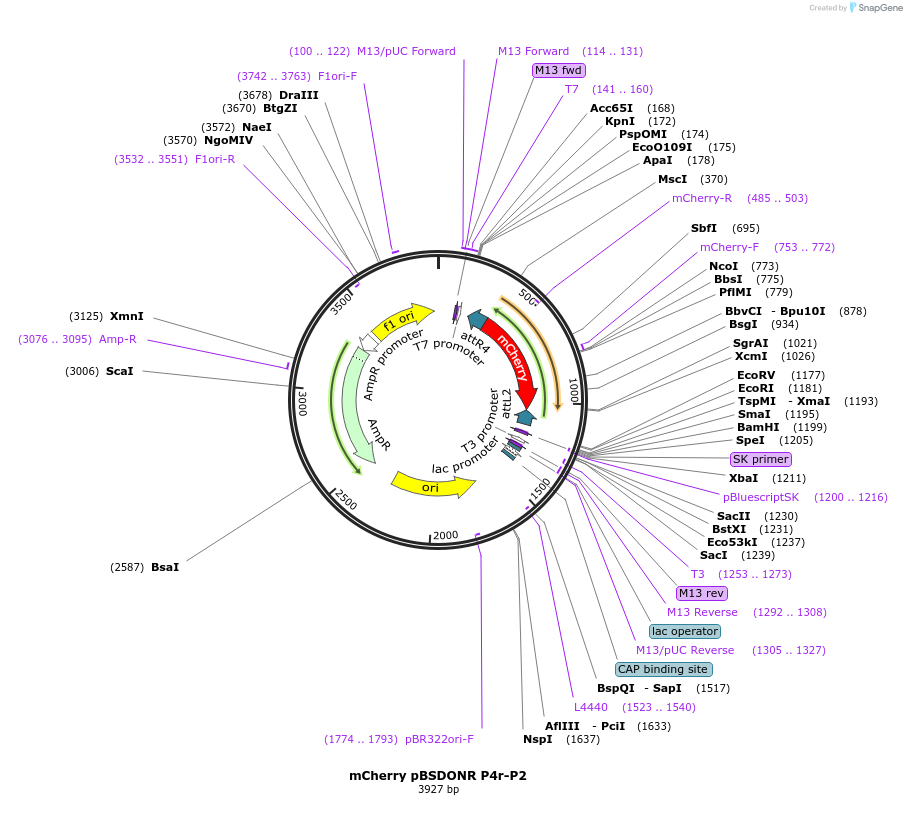
mCherry pBSDONR P4r-P2
Plasmid#121528PurposeFor making C-terminal mCherry fusions using multisite Gateway cloningDepositorInsertmCherry
ExpressionBacterialAvailable SinceAug. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
pET15b-At 14-3-3 iota
Plasmid#44937DepositorAvailable SinceMay 14, 2013AvailabilityAcademic Institutions and Nonprofits only -
pcDNA-FLAG-Nv-NF-kB-Ser
Plasmid#27259DepositorInsertNv-NF-kB
TagsFLAGExpressionMammalianMutation"Ser allele" = Serine at aa 67Available SinceFeb. 1, 2011AvailabilityAcademic Institutions and Nonprofits only -
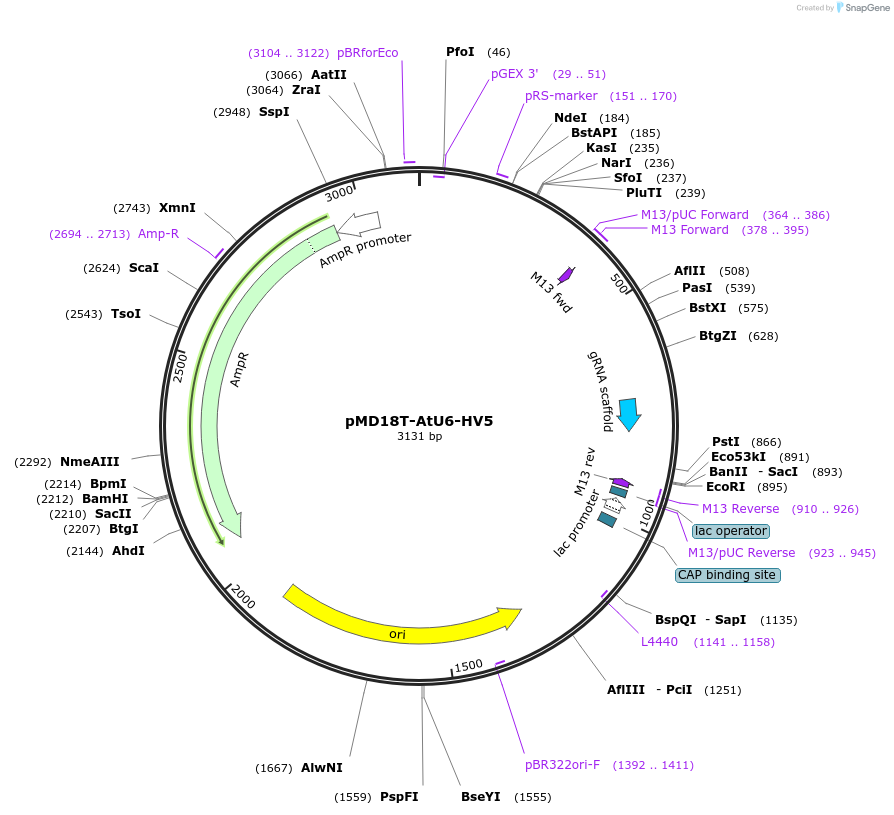
pMD18T-AtU6-HV5
Plasmid#239470PurposeEncodes a gRNA expression cassette driven by the AtU6 promoter.DepositorInsertU6-gRNA
UseCRISPRExpressionPlantAvailable SinceDec. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
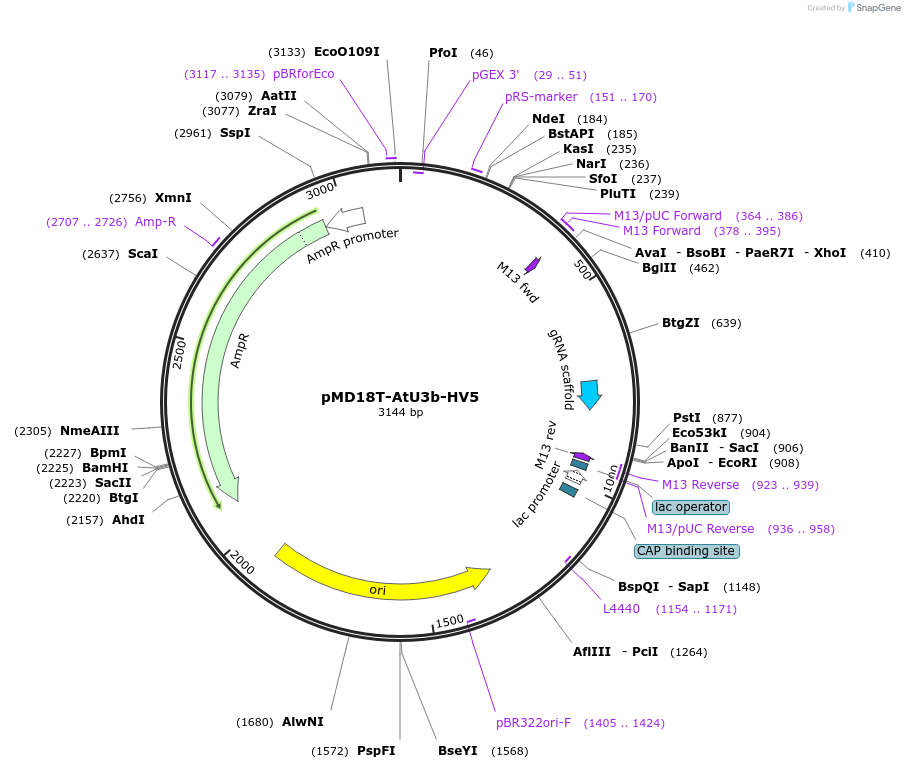
pMD18T-AtU3b-HV5
Plasmid#239477PurposeEncodes a gRNA expression cassette driven by the AtU3b promoter.DepositorInsertU3b-gRNA
UseCRISPRExpressionPlantAvailable SinceDec. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
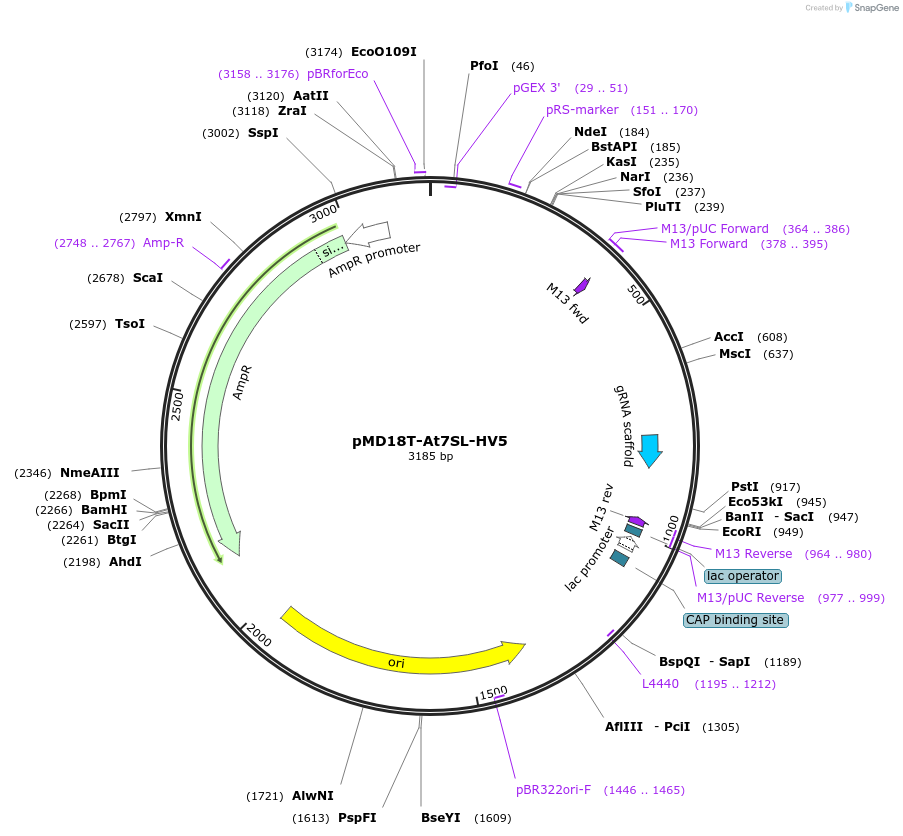
pMD18T-At7SL-HV5
Plasmid#239478PurposeEncodes a gRNA expression cassette driven by the At7SL promoter.DepositorInsert7SL-gRNA
UseCRISPRExpressionPlantAvailable SinceDec. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
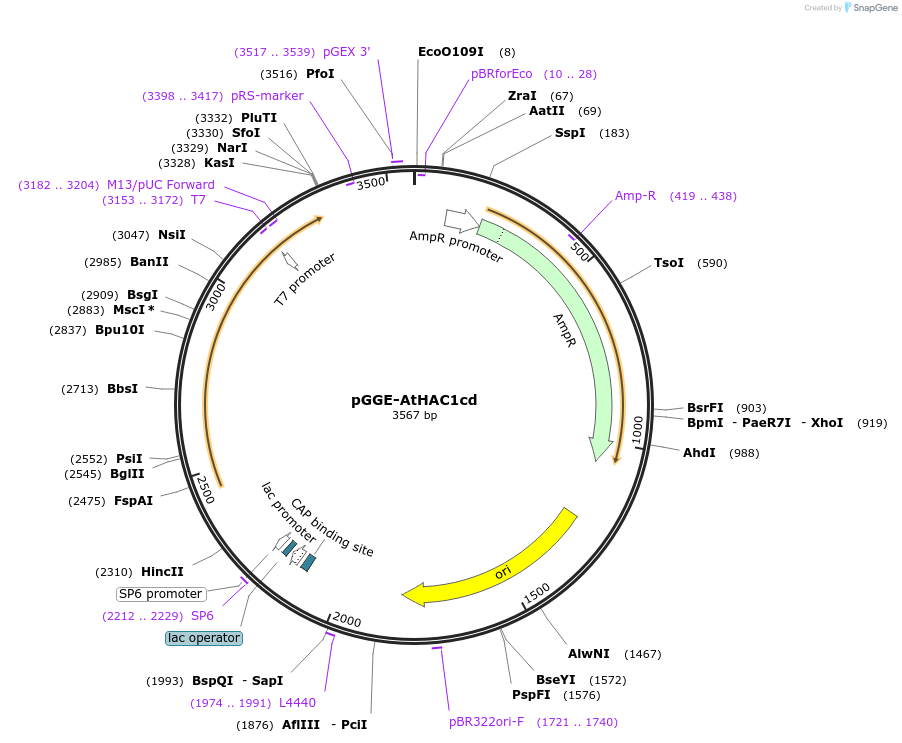
pGGE-AtHAC1cd
Plasmid#246809PurposeA GreenGate entry vector containing coding region of AtHAC1 catalytic domainDepositorInsertAtHAC1cd (HAC1 Mustard Weed)
UseGreengate cloning entry vectorAvailable SinceDec. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
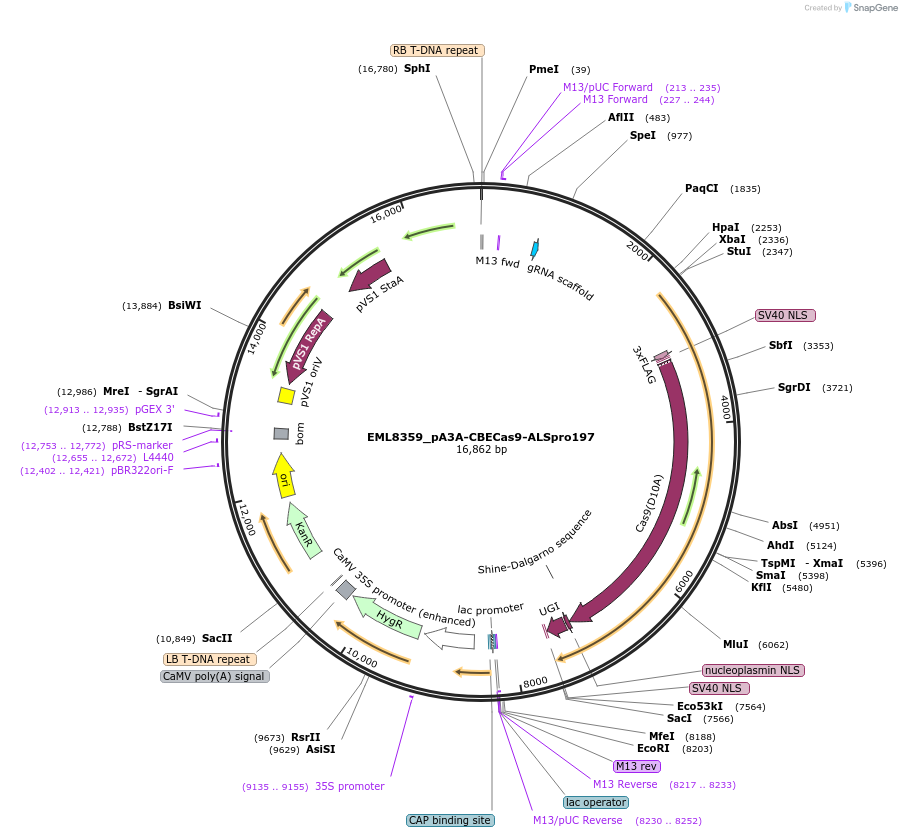
EML8359_pA3A-CBECas9-ALSpro197
Plasmid#234353PurposeBase editor targets to ALS Pro197 locusDepositorInsertALS
UseCRISPRExpressionPlantAvailable SinceNov. 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
pEPDG1CB0004
Plasmid#227542PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CaTXSS_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
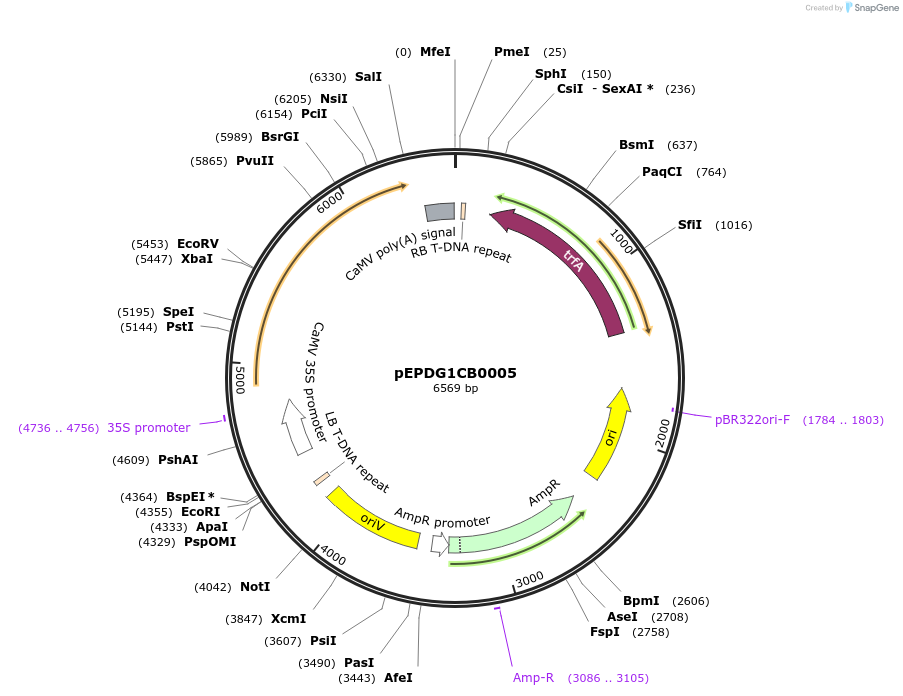
pEPDG1CB0005
Plasmid#227548PurposeLevel 1 - Position 1 with 35S promoter and terminator for plant expressionDepositorInsert35Sshort_TMV_CYP716A392a_35S
ExpressionPlantMutationBsaI sites removed by silent mutationPromoter35Sshort_TMV (pICH51277)Available SinceJuly 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
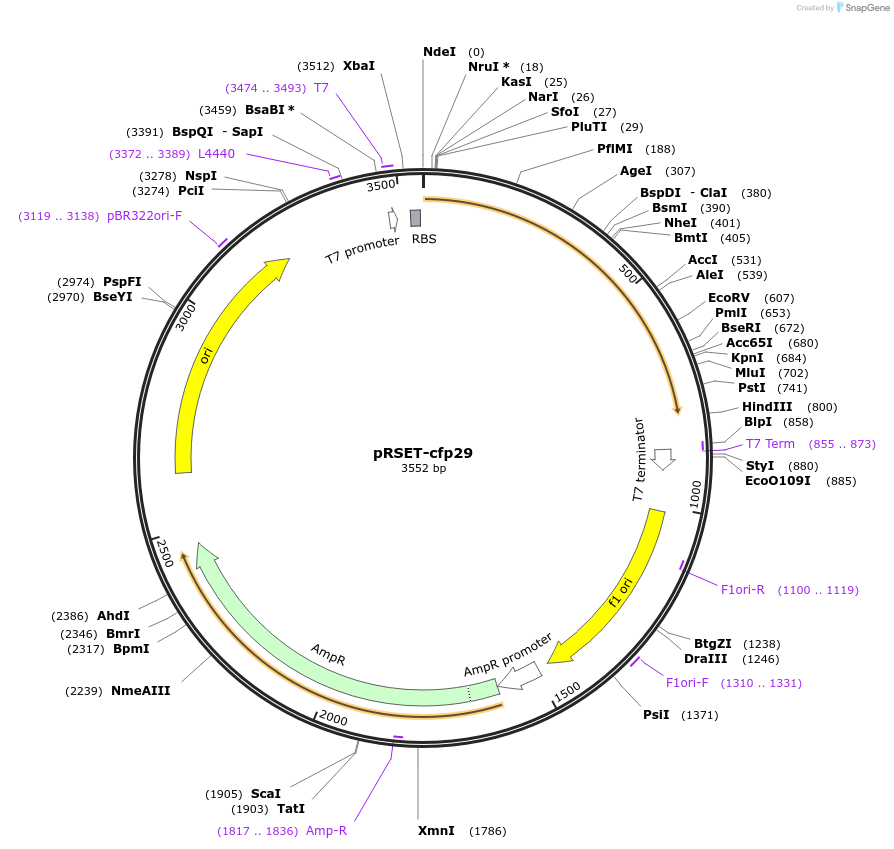
pRSET-cfp29
Plasmid#233651PurposeMycobacterium tuberculosis Encapsulin CFP29 expression plasmid in E. coliDepositorInsertEncapsulin
ExpressionBacterialPromoterT7 promoterAvailable SinceJune 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
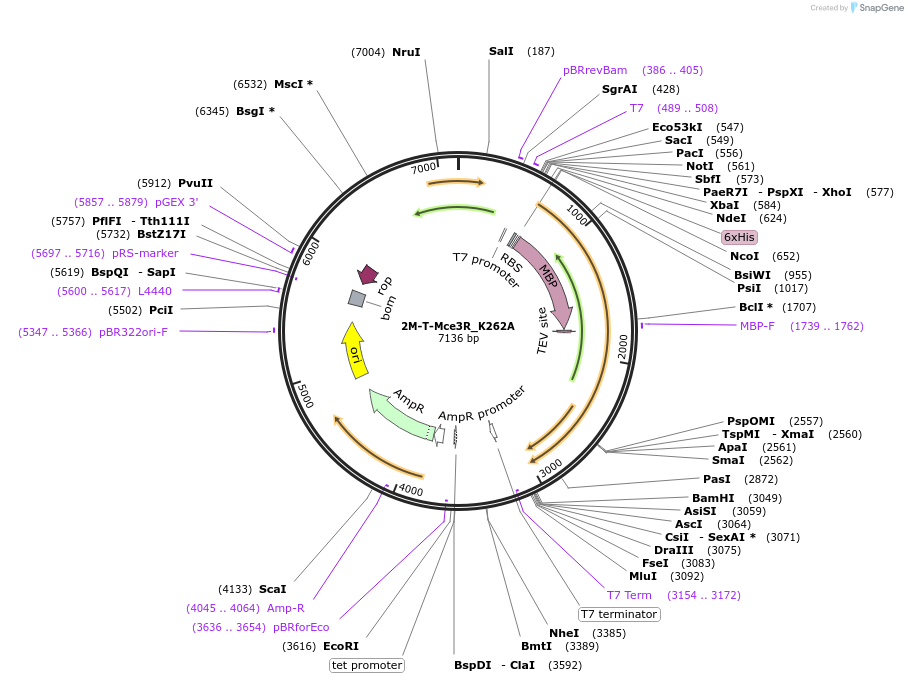
2M-T-Mce3R_K262A
Plasmid#225728PurposeTo express Mce3R mutant K262A in E. coliDepositorInsertmce3R
Tags6xHis-MBPExpressionBacterialMutationK262AAvailable SinceJan. 17, 2025AvailabilityAcademic Institutions and Nonprofits only