We narrowed to 9,586 results for: Coli
-
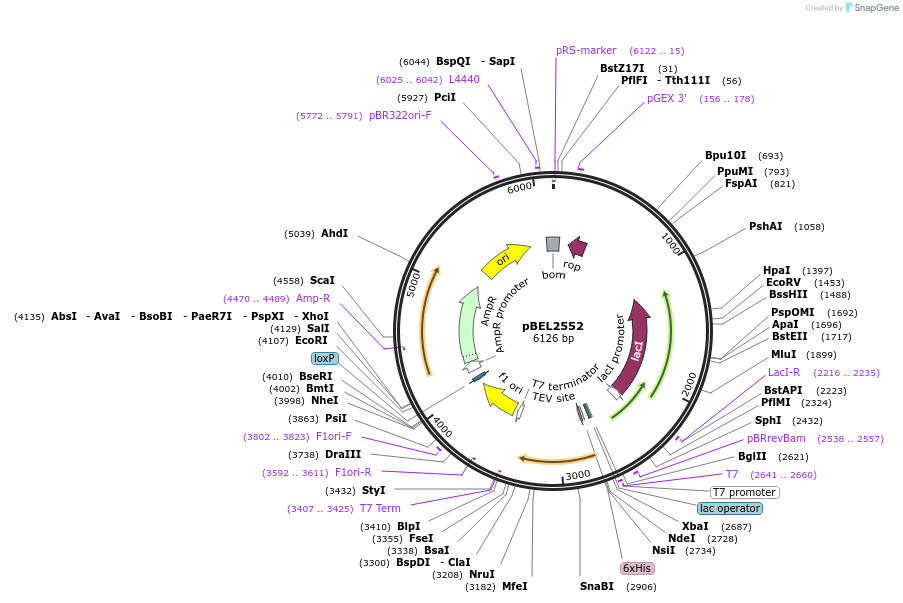
Plasmid#195670PurposeExpression/purification of 6xHis-TEV-MlaC(A168R)DepositorInsertMlaC
ExpressionBacterialMutation6xHis-TEV-MlaC(A168R)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2553
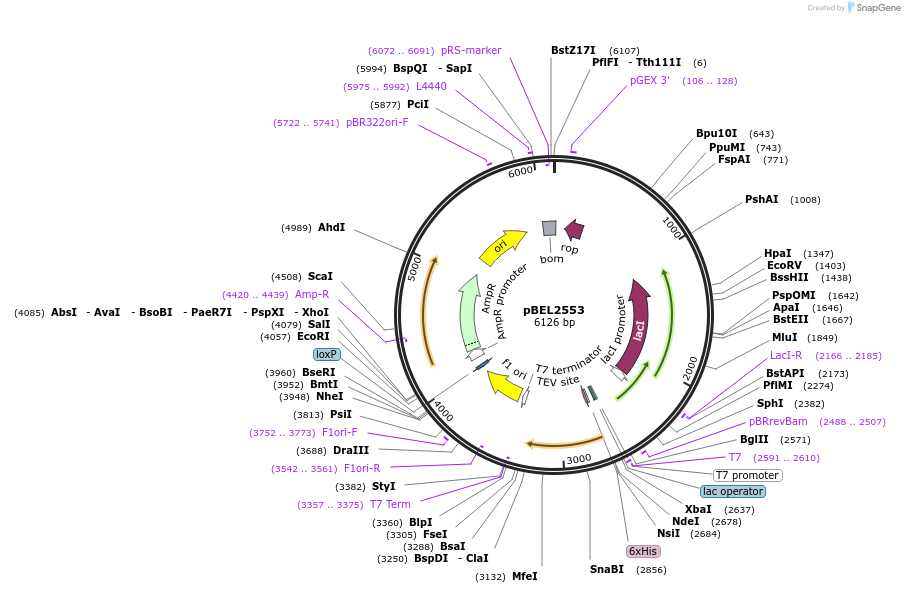
Plasmid#195671PurposeExpression/purification of 6xHis-TEV-MlaC(D165I)DepositorInsertMlaC
ExpressionBacterialMutation6xHis-TEV-MlaC(D165I)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2554
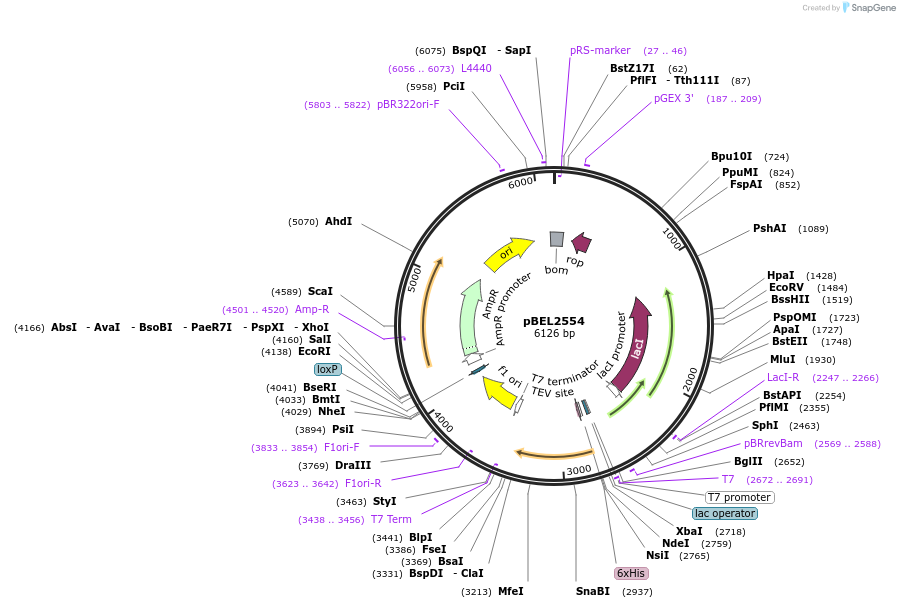
Plasmid#195672PurposeExpression/purification of 6xHis-TEV-MlaC(E169R)DepositorInsertMlaC
ExpressionBacterialMutation6xHis-TEV-MlaC(E169R)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2522
Plasmid#195644PurposeComplementation of MlaC(A108R)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(A108R)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
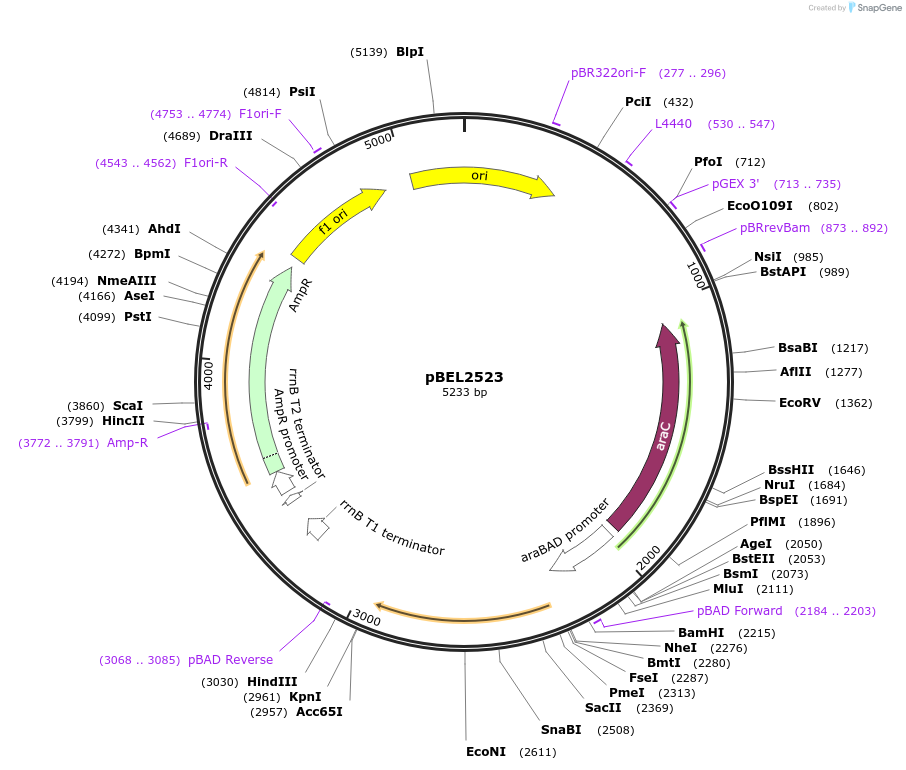
pBEL2523
Plasmid#195645PurposeComplementation of MlaC(A163R)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(A163R)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
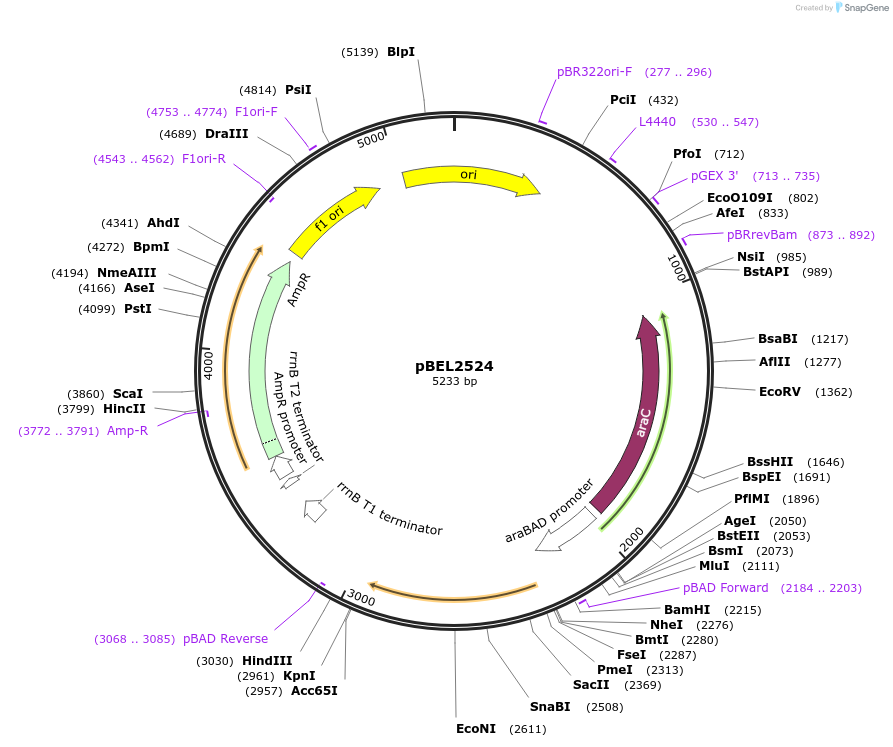
pBEL2524
Plasmid#195646PurposeComplementation of MlaC(A168R)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(A168R)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
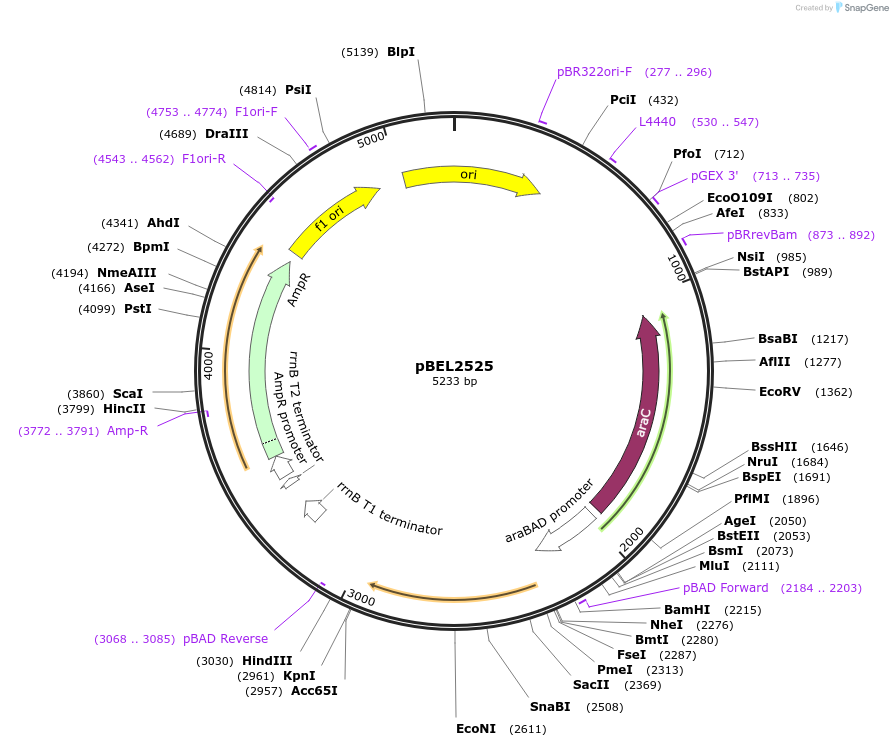
pBEL2525
Plasmid#195647PurposeComplementation of MlaC(D165I)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(D165I)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
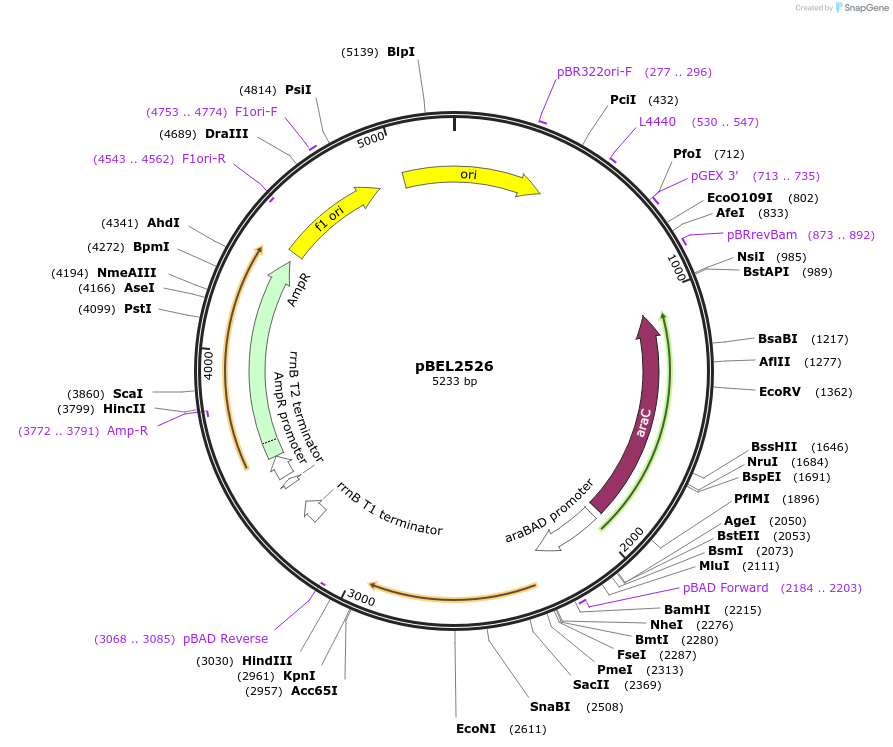
pBEL2526
Plasmid#195648PurposeComplementation of MlaC(E169R)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(E169R)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2527
Plasmid#195649PurposeComplementation of MlaC(G170K)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(G170K)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2528
Plasmid#195650PurposeComplementation of MlaC(I130D)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(I130D)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
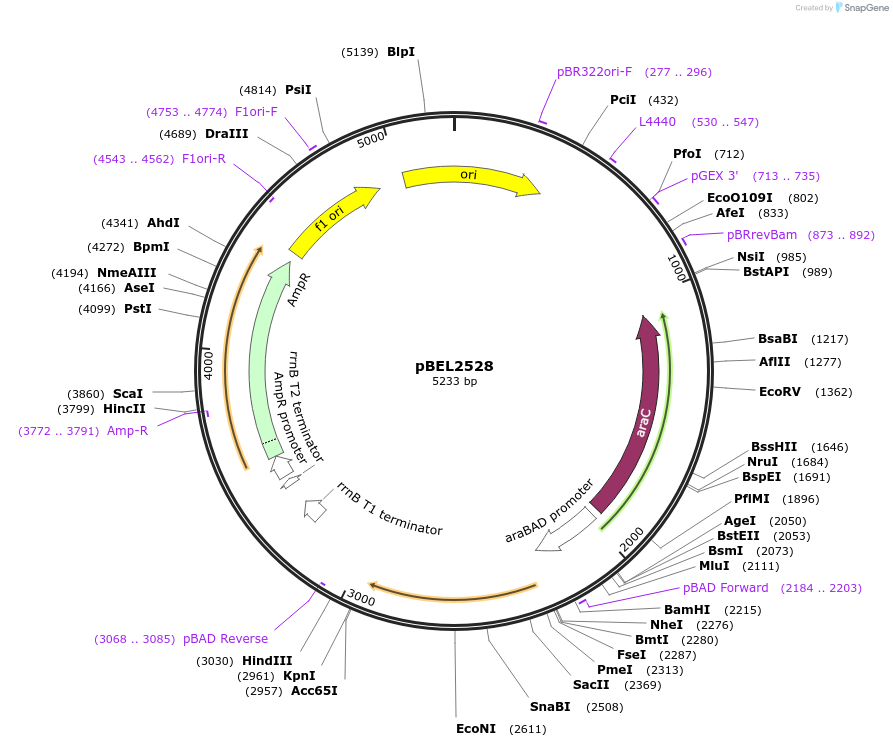
pBEL2530
Plasmid#195651PurposeComplementation of MlaC(I137D)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(I137D)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
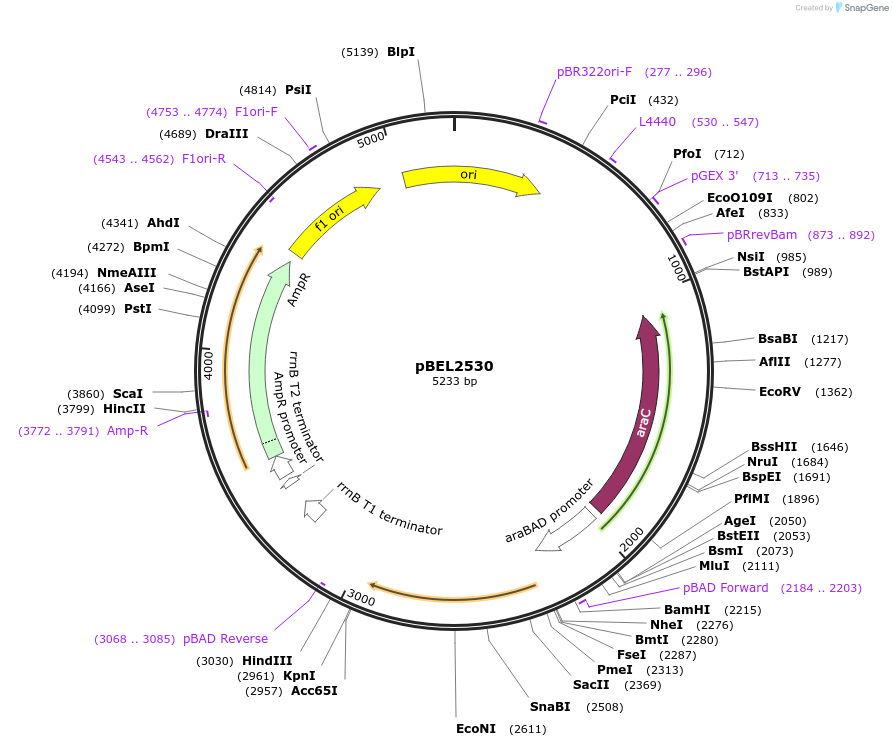
pBEL2531
Plasmid#195652PurposeComplementation of MlaC(L76D)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(L76D)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
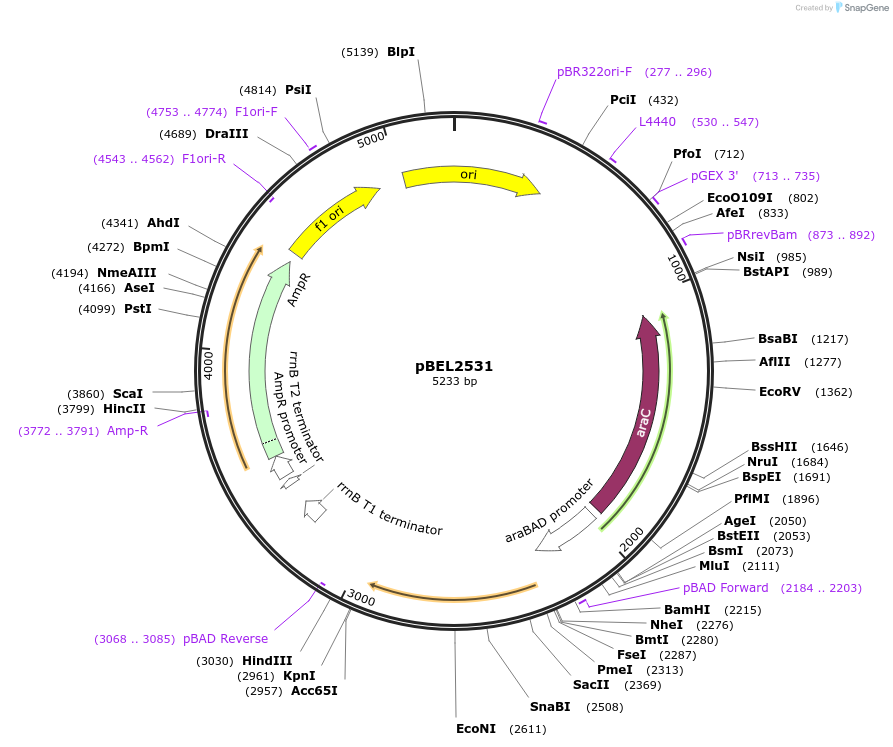
pBEL2532
Plasmid#195653PurposeComplementation of MlaC(N155A)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(N155A)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
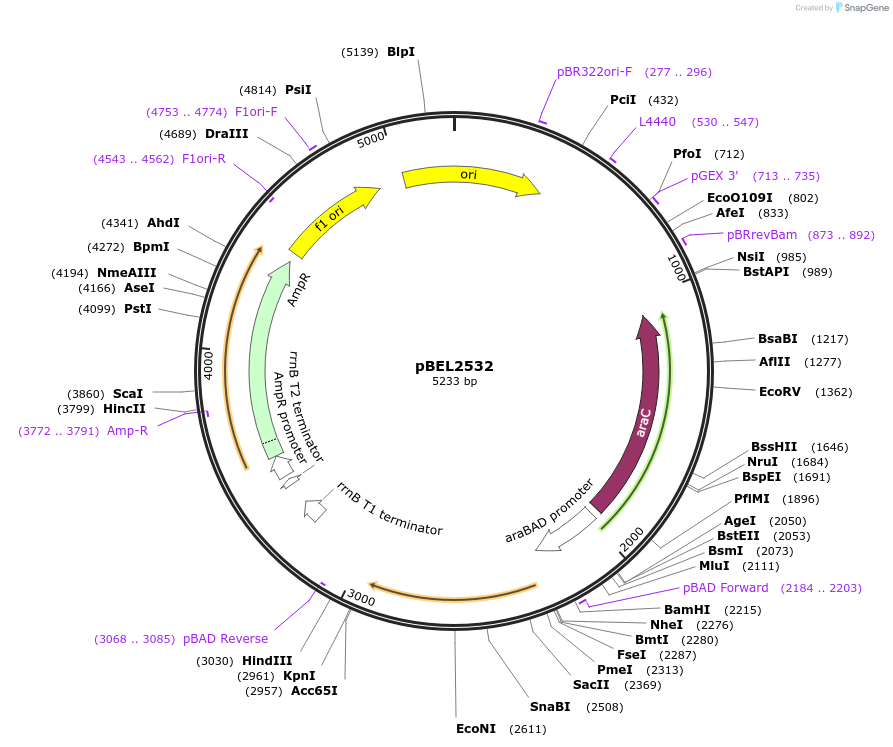
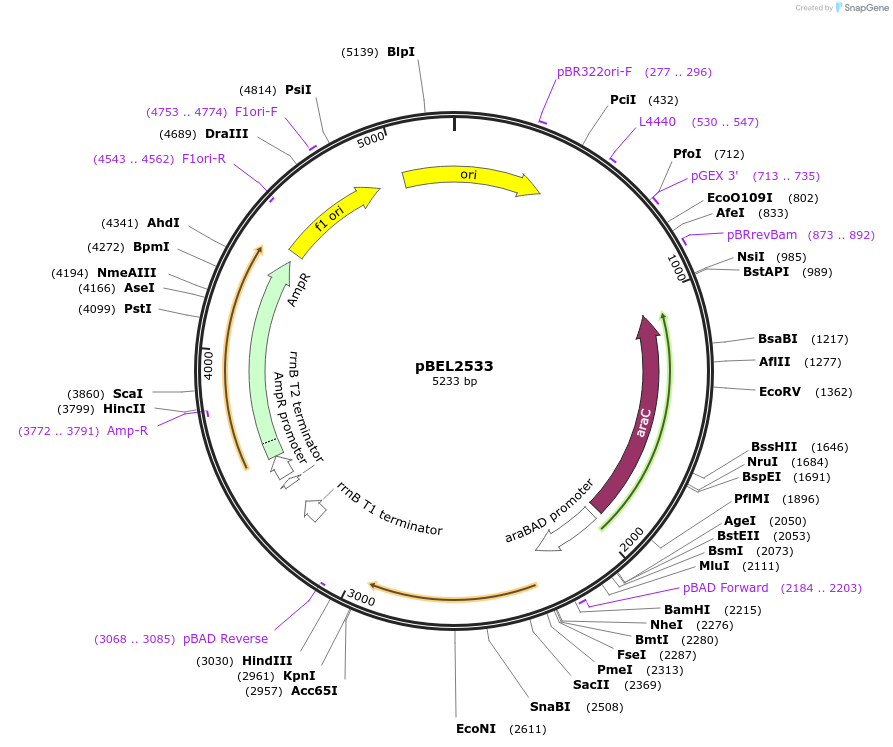
pBEL2533
Plasmid#195654PurposeComplementation of MlaC(N179E)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(N179E)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
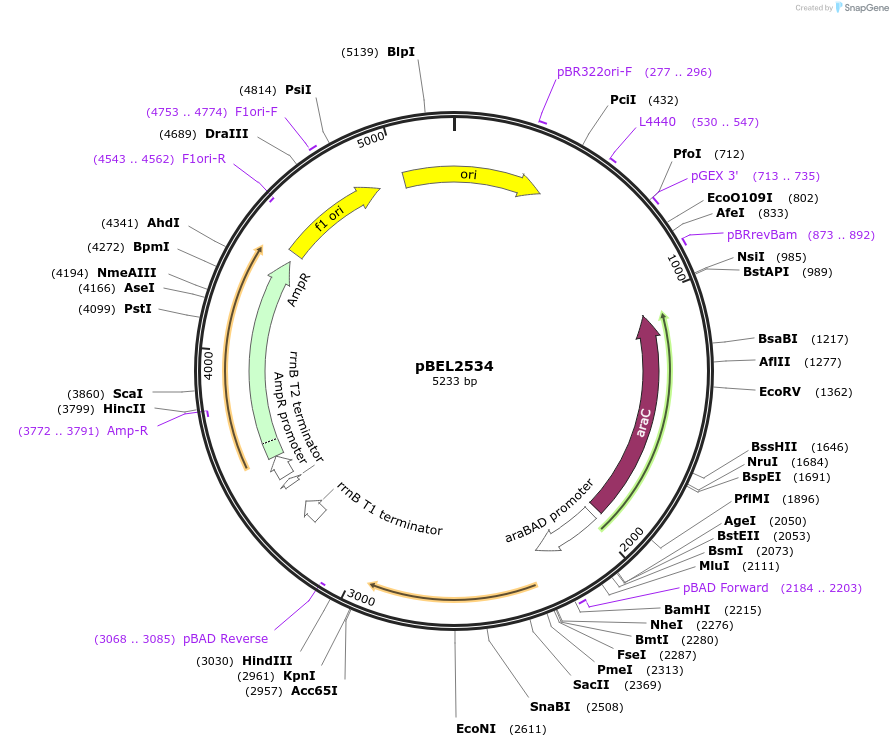
pBEL2534
Plasmid#195655PurposeComplementation of MlaC(Q115A)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(Q115A)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
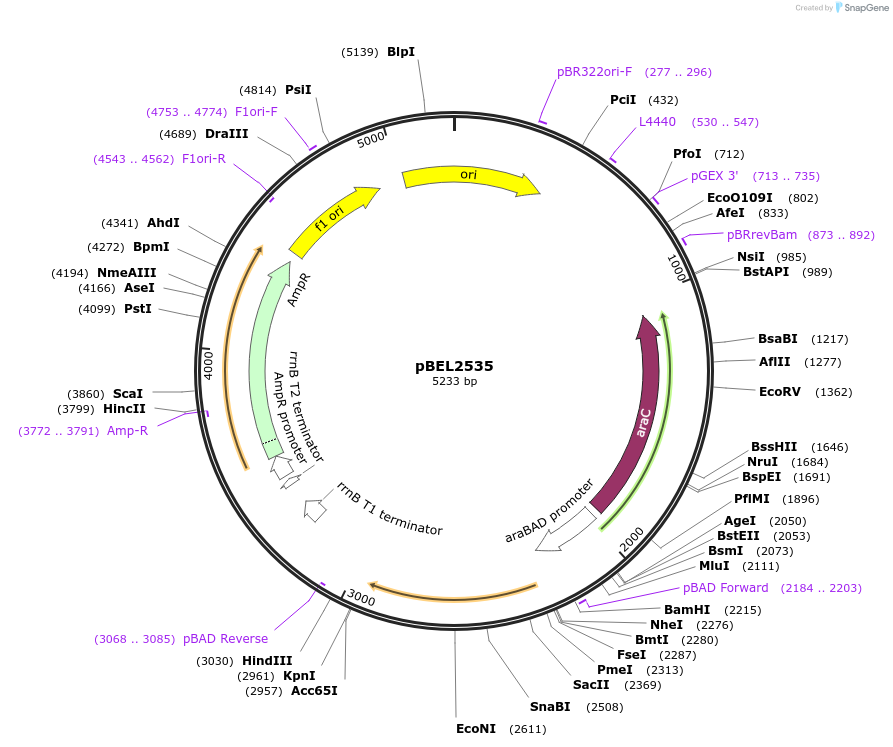
pBEL2535
Plasmid#195656PurposeComplementation of MlaC(R134E)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(R134E)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
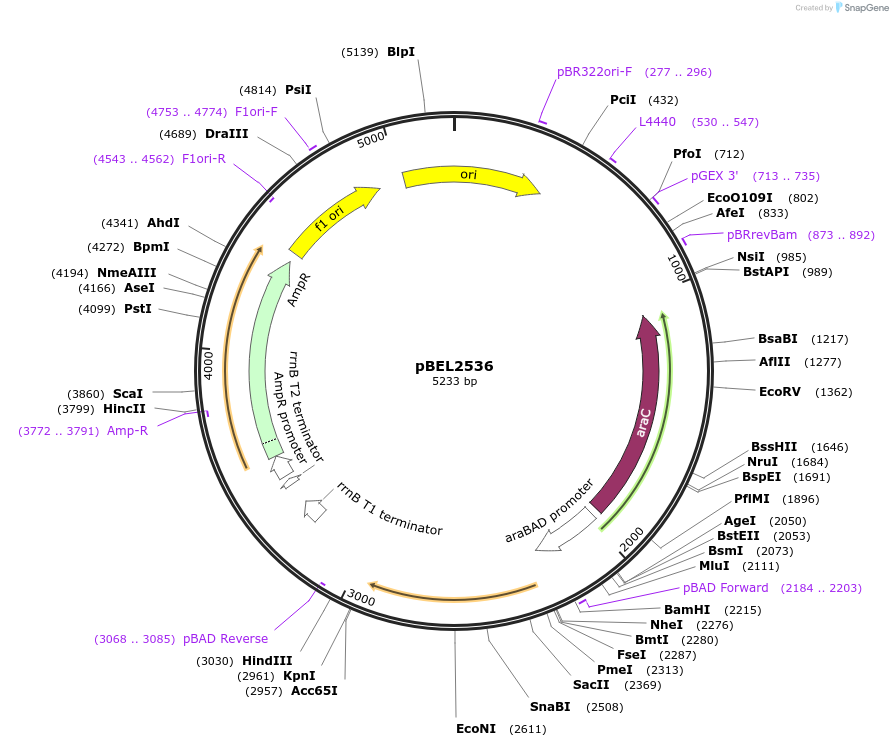
pBEL2536
Plasmid#195657PurposeComplementation of MlaC(R147D)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(R147D)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
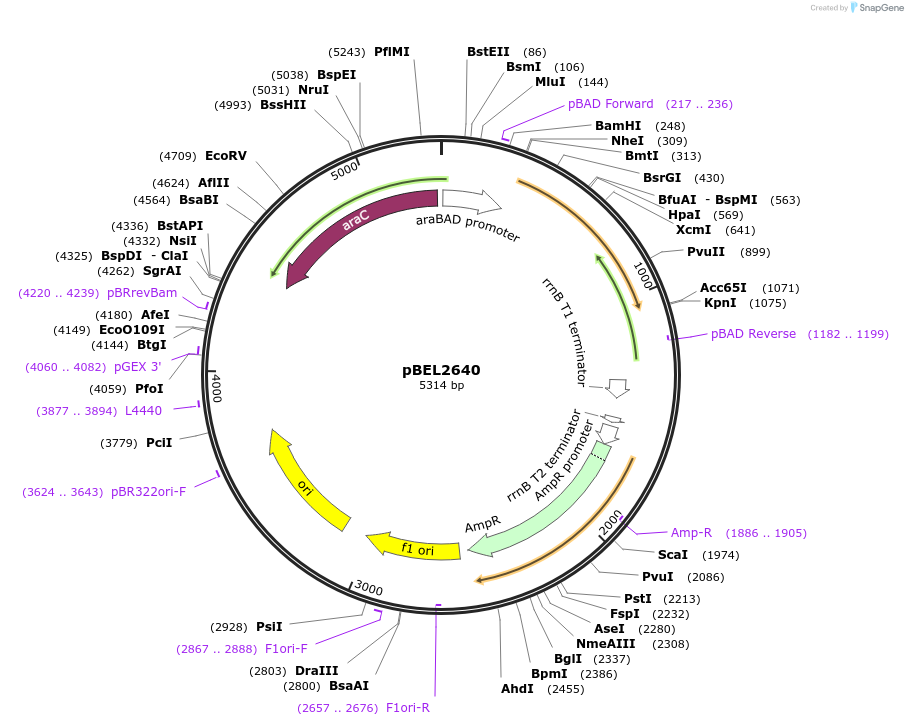
pBEL2640
Plasmid#195629PurposeComplementation of MlaA(L245N)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(L245N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
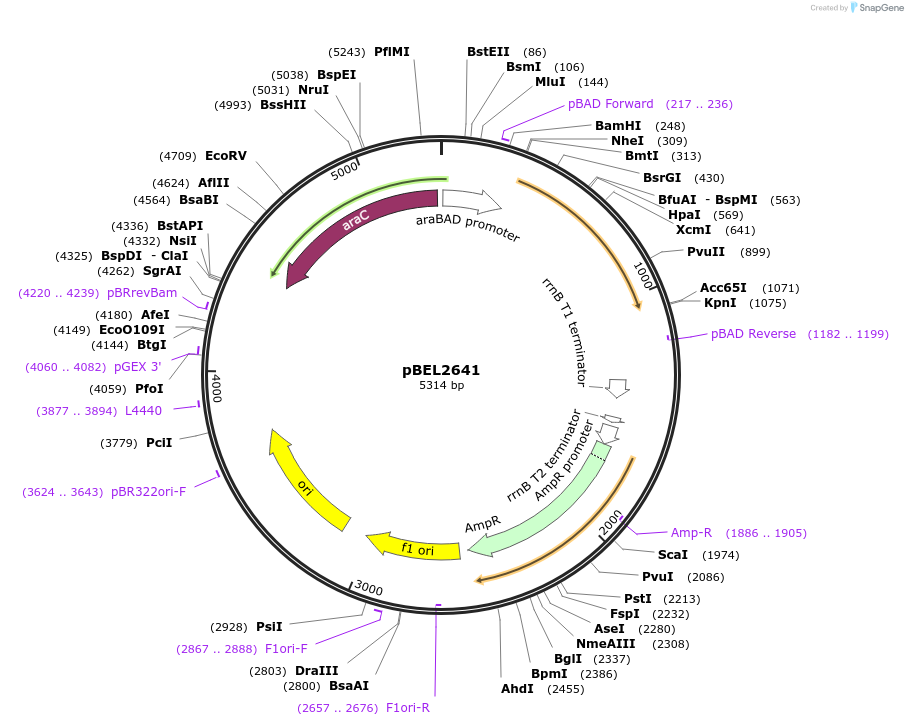
pBEL2641
Plasmid#195630PurposeComplementation of MlaA(I248N)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(I248N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
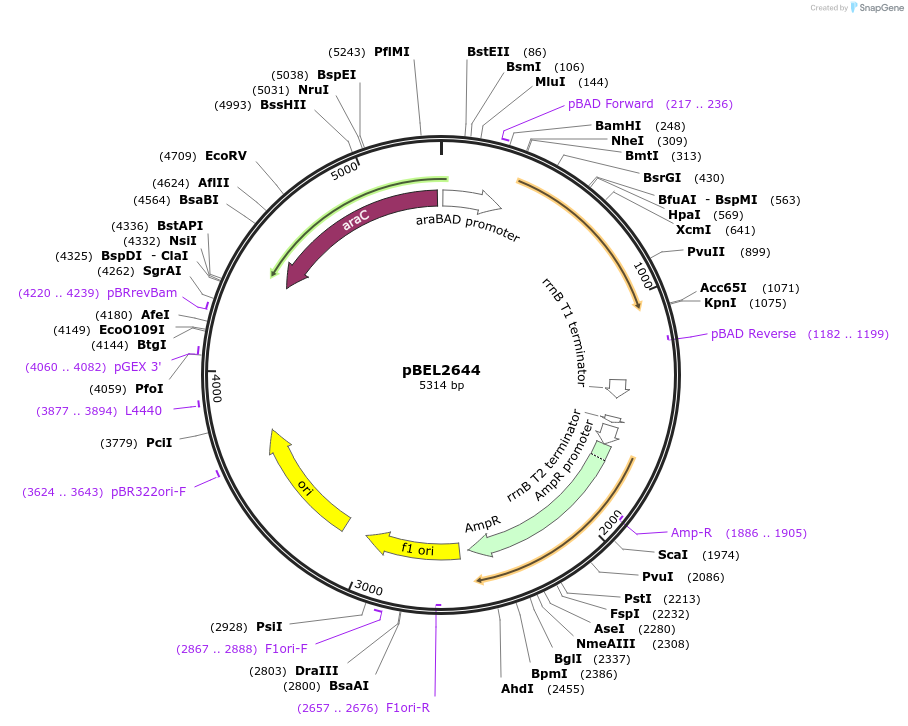
pBEL2644
Plasmid#195631PurposeComplementation of MlaA(D198N)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(D198N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
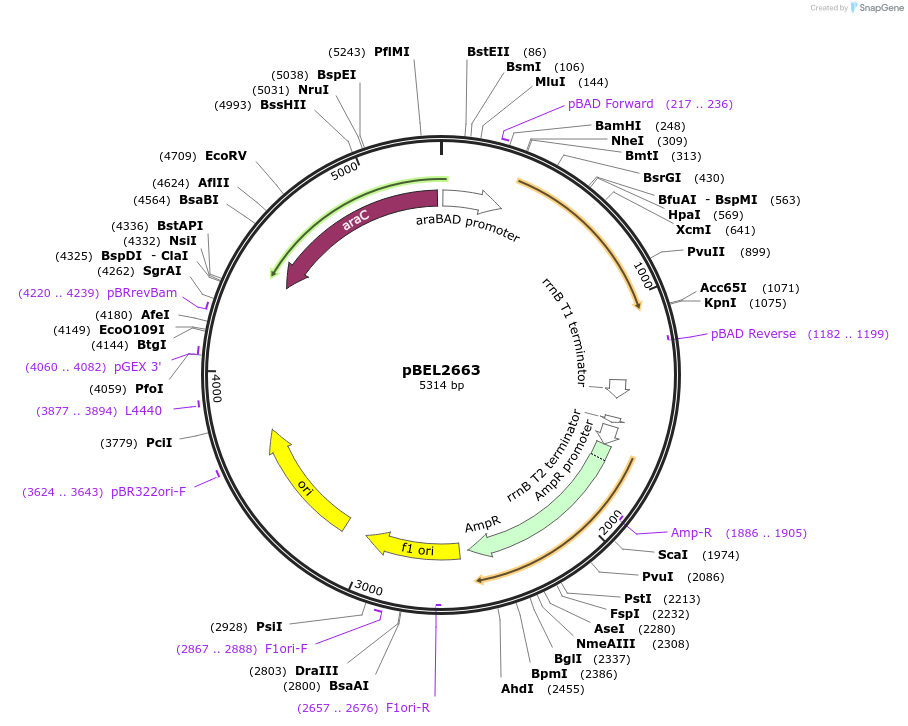
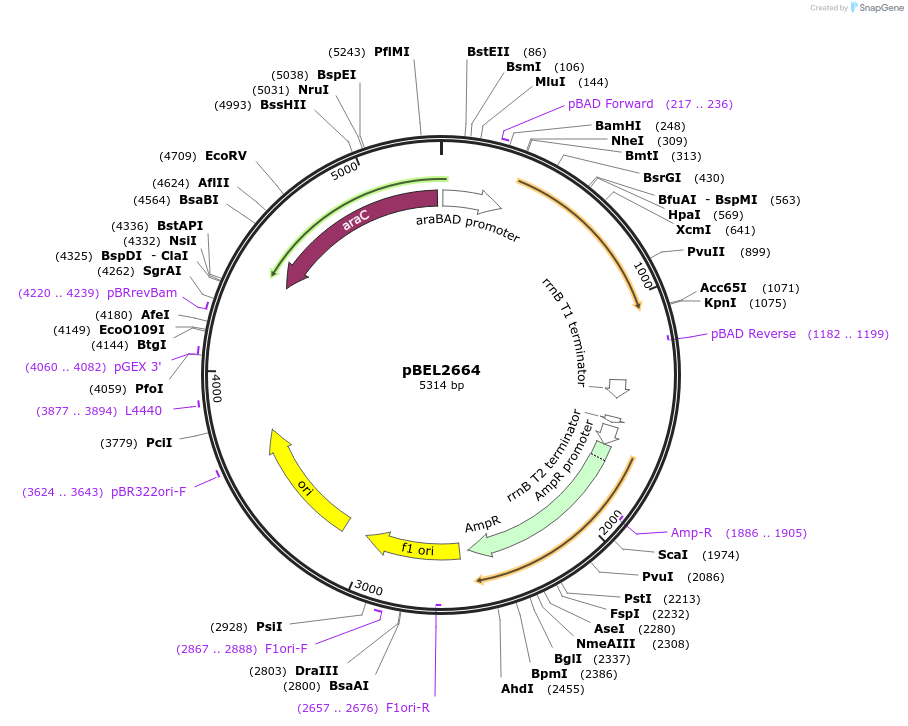
pBEL2663
Plasmid#195632PurposeComplementation of MlaA(F223N)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(F223N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2664
Plasmid#195633PurposeComplementation of MlaA(L230N)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(L230N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2665
Plasmid#195634PurposeComplementation of MlaA(delta244-251)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(delta244-251)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
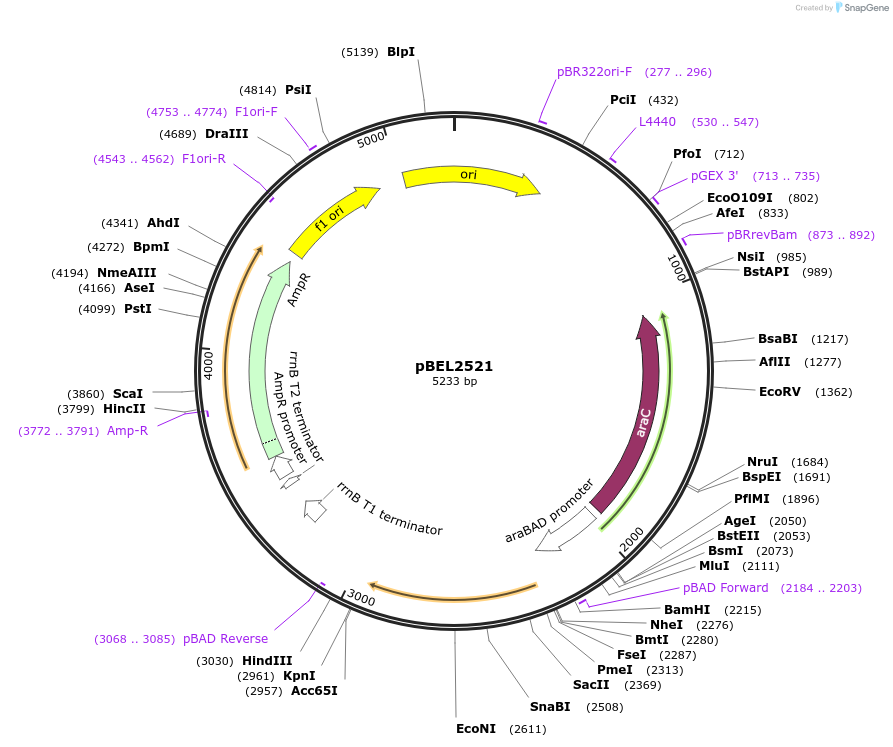
pBEL2521
Plasmid#195643PurposeComplementation of MlaC(A34R)DepositorInsertMlaC
ExpressionBacterialMutationMlaC(A34R)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL2512
Plasmid#195626PurposeComplementation of MlaA(delta238-251)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(delta238-251)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
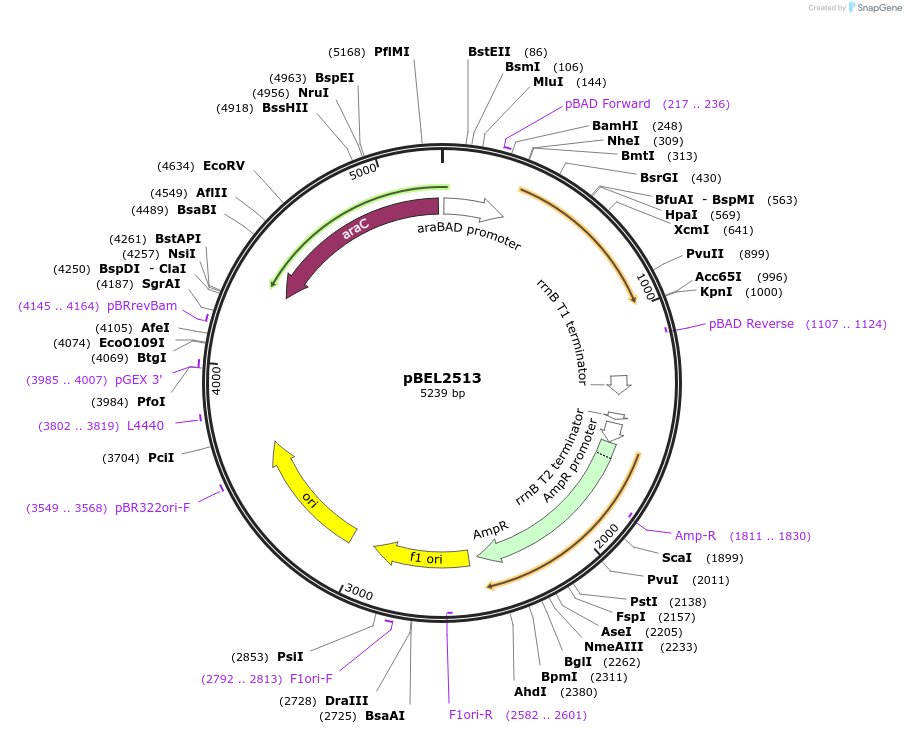
pBEL2513
Plasmid#195627PurposeComplementation of MlaA(delta227-251)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(delta227-251)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
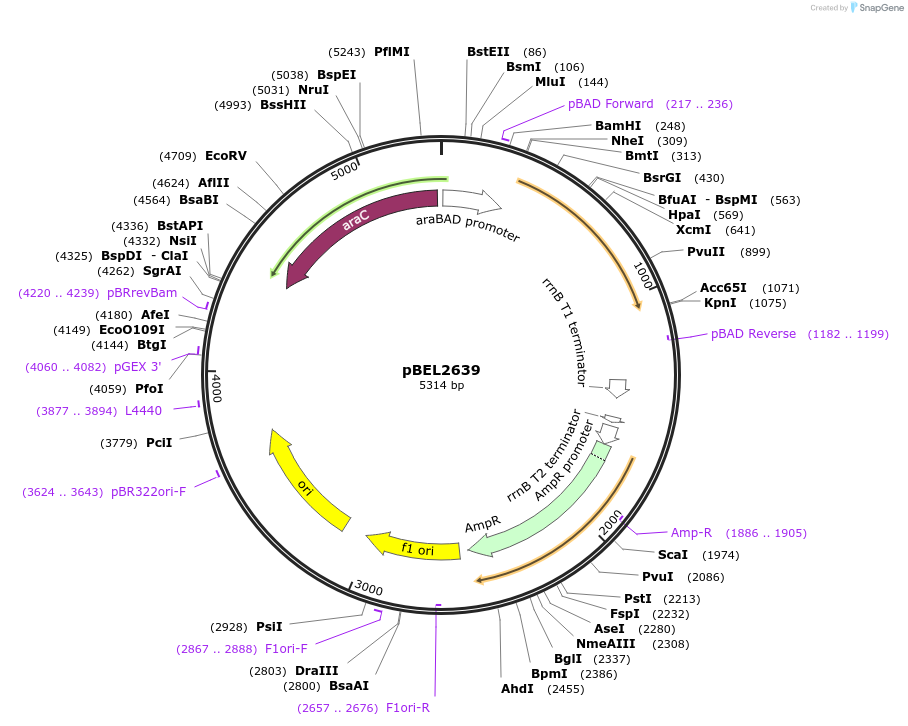
pBEL2639
Plasmid#195628PurposeComplementation of MlaA(I241N)DepositorInsertMlaA
ExpressionBacterialMutationMlaA(I241N)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
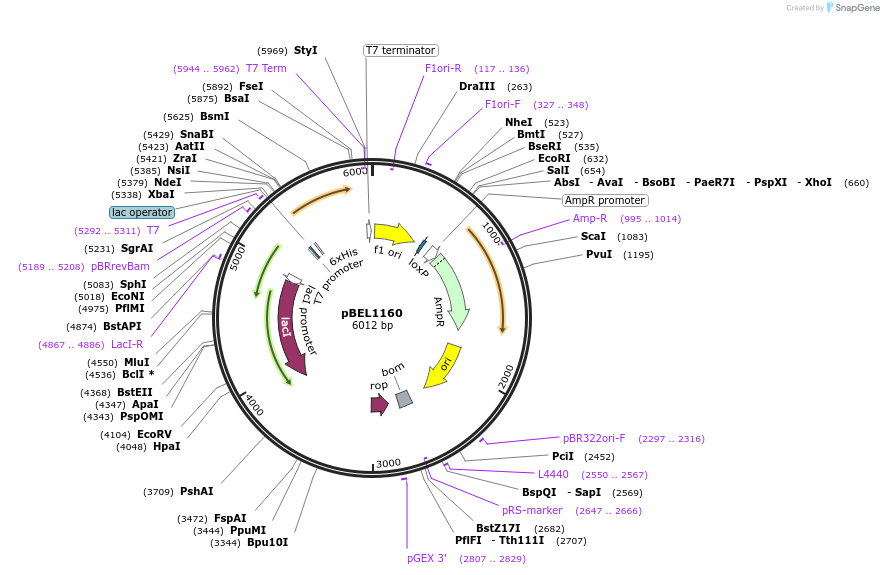
pBEL1160
Plasmid#196025PurposeExpression/purification of 6xHis-TEV-MlaDDepositorInsertMlaD
ExpressionBacterialAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
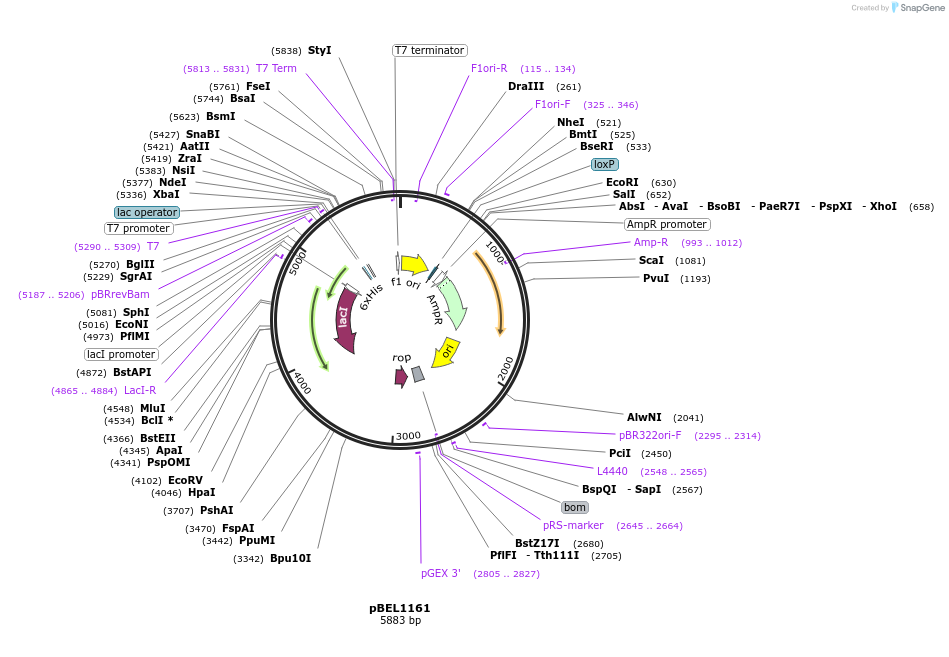
pBEL1161
Plasmid#196026PurposeExpression/purification of 6xHis-TEV-MlaDdelta141-183DepositorInsertMlaD
ExpressionBacterialMutation6xHis-TEV-MlaDdelta141-183Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
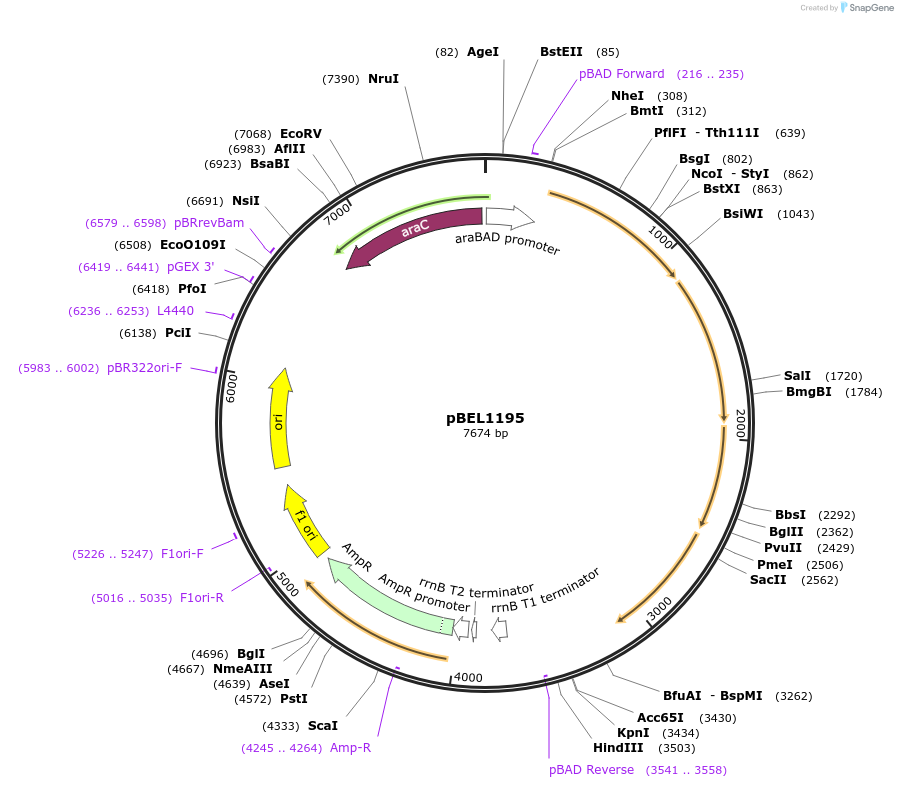
pBEL1195
Plasmid#196027PurposeComplementation of MlaF-MlaE-MlaD-MlaC-MlaBDepositorInsertMlaFEDCB
ExpressionBacterialAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
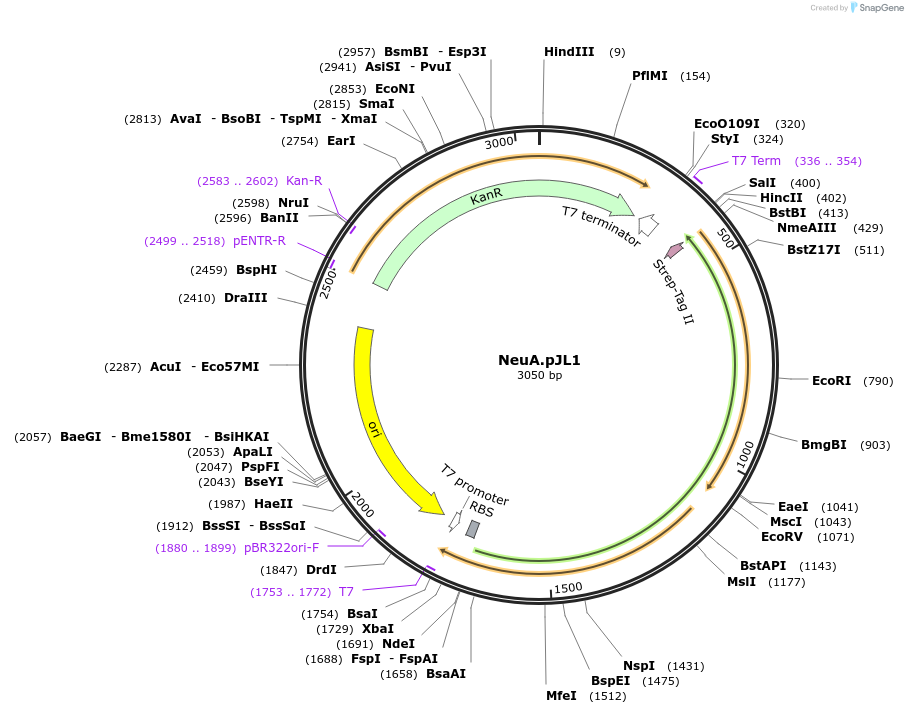
NeuA.pJL1
Plasmid#199115PurposeIn vitro expression of N-acylneuraminate cytodiylyltransfease (NeuA), which conjugates sialic acid and CTP to make activated sugar donor, CMP-sialic acid (see CSTI.PJL1 and PdST6.PJL1)DepositorInsertN-acylneuraminate cytodiylyltransfease (NeuA)
TagsStrep-tagExpressionBacterialPromoterT7Available SinceApril 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
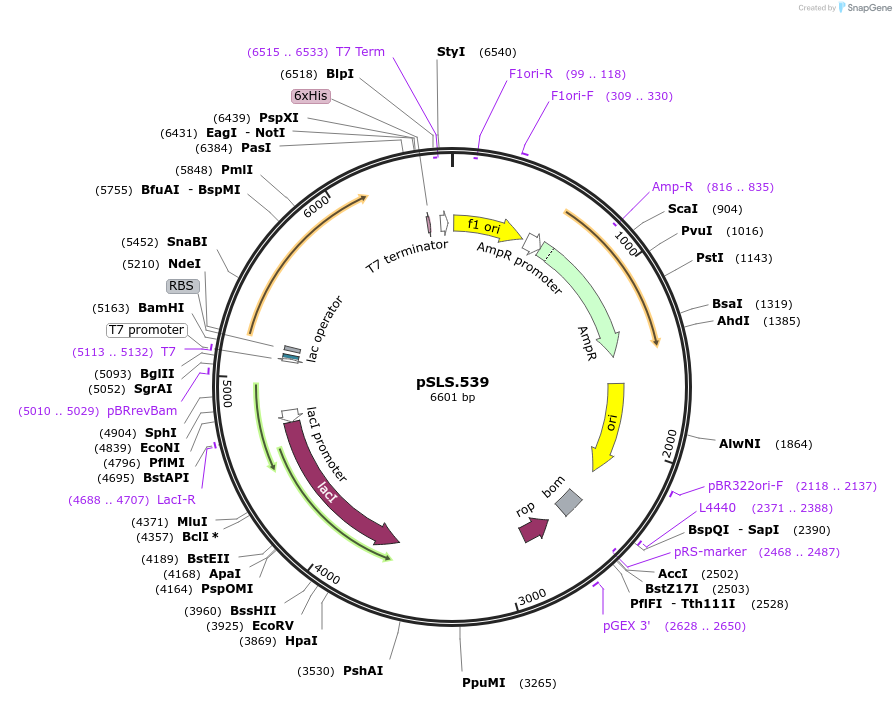
pSLS.539
Plasmid#184958PurposeTest effect of a1/a2 length on editing fabH using retron recombineeringDepositorInsertEco1 RT and recombineering ncRNA, fabH G954A, a1/a2 length: 12
ExpressionBacterialMutationfabH donor G954APromoterT7/lacAvailable SinceJan. 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
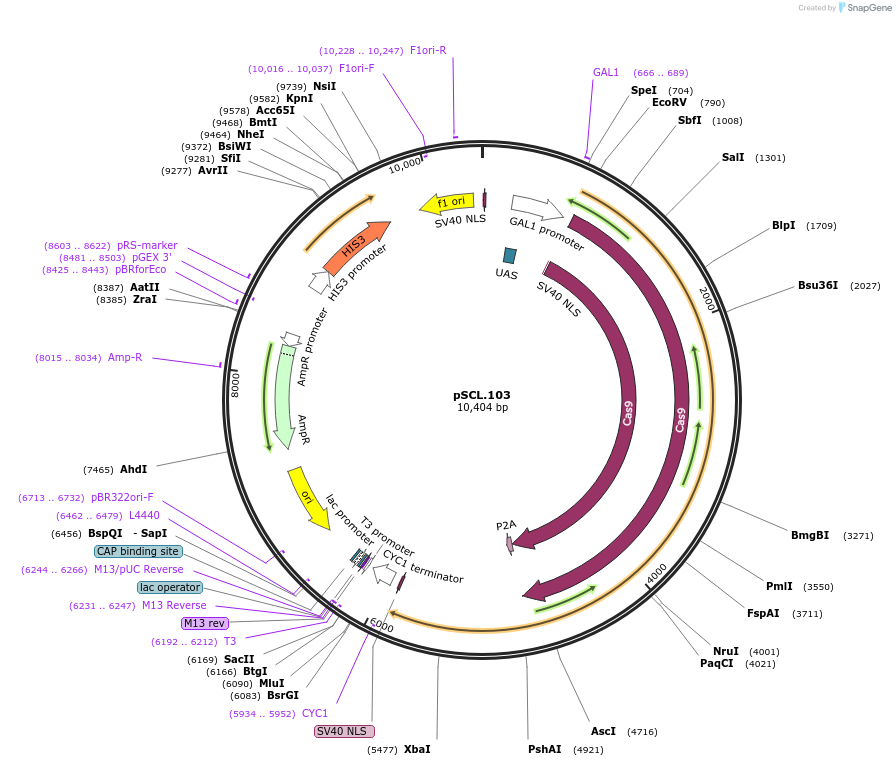
pSCL.103
Plasmid#184988PurposeExpress Cas9-P2A-Eco1RTDepositorInsertCas9-P2A-Eco1RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
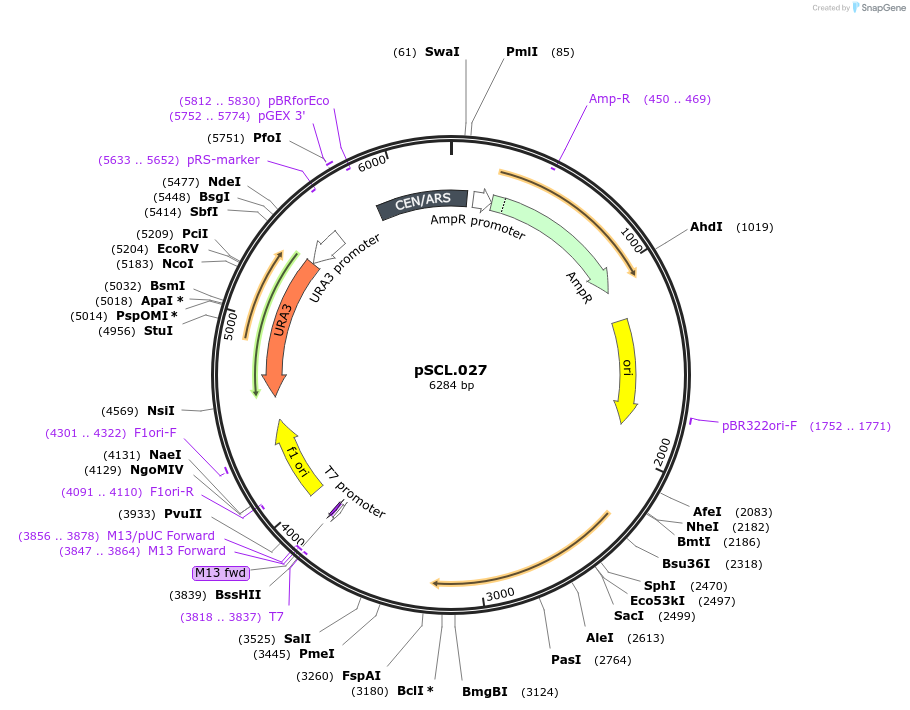
pSCL.027
Plasmid#184964PurposeExpress -Eco1 RT and ncRNA in yeastDepositorInsertEco1: RT and ncRNA(wt), a1/a2 length: 12
ExpressionYeastMutationhuman codon optimized RTPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
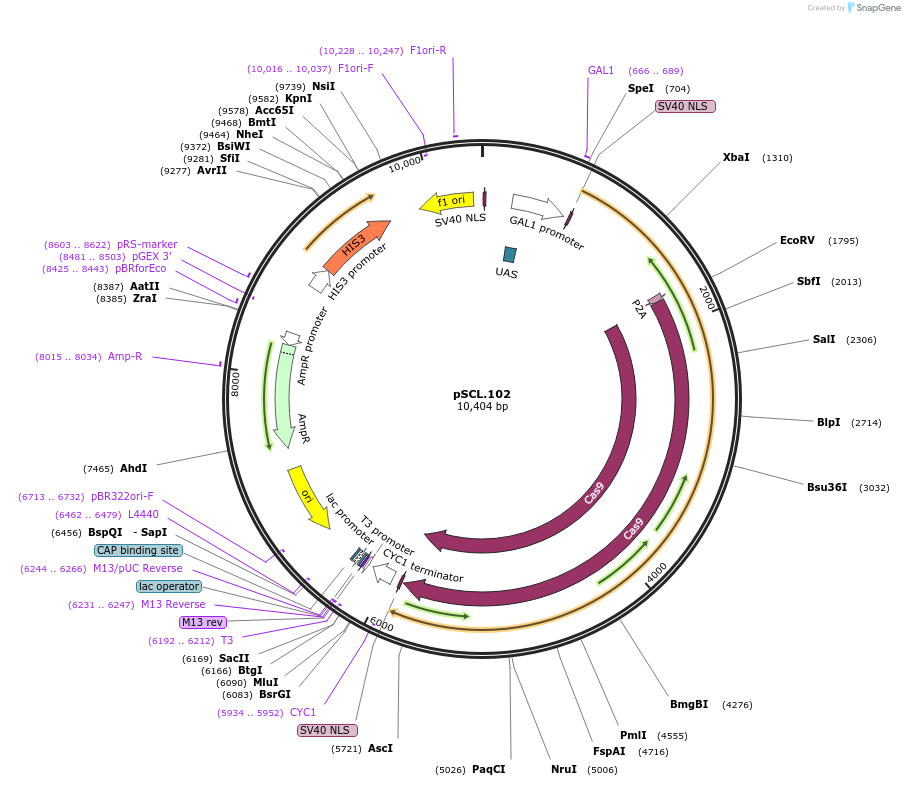
pSCL.102
Plasmid#184987PurposeExpress -Eco1 RT-P2A-Cas9DepositorInsertEco1RT-P2A-Cas9
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
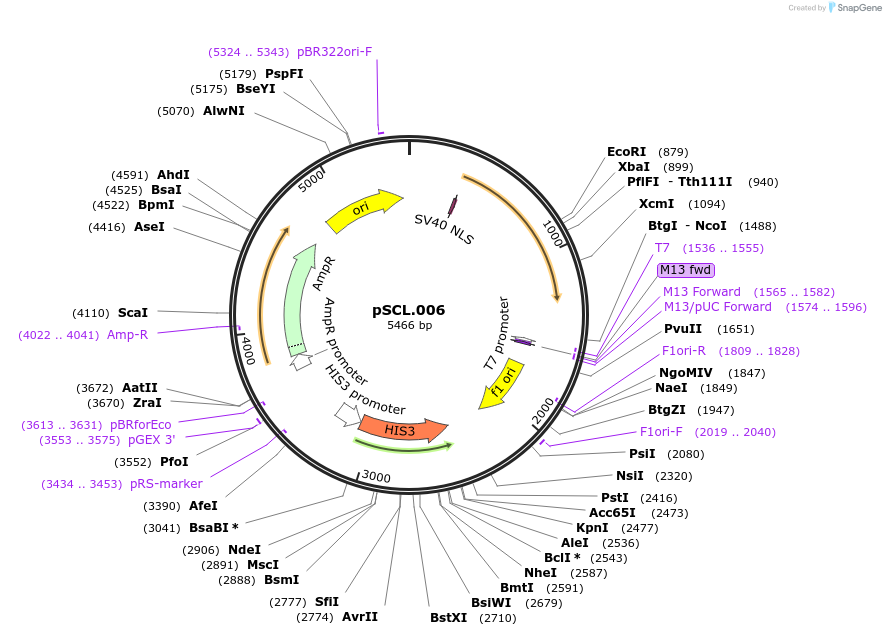
pSCL.006
Plasmid#184970PurposeExpress -Eco1 RT in yeastDepositorInsertIntegrating inducible cassette: Eco1 RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
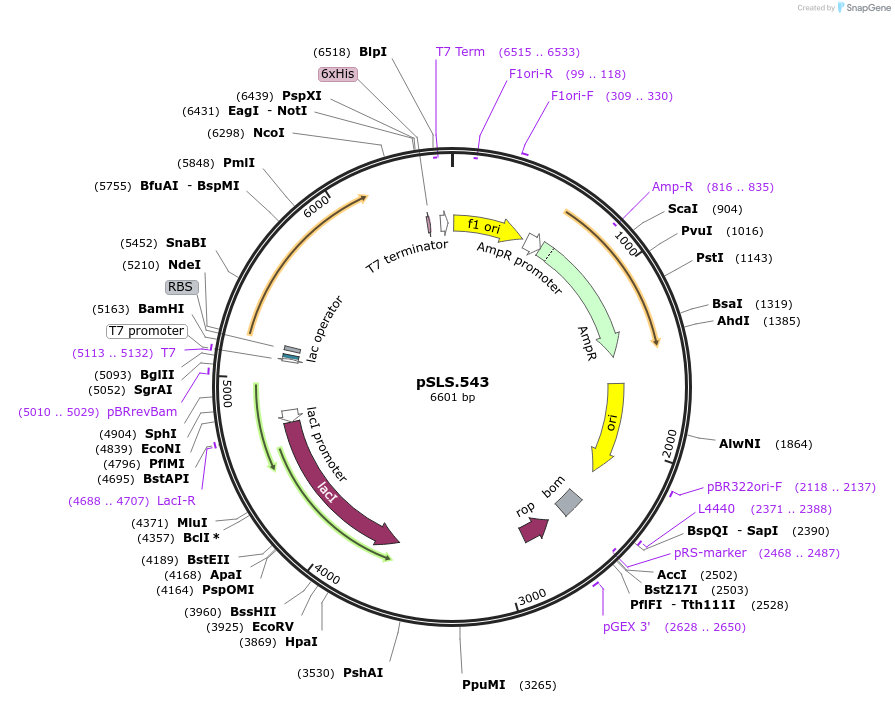
pSLS.543
Plasmid#184960PurposeTest effect of a1/a2 length on editing murF using retron recombineeringDepositorInsertEco1 RT and recombineering ncRNA, murF G1359A, a1/a2 length: 12
ExpressionBacterialMutationmurF donor G1359APromoterT7/lacAvailable SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
pSLS.545
Plasmid#184962PurposeTest effect of a1/a2 length on editing priB using retron recombineeringDepositorInsertEco1 RT and recombineering ncRNA, priB G1069A, a1/a2 length: 12
ExpressionBacterialMutationpriB donor G1069APromoterT7/lacAvailable SinceNov. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
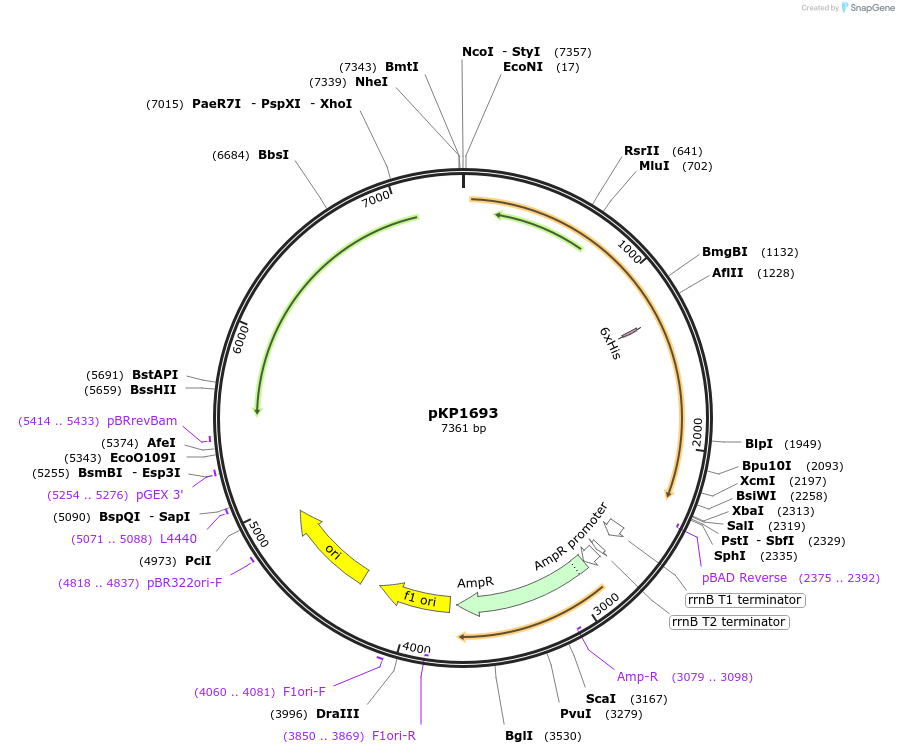
pKP1693
Plasmid#167595PurposeExpress FepA with His6 tag between residues 393 and 394 of the mature amino acid sequence . Please note that mutation numbers are not based on start codon of insert.DepositorInsertFepA
ExpressionBacterialMutationHis6 tag inserted after residue 393 in the mature…Available SinceJuly 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
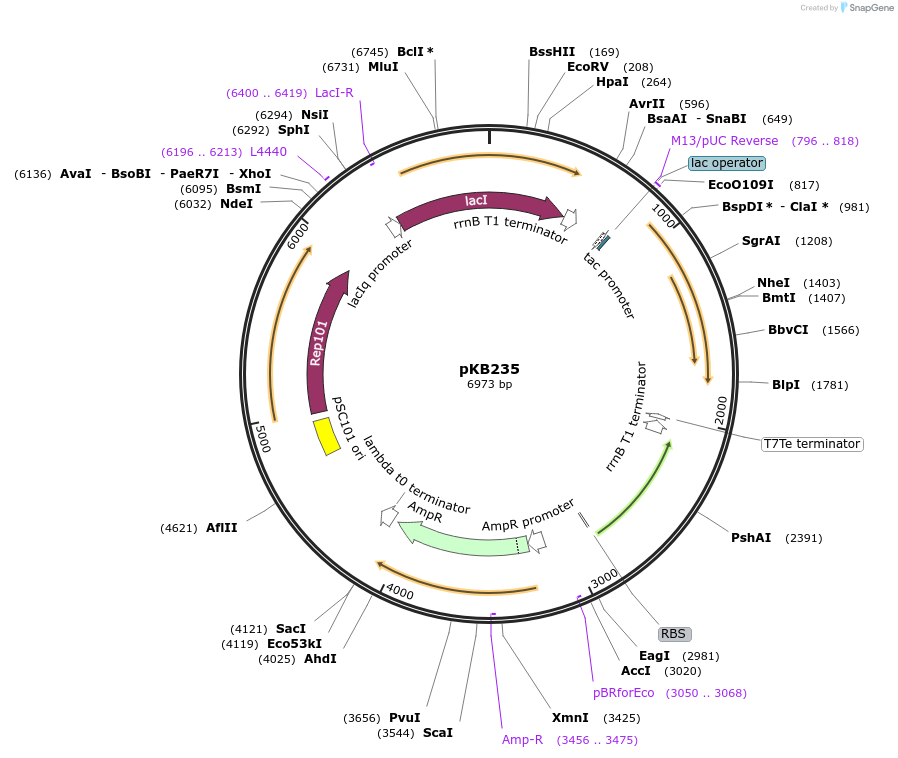
pKB235
Plasmid#170024PurposeEncodes LacI-controlled expression of surface-displayed CCL20 and PvirK-mNeonGreen reporterDepositorArticleInsertsUseSynthetic BiologyTagsLpp, OmpA, and TetherExpressionBacterialPromoterPtac and PvirKAvailable SinceJuly 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
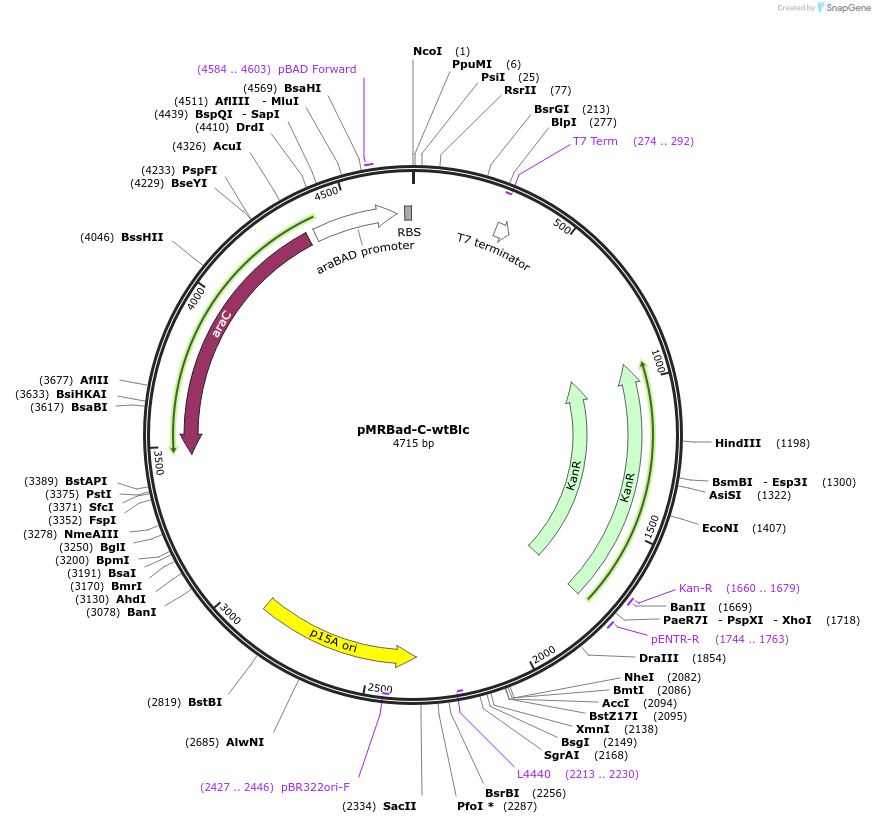
pMRBad-C-wtBlc
Plasmid#168257PurposeC-fragment. This plasmid can be co-transformed with either Plasmid 168259: pET11a-N-wtBlc or Plasmid 168472: pET11a-N-DiB2 to obtain wtBlc‐split or DiB2‐split protein, correspondinglyDepositorInsertC-fragment of wtBlc
ExpressionBacterialPromoteraraBADAvailable SinceMay 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
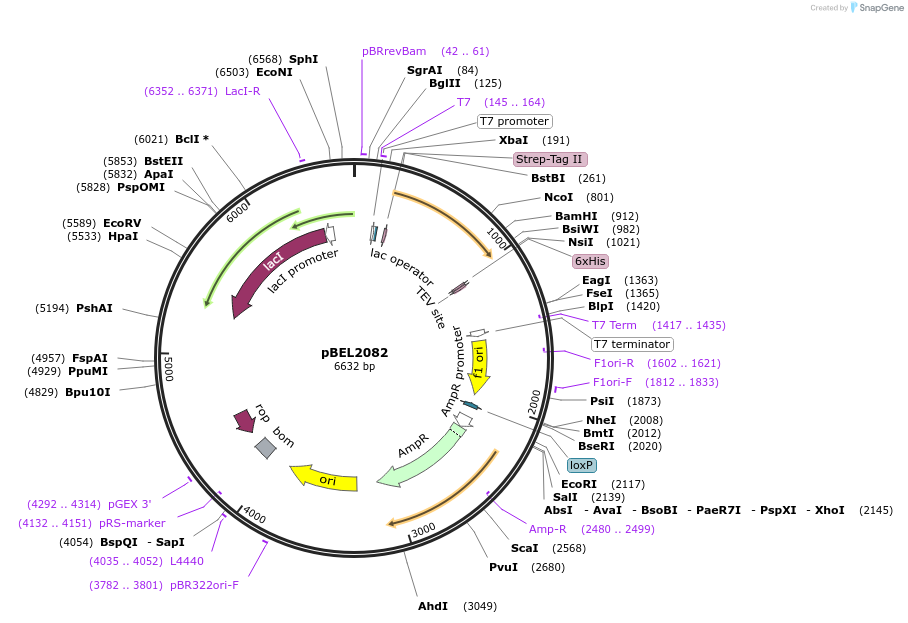
pBEL2082
Plasmid#155162PurposeExpresses (Strep-MlaF_Δ[248-269])-(6xHis-TEV-MlaB)DepositorInsert(MlaFΔ[248-269])-(6xHis-TEV-MlaB)
ExpressionBacterialMutationdeleted amino acids 248-269 in MlaFAvailable SinceDec. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
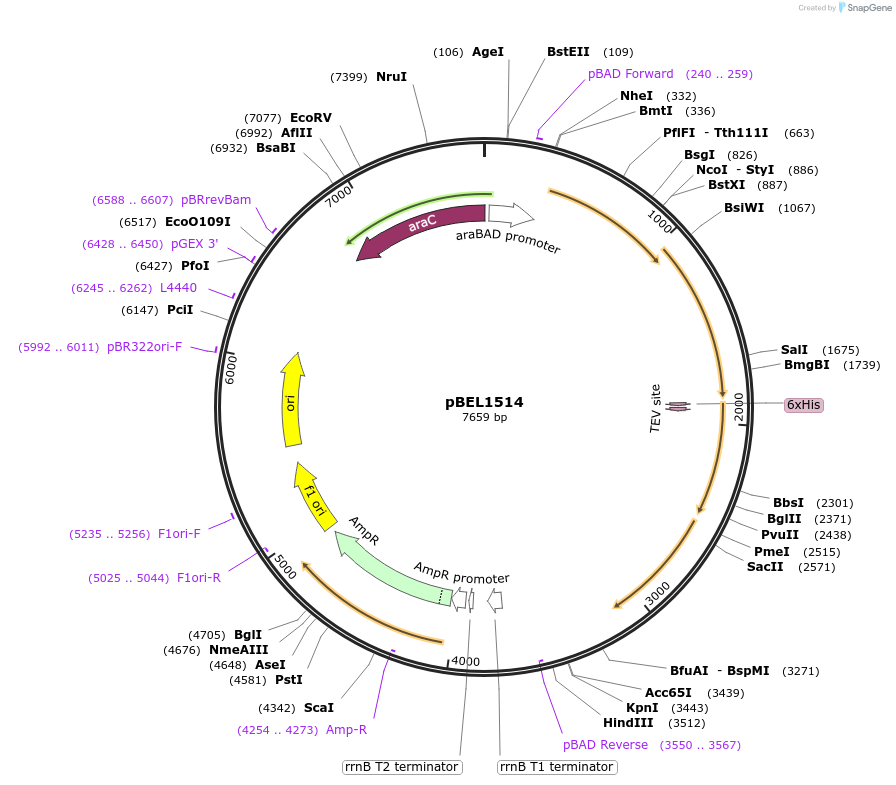
pBEL1514
Plasmid#155147PurposeExpresses MlaFEDCB operon with MlaF C-terminus deleted [1-246], His-tag on MlaDDepositorInsertmlaF(1-246)-mlaE-[6xHis-2xQH-TEV-mlaD]-mlaCB operon
PromoteraraBADAvailable SinceDec. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
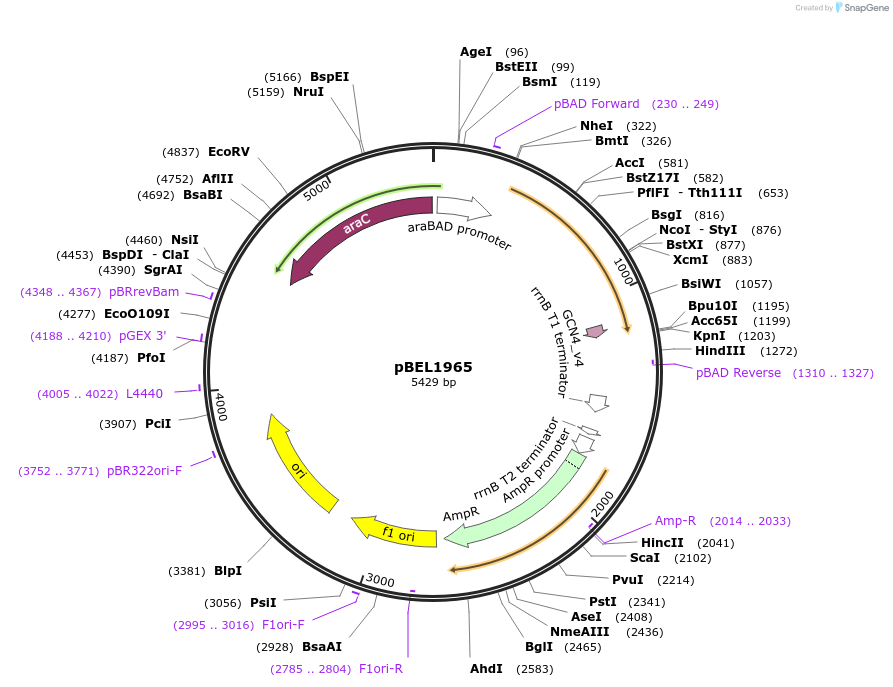
pBEL1965
Plasmid#155152PurposeExpresses MlaF(1-246)-(6 aa linker)-GCN4DepositorInsertmlaF(1-246)-GCN4_Dimerization_Domain
Mutationdeleted amino acids 247-269 in MlaF; added dimeri…PromoteraraBADAvailable SinceDec. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
pBEL1966
Plasmid#155153PurposeExpresses MlaF(1-246)-(10 aa linker)-GCN4DepositorInsertmlaF(1-246)-GCN4_Dimerization_Domain
Mutationdeleted amino acids 247-269 in MlaF; added dimeri…PromoteraraBADAvailable SinceDec. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
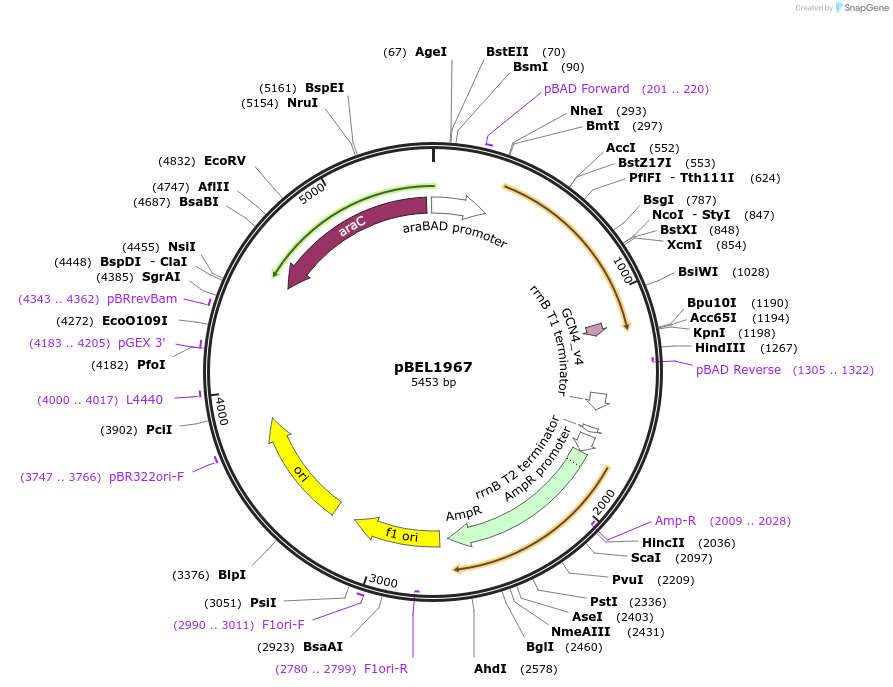
pBEL1967
Plasmid#155154PurposeExpresses MlaF(1-246)-(14 aa linker)-GCN4DepositorInsertmlaF(1-246)-GCN4_Dimerization_Domain
Mutationdeleted amino acids 247-269 in MlaF; added dimeri…PromoteraraBADAvailable SinceDec. 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
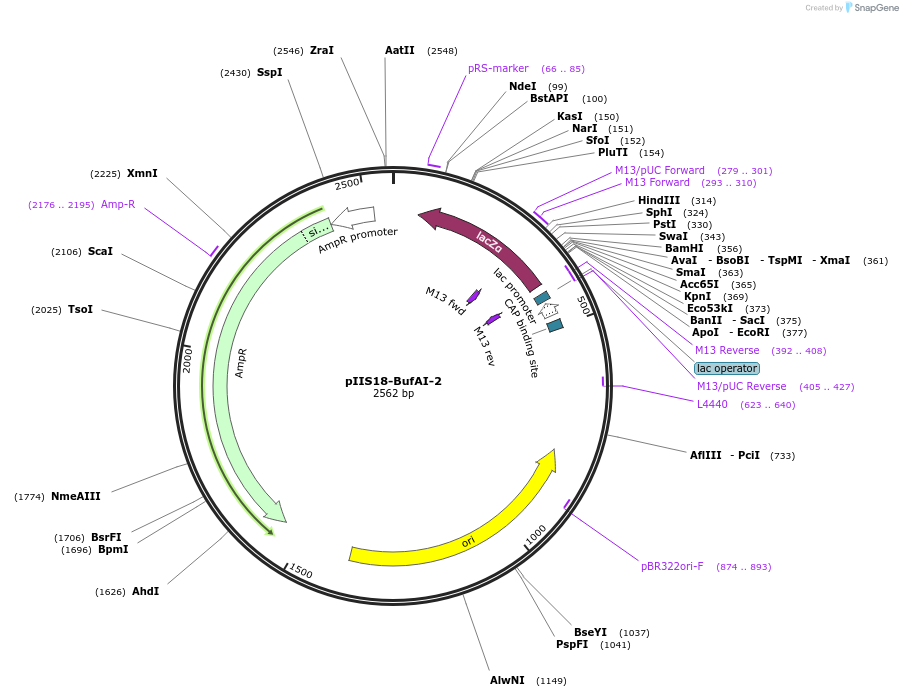
pIIS18-BufAI-2
Plasmid#128651PurposeEntry vector useful for golden gate or other seamless cloning technique based on Type IIS restriction enzymesDepositorTypeEmpty backboneUseUnspecifiedAvailable SinceAug. 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
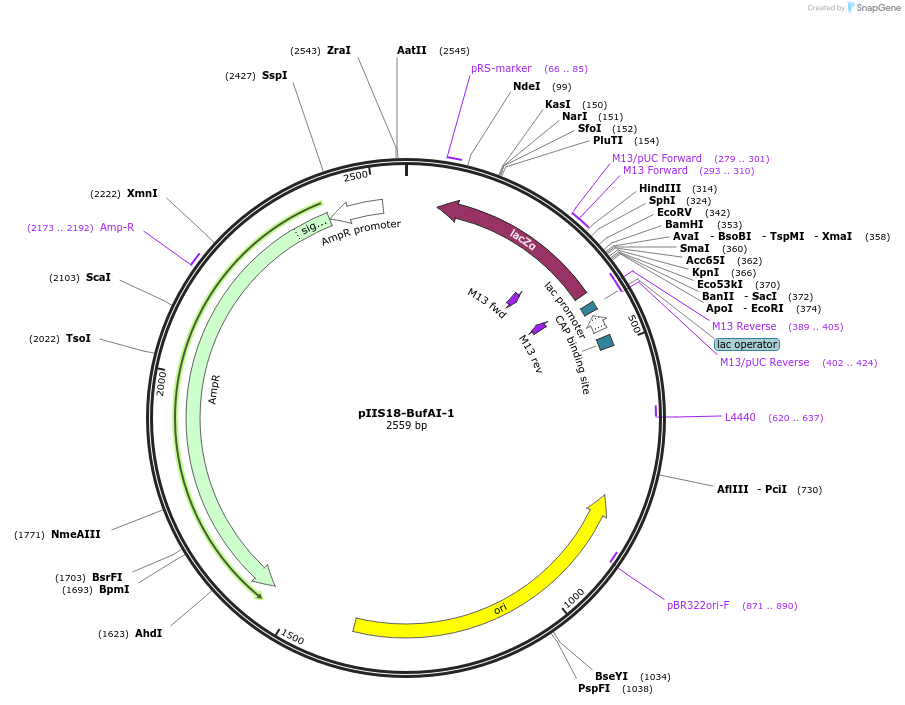
pIIS18-BufAI-1
Plasmid#128650PurposeEntry vector useful for golden gate or other seamless cloning technique based on Type IIS restriction enzymesDepositorTypeEmpty backboneUseUnspecifiedAvailable SinceAug. 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
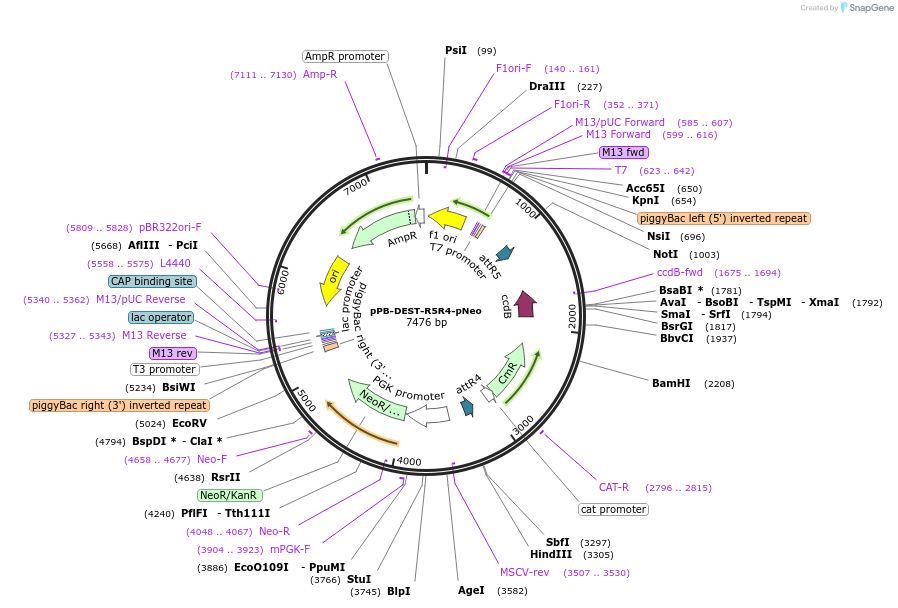
pPB-DEST-R5R4-pNeo
Plasmid#141058PurposeMutisite Gateway mediated vector constructionDepositorInsertNeomycin resistance gene
ExpressionMammalianAvailable SinceJuly 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
pHCKan-yibDp-cimA3.7
Plasmid#134595PurposeLow Phosphate inducible mutant citramalate synthaseDepositorInsertcimA3.7
ExpressionBacterialMutationI47V, E114V, H126Q, T204A, L238SPromoteryibDpAvailable SinceMay 5, 2020AvailabilityAcademic Institutions and Nonprofits only