We narrowed to 42,020 results for: ids
-
Plasmid#54948PurposeLocalization: Golgi TGN, Excitation: 568, Emission: 592DepositorAvailable SinceAug. 7, 2014AvailabilityAcademic Institutions and Nonprofits only
-
mPlum-LC-Myosin-N-7
Plasmid#55983PurposeLocalization: Light Chain Myosin, Excitation: 590, Emission: 649DepositorAvailable SinceFeb. 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
mRuby-Rab4a-7
Plasmid#55878PurposeLocalization: Ras GTPase, Excitation: 558, Emission: 605DepositorAvailable SinceMarch 4, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
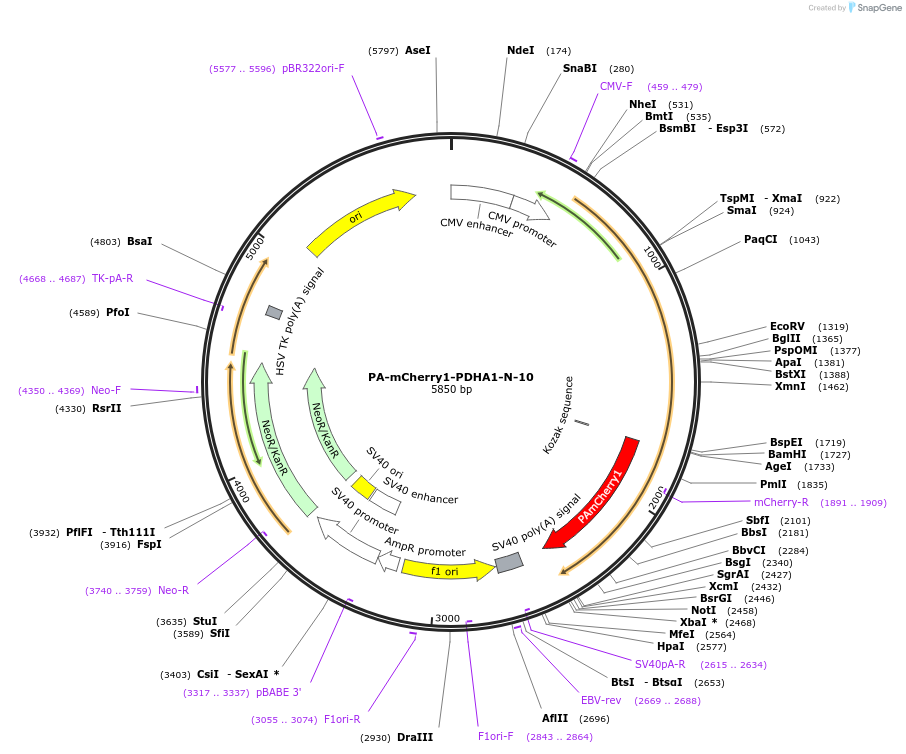
PA-mCherry1-PDHA1-N-10
Plasmid#57193PurposeLocalization: Mitochondria, Excitation: 400 / 504, Emission: 515 / 517DepositorAvailable SinceJan. 13, 2015AvailabilityAcademic Institutions and Nonprofits only -
mTagRFP-T-Rab4a-7
Plasmid#58025PurposeLocalization: Ras GTPase, Excitation: 555, Emission: 584DepositorAvailable SinceNov. 4, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
mRuby-Vinculin-23
Plasmid#55885PurposeLocalization: Focal Adhesions, Excitation: 558, Emission: 605DepositorAvailable SinceMarch 4, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
mEmerald-PTK2-N-10
Plasmid#54241PurposeLocalization: Focal Adhesion Kinase, Excitation: 487, Emission: 509DepositorAvailable SinceJuly 10, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
mEmerald-TPX2-N-10
Plasmid#54285PurposeLocalization: Centrosomes, Excitation: 487, Emission: 509DepositorAvailable SinceJuly 25, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
mCherry-Zyxin-C-13
Plasmid#55167PurposeLocalization: Focal Adhesions, Excitation: 587, Emission: 610DepositorInsertZyxin (ZYX Human)
TagsmCherryExpressionMammalianMutationS11P, G42R and E62DPromoterCMVAvailable SinceSept. 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
Nup54-mEGFP494(0)
Plasmid#163426PurposeMammalian expression of Nup54 with a mEGFP (missing the first 5 amino acids) at amino acid position 494 with a rigid alpha helix of 5 amino acids at the carboxyl end of the mEGFPDepositorAvailable SinceJan. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
EBFP2-Vinculin-23
Plasmid#55252PurposeLocalization: Focal Adhesions, Excitation: 383, Emission: 448DepositorAvailable SinceOct. 22, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
mEmerald-Tensin1-N-18
Plasmid#54275PurposeLocalization: Focal Adhesions, Excitation: 487, Emission: 509DepositorAvailable SinceJune 16, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
mEmerald-Tropomyosin-C-14
Plasmid#54289PurposeLocalization: Actin, Excitation: 487, Emission: 509DepositorAvailable SinceJuly 10, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
mGeos-M-PDHA1-N-10
Plasmid#57531PurposeLocalization: Mitochondria, Excitation: 503 / 569, Emission: 514 / 581DepositorAvailable SinceJan. 16, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
mOrange2-H4-23
Plasmid#57964PurposeLocalization: Nucleus/Histones, Excitation: 549, Emission: 565DepositorAvailable SinceOct. 24, 2014AvailabilityAcademic Institutions and Nonprofits only -
mKOet-H4-23
Plasmid#57926PurposeLocalization: Nucleus/Histones, Excitation: 548, Emission: 561DepositorAvailable SinceOct. 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
Dendra2-SiT-N-15
Plasmid#57739PurposeLocalization: Golgi TGN, Excitation: 490 / 553, Emission: 507 / 573DepositorAvailable SinceJan. 6, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
mWasabi-Vinculin-23
Plasmid#56512PurposeLocalization: Focal Adhesions, Excitation: 493, Emission: 509DepositorInsertVinculin (VCL Human)
TagsmWasabiExpressionMammalianMutationV234L in Vinculin.PromoterCMVAvailable SinceJan. 13, 2015AvailabilityAcademic Institutions and Nonprofits only -
mEos2-Moesin-N-14
Plasmid#57402PurposeLocalization: Cytoskeleton/Membrane, Excitation: 505 / 569, Emission: 516 / 581DepositorAvailable SinceFeb. 5, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
mPA-GFP-VASP-5
Plasmid#57161PurposeLocalization: Actin/Focal Adhesions, Excitation: 400 / 504, Emission: 515 / 517DepositorInsertVASP
TagsmPA-GFPExpressionMammalianMutationA209T in VASPPromoterCMVAvailable SinceJan. 6, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits