We narrowed to 9,536 results for: pho
-
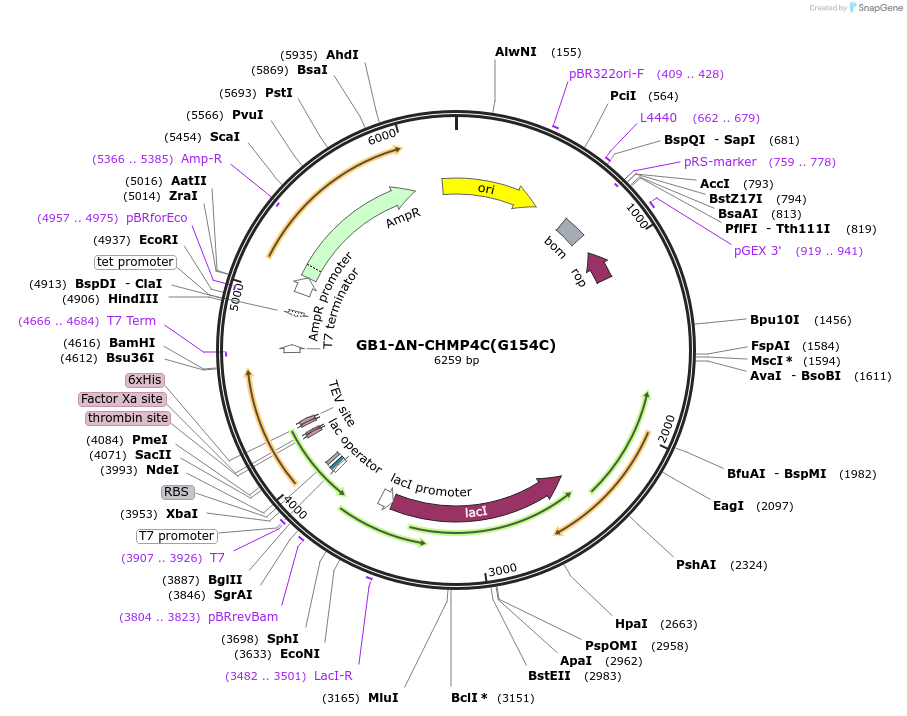
Plasmid#190783PurposeCodon-optimized bacterial expression plasmid. Expresses GB1-His6-TEV-CHMP4C(121-233, G154C)DepositorInsertGB1-His6-TEV-CHMP4C(121-233, G154C) (CHMP4C Human)
TagsGB1-His6-TEVExpressionBacterialMutationG154CAvailable SinceJune 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
GB1-ΔN-CHMP4C(M165C)
Plasmid#190784PurposeCodon-optimized bacterial expression plasmid. Expresses GB1-His6-TEV-CHMP4C(121-233, M165C)DepositorInsertGB1-His6-TEV-CHMP4C(121-233, M165C) (CHMP4C Human)
TagsGB1-His6-TEVExpressionBacterialMutationM165CAvailable SinceJune 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
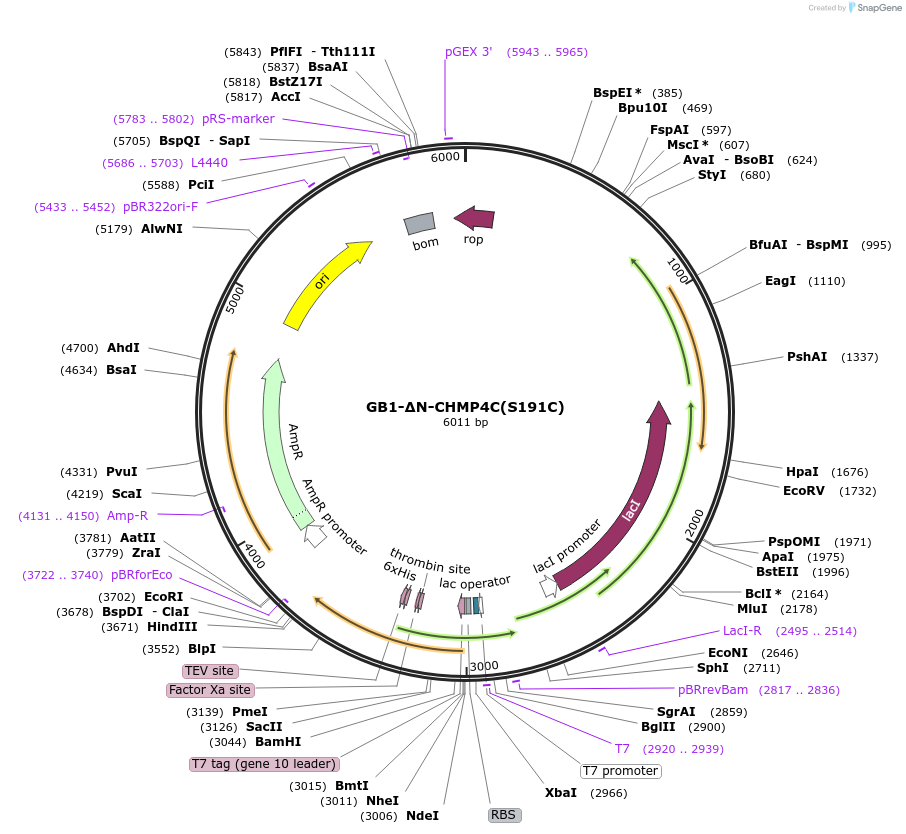
GB1-ΔN-CHMP4C(S191C)
Plasmid#180027PurposeCodon-optimized bacterial expression plasmid. Expresses GB1-His6-TEV-CHMP4C(121-233, S191C).DepositorInsertGB1-His6-TEV-CHMP4C(121-233, S191C) (CHMP4C Human)
TagsGB1-His6-TEVExpressionBacterialMutationS191CAvailable SinceJune 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
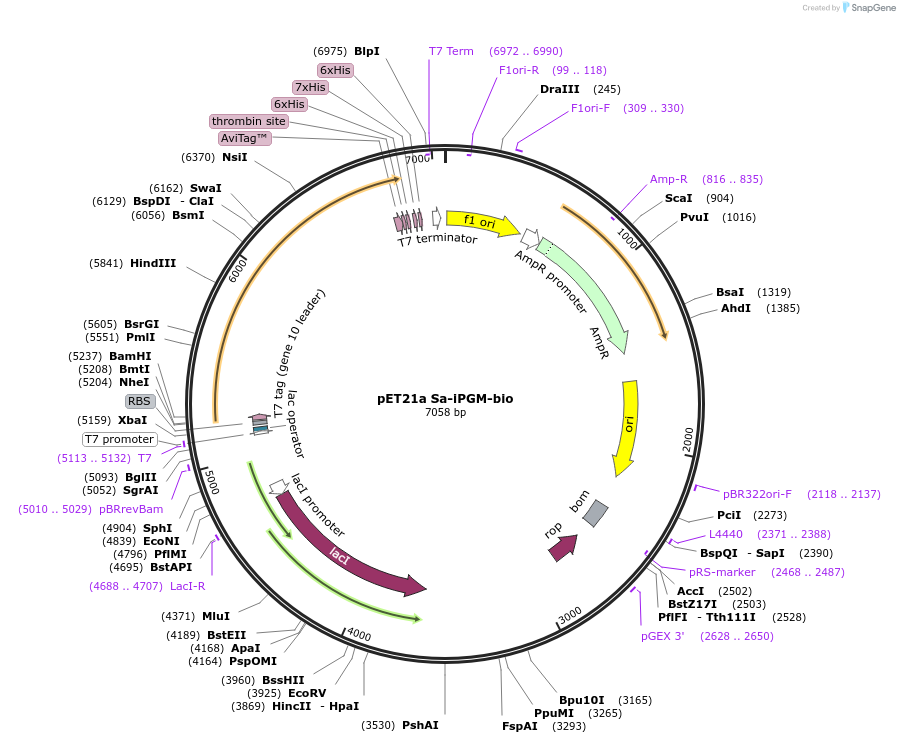
pET21a Sa-iPGM-bio
Plasmid#179523PurposeExpresses S. aureus iPGM containing a C-terminal biotinylation site and His tag in E. coliDepositorInsertS. aureus iPGM bio-6His
TagsHis tag and biotinylation sequenceExpressionBacterialPromoterT7Available SinceSept. 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
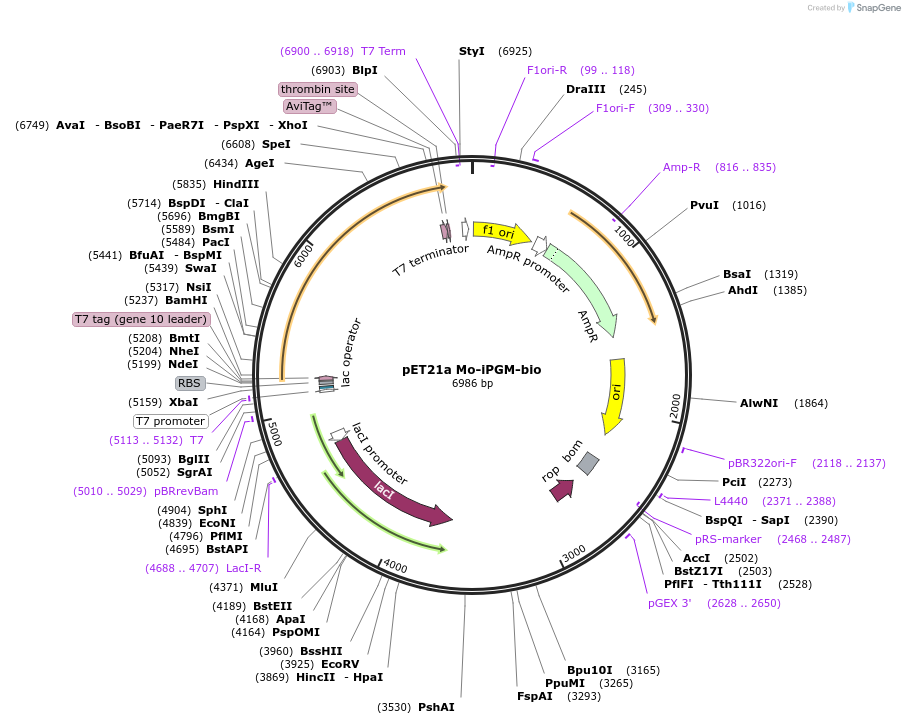
pET21a Mo-iPGM-bio
Plasmid#179614PurposeExpresses M. orale iPGM containing a C-terminal biotinylation site and His tag in E. coliDepositorInsertM. orale iPGM bio-6His
TagsHis tag and biotinylation sequenceExpressionBacterialPromoterT7Available SinceSept. 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
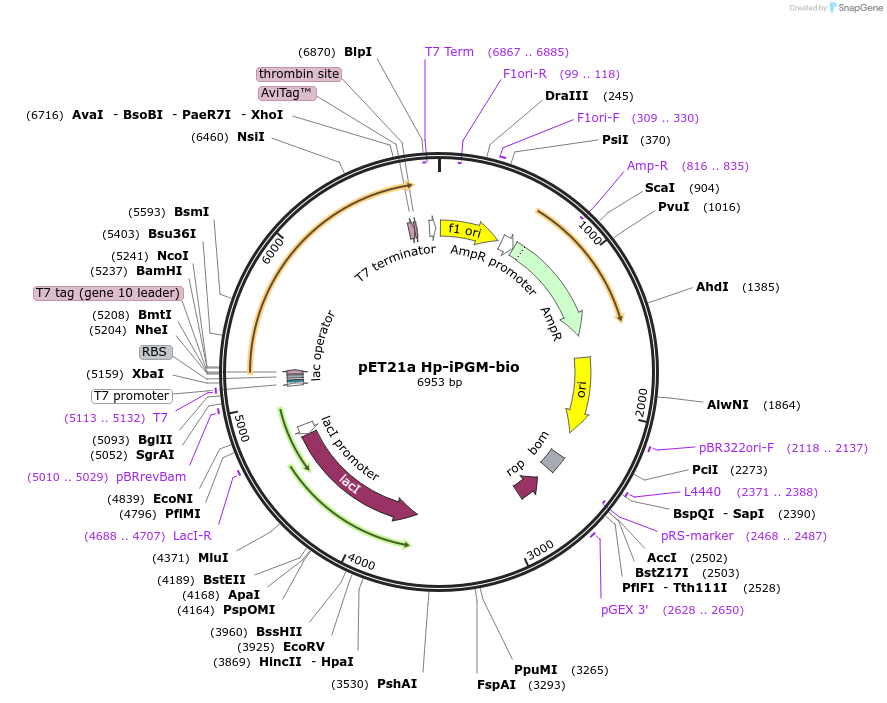
pET21a Hp-iPGM-bio
Plasmid#179615PurposeExpresses H. pylori iPGM containing a C-terminal biotinylation site and His tag in E. coliDepositorInsertH. pylori iPGM bio-6His
TagsHis tag and biotinylation sequenceExpressionBacterialPromoterT7Available SinceSept. 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
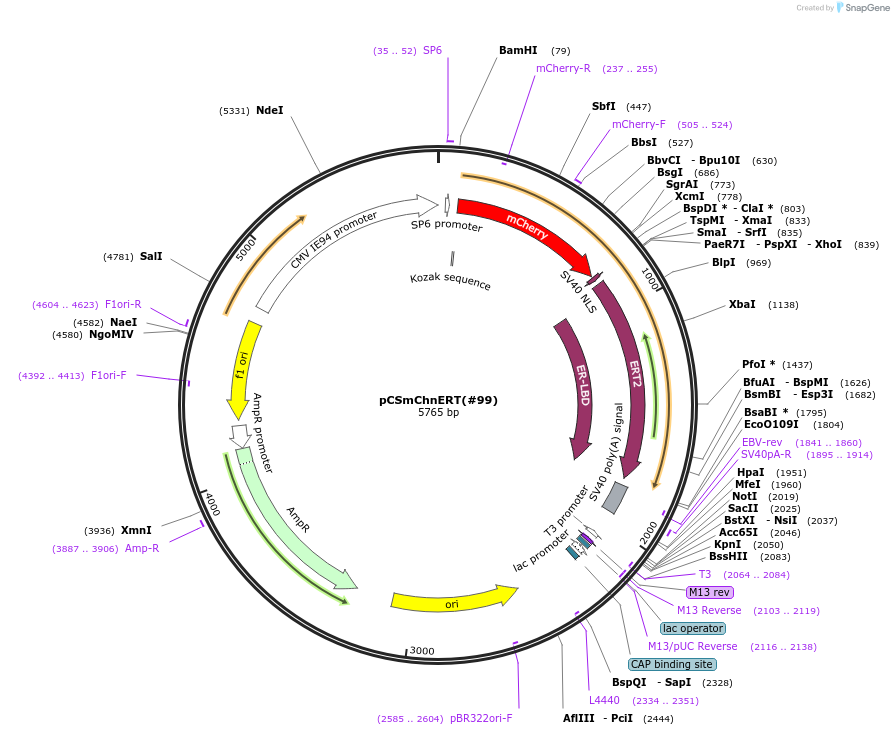
pCSmChnERT(#99)
Plasmid#184063Purposecyclofen-inducible mCherry nuclear relocation via mammalian cell transfection or mRNA synthesisDepositorInsertmCherry-nls-ERT2
UseSynthetic BiologyExpressionMammalianMutationERT2: G400V, M543A, L544A, deleted 1-281;PromotersCMV+SP6Available SinceAug. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
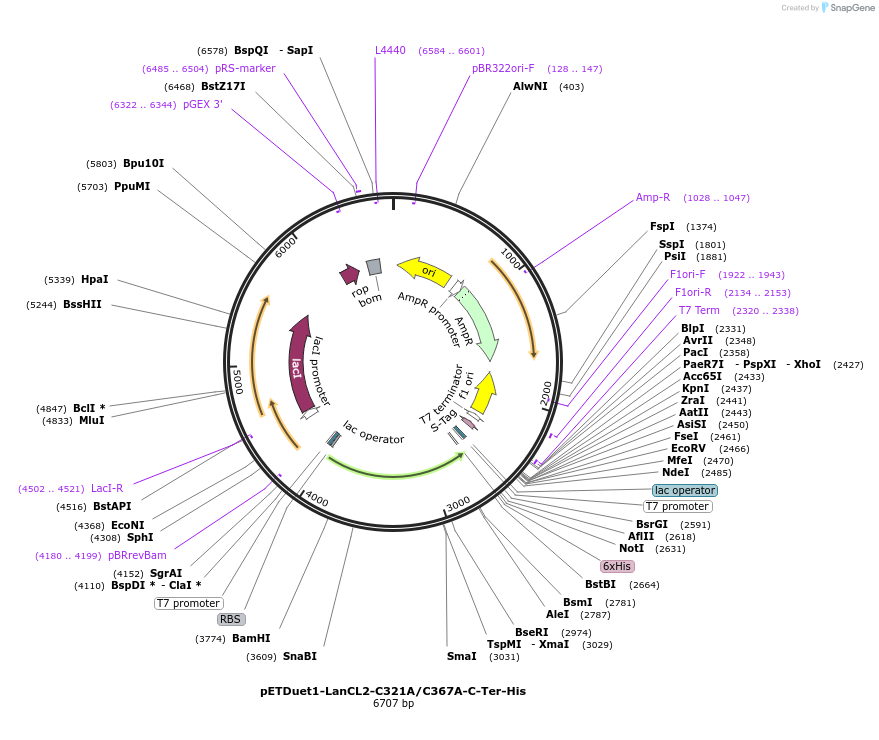
pETDuet1-LanCL2-C321A/C367A-C-Ter-His
Plasmid#154188PurposeTo express LanCL2-C321A/C367A in bacterial cellsDepositorInsertLanCL2
TagsHexahistidineExpressionBacterialMutationCys321->Ala321; Cys367->Ala367Available SinceAug. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
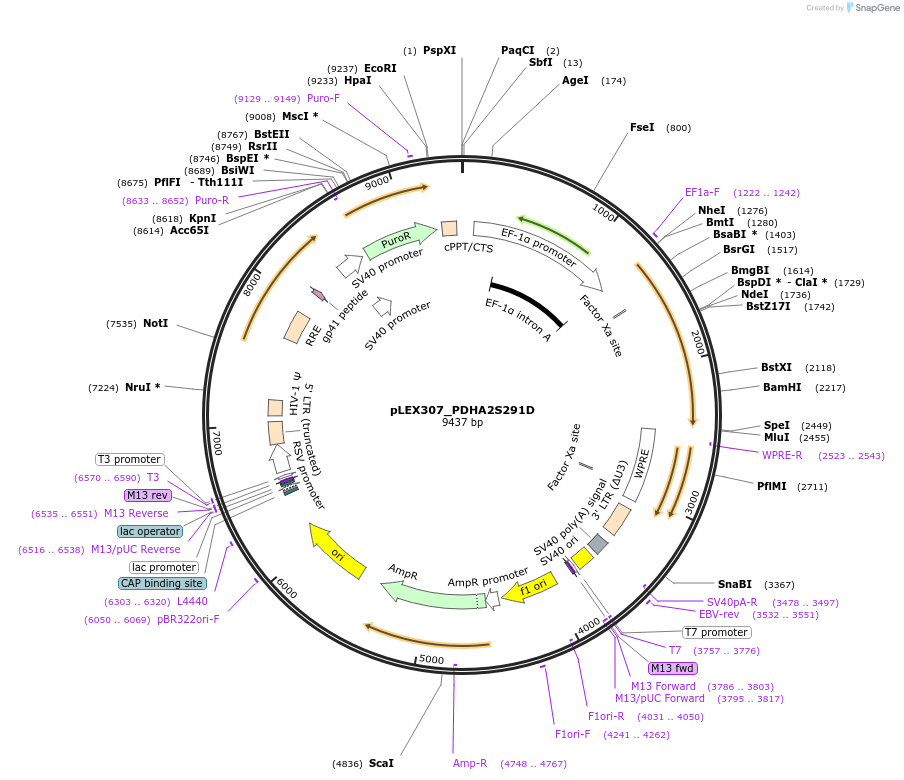
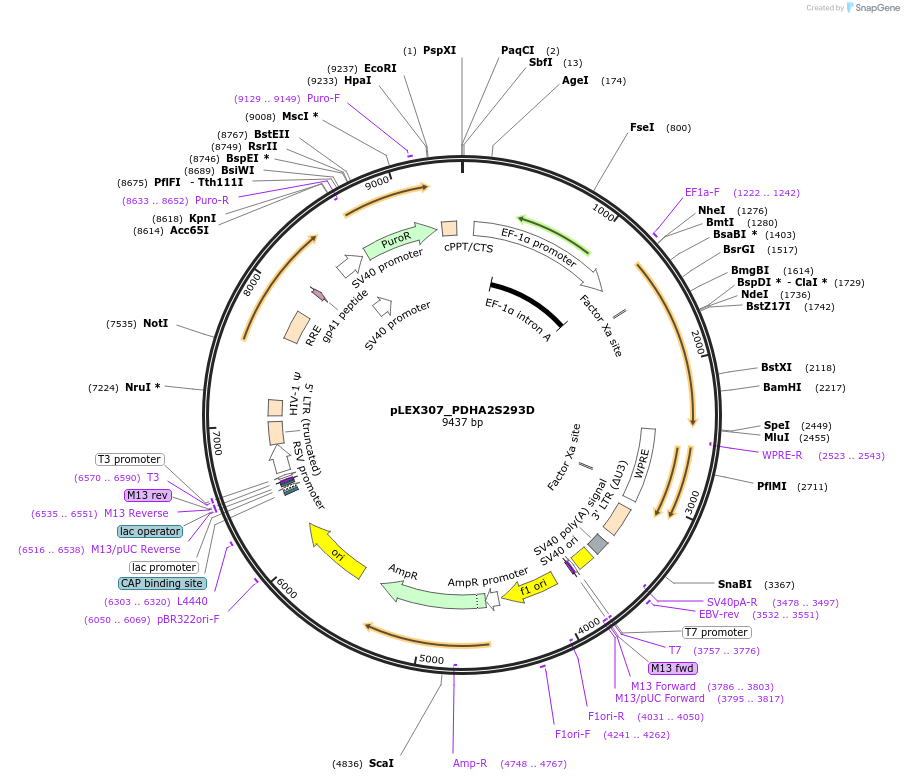
pLEX307_PDHA2S291D
Plasmid#115193PurposeLentiviral transduction and expression of PDHA2S291D into any mammalian cellDepositorAvailable SinceJuly 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLEX307_PDHA2S293D
Plasmid#115194PurposeLentiviral transduction and expression of PDHA2S293D into any mammalian cellDepositorAvailable SinceJuly 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
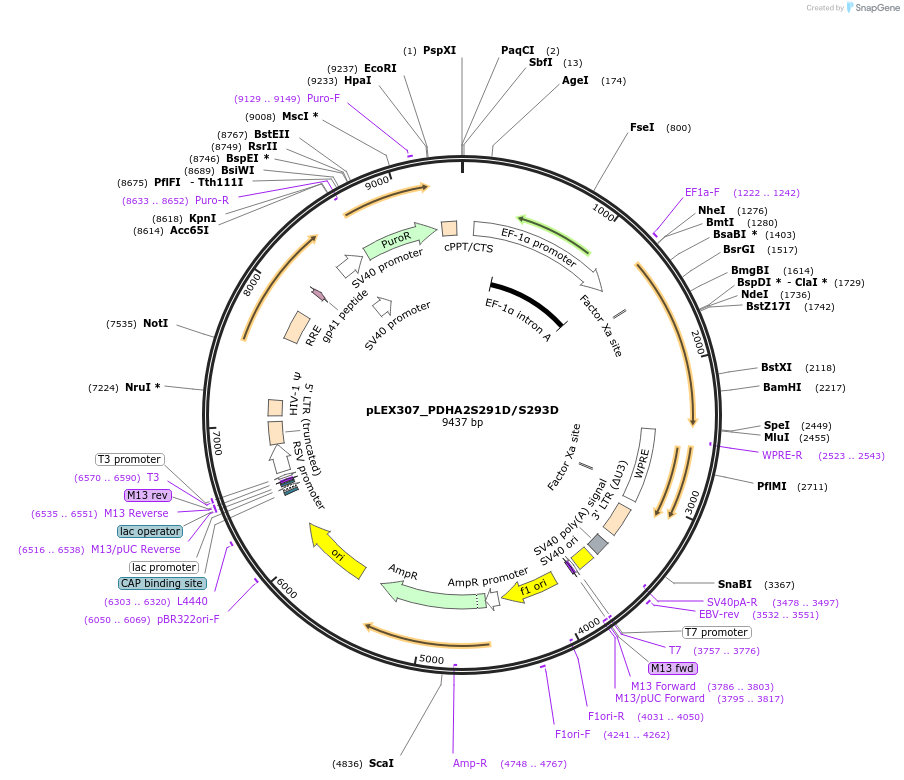
pLEX307_PDHA2S291D/S293D
Plasmid#115195PurposeLentiviral transduction and expression of PDHA2S291D/S293D into any mammalian cellDepositorAvailable SinceJuly 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
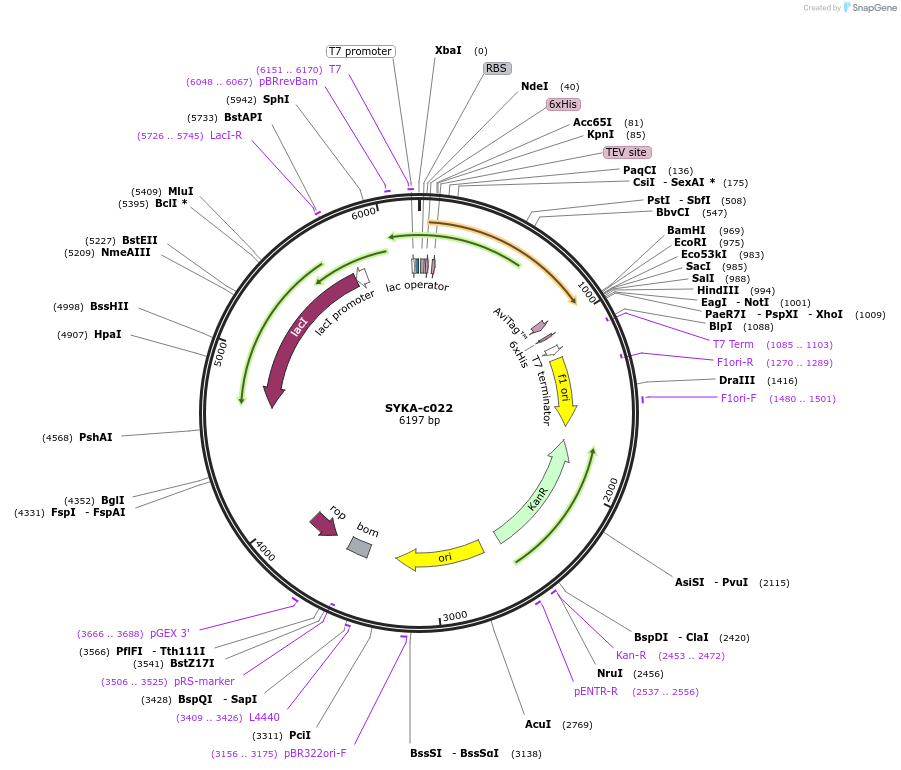
SYKA-c022
Plasmid#175490PurposeProtein expression in bacterial cells. Tandem SH2 domains, M6-N269. Can be biotinylated.DepositorAvailable SinceJan. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
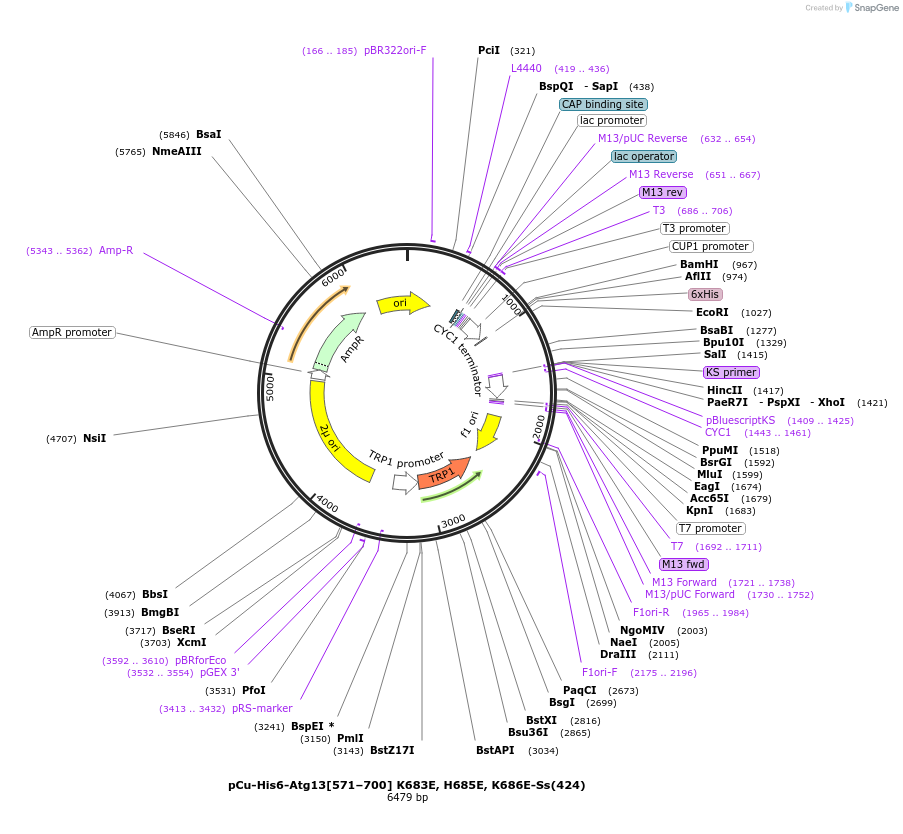
pCu-His6-Atg13[571–700] K683E, H685E, K686E-Ss(424)
Plasmid#166855PurposeExpresses 6xHis tagged ATG13 [571–700] fragment mutated in K683E, H685E, K686E with 14 scramble amino acid sequence DIKLIDTVDLESCN under a Cu promoter in yeastDepositorInsertAtg13[571–700] (ATG13 Budding Yeast)
Tags6xHisExpressionYeastMutationK683E, H685E, K686EPromoterpCuAvailable SinceApril 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
pCu-His6-Atg13[571–700] F641E, I645E-Ss(424)
Plasmid#166854PurposeExpresses 6xHis tagged ATG13 [571–700] fragment mutated in F641E, I645E with 14 scramble amino acid sequence DIKLIDTVDLESCN under a Cu promoter in yeastDepositorInsertAtg13[571–700] (ATG13 Budding Yeast)
Tags6xHisExpressionYeastMutationF641E, I645EPromoterpCuAvailable SinceApril 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
pCu-His6-Atg13[571–700] F641G, I645G-Ss(424)
Plasmid#166853PurposeExpresses 6xHis tagged ATG13 [571–700] fragment mutated in F641G, I645G with 14 scramble amino acid sequence DIKLIDTVDLESCN under a Cu promoter in yeastDepositorInsertAtg13[571–700] (ATG13 Budding Yeast)
Tags6xHisExpressionYeastMutationF641G, I645GPromoterpCuAvailable SinceApril 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
CPH3400 (YCp LEU2 Rpb1 A1529_G1534delInsLEVLFQGP)
Plasmid#91806PurposeYeast expression of S. cerevisiae Rpb1 with an internal PreScission protease siteDepositorInsertRPO21 (RPO21 Budding Yeast)
ExpressionYeastMutationResidues A1529-G1534 replaced with a PreScission …PromoterEndogenous Rpb1 promoterAvailable SinceMarch 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
CPH3477 (YCp LEU2 Rpb1 P1455_E1456InsLEVLFQGP)
Plasmid#91807PurposeYeast expression of S. cerevisiae Rpb1 with an internal PreScission protease siteDepositorInsertRPO21 (RPO21 Budding Yeast)
ExpressionYeastMutationPreScission Protease site (LEVLFQGP) inserted bet…PromoterEndogenous Rpb1 promoterAvailable SinceMarch 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
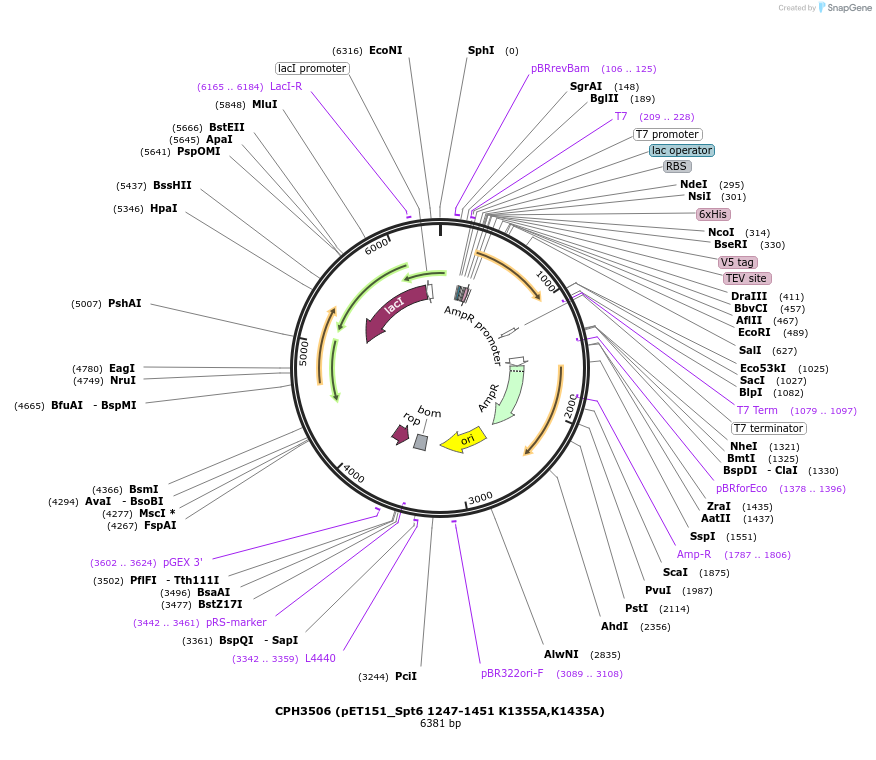
CPH3506 (pET151_Spt6 1247-1451 K1355A,K1435A)
Plasmid#91818PurposeBacterial expression of residues 1247-1451 from S. cerevisiae Spt6 with K1435A and K1355A mutationsDepositorInsertSPT6 (SPT6 Budding Yeast)
Tags6xHisExpressionBacterialMutationchanged lysine 1355 and lysine 1435 to alanines; …PromoterT7Available SinceMarch 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
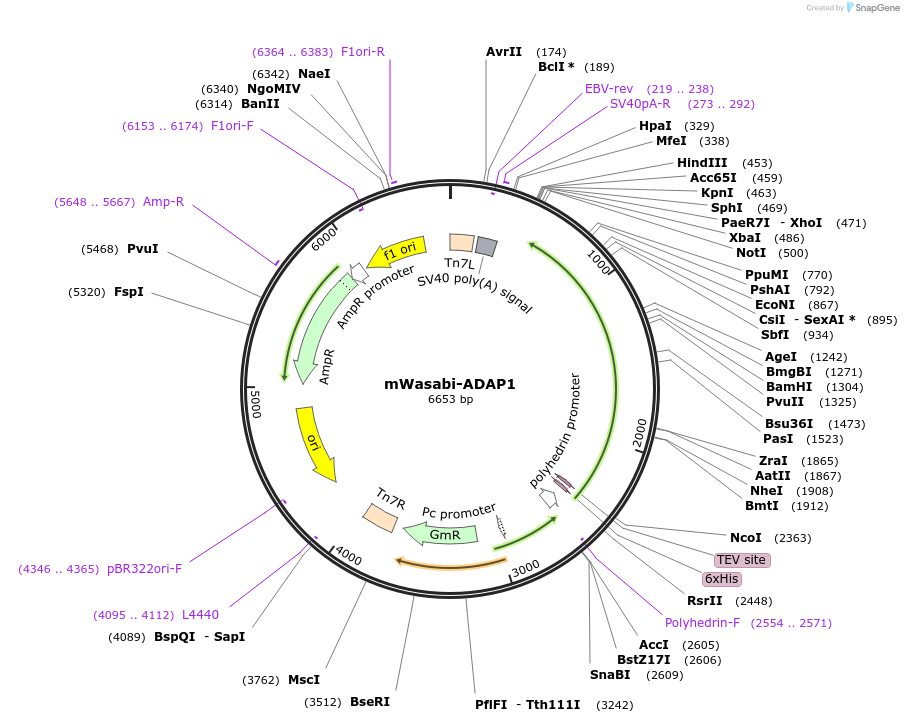
mWasabi-ADAP1
Plasmid#163906PurposeFull length mouse ADAP1 with N terminal fluorescent tag for purification from insect cellsDepositorAvailable SinceMarch 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
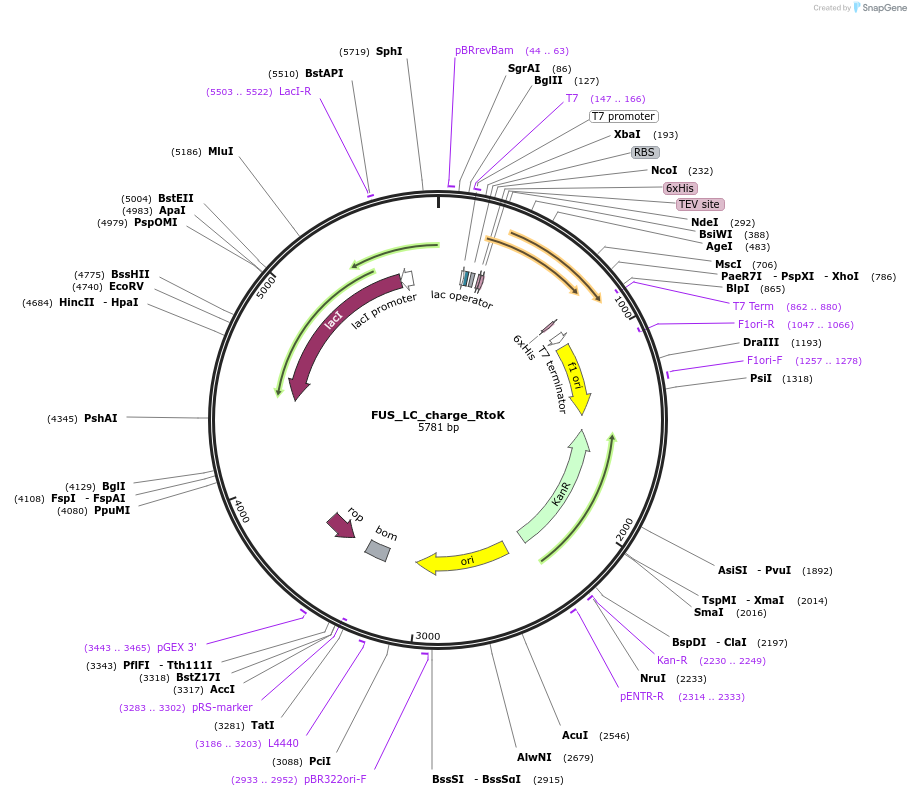
FUS_LC_charge_RtoK
Plasmid#139127Purposeexpress FUS LC with hnRNPA2-like charge patterning where all R are changed to KDepositorInsertFUS (FUS Human)
ExpressionBacterialMutationAdd K and D to FUS LC so it has the same charge p…Available SinceDec. 7, 2020AvailabilityAcademic Institutions and Nonprofits only