We narrowed to 9,847 results for: try
-
Plasmid#237480PurposeEncodes full-length SARS-CoV-2 Spike protein (Belgium/GHB-03021 isolate), featuring the E484D substitution, which likely emerged during passaging and enhances TMEM106B binding.DepositorInsertSARS-CoV-2 Spike protein (Belgium/GHB-03021 isolate)
UseLentiviralTagsIRES-mNeonGreen-NES/PKI and P2A-BlasticidinExpressionMammalianMutationS484D + S813I + deleted amino acids 68-76 + 676-6…PromoterCMVAvailable SinceJuly 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
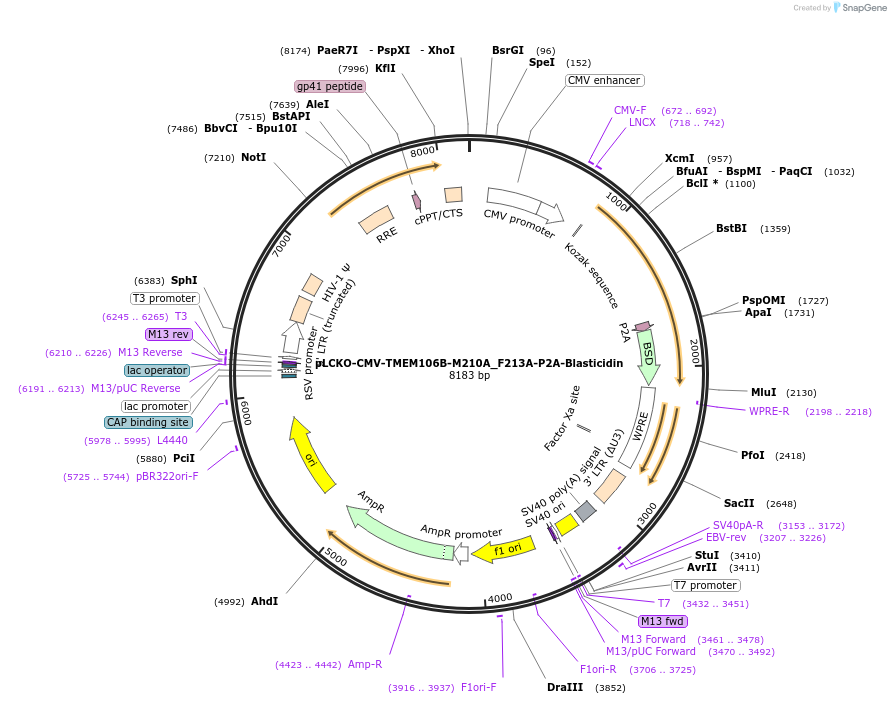
pLCKO-CMV-TMEM106B-M210A_F213A-P2A-Blasticidin
Plasmid#237479PurposeEncodes full-length human TMEM106B. M210A/F213A mutations block S1 binding and SARS-CoV-2 Belgium/GHB-03021/2020 TMEM106B mediated infection, without affecting TMEM106B expression or localization.DepositorInsertTMEM106B (TMEM106B Human)
UseLentiviralTagsP2A-BlasticidinExpressionMammalianMutationM210A + F213APromoterCMVAvailable SinceJune 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
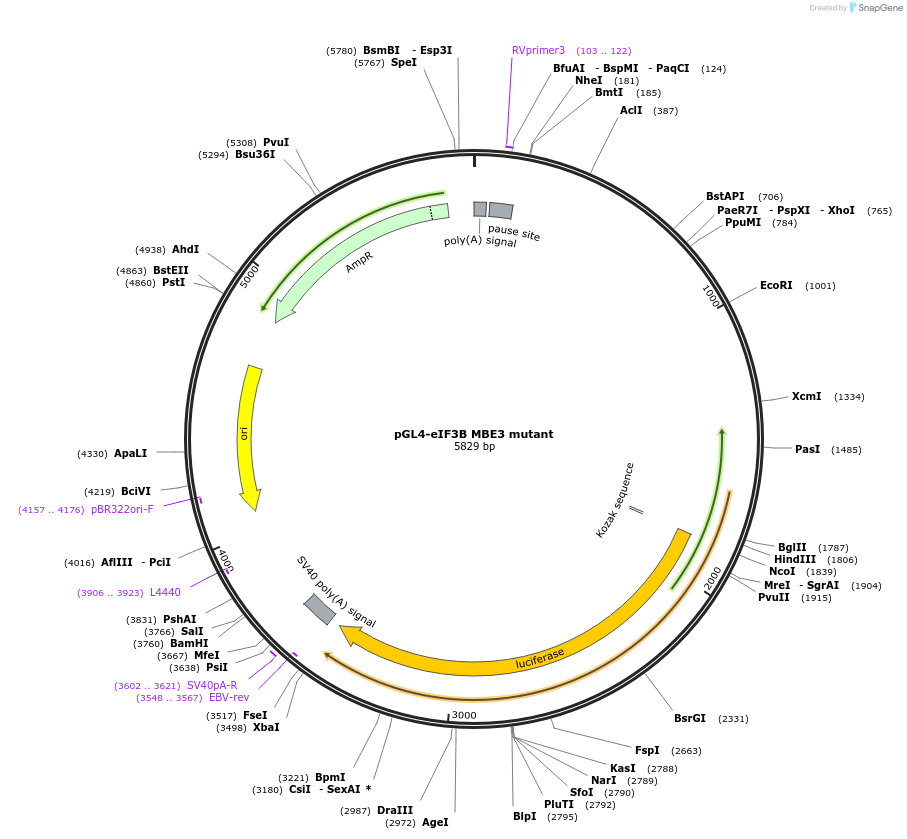
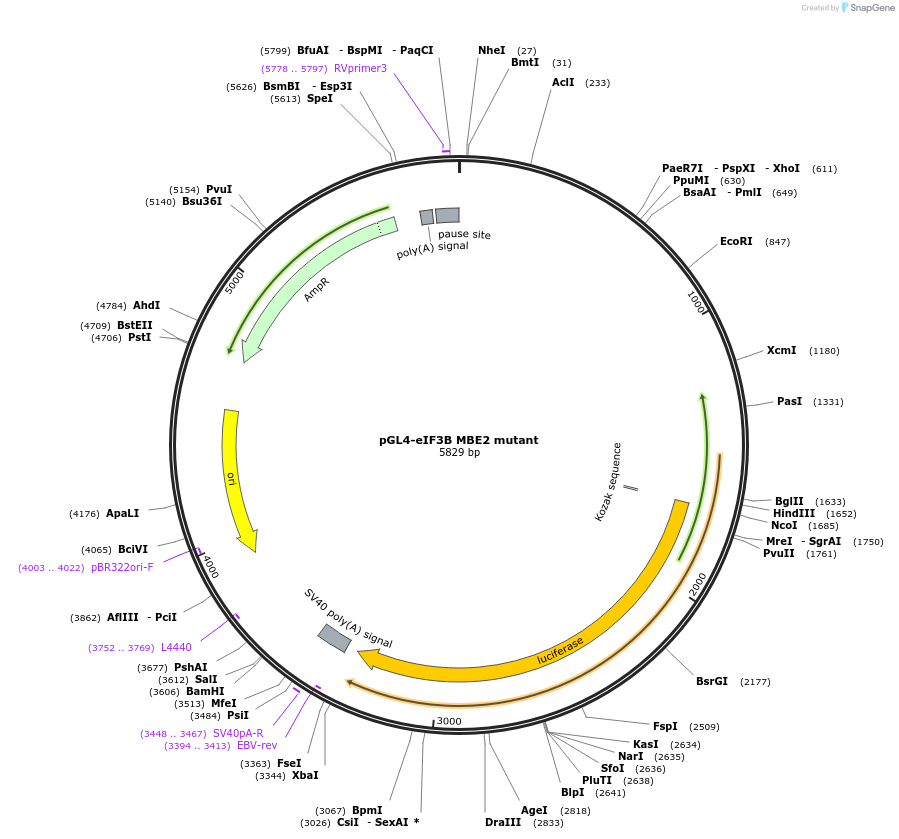
pGL4-eIF3B MBE3 mutant
Plasmid#133310PurposeExpression of luciferase driven by eIF3B promoter region mutated at the E-box binding motif 3DepositorInserteIF3B promoter MBE3 mutant (EIF3B Human)
UseLuciferaseExpressionMammalianMutationccacgtgacc changed to cAaAAAAaccPromotereIF3BAvailable SinceOct. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
pGL4-eIF3B MBE2 mutant
Plasmid#133309PurposeExpression of luciferase driven by eIF3B promoter region mutated at the E-box binding motif 2DepositorInserteIF3B promoter MBE2 mutant (EIF3B Human)
UseLuciferaseExpressionMammalianMutationgccacatgcacc changed to gcAaAaAAcaccPromotereIF3BAvailable SinceOct. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
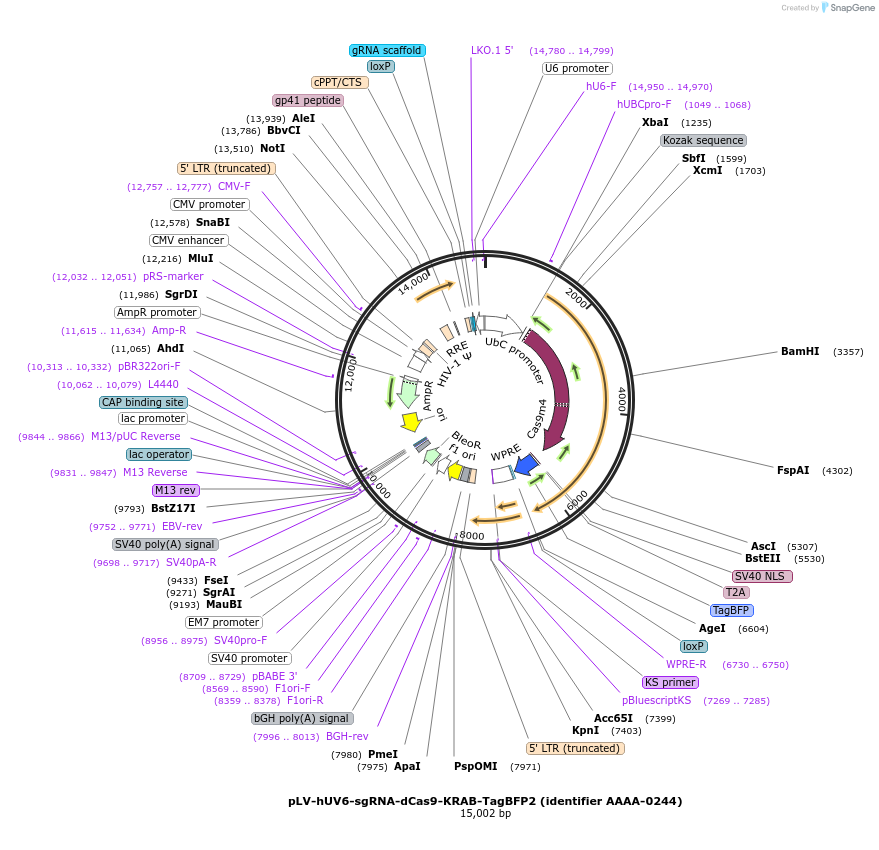
pLV-hUV6-sgRNA-dCas9-KRAB-TagBFP2 (identifier AAAA-0244)
Plasmid#202553PurposeNontargeting control vector with TagBFP2DepositorInsertHumanized dCas9-KRAB-TagBFP2 (tag blue fluorescent protein 2) (TRIM28 S. Pyogenes (dCas9), H. sapiens (KRAB), Entacmaea quadricolor (TagBFP2))
UseCRISPR and LentiviralTags3xFlag (N-terminus of Cas9-KRAB), T2A-TagBFP2MutationPuromycin-resistance gene in the pLV hU6-sgRNA hU…Available SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
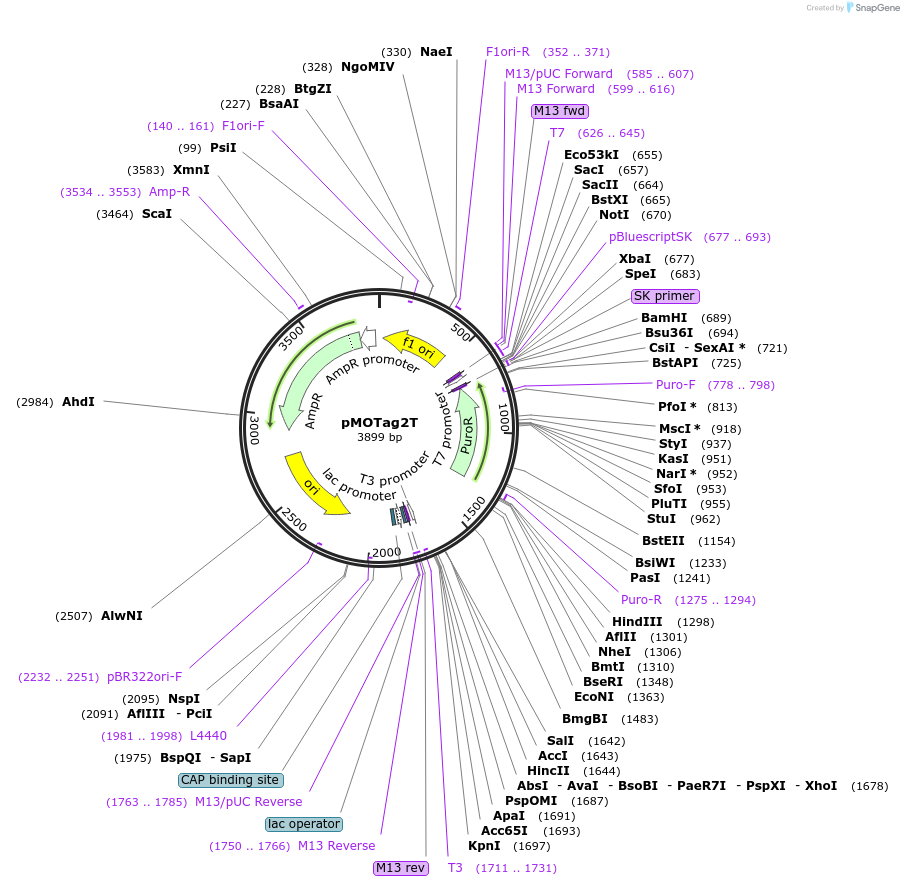
pMOTag2T
Plasmid#26295DepositorTypeEmpty backboneUseTrypanosome in-situ tagging vectorTagsTy-1Available SinceSept. 16, 2010AvailabilityAcademic Institutions and Nonprofits only -
E. coli K-12 MG1655_rLP5(nth-ydgR)
Bacterial Strain#110245PurposeE. coli K-12 MG1655 with Landing Pad engineered into the nth-ydgR intergenic region. The Landing Pad consists of floxed mCherry,Chlor that are cotranscribed.DepositorBacterial ResistanceChloramphenicolAvailable SinceMarch 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
TB205 △trpC
Bacterial Strain#230038PurposeFluorescently labelled (mCherry) E. coli, auxotrophic for tryptophanDepositorBacterial ResistanceNoneAvailable SinceJan. 31, 2025AvailabilityAcademic Institutions and Nonprofits only -
TB204 △trpC
Bacterial Strain#230037PurposeFluorescently labelled (GFP) E. coli, auxotrophic for tryptophanDepositorBacterial ResistanceNoneAvailable SinceJan. 31, 2025AvailabilityAcademic Institutions and Nonprofits only -
E. coli K-12 MG1655_fLP4(essQ-cspB)
Bacterial Strain#110244PurposeE. coli K-12 MG1655 with Landing Pad engineered into the essQ-cspB intergenic region. The Landing Pad consists of floxed mCherry,Chlor that are cotranscribed.DepositorBacterial ResistanceChloramphenicolAvailable SinceMarch 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
E. coli K-12 MG1655_rLP6(ygcE-ygcF)
Bacterial Strain#110246PurposeE. coli K-12 MG1655 with Landing Pad engineered into the ygcE-ygcF intergenic region. The Landing Pad consists of floxed mCherry,Chlor that are cotranscribed.DepositorBacterial ResistanceChloramphenicolAvailable SinceMarch 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
E. coli K-12 MG1655_rLP1(atpI-gidB)
Bacterial Strain#110241PurposeE. coli K-12 MG1655 with Landing Pad engineered into the atpl-gidB intergenic region. The Landing Pad consists of floxed mCherry,Chlor that are cotranscribed.DepositorBacterial ResistanceChloramphenicolAvailable SinceMarch 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
E. coli K-12 MG1655_fLP3(ybbD-ylbG)
Bacterial Strain#110243PurposeE. coli K-12 MG1655 with Landing Pad engineered into the ybbD-ylbG intergenic region. The Landing Pad consists of floxed mCherry,Chlor that are cotranscribed.DepositorBacterial ResistanceChloramphenicolAvailable SinceMarch 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
E. coli K-12 MG1655_fLP2(yieN-trkD)
Bacterial Strain#110242PurposeE. coli K-12 MG1655 with Landing Pad engineered into the yieN-trkD intergenic region. The Landing Pad consists of floxed mCherry,Chlor that are cotranscribed.DepositorBacterial ResistanceChloramphenicolAvailable SinceMarch 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
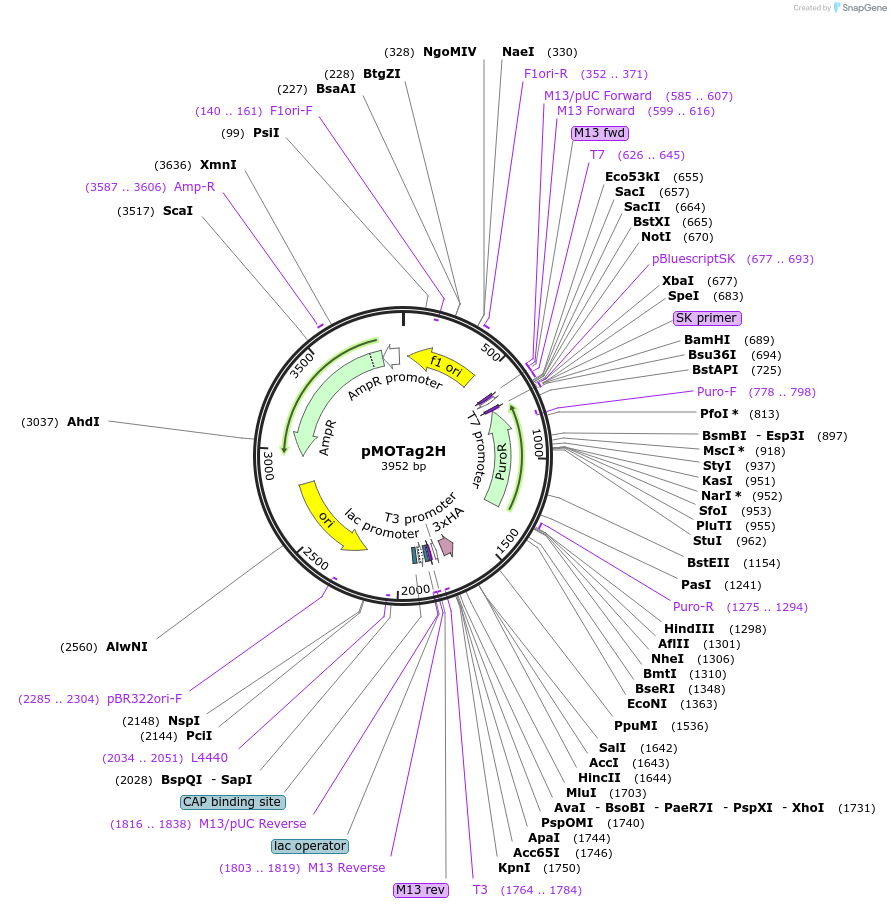
pMOTag2H
Plasmid#26296DepositorTypeEmpty backboneUseTrypanosome in-situ tagging vectorTagsTriple HAAvailable SinceSept. 16, 2010AvailabilityAcademic Institutions and Nonprofits only -
pMOTag5H
Plasmid#26299DepositorTypeEmpty backboneUseTrypanosome in-situ tagging vectorTagstriple HAAvailable SinceSept. 16, 2010AvailabilityAcademic Institutions and Nonprofits only -
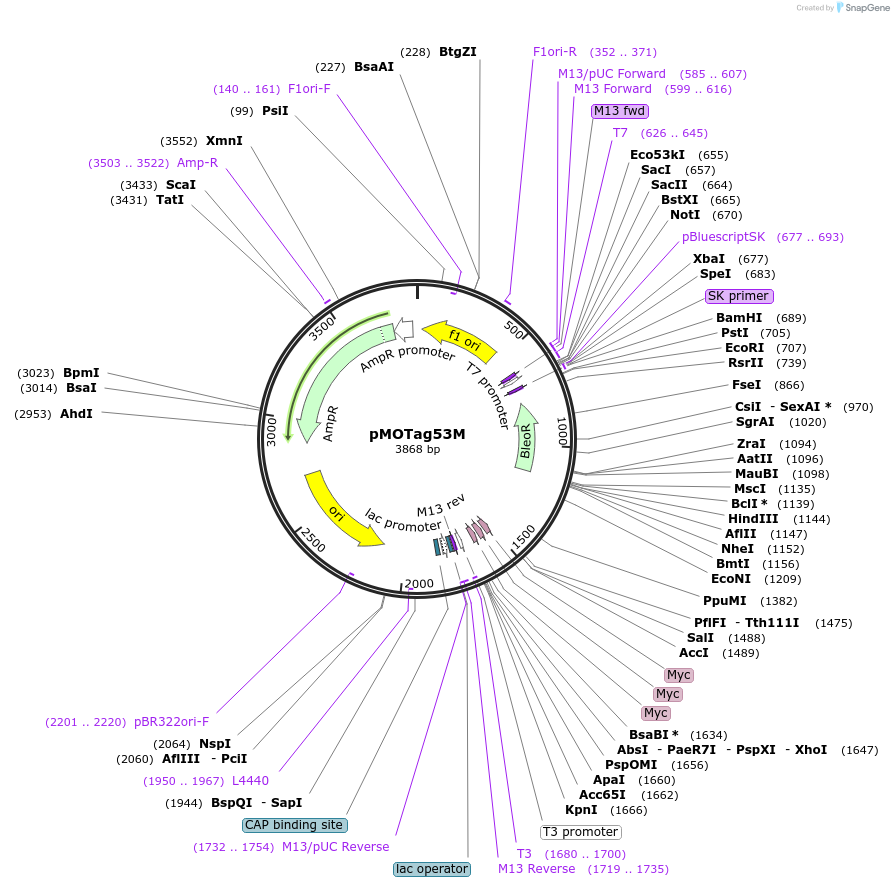
pMOTag53M
Plasmid#26297DepositorTypeEmpty backboneUseTrypanosome in-situ tagging vectorTagstriple cMycAvailable SinceSept. 16, 2010AvailabilityAcademic Institutions and Nonprofits only -
ZM2
Plasmid#32784DepositorInsertT7rnap
UseCre/Lox; Floxed targeting vector for trypanosomeAvailable SinceDec. 13, 2011AvailabilityAcademic Institutions and Nonprofits only -
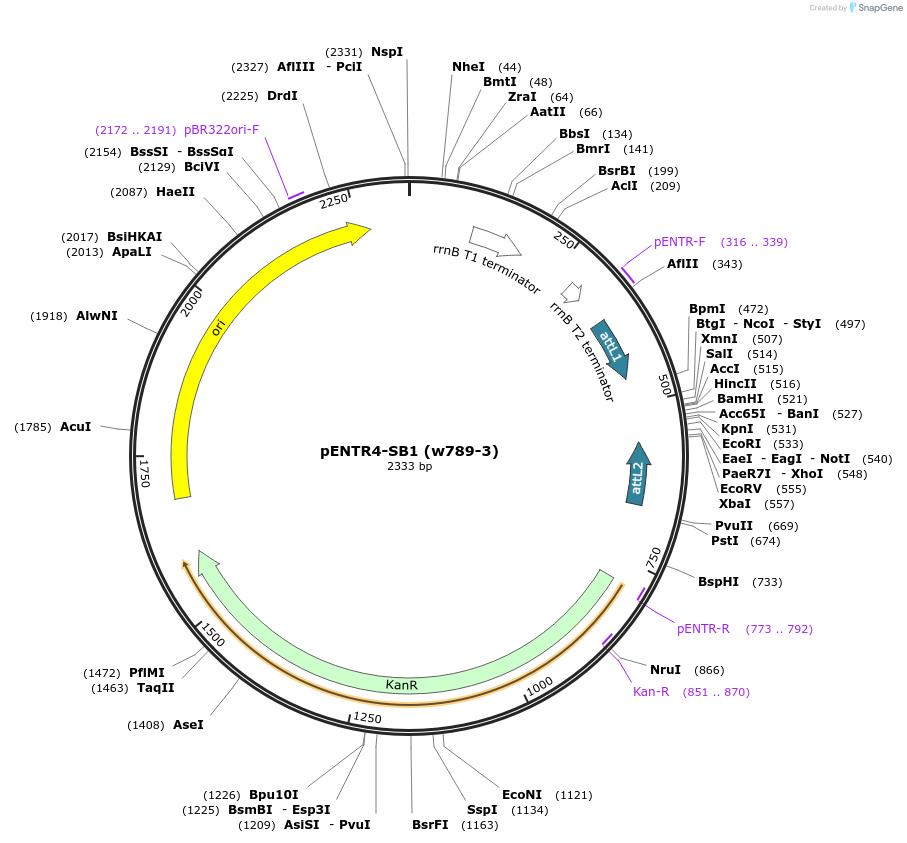
pENTR4-SB1 (w789-3)
Plasmid#26956DepositorTypeEmpty backboneUseEntry vectorTagsSB1Available SinceApril 27, 2011AvailabilityAcademic Institutions and Nonprofits only -
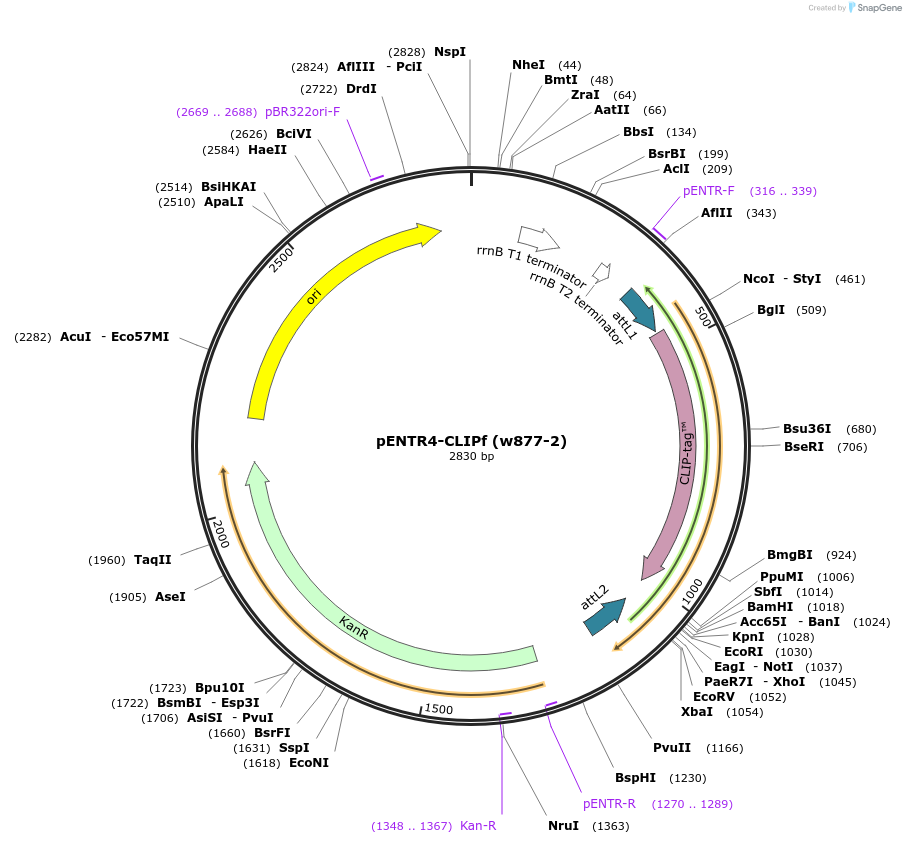
pENTR4-CLIPf (w877-2)
Plasmid#29650DepositorTypeEmpty backboneUseEntry vectorTagsCLIPfAvailable SinceJuly 6, 2011AvailabilityAcademic Institutions and Nonprofits only