We narrowed to 13,323 results for: BASE
-
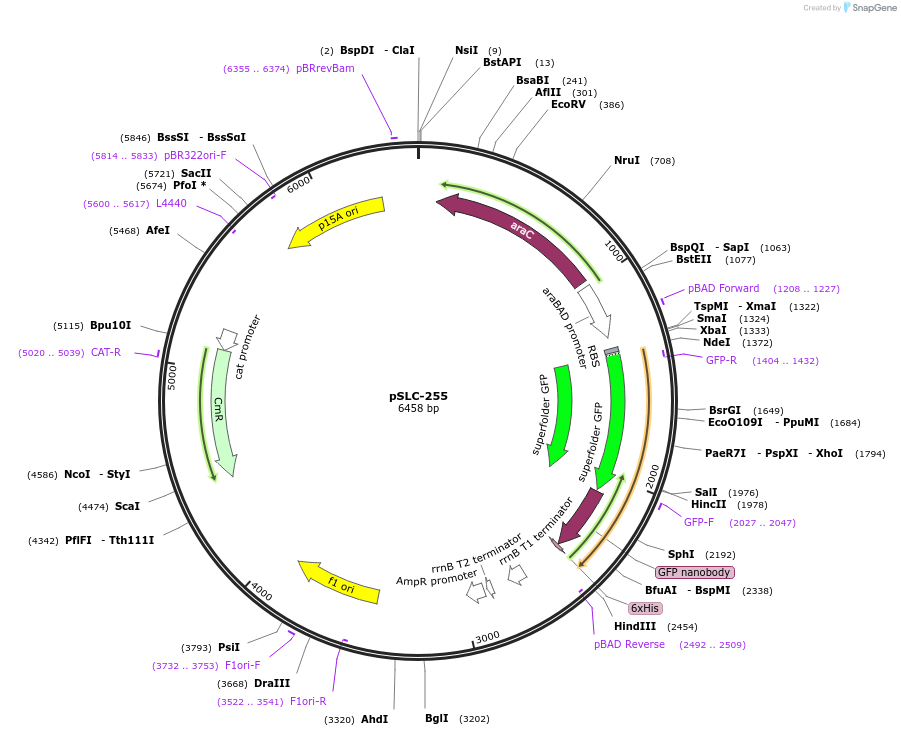
Plasmid#124292PurposeExpression of vsfGFP-9. This construct improves brightness of sfGFP by fusing a GFP-specific single domain antibody to it that creates a monomeric fluorophore.DepositorInsertvsfGFP-9
ExpressionBacterialAvailable SinceSept. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
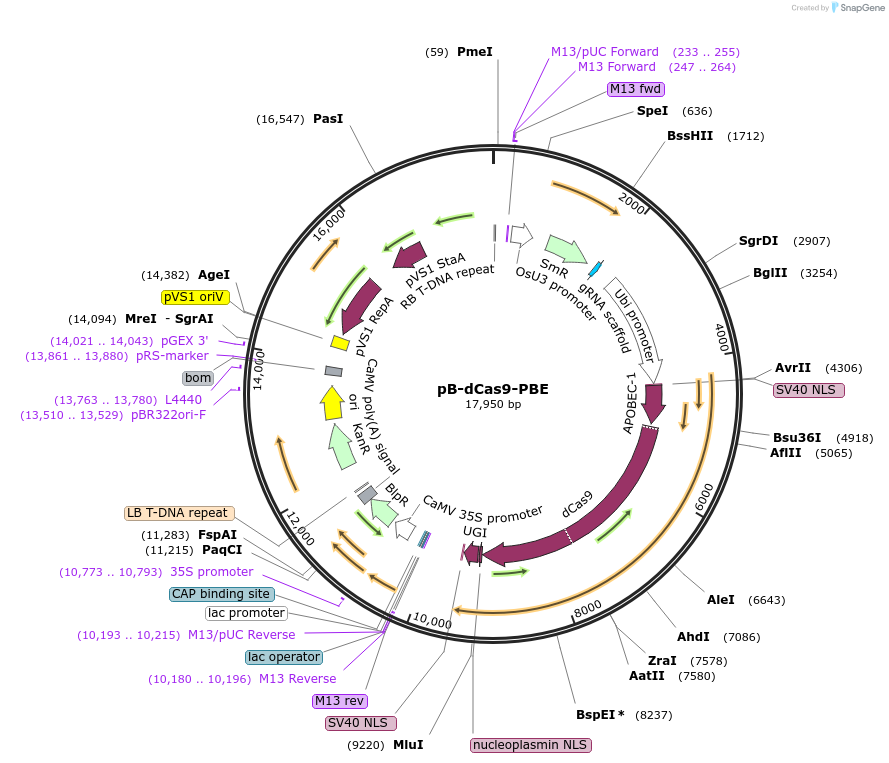
pB-dCas9-PBE
Plasmid#98159PurposeExpresses CRISPR base editor in maizeDepositorInsertNLS-APOBEC1-XTEN-dCas9-UGI-NLS
UseMaizeExpressionPlantPromoterUbiAvailable SinceAug. 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
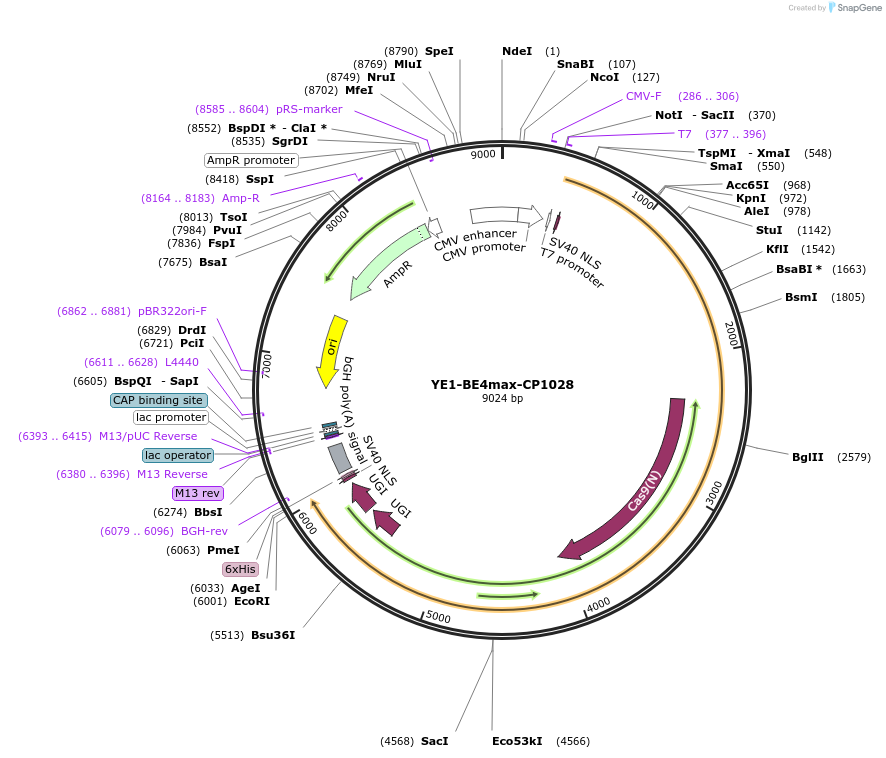
YE1-BE4max-CP1028
Plasmid#138160PurposeC-to-T base editorDepositorInsertrAPOBEC1(YE1)-nSpCas9 (CP1028) -UGI-UGI
TagsNLSExpressionMammalianMutationWithin rAPOBEC1 W90Y + R126E; within Cas9 (CP1028…PromoterCMVAvailable SinceFeb. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
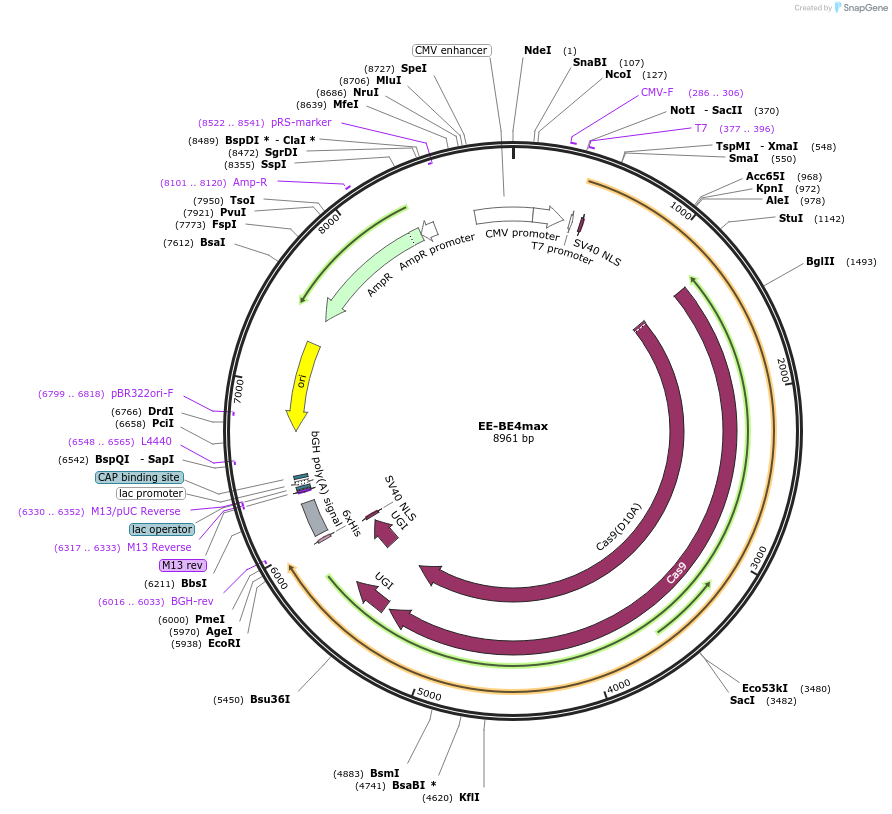
EE-BE4max
Plasmid#138158PurposeC-to-T base editorDepositorInsertrAPOBEC1(EE)-nSpCas9-UGI-UGI
TagsNLSExpressionMammalianMutationWithin rAPOBEC1 R126E + R132E; within Cas9 D10APromoterCMVAvailable SinceFeb. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
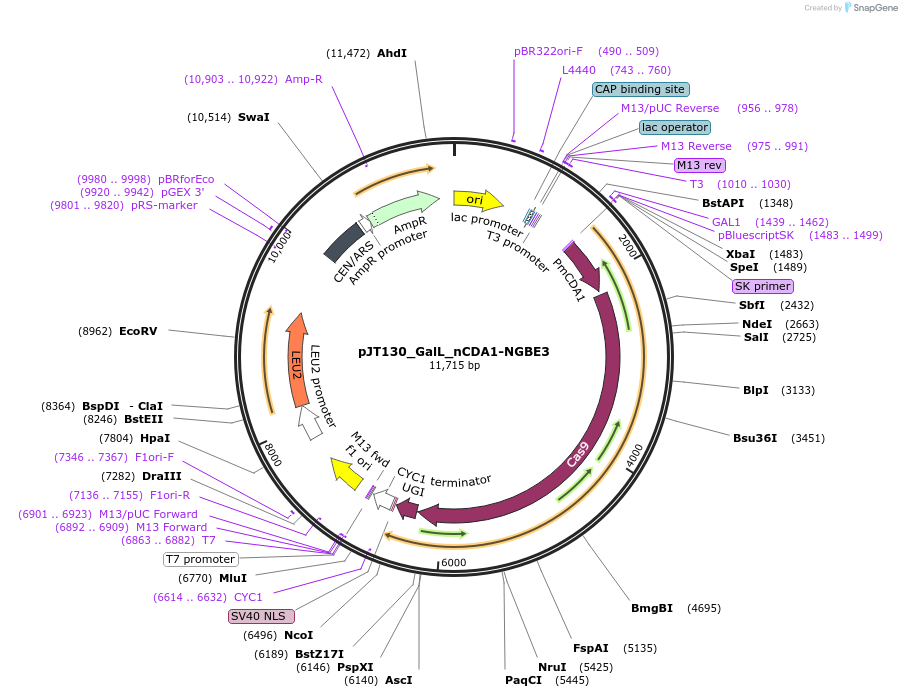
pJT130_GalL_nCDA1-NGBE3
Plasmid#145094PurposeExpressing base editor nCDA1-NGBE3 in yeast cellsDepositorInsertnCDA1-NGBE3
UseCRISPRExpressionYeastMutationspCas9(D10A/L1111R/D1135V/G1218R/E1219F/A1322R/R1…PromoterGalLAvailable SinceJuly 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
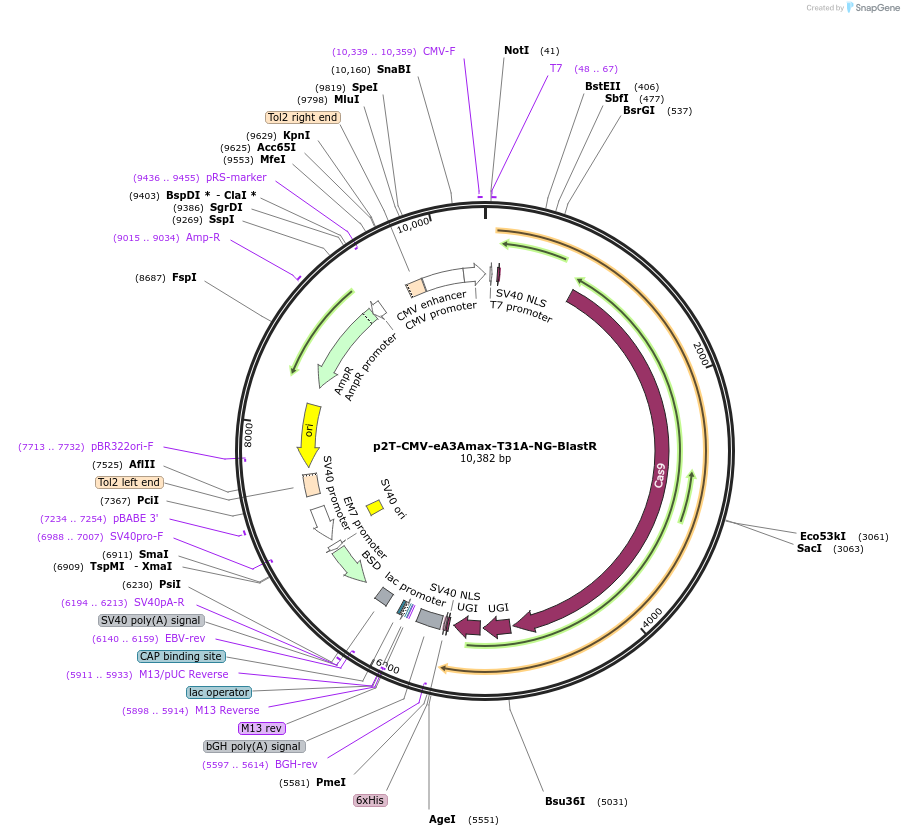
p2T-CMV-eA3Amax-T31A-NG-BlastR
Plasmid#153000PurposeC-to-T/G/A base editorDepositorInserteA3A-BE4 T31A-nSpCas9-UGI-UGI
UseCRISPRExpressionMammalianPromoterCMVAvailable SinceJuly 24, 2020AvailabilityAcademic Institutions and Nonprofits only -
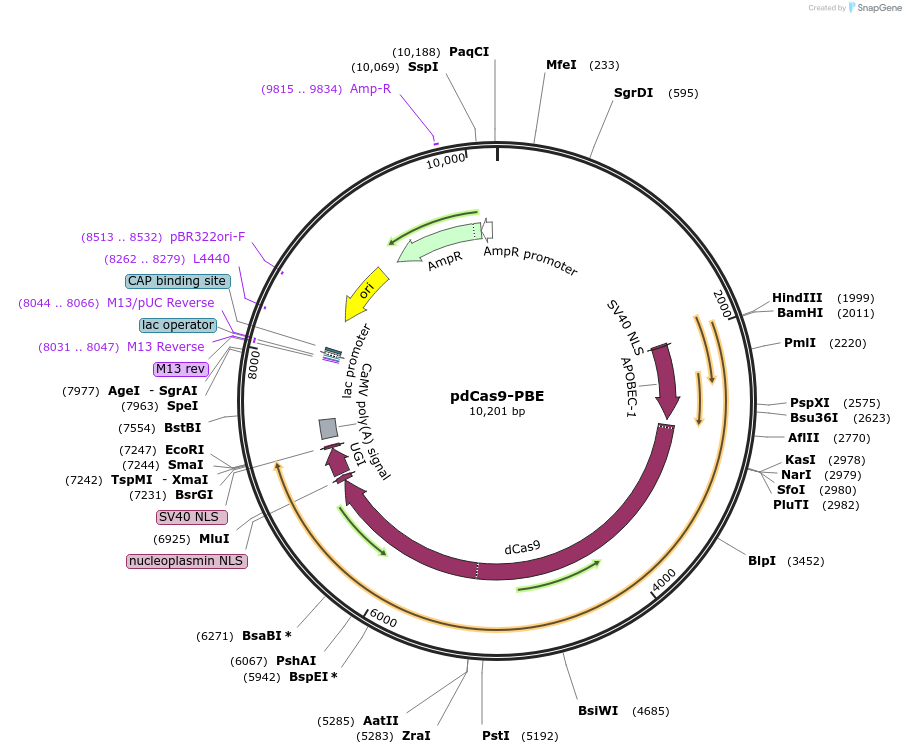
pdCas9-PBE
Plasmid#98161PurposeExpresses CRISPR base editor in protoplastsDepositorInsertNLS-APOBEC1-XTEN-dCas9-UGI-NLS
ExpressionPlantPromoterUbiAvailable SinceJuly 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
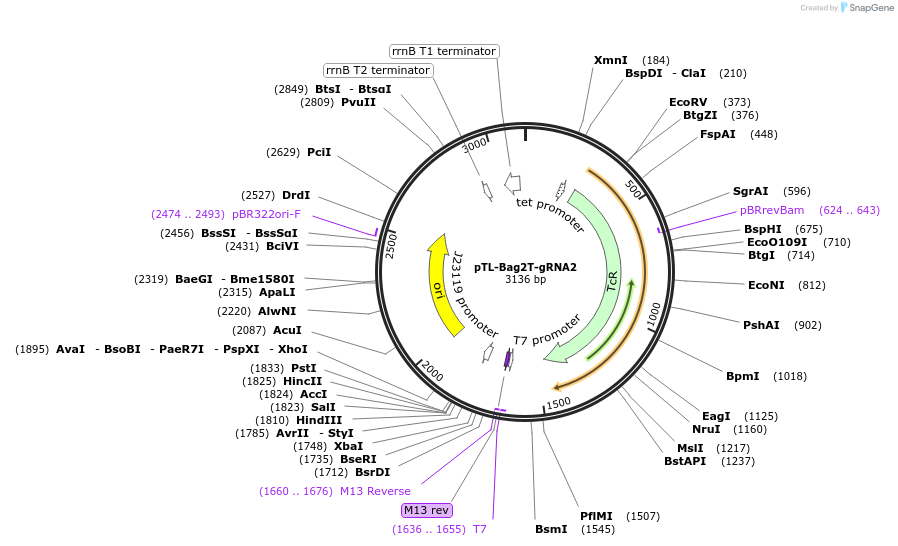
pTL-Bag2T-gRNA2
Plasmid#196253PurposePlasmid for cloning the second CRISPR-Cas9 guide RNA for guide RNAsDepositorInsertCRISPR-Cas9 guide RNA scaffold and multiple cloning site
ExpressionBacterialPromoterJ23119 promoterAvailable SinceFeb. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
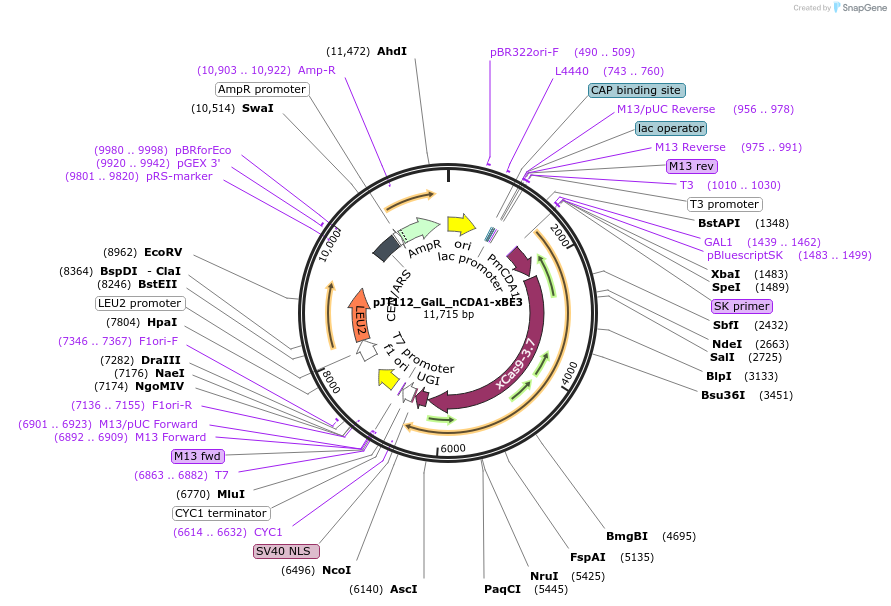
pJT112_GalL_nCDA1-xBE3
Plasmid#141318PurposeExpressing base editor nCDA1-xBE3 in yeast cellsDepositorInsertnCDA1-xBE3
UseCRISPRExpressionYeastMutationspCas9(D10A, A262T, R324L, S409I, E480K, E543D, M…PromoterGalLAvailable SinceJuly 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
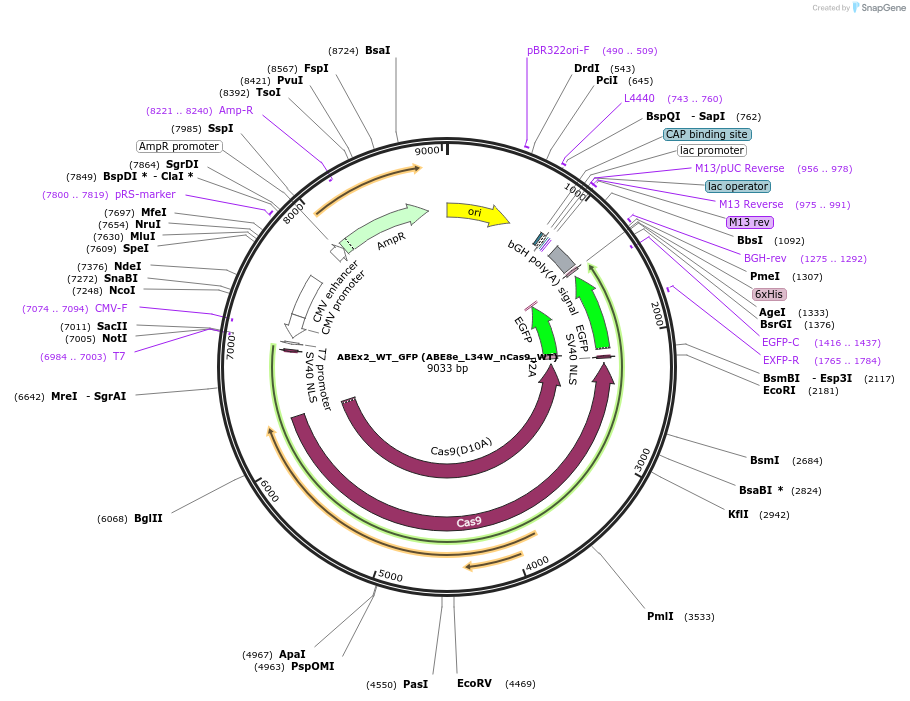
ABEx2_WT_GFP (ABE8e_L34W_nCas9_WT)
Plasmid#251549PurposeMammalian expression plasmid encoding an nCas9-WT adenine base editor (ABE8e) carrying the L34W mutation in the deaminase domainDepositorInsertecTadA(8e)_L34W-nCas9_WT
UseCRISPRExpressionMammalianMutationABEx2 L34WAvailable SinceFeb. 3, 2026AvailabilityAcademic Institutions and Nonprofits only -
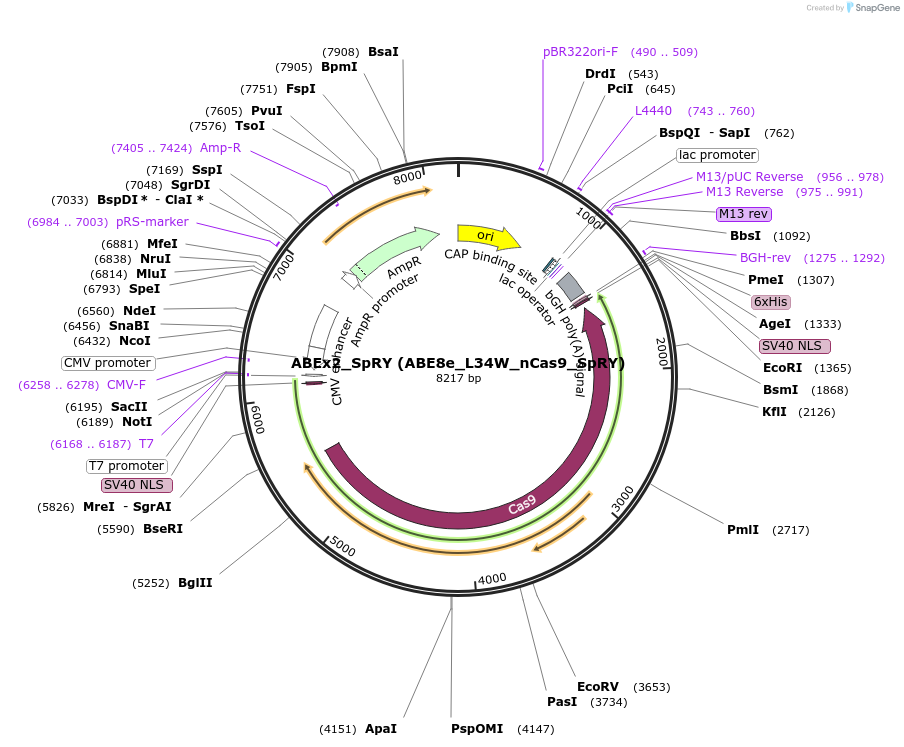
ABEx2_SpRY (ABE8e_L34W_nCas9_SpRY)
Plasmid#251545PurposeMammalian expression plasmid encoding an nCas9-SpRY adenine base editor (ABE8e) carrying the L34W mutation in the deaminase domainDepositorInsertecTadA(8e)_L34W-nCas9_SpRY
UseCRISPRExpressionMammalianMutationABEx2 L34WAvailable SinceFeb. 3, 2026AvailabilityAcademic Institutions and Nonprofits only -
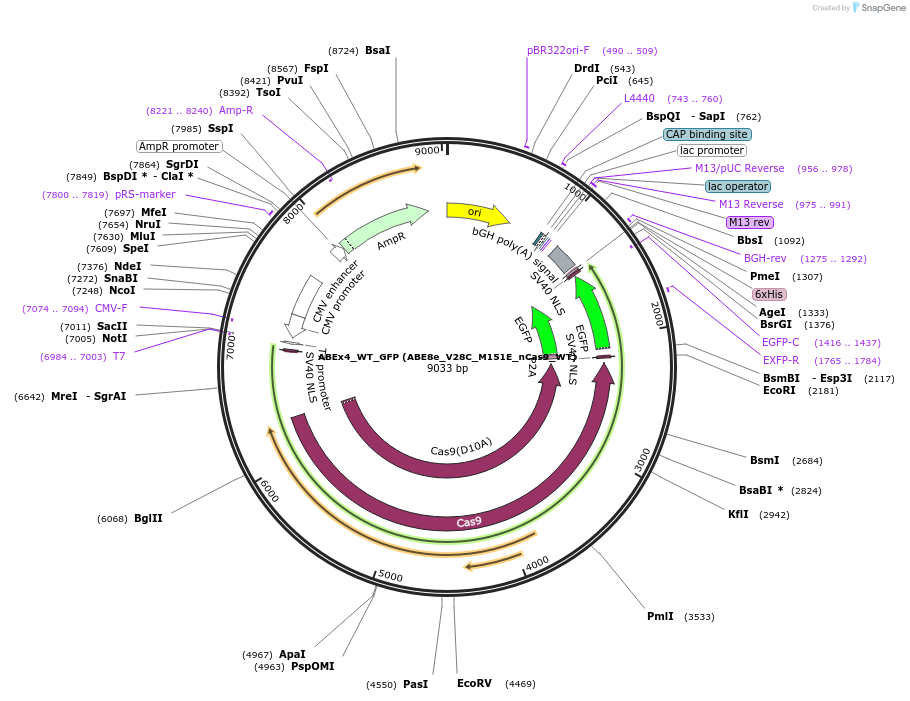
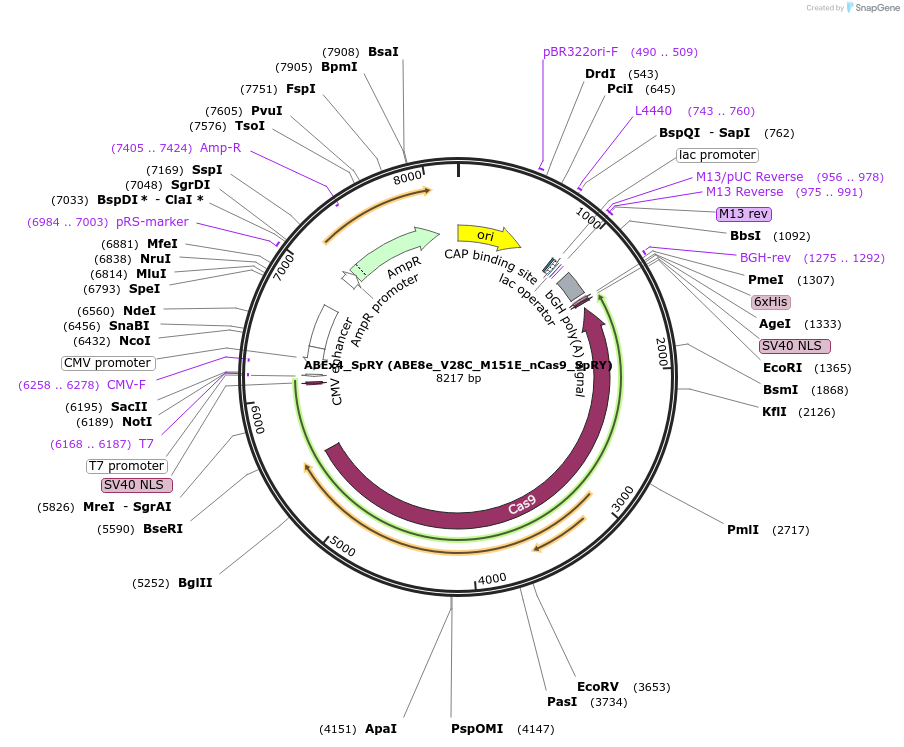
ABEx4_WT_GFP (ABE8e_V28C_M151E_nCas9_WT)
Plasmid#251551PurposeMammalian expression plasmid encoding an nCas9-WT adenine base editor (ABE8e) carrying the V28C and M151E mutations in the deaminase domainDepositorInsertecTadA(8e)_V28C_M151E-nCas9_WT
UseCRISPRExpressionMammalianMutationABEx4 V28C_M151EAvailable SinceFeb. 3, 2026AvailabilityAcademic Institutions and Nonprofits only -
ABEx4_SpRY (ABE8e_V28C_M151E_nCas9_SpRY)
Plasmid#251547PurposeMammalian expression plasmid encoding an nCas9-SpRY adenine base editor (ABE8e) carrying the V28C and M151E mutations in the deaminase domainDepositorInsertecTadA(8e)_V28C_M151E-nCas9_SpRY
UseCRISPRExpressionMammalianMutationABEx4 V28C_M151EAvailable SinceFeb. 3, 2026AvailabilityAcademic Institutions and Nonprofits only -
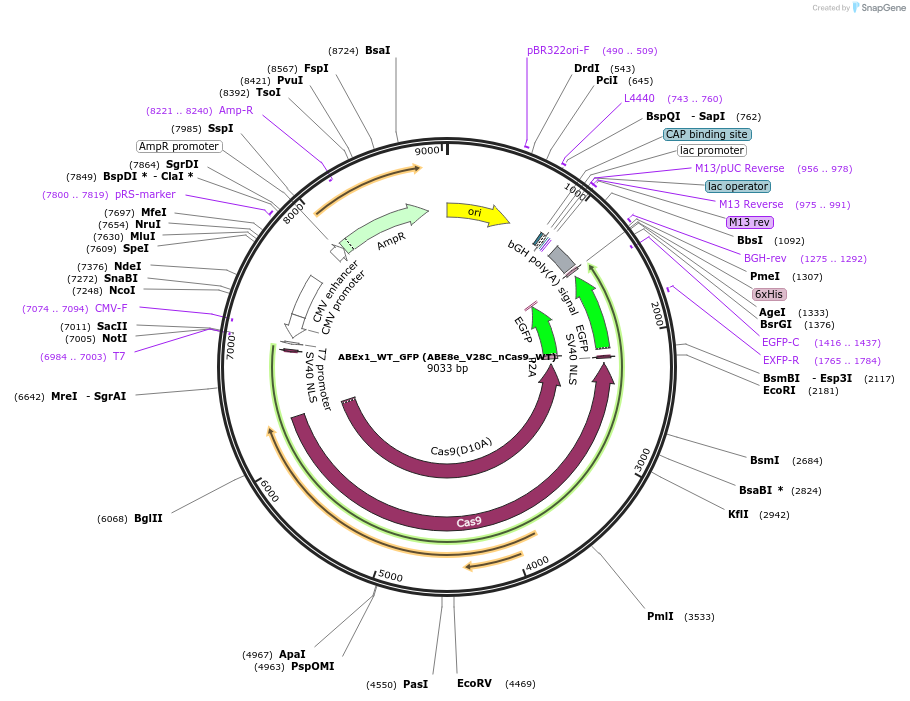
ABEx1_WT_GFP (ABE8e_V28C_nCas9_WT)
Plasmid#251548PurposeMammalian expression plasmid encoding an nCas9-WT adenine base editor (ABE8e) carrying the V28C mutation in the deaminase domainDepositorInsertecTadA(8e)_V28C-nCas9_WT
UseCRISPRExpressionMammalianMutationABEx1 V28CAvailable SinceFeb. 3, 2026AvailabilityAcademic Institutions and Nonprofits only -
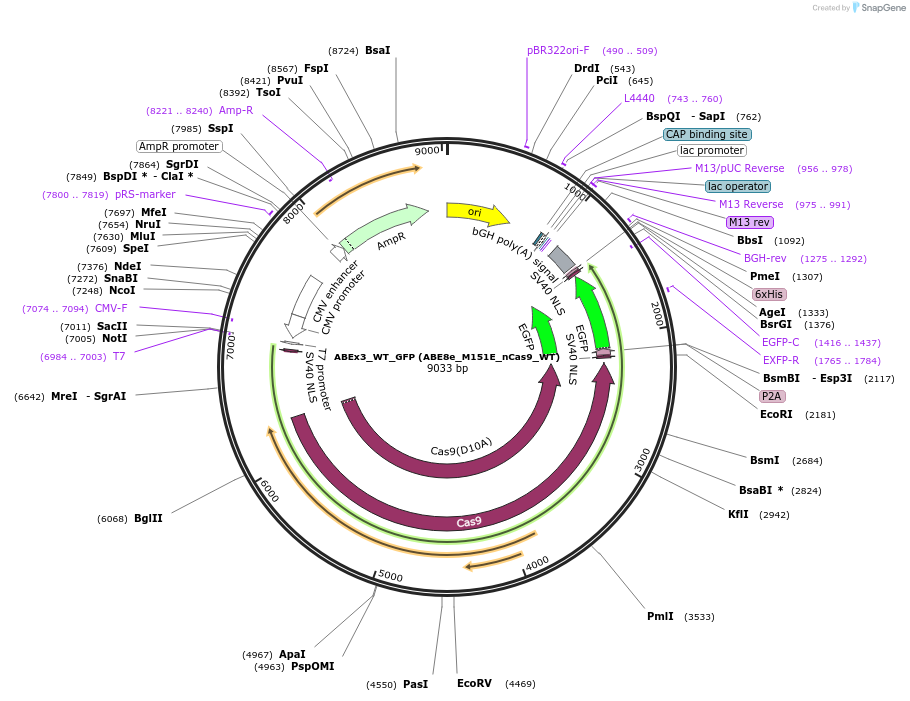
ABEx3_WT_GFP (ABE8e_M151E_nCas9_WT)
Plasmid#251550PurposeMammalian expression plasmid encoding an nCas9-WT adenine base editor (ABE8e) carrying the M151E mutation in the deaminase domainDepositorInsertecTadA(8e)_M151E-nCas9_WT
UseCRISPRExpressionMammalianMutationABEx3 M151EAvailable SinceFeb. 3, 2026AvailabilityAcademic Institutions and Nonprofits only -
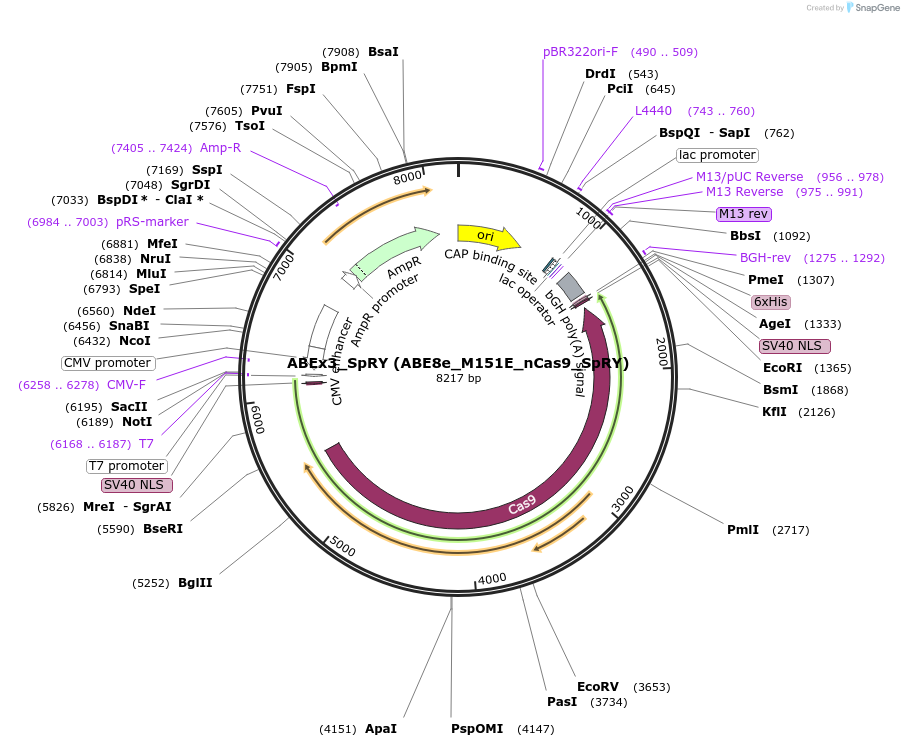
ABEx3_SpRY (ABE8e_M151E_nCas9_SpRY)
Plasmid#251546PurposeMammalian expression plasmid encoding an nCas9-SpRY adenine base editor (ABE8e) carrying the M151E mutation in the deaminase domainDepositorInsertecTadA(8e)_M151E-nCas9_SpRY
UseCRISPRExpressionMammalianMutationABEx3 M151EAvailable SinceFeb. 3, 2026AvailabilityAcademic Institutions and Nonprofits only -
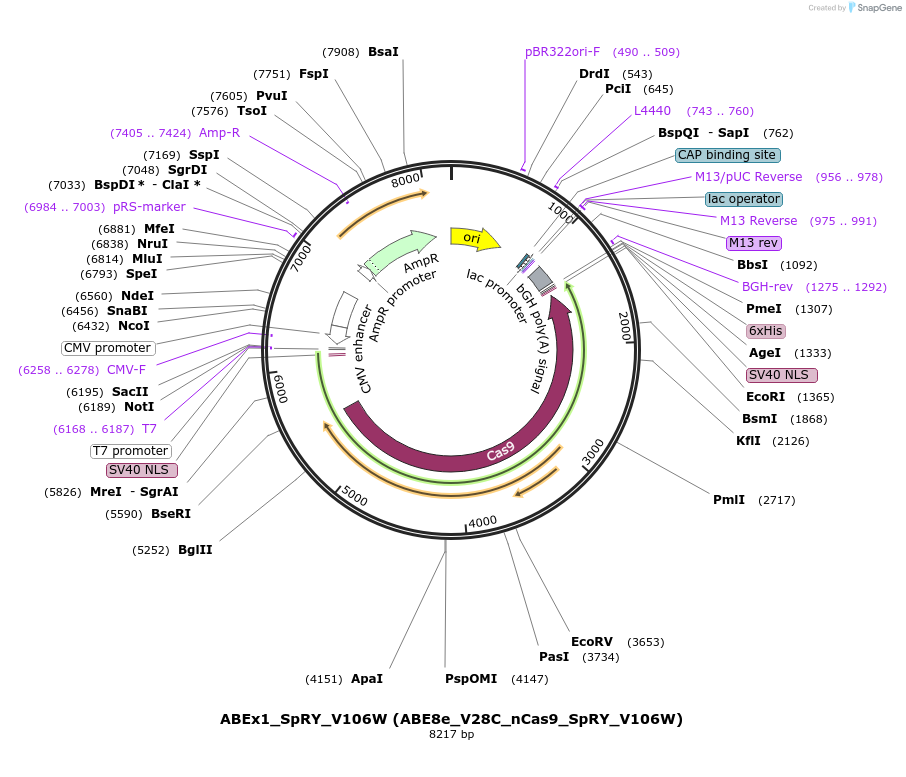
ABEx1_SpRY_V106W (ABE8e_V28C_nCas9_SpRY_V106W)
Plasmid#251552PurposeMammalian expression plasmid encoding an nCas9-SpRY_V106W adenine base editor (ABE8e) carrying the V28C mutation in the deaminase domainDepositorInsertecTadA(8e)_V28C-nCas9_SpRY_V106W
UseCRISPRExpressionMammalianMutationABEx4 V28C_M151EAvailable SinceFeb. 3, 2026AvailabilityAcademic Institutions and Nonprofits only -
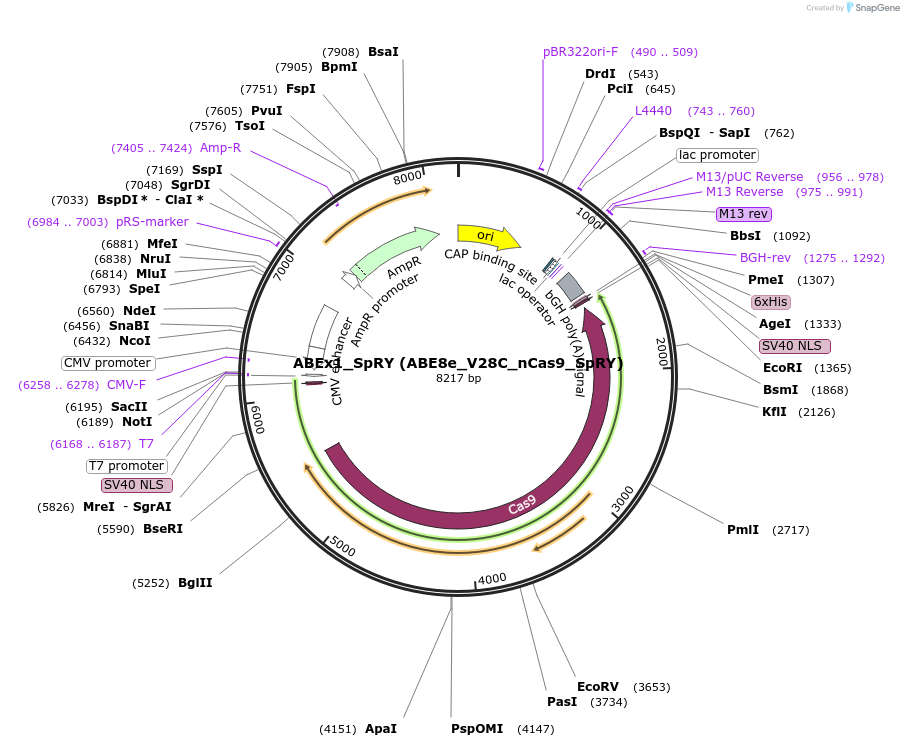
ABEx1_SpRY (ABE8e_V28C_nCas9_SpRY)
Plasmid#251544PurposeMammalian expression plasmid encoding an nCas9-SpRY adenine base editor (ABE8e) carrying the V28C mutation in the deaminase domainDepositorInsertecTadA(8e)_V28C-nCas9_SpRY
UseCRISPRExpressionMammalianMutationABEx1 V28CAvailable SinceFeb. 3, 2026AvailabilityAcademic Institutions and Nonprofits only -
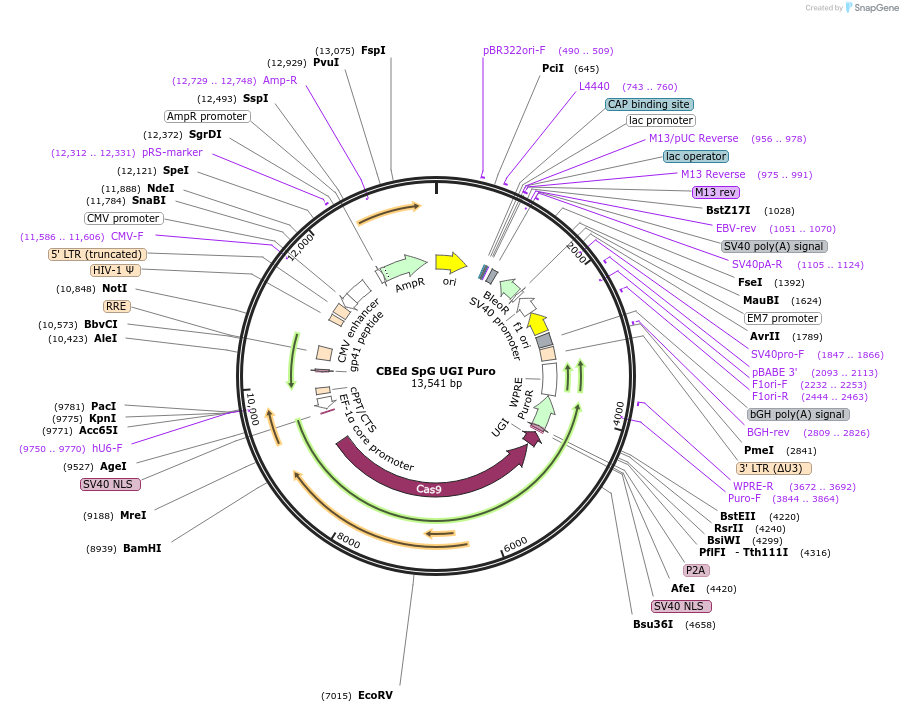
CBEd SpG UGI Puro
Plasmid#235045PurposeCytosine base editor d with SpG nicking Cas9 makes C > T editsDepositorInsertCBEd, nCas9
UseCRISPR and LentiviralTagsP2A-PuroRPromoterEF1a coreAvailable SinceJan. 7, 2026AvailabilityAcademic Institutions and Nonprofits only -
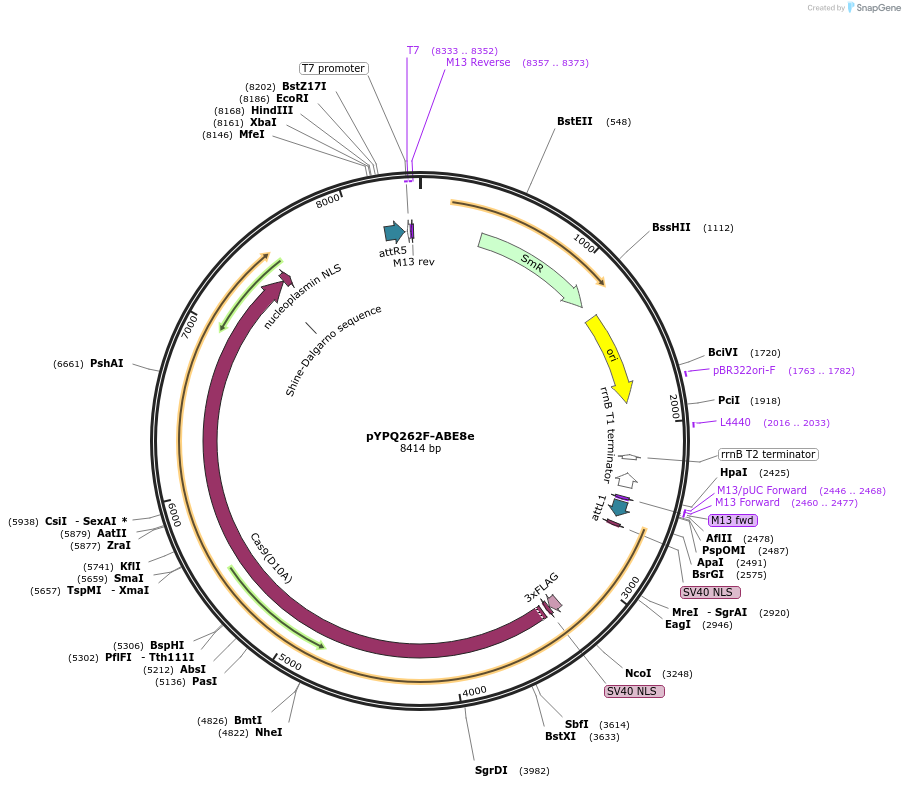
pYPQ262F-ABE8e
Plasmid#199179PurposeGateway entry clone (attL1 & attR5) for CRISPR-zSpCas9 ABE8e-HF mediated A-G base editingDepositorInsertABE8e(V106W)-zSpCas9(D10A)
UseCRISPR; Gateway compatible abe8e(v106w)-zspcas9(d…TagsNLSExpressionPlantAvailable SinceMarch 4, 2024AvailabilityAcademic Institutions and Nonprofits only