We narrowed to 1,744 results for: SUA
-
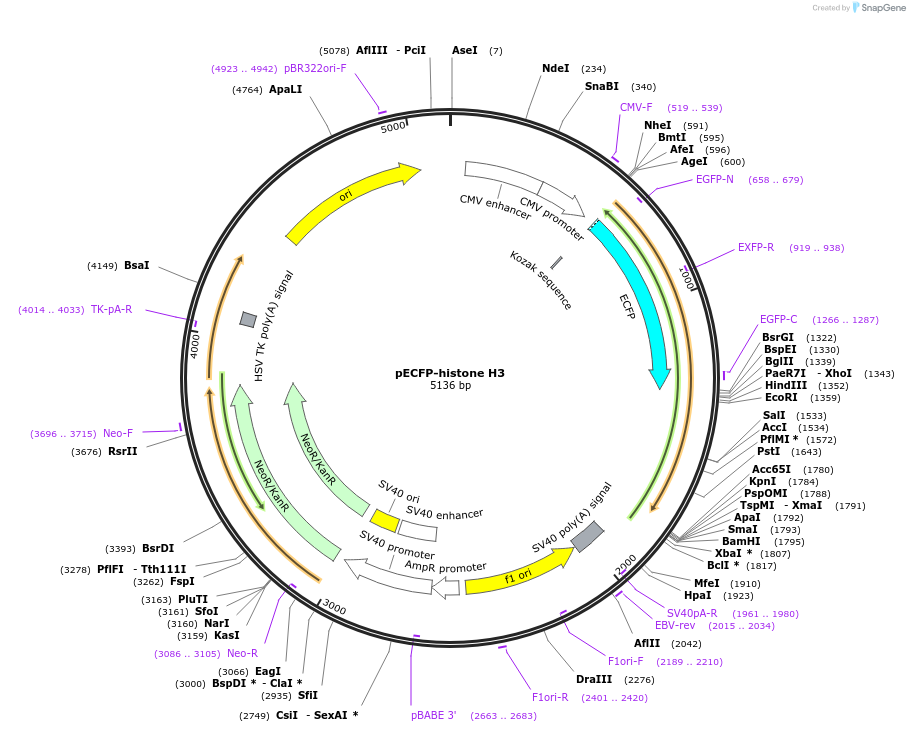
Plasmid#179698Purposevisualization of nucleus/chromosomeDepositorAvailable SinceMarch 9, 2022AvailabilityAcademic Institutions and Nonprofits only
-
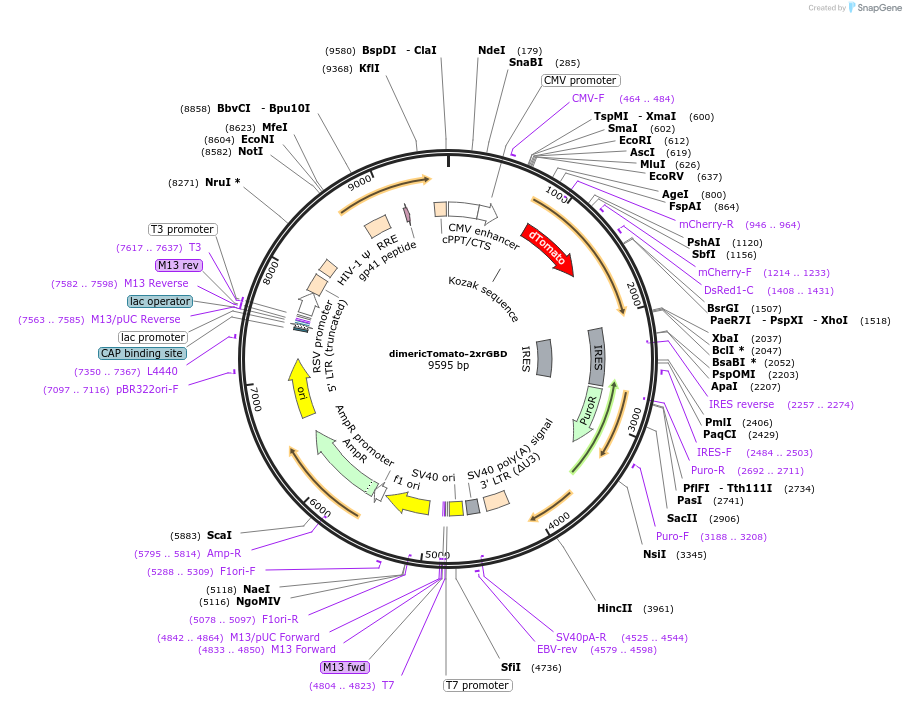
dimericTomato-2xrGBD
Plasmid#176098PurposeVisualization of active RhoADepositorAvailable SinceOct. 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
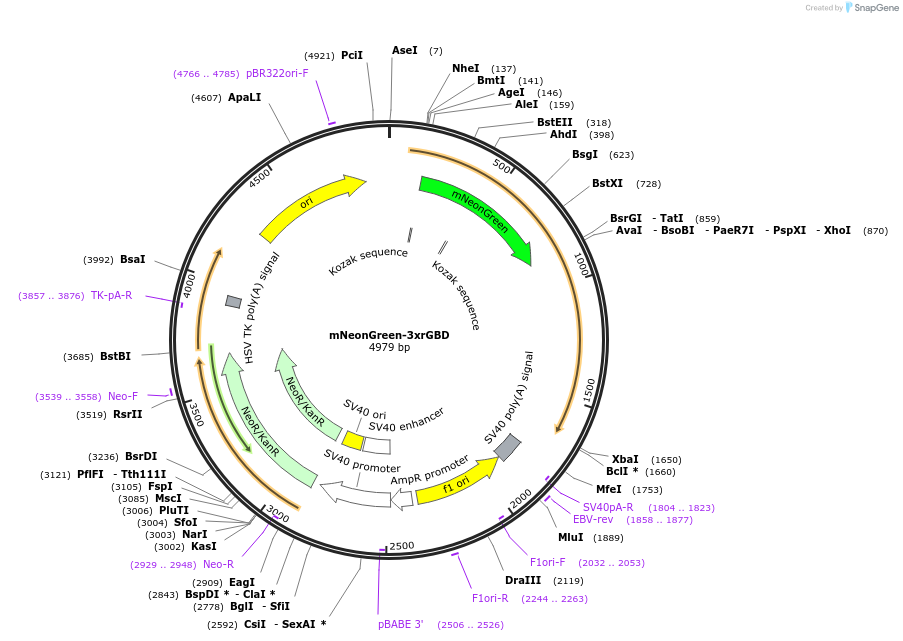
mNeonGreen-3xrGBD
Plasmid#176091PurposeVisualization of active RhoDepositorAvailable SinceOct. 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
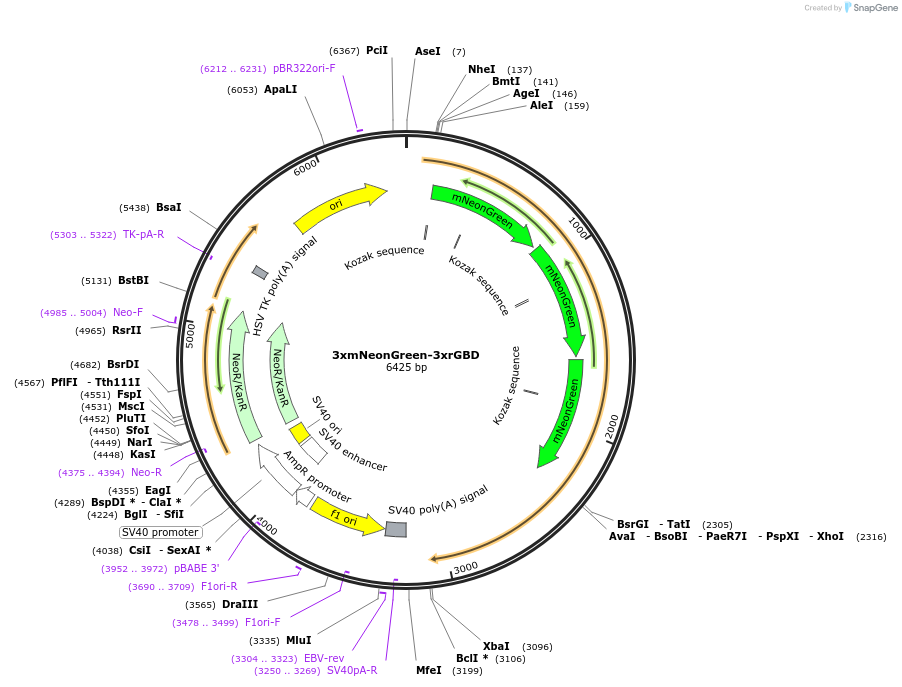
3xmNeonGreen-3xrGBD
Plasmid#176101PurposeVisualization of active RhoDepositorAvailable SinceOct. 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
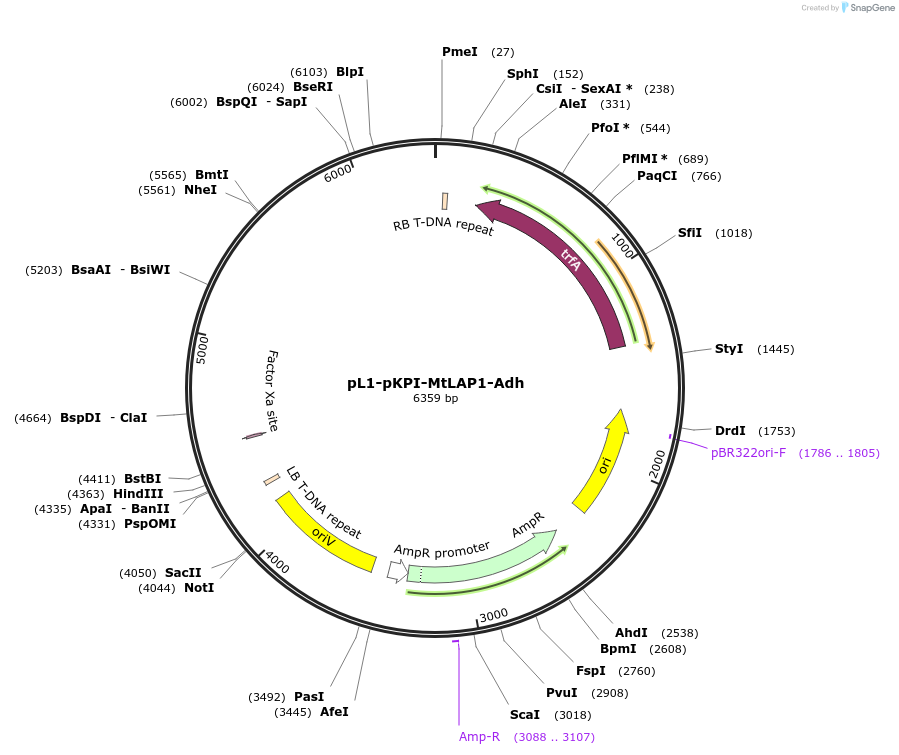
pL1-pKPI-MtLAP1-Adh
Plasmid#193520PurposeAnthocyanin pigmentation based visual marker for arbuscular mycorrhizal fungal colonization of Medicago truncatula rootsDepositorInsertMtLAP1
ExpressionPlantPromoterMedtr8g059790(KPI)Available SinceJune 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
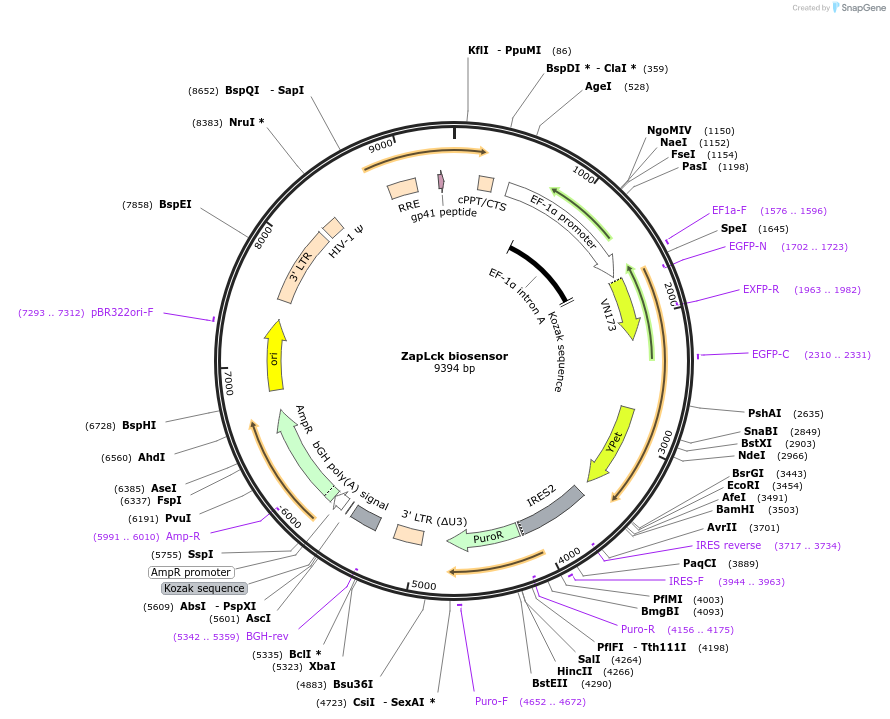
ZapLck biosensor
Plasmid#131584PurposeLck FRET Biosensor. To visualize Lck kinase activity in mammalian cells by FRETDepositorInsertZapLck biosensor
UseLentiviralAvailable SinceSept. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
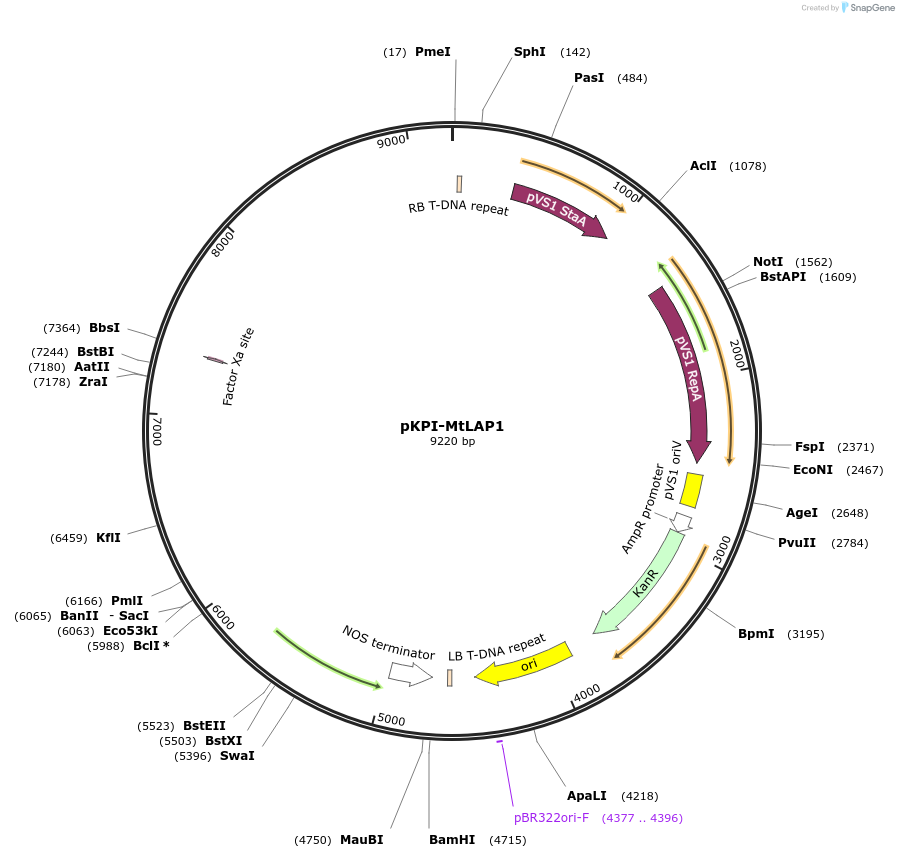
pKPI-MtLAP1
Plasmid#193521PurposeAnthocyanin pigmentation based visual marker for arbuscular mycorrhizal fungal colonization of Medicago truncatula rootsDepositorInsertsMtLAP1
DsRed
ExpressionPlantPromoterATUBI10 and Medtr8g059790(KPI)Available SinceJune 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
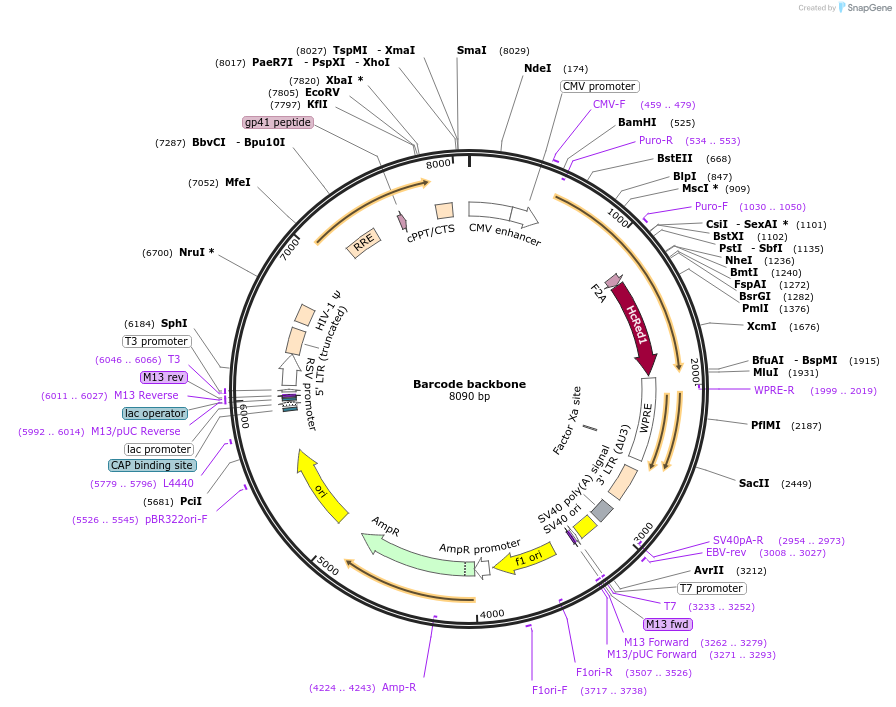
Barcode backbone
Plasmid#158665PurposeThis plasmid serves as the backbone for the visual barcodes described in our paper. It has a CMV-Puro-F2A backbone to which we insert a fluorescent protein cellular localization combinationDepositorTypeEmpty backboneUseLentiviralTagshcREDExpressionMammalianAvailable SinceMay 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
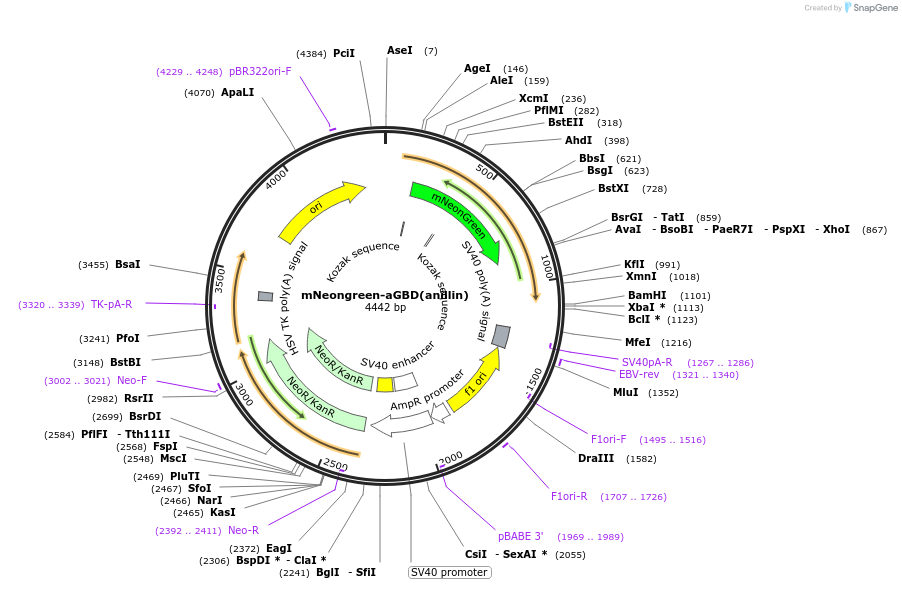
mNeongreen-aGBD(anillin)
Plasmid#129633PurposeVisualization of active RhoADepositorInsertRhoA binding domain of anillin (712-786)
ExpressionMammalianPromoterCMVdelAvailable SinceFeb. 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
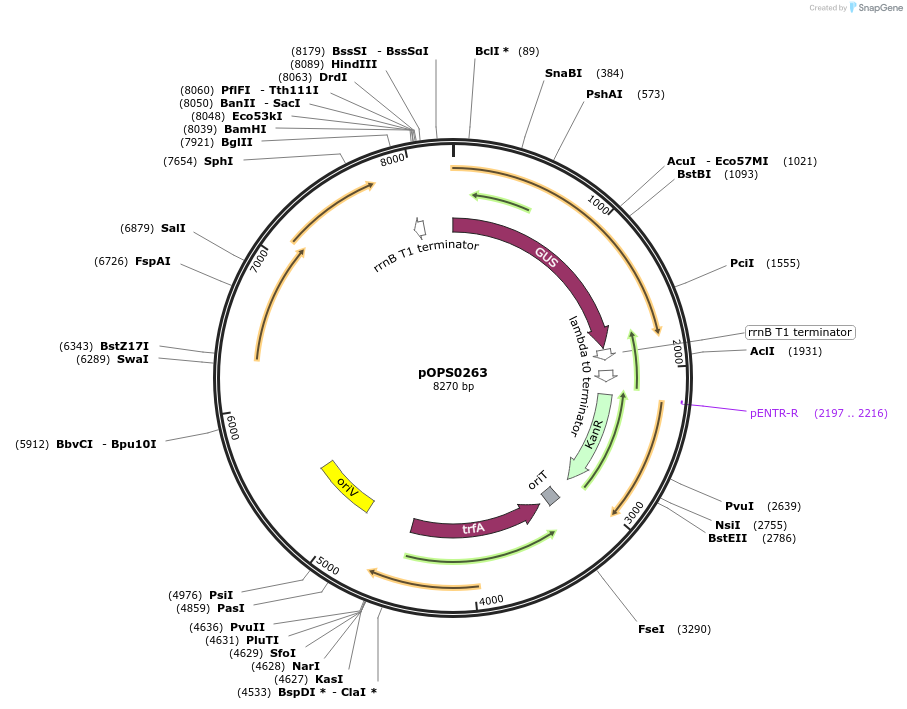
pOPS0263
Plasmid#133228PurposeReporter plasmid for R. leguminosarum suitable for experiments in environments where there is no antibiotic selection present.DepositorInsertconstructed with PsNifH (pOGG043), gusA (pOGG083) and T-pharma (pOGG003)
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceApril 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
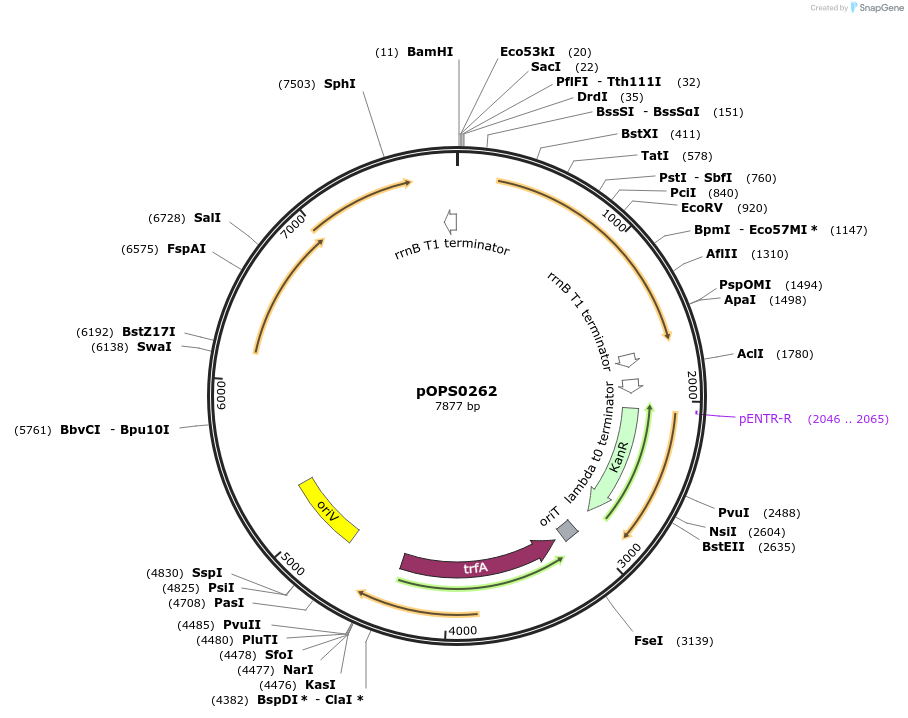
pOPS0262
Plasmid#133227PurposeReporter plasmid for R. leguminosarum suitable for experiments in environments where there is no antibiotic selection present.DepositorInsertconstructed with PsNifH (pOGG043), celB (pOGG050) and T-pharma (pOGG003)
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceApril 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
SEPTOPO
Plasmid#64944Purposesource of SEP (Super Ecliptic pHluorin) sequenceDepositorInsertSEP
ExpressionBacterialPromoterlacAvailable SinceJune 11, 2015AvailabilityAcademic Institutions and Nonprofits only -
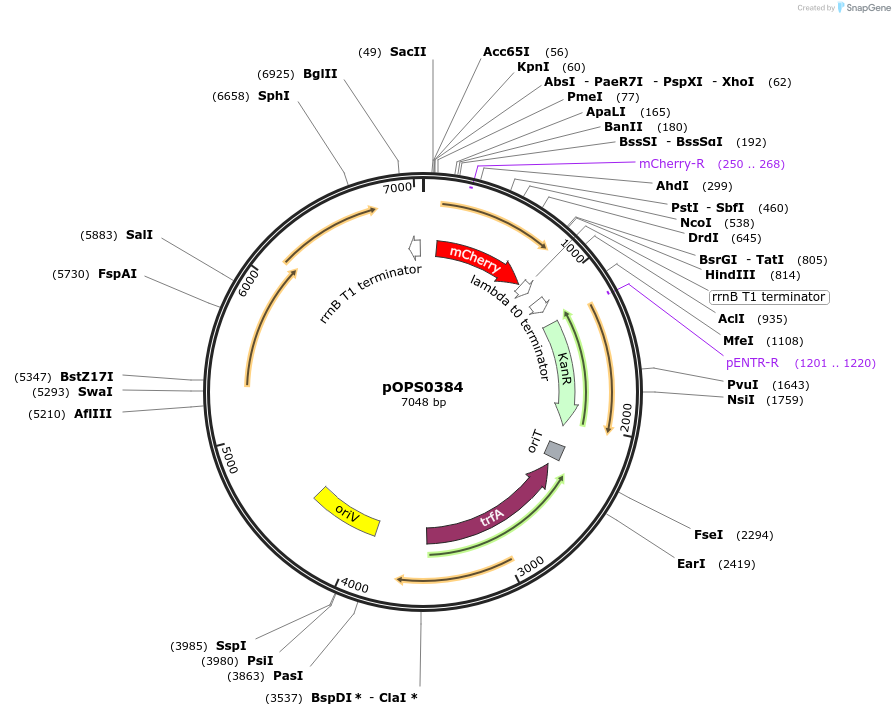
pOPS0384
Plasmid#133234PurposeReporter plasmid for R. leguminosarum suitable for experiments in environments where there is no antibiotic selection present.DepositorInsertconstructed with NoP (pOGG113), mCherry (EC15071) and T-pharma (pOGG003)
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceApril 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
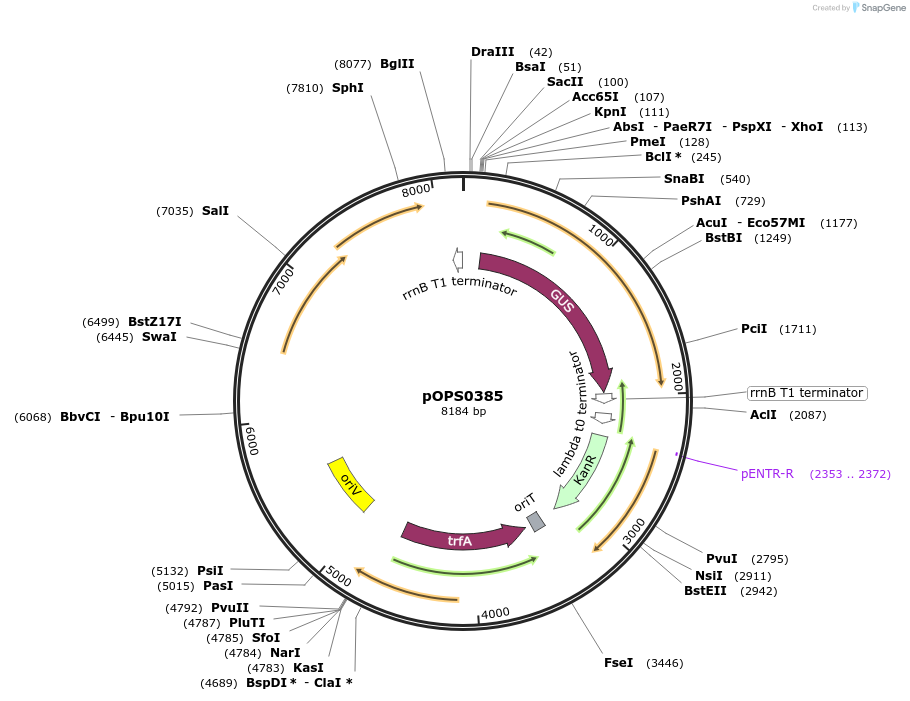
pOPS0385
Plasmid#133235PurposeReporter plasmid for R. leguminosarum suitable for experiments in environments where there is no antibiotic selection present.DepositorInsertconstructed with NoP (pOGG113), gusA (pOGG083) and T-pharma (pOGG003)
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceApril 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
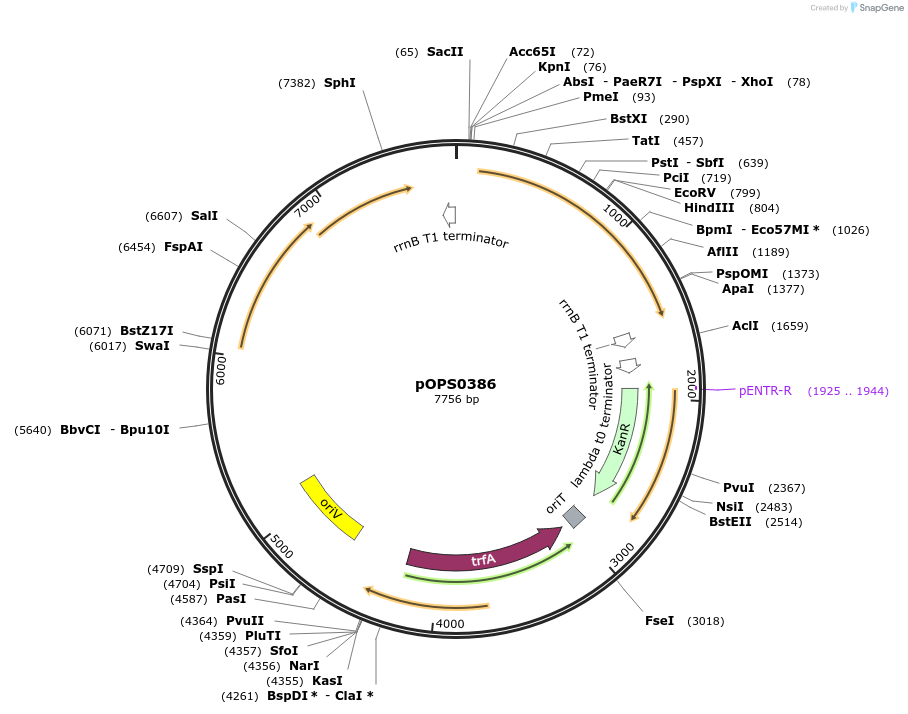
pOPS0386
Plasmid#133236PurposeReporter plasmid for R. leguminosarum suitable for experiments in environments where there is no antibiotic selection present.DepositorInsertconstructed with NoP (pOGG113), celB (pOGG050) and T-pharma (pOGG003)
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceApril 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
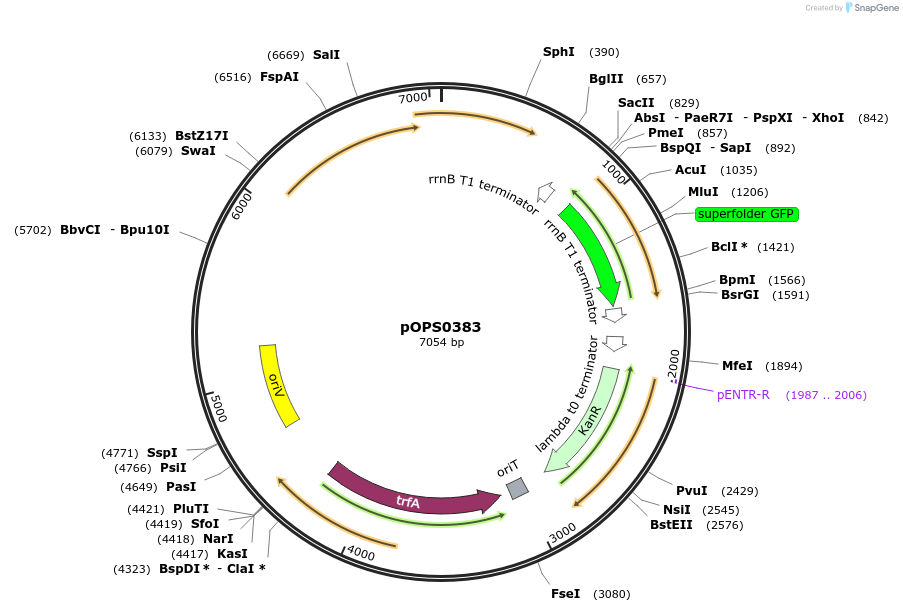
pOPS0383
Plasmid#133233PurposeReporter plasmid for R. leguminosarum suitable for experiments in environments where there is no antibiotic selection present.DepositorInsertconstructed with NoP (pOGG113), sfGFP (pOGG037) and T-pharma (pOGG003)
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceApril 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
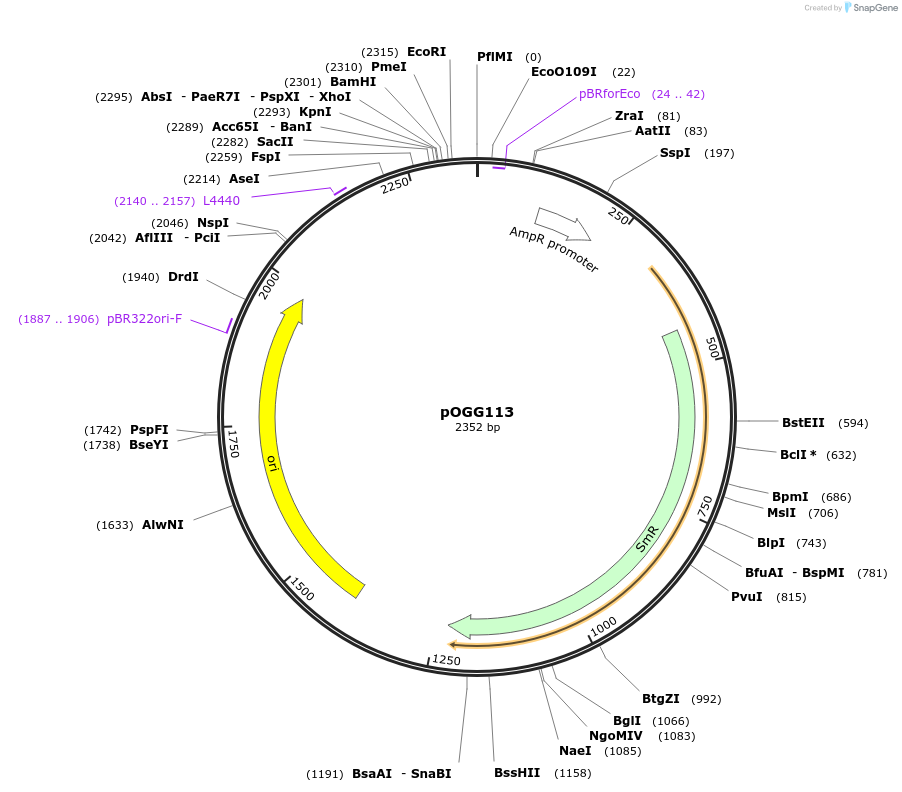
pOGG113
Plasmid#133124PurposepL0M-PU-NoP, promotorless region used as negative controlDepositorInsertNoP promotorless region from plasmid pIJJ11268
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
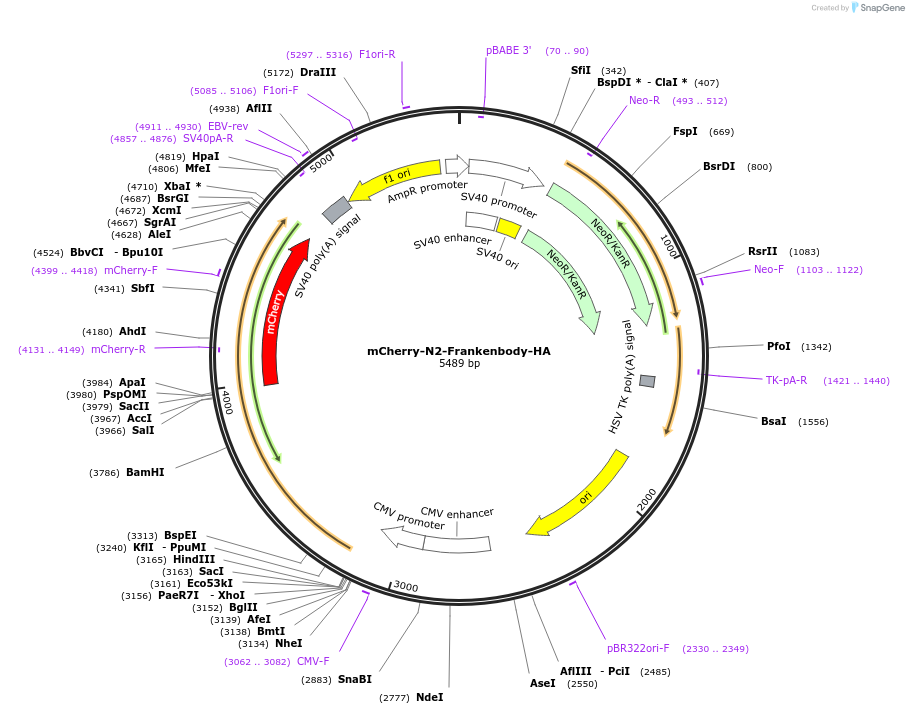
mCherry-N2-Frankenbody-HA
Plasmid#175292PurposeVisualize endogeneous loci coupled with ZF probes harboring 10xHA tagDepositorInsertanti-HA frankenbody fused with mCherry
TagsmCherryExpressionMammalianAvailable SinceOct. 7, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
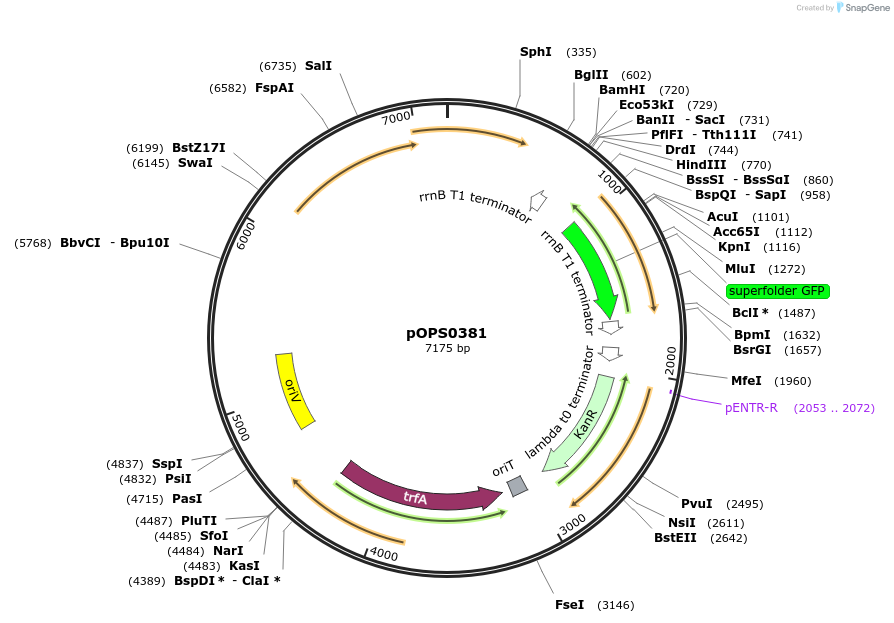
pOPS0381
Plasmid#133231PurposeReporter plasmid for R. leguminosarum suitable for experiments in environments where there is no antibiotic selection present.DepositorInsertconstructed with constitutive promoter PsNifH (pOGG043), sfGFP (pOGG037) and T-pharma (pOGG003)
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceApril 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
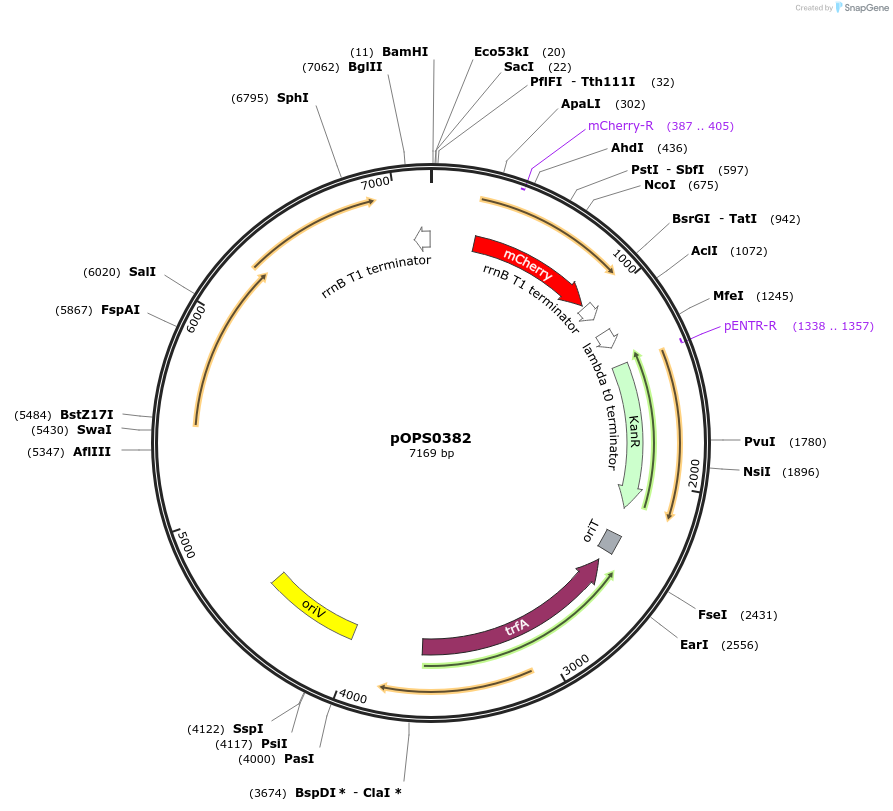
pOPS0382
Plasmid#133232PurposeReporter plasmid for R. leguminosarum suitable for experiments in environments where there is no antibiotic selection present.DepositorInsertconstructed with constitutive promoter PsNifH (pOGG043), mCherry (EC15071) and T-pharma (pOGG003)
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceApril 8, 2020AvailabilityAcademic Institutions and Nonprofits only