We narrowed to 13,079 results for: ache
-
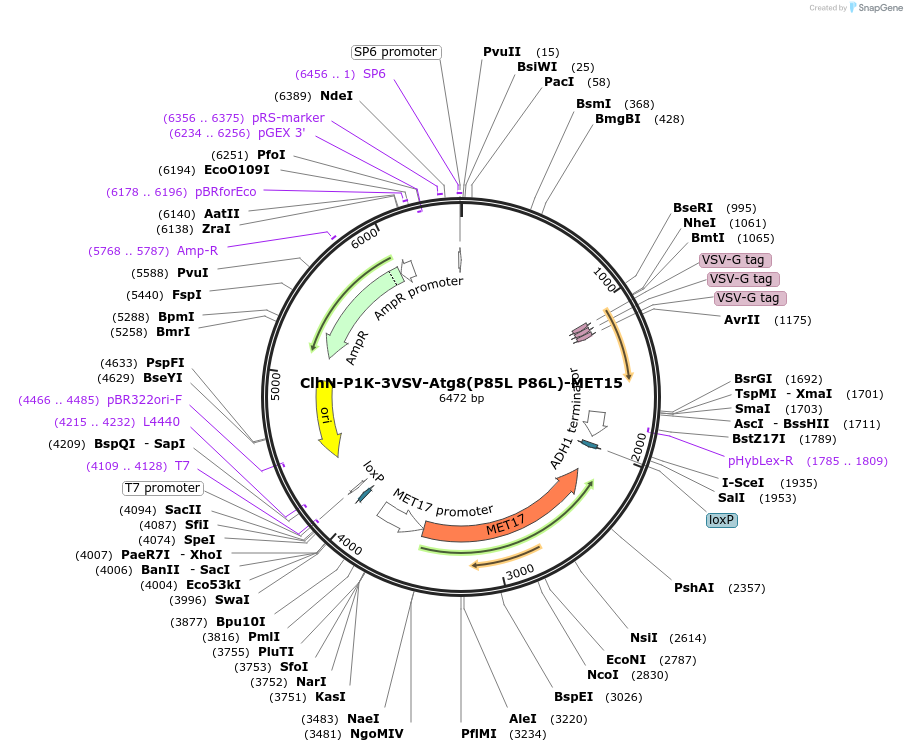
Plasmid#207050PurposeExpression of 3VSV-Atg8 point mutant. Uses auxotrophic marker MET15(Saccharomyces cerevisiae).DepositorInsertATG8
Tags3xVSVExpressionYeastMutationP85L P86LAvailable SinceMarch 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
ClhN-P1K-3VSV-Atg8(T56A R65A T87A)-MET15
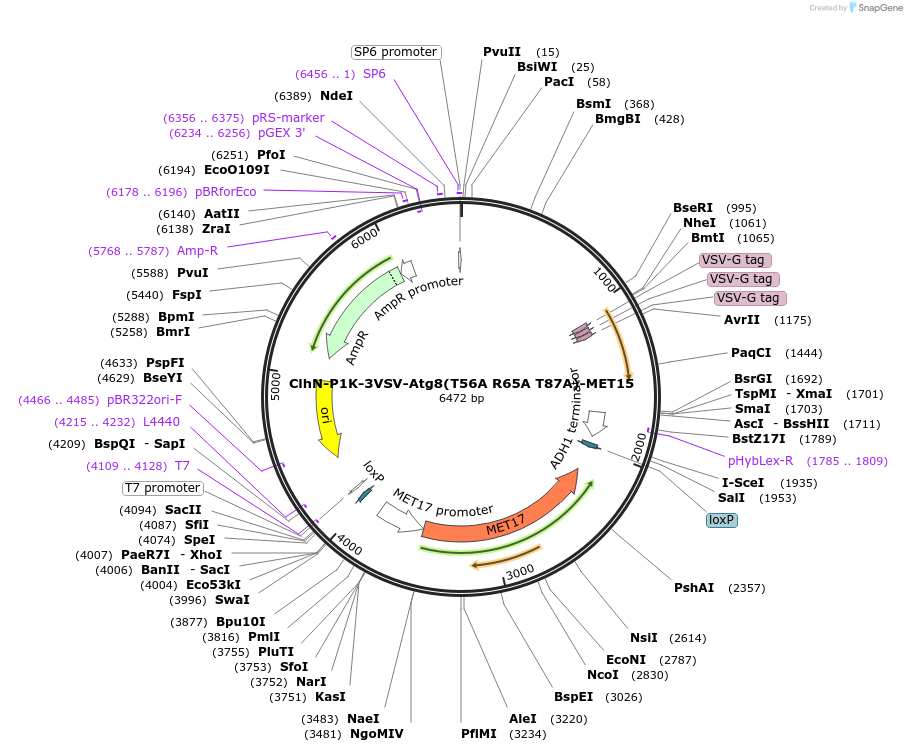
Plasmid#207051PurposeExpression of 3VSV-Atg8 point mutant. Uses auxotrophic marker MET15(Saccharomyces cerevisiae).DepositorInsertATG8
Tags3xVSVExpressionYeastMutationT56A R65A T87AAvailable SinceMarch 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
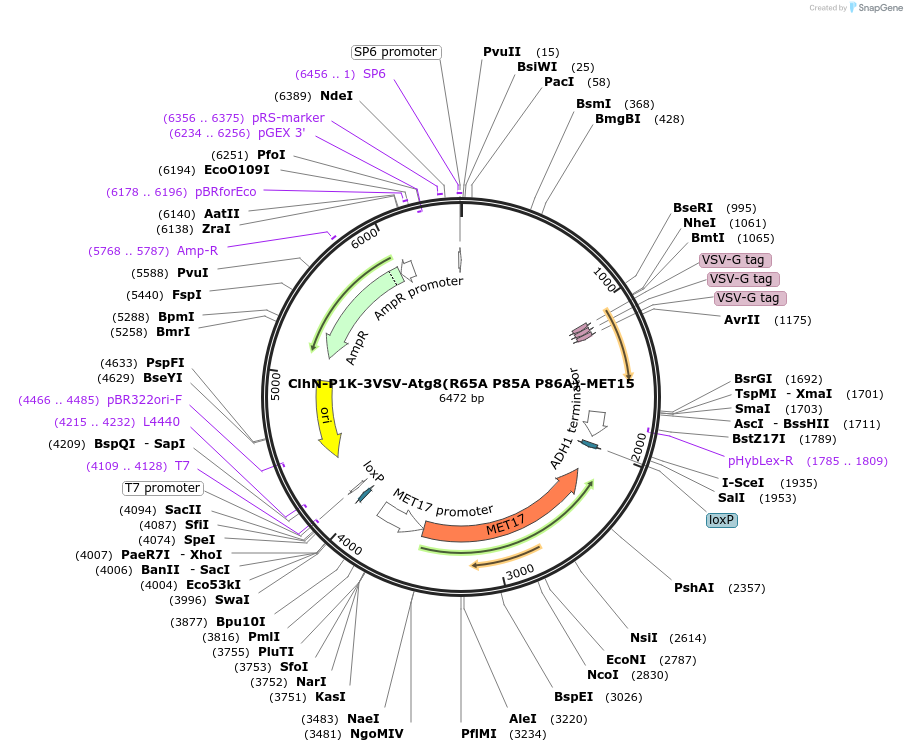
ClhN-P1K-3VSV-Atg8(R65A P85A P86A)-MET15
Plasmid#207052PurposeExpression of 3VSV-Atg8 point mutant. Uses auxotrophic marker MET15(Saccharomyces cerevisiae).DepositorInsertATG8
Tags3xVSVExpressionYeastMutationR65A P85A P86AAvailable SinceMarch 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
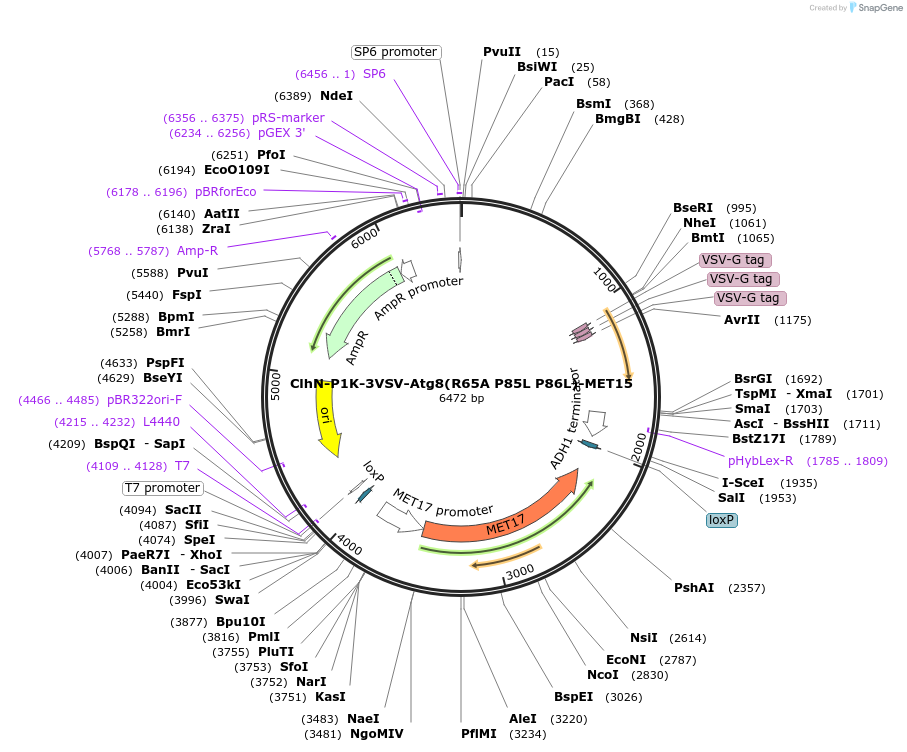
ClhN-P1K-3VSV-Atg8(R65A P85L P86L)-MET15
Plasmid#207053PurposeExpression of 3VSV-Atg8 point mutant. Uses auxotrophic marker MET15(Saccharomyces cerevisiae).DepositorInsertATG8
Tags3xVSVExpressionYeastMutationR65A P85L P86LAvailable SinceMarch 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
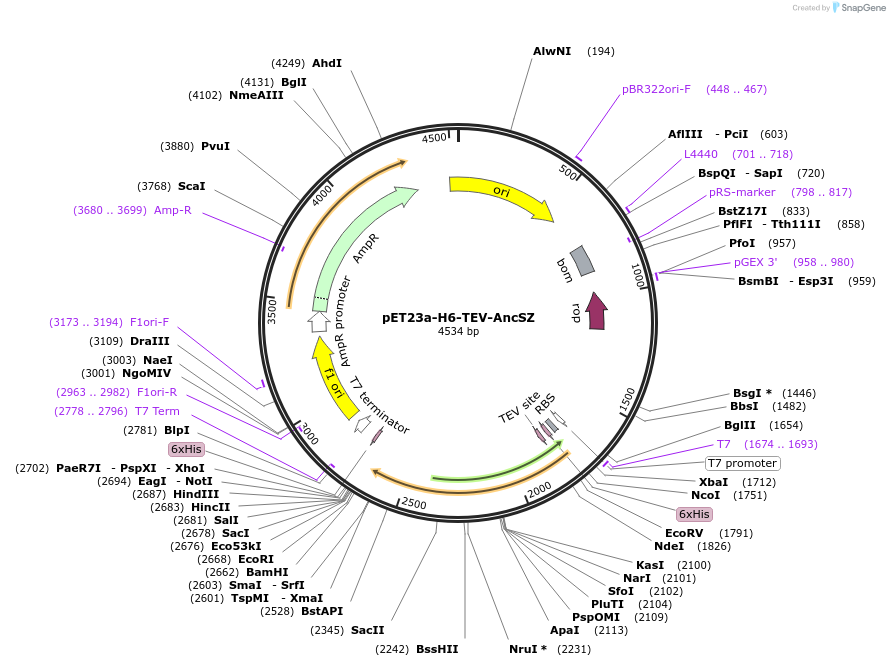
pET23a-H6-TEV-AncSZ
Plasmid#214235PurposeBacterial expression construct of AncSZ, an kinase domain engineered through ancestorial reconstruction of the kinase domains of both SYK and ZAP-70. For in vitro protein tyrosine phosphorylationDepositorInsertSyk kinase (SYK Human)
Tags6xHisExpressionBacterialMutationSequence gained through ancestral reconstruction …PromoterT7Available SinceFeb. 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
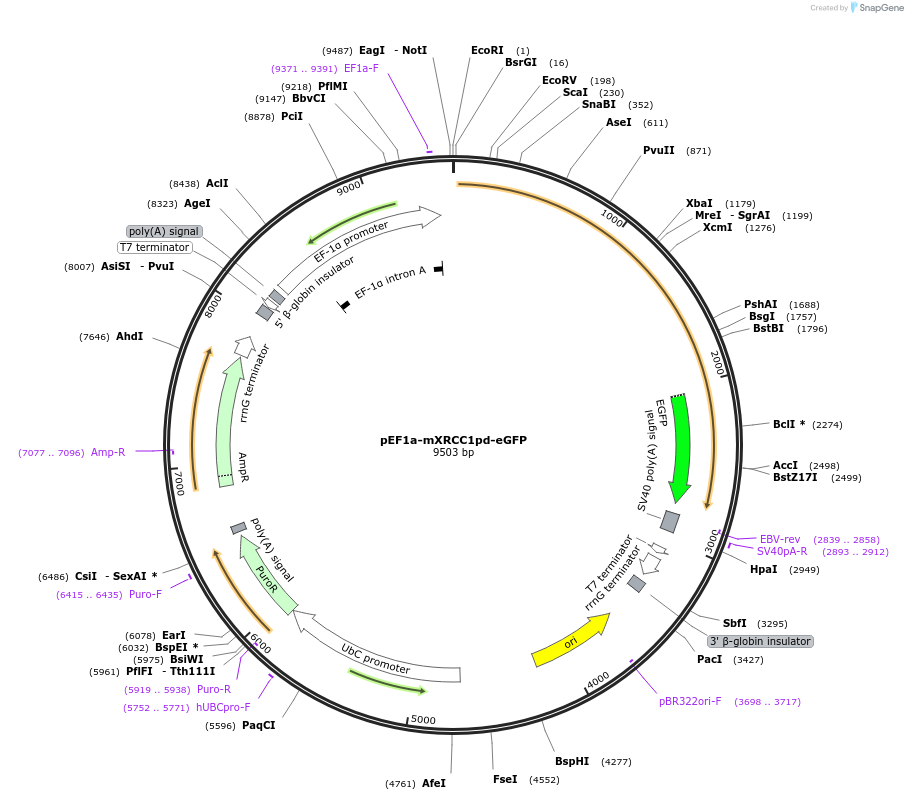
pEF1a-mXRCC1pd-eGFP
Plasmid#206035PurposeMammalian expression of PAR-deficient mXRCC1pd coupled to eGFP under the control of a EF1a promoterDepositorInsertmXRCC1pd
TagseGFPExpressionMammalianMutationS105A, S186A, K188A, S195A, S221A, S222A,S238A, S…PromoterEf1aAvailable SinceJan. 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
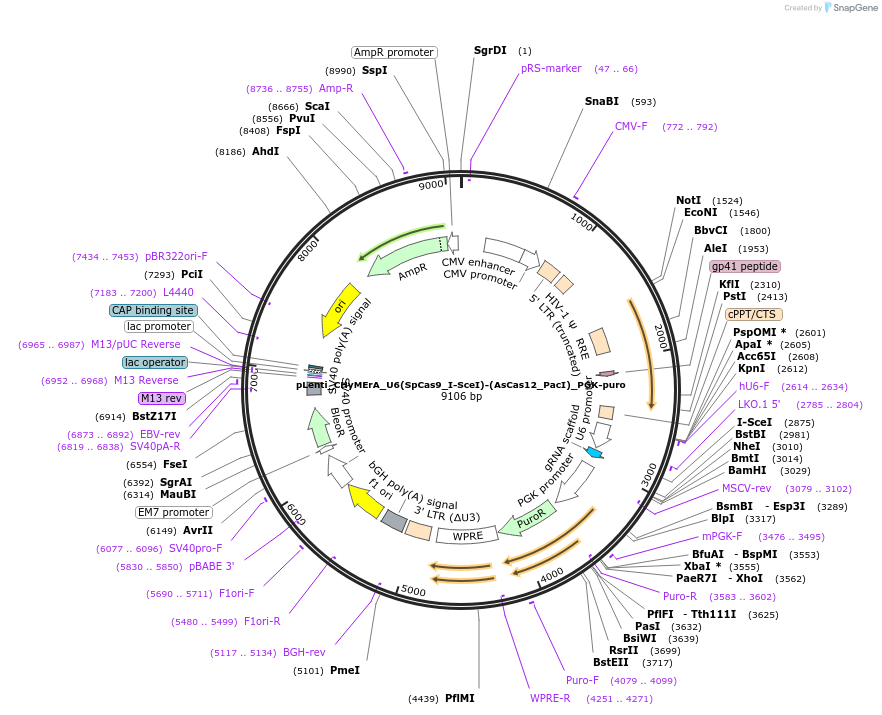
pLenti_CHyMErA_U6(SpCas9_I-SceI)-(AsCas12_PacI)_PGK-puro
Plasmid#189634PurposeLentiviral expression of single SpCas9 and AsCas12a gRNAs for generating combinatorial CHyMErA 3Cs librariesDepositorInserthU6 Cas9-Cas12a gRNA cassette, PGK puro cassette
UseCRISPR and LentiviralExpressionMammalianAvailable SinceNov. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
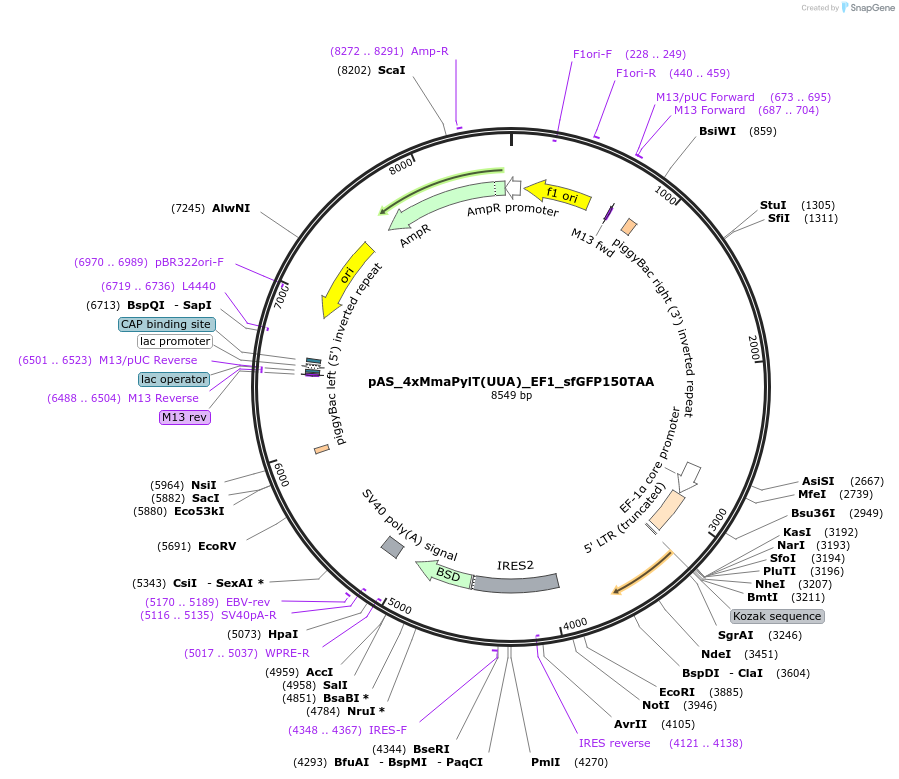
pAS_4xMmaPylT(UUA)_EF1_sfGFP150TAA
Plasmid#174897Purposeochre suppression reporter sfGFP150TAA expression, with MmaPylT(UUA) ochre suppressor tRNA cassetteDepositorInsertsf GFP
ExpressionMammalianMutation150TAA in sfGFPPromoterEF1Available SinceNov. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
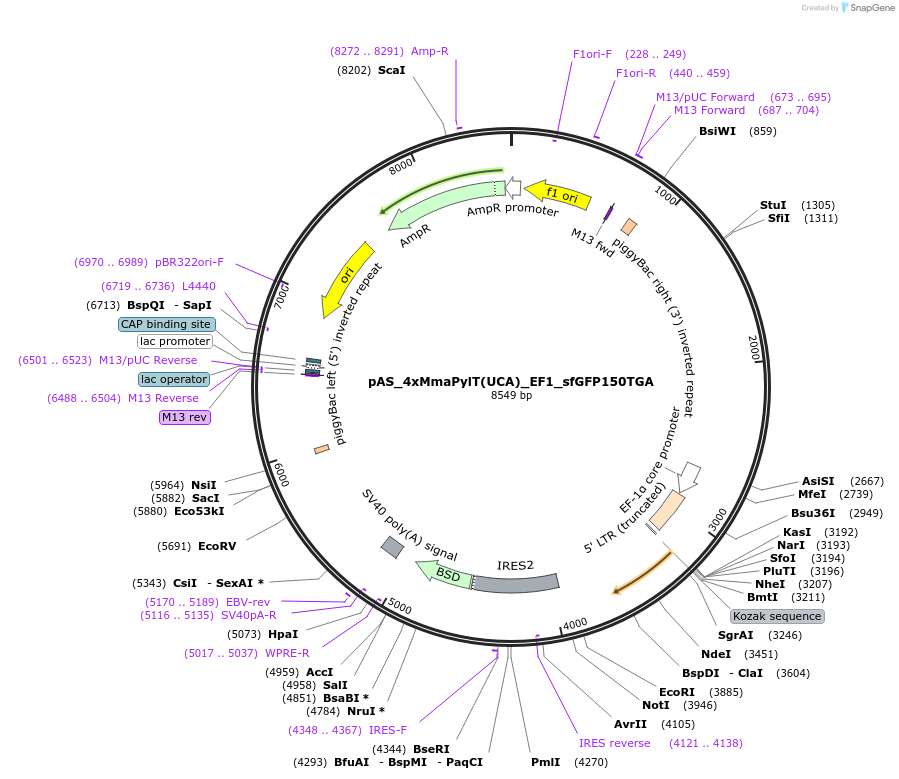
pAS_4xMmaPylT(UCA)_EF1_sfGFP150TGA
Plasmid#174898Purposeopal suppression reporter sfGFP150TGA expression, with MmaPylT(UCA) opal suppressor tRNA cassetteDepositorInsertsfGFP
ExpressionMammalianMutation150TGA in sfGFPPromoterEF1Available SinceNov. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
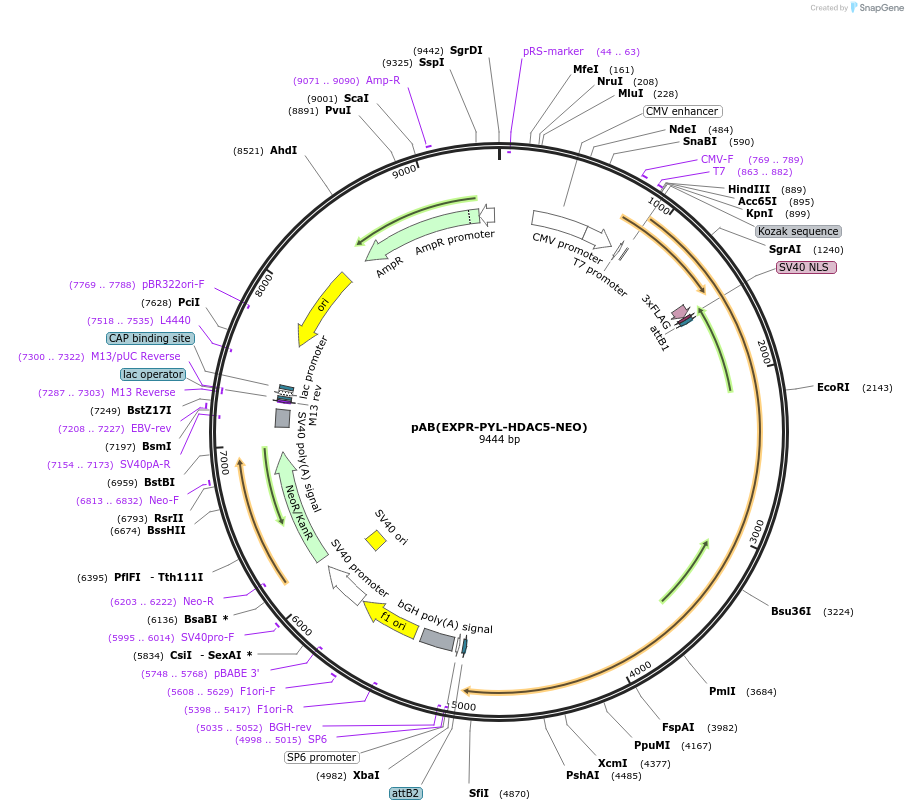
pAB(EXPR-PYL-HDAC5-NEO)
Plasmid#114395PurposeFor PYL-HDAC5 expressionDepositorInsertPYL-HDAC5
Tags3xFlag-NLS (internal)ExpressionMammalianMutationD593E compared with NCBI reference NP_005465.2PromoterCMVAvailable SinceAug. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
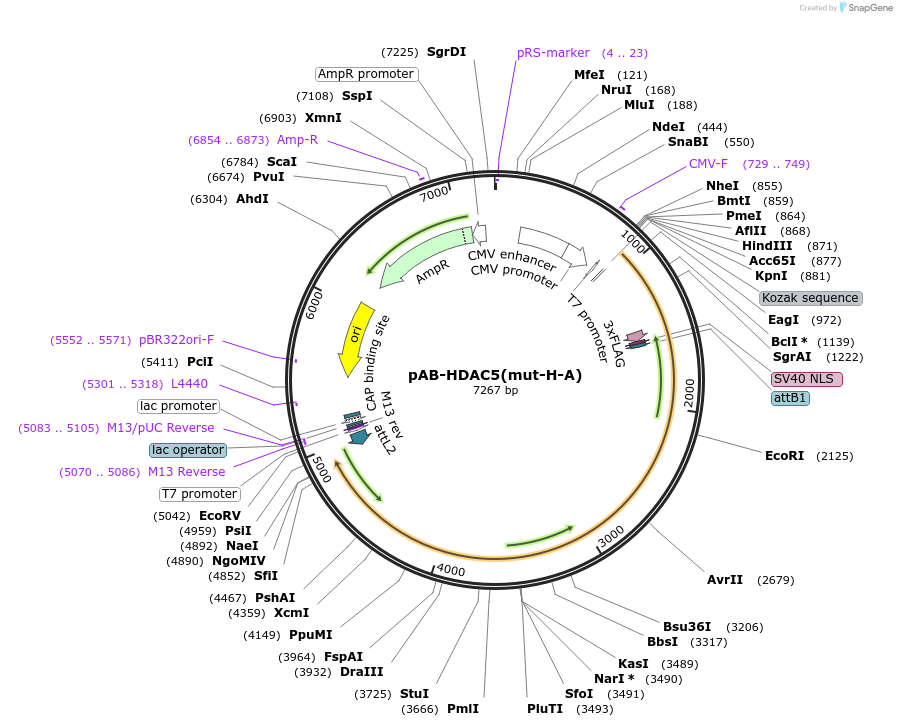
pAB-HDAC5(mut-H-A)
Plasmid#114397PurposeFor PYL-HDAC5 H1006A expressionDepositorInsertPYL-HDAC5 H1006A
Tags3xFlag-NLS (internal)ExpressionMammalianMutationD593E compared to NCBI ref NP_005465.2PromoterCMVAvailable SinceAug. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
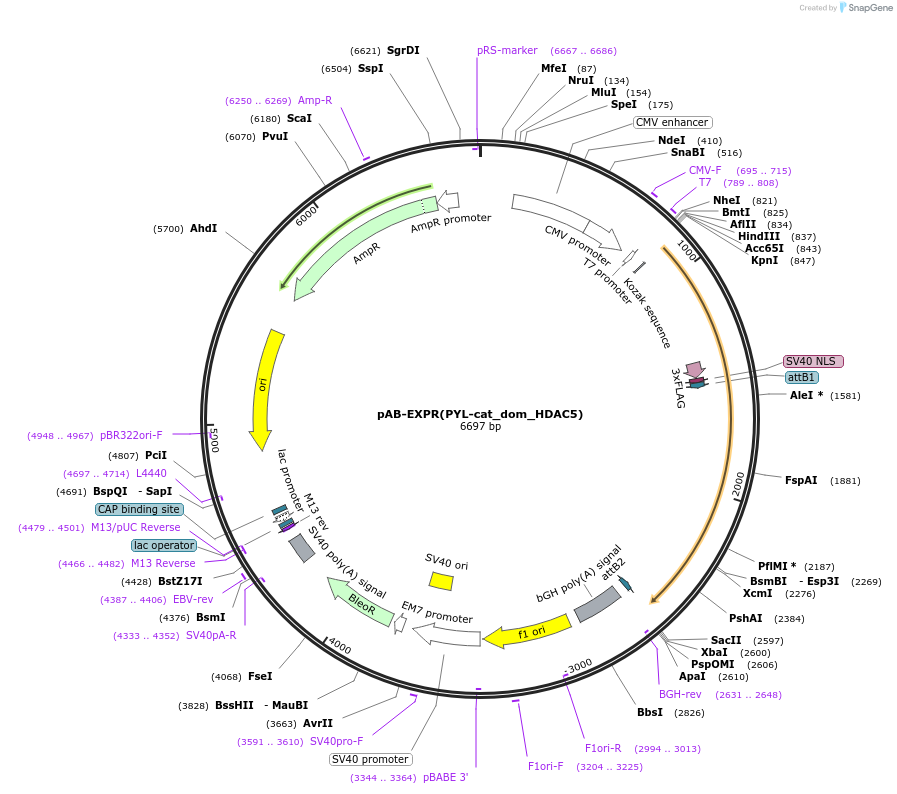
pAB-EXPR(PYL-cat_dom_HDAC5)
Plasmid#114401PurposeFor PYL-HDAC5 catalytic domain expressionDepositorInsertPYL-HDAC5 catalytic domain
Tags3xFlag-NLS (internal)ExpressionMammalianMutationS220F and the deletion of M221PromoterCMVAvailable SinceAug. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAB-EXPR(PYL-Nterm_dom_HDAC5)
Plasmid#114402PurposeFor PYL-HDAC5 N terminal domain expressionDepositorInsertPYL-HDAC5 N terminal domain
Tags3xFlag-NLS (internal)ExpressionMammalianMutationD593E compared to NCBI ref NP_005465.2PromoterCMVAvailable SinceAug. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
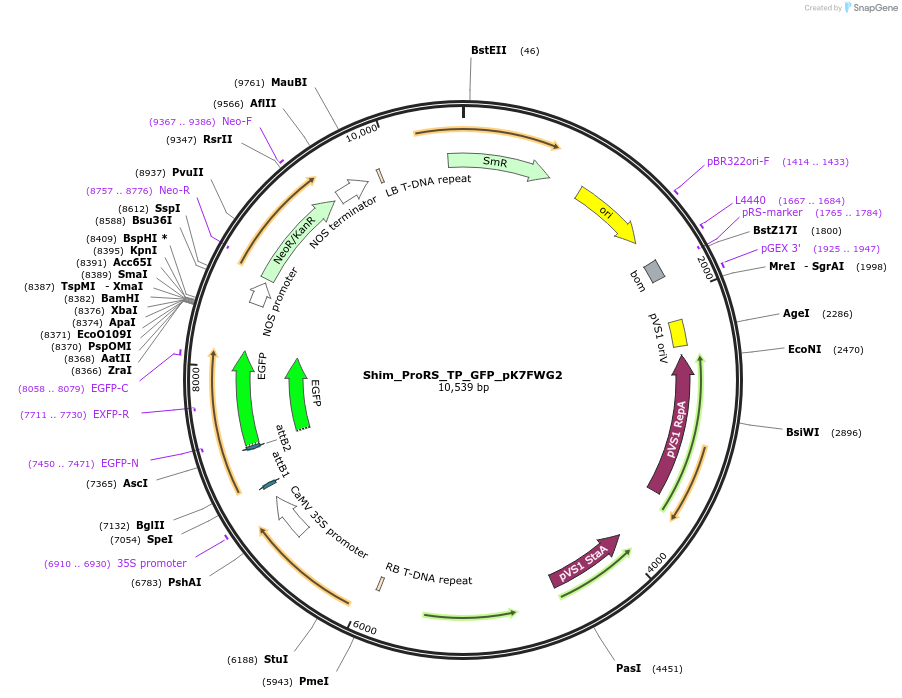
Shim_ProRS_TP_GFP_pK7FWG2
Plasmid#202651PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from putative cytosolic ProRS
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
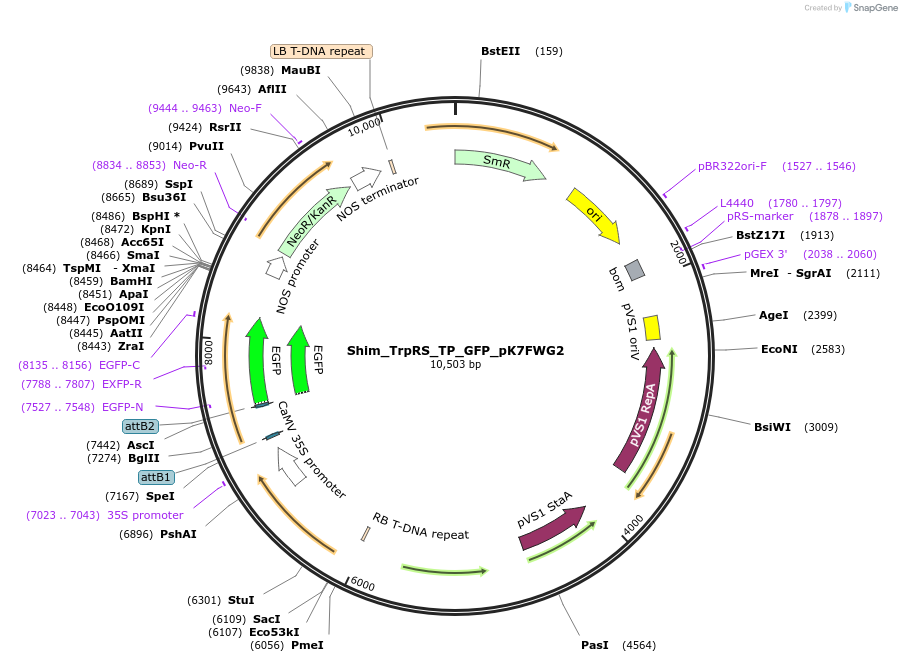
Shim_ThrRS_TP_GFP_pK7FWG2
Plasmid#202652PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from putative cytosolic ThrRS
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
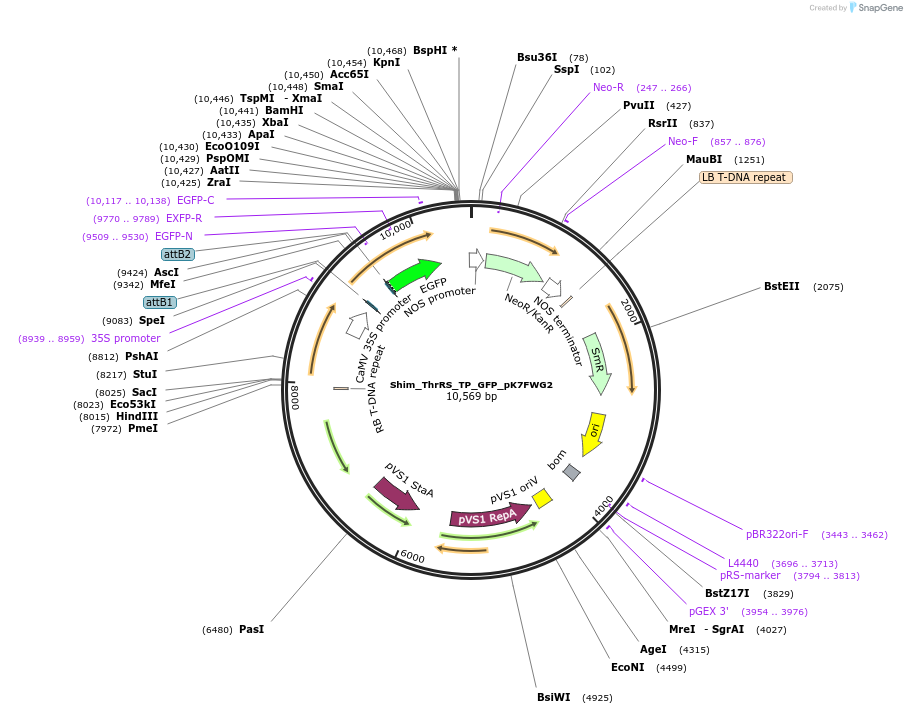
Shim_TrpRS_TP_GFP_pK7FWG2
Plasmid#202653PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from putative cytosolic TrpRS
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
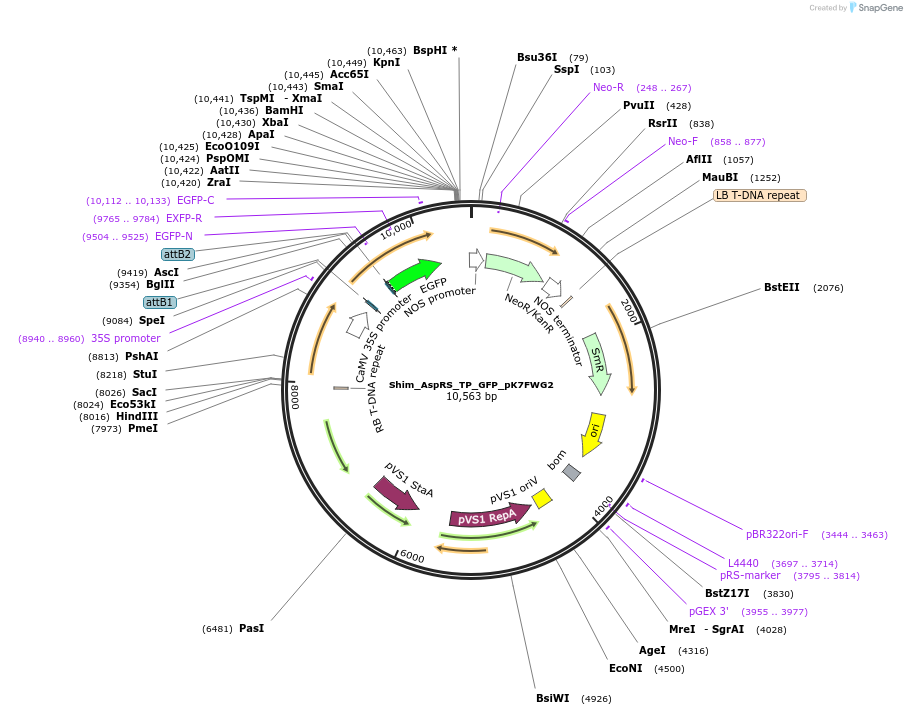
Shim_AspRS_TP_GFP_pK7FWG2
Plasmid#202647PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from putative cytosolic AspRS
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
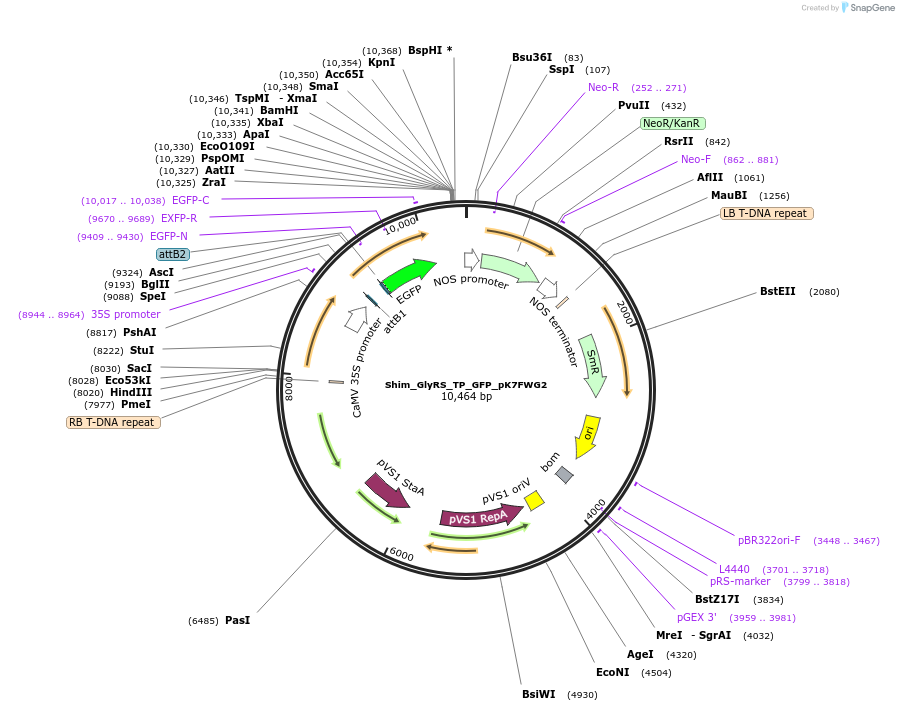
Shim_GlyRS_TP_GFP_pK7FWG2
Plasmid#202648PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from putative cytosolic GlyRS
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
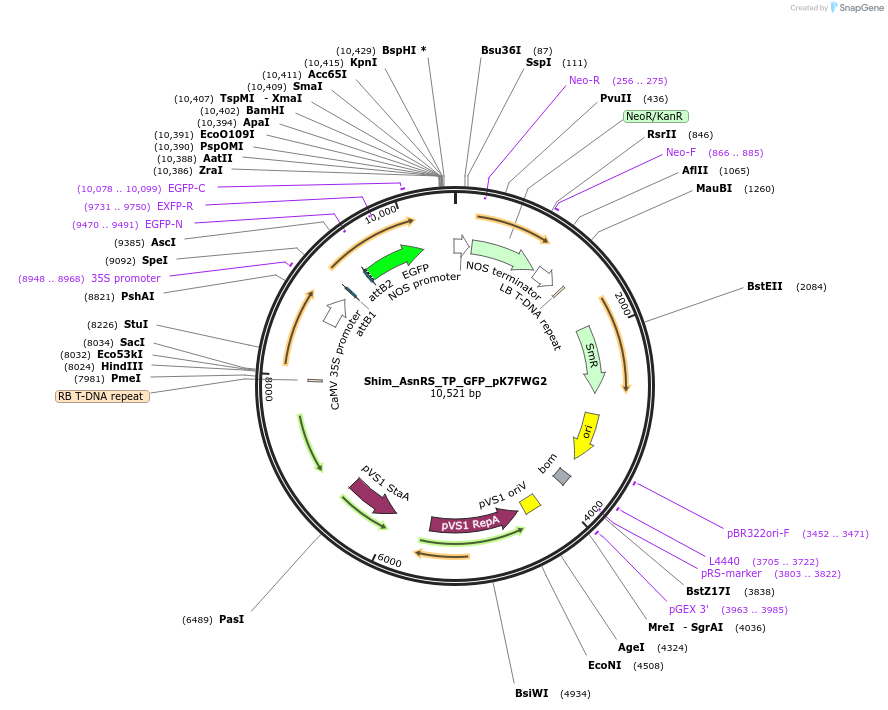
Shim_AsnRS_TP_GFP_pK7FWG2
Plasmid#202649PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from putative cytosolic AsnRS
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
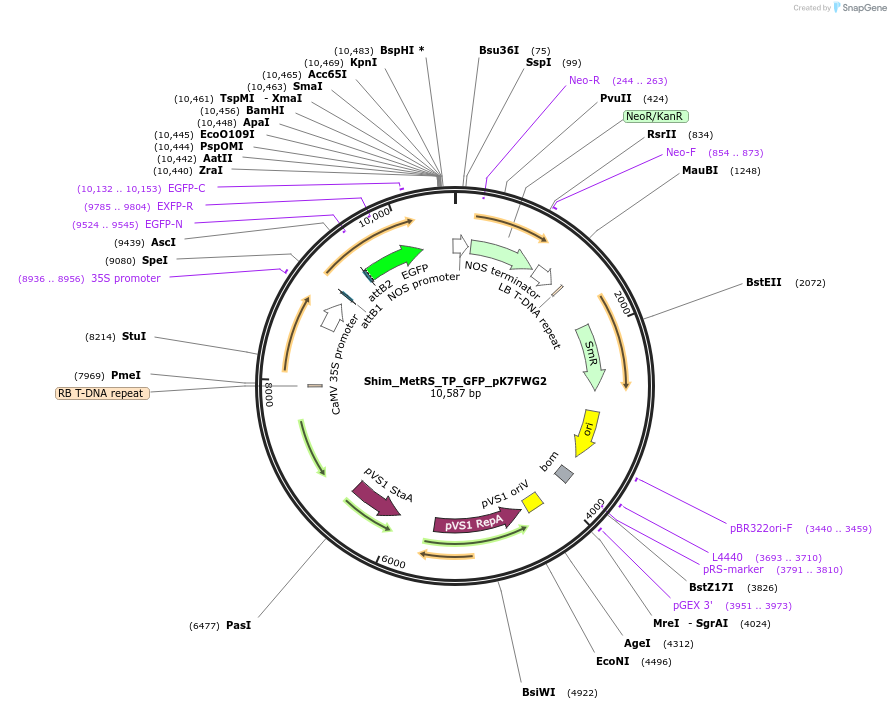
Shim_MetRS_TP_GFP_pK7FWG2
Plasmid#202650PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from putative cytosolic MetRS
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits