We narrowed to 13,849 results for: 109
-
-
-
-
-
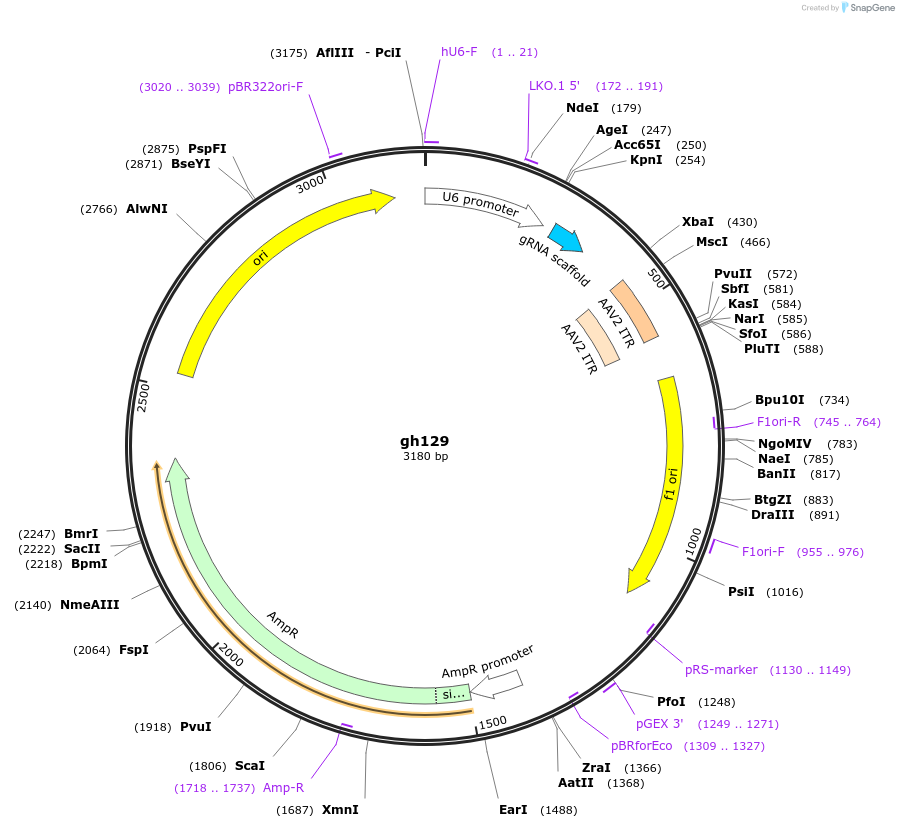
gh129
Plasmid#106799Purposeexpression of gRNA targeting CSGALNACT1DepositorInsertCSGALNACT1 (CSGALNACT1 Human)
ExpressionMammalianAvailable SinceNov. 9, 2018AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
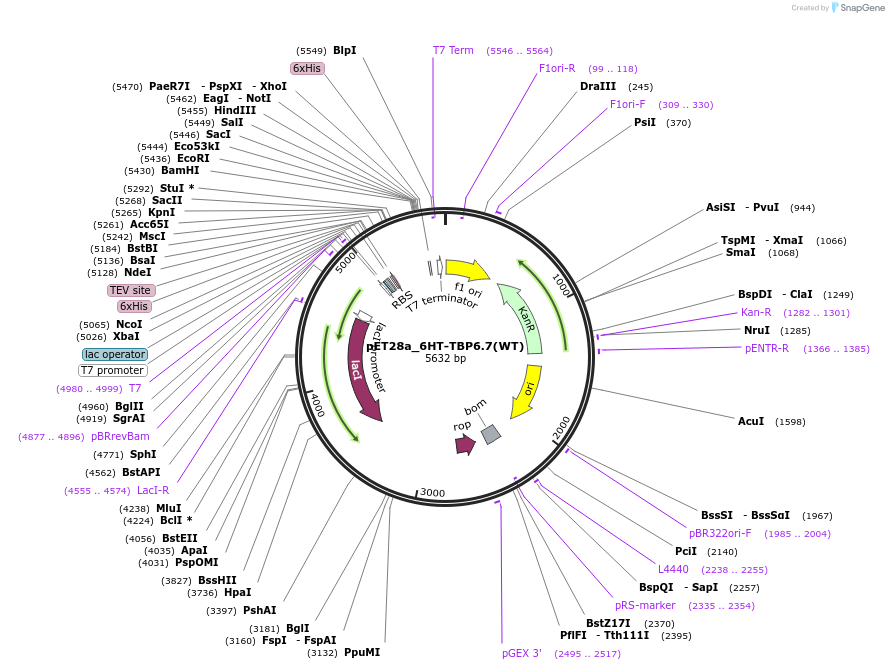
pET28a_6HT-TBP6.7(WT)
Plasmid#112720PurposeWild-type TBP6.7 was one of the first evolved TAR-binding proteins to bind with exceptional affinity. It was this protein that was used to obtain co-crystals with WT TAR RNA.DepositorInsert6His-TEV-TBP6.7(WT)
Tags6His-TEVExpressionBacterialPromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
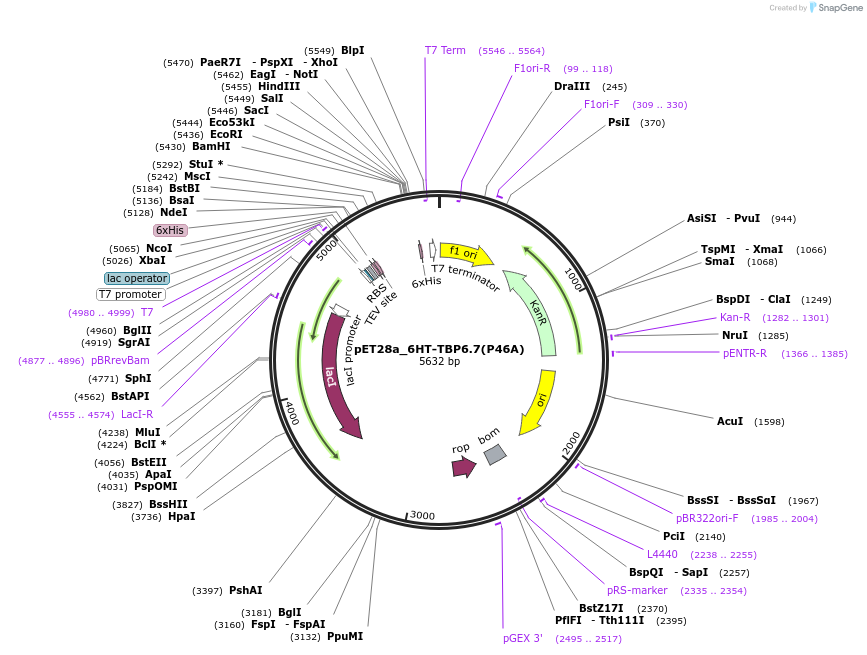
pET28a_6HT-TBP6.7(P46A)
Plasmid#112721PurposeThis mutation (P46A) marks the first residue within the evolved region of U1A.DepositorInsert6His-TEV-TBP6.7(P46A)
Tags6His-TEVExpressionBacterialMutationP46APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
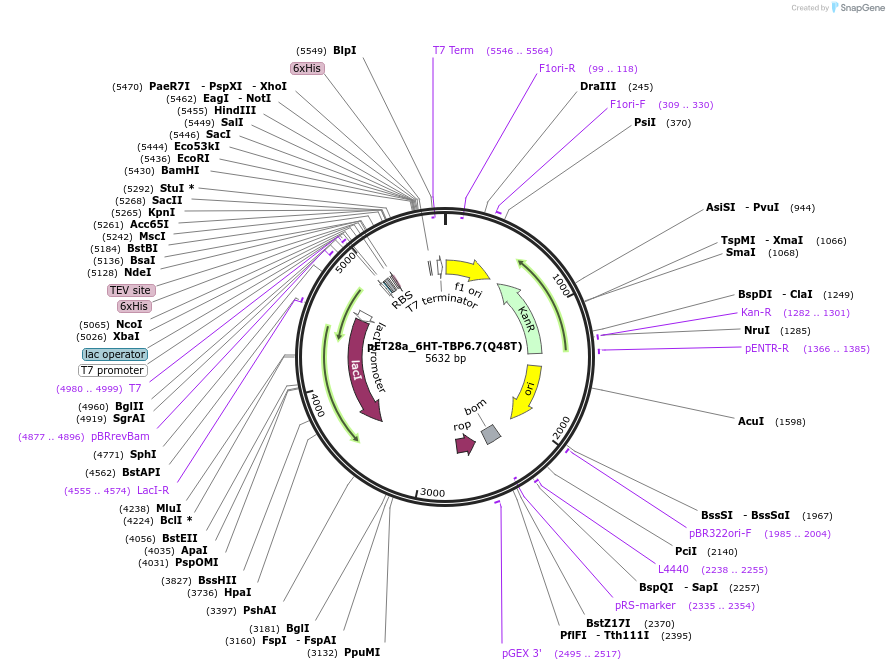
pET28a_6HT-TBP6.7(Q48T)
Plasmid#112725PurposeThis mutant of TBP6.7 binds to TAR very similarly to TBP6.7(WT), with only 1.4-fold reduced Kd.DepositorInsert6His-TEV-TBP6.7(Q48T)
Tags6His-TEVExpressionBacterialMutationQ48TPromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
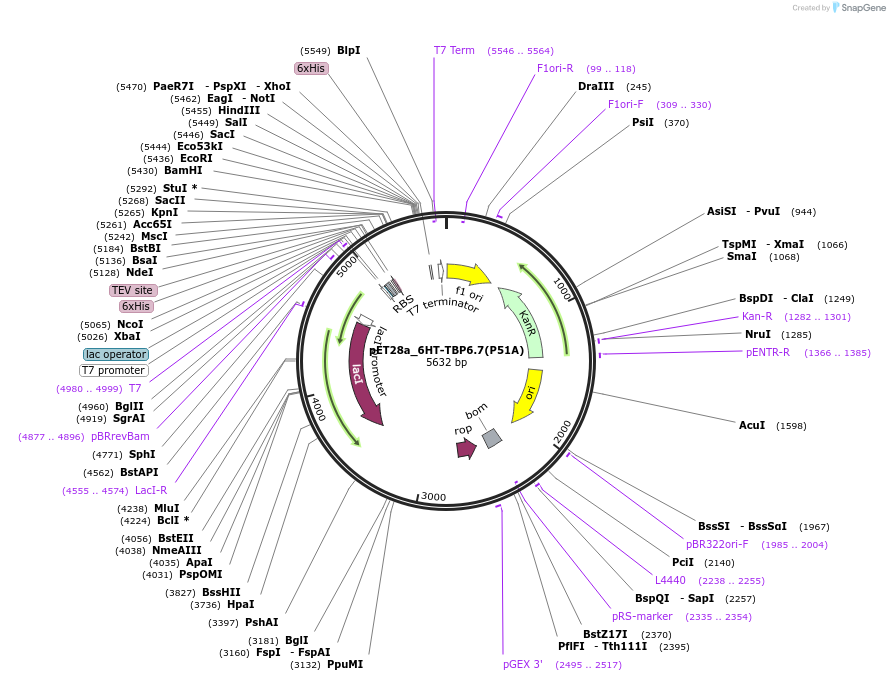
pET28a_6HT-TBP6.7(P51A)
Plasmid#112728PurposeSimilar to P46, P51 was selected and is present in all evolved TBPs. Mutating this position into Ala has a modest effect on binding.DepositorInsert6His-TEV-TBP6.7(P51A)
Tags6His-TEVExpressionBacterialMutationP51APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
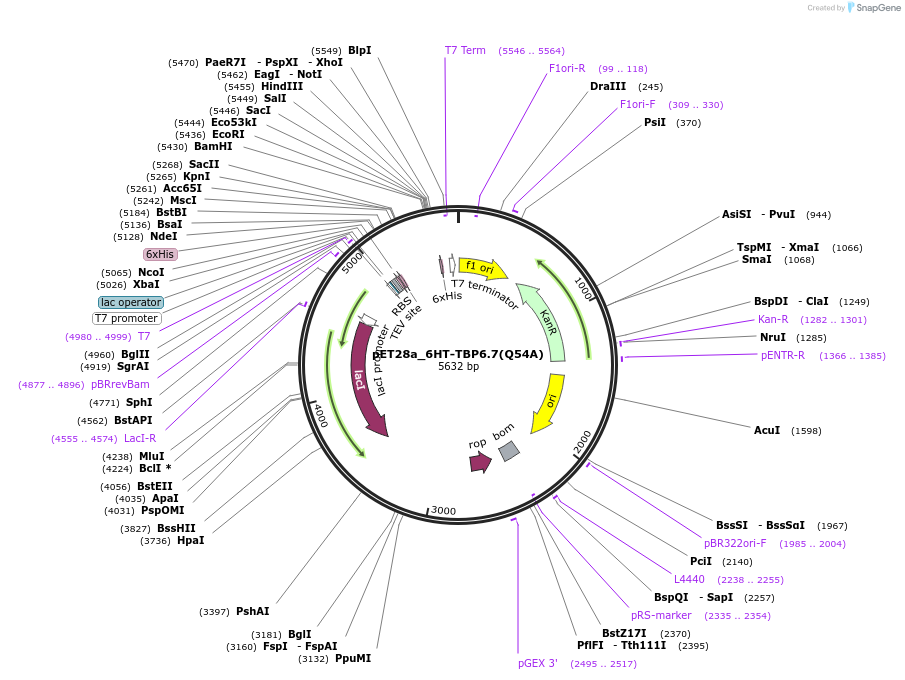
pET28a_6HT-TBP6.7(Q54A)
Plasmid#112730PurposeAlthough not evolved, this residue within TBPs participates in stabilizing the polypeptide loop responsible for RNA recognition.DepositorInsert6His-TEV-TBP6.7(Q54A)
Tags6His-TEVExpressionBacterialMutationQ54APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
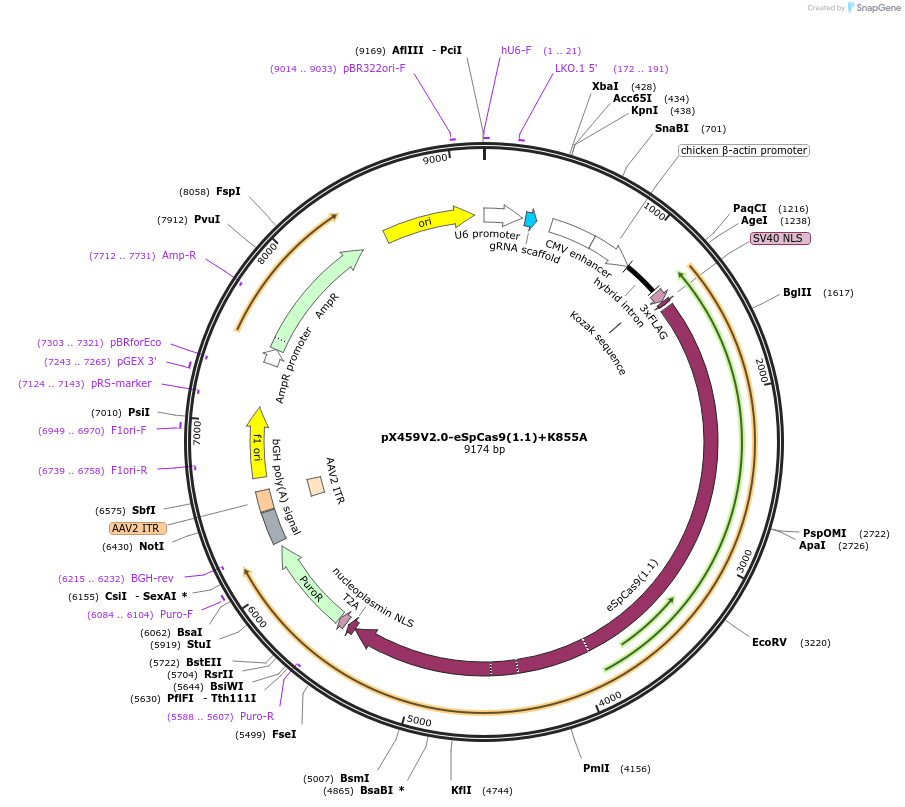
pX459V2.0-eSpCas9(1.1)+K855A
Plasmid#108298PurposepX459 V2.0 (Plasmid #62988) with the K848A, K855A, K1003A, and R1060A mutationsDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceMay 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
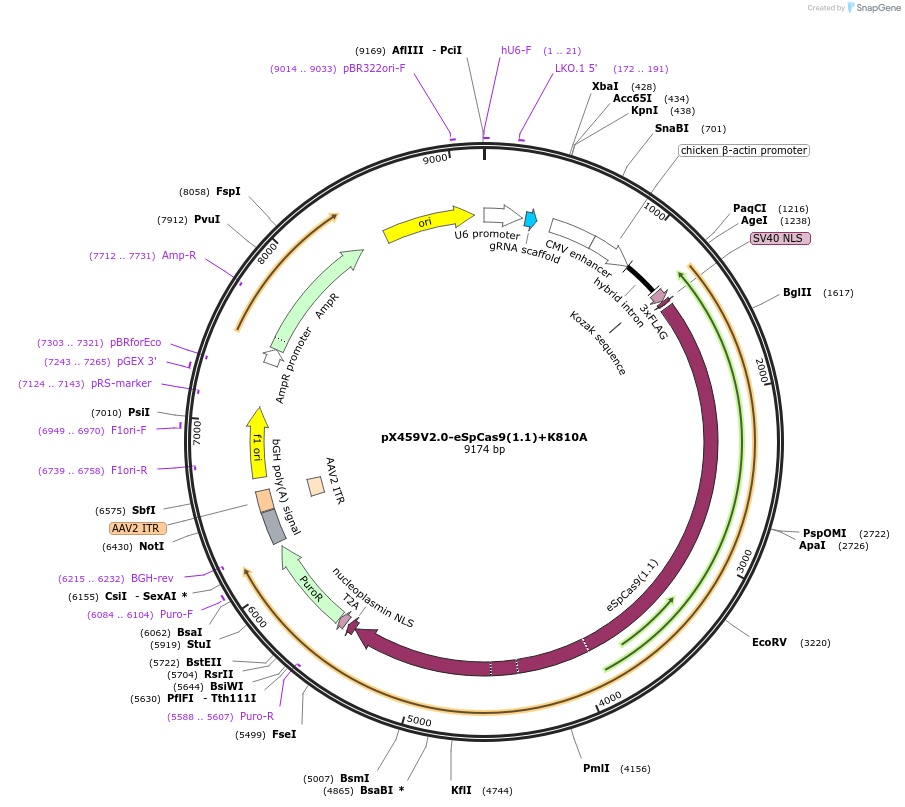
pX459V2.0-eSpCas9(1.1)+K810A
Plasmid#108297PurposepX459 V2.0 (Plasmid #62988) with the K810A, K848A, K1003A, and R1060A mutationsDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceMay 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
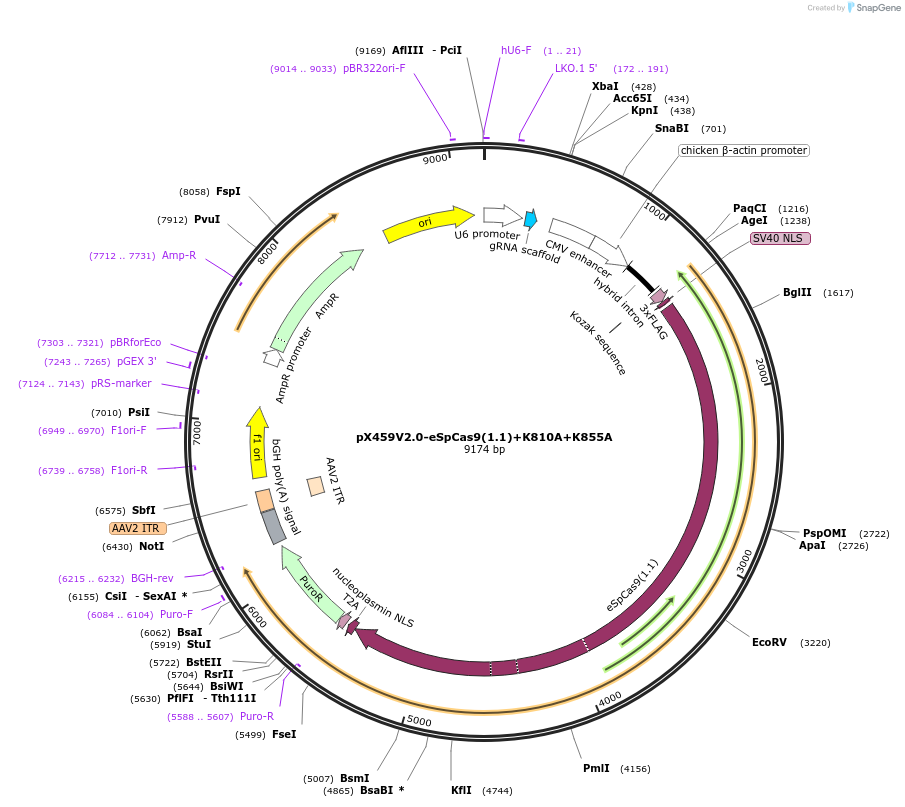
pX459V2.0-eSpCas9(1.1)+K810A+K855A
Plasmid#108299PurposepX459 V2.0 (Plasmid #62988) with the K810A, K848A, K855A, K1003A, and R1060A mutationsDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceMay 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
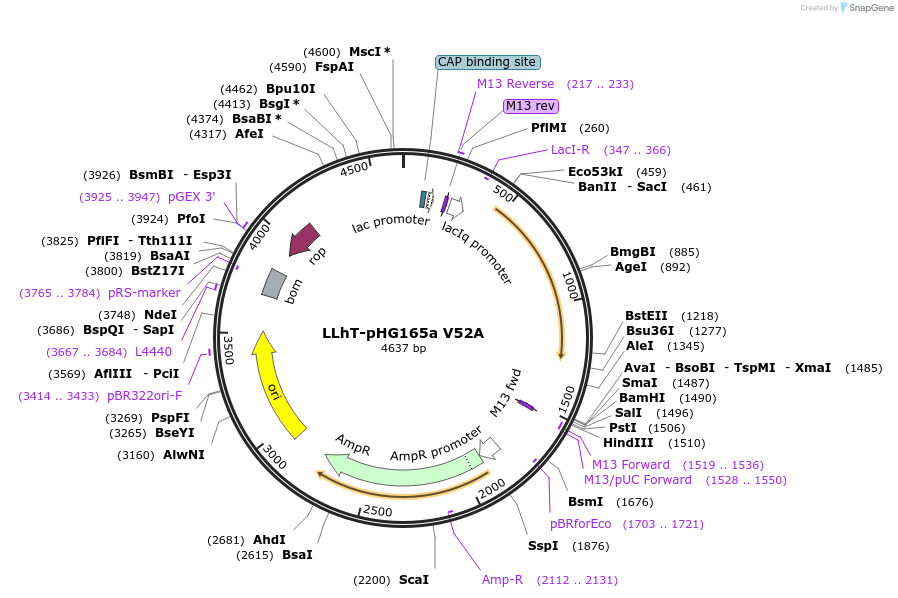
LLhT-pHG165a V52A
Plasmid#90050PurposeConstitutive expression of a chimeric LacI:TreR transcription repressorDepositorInsertLLhT/V52A
ExpressionBacterialMutationV52APromoterIqAvailable SinceJune 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
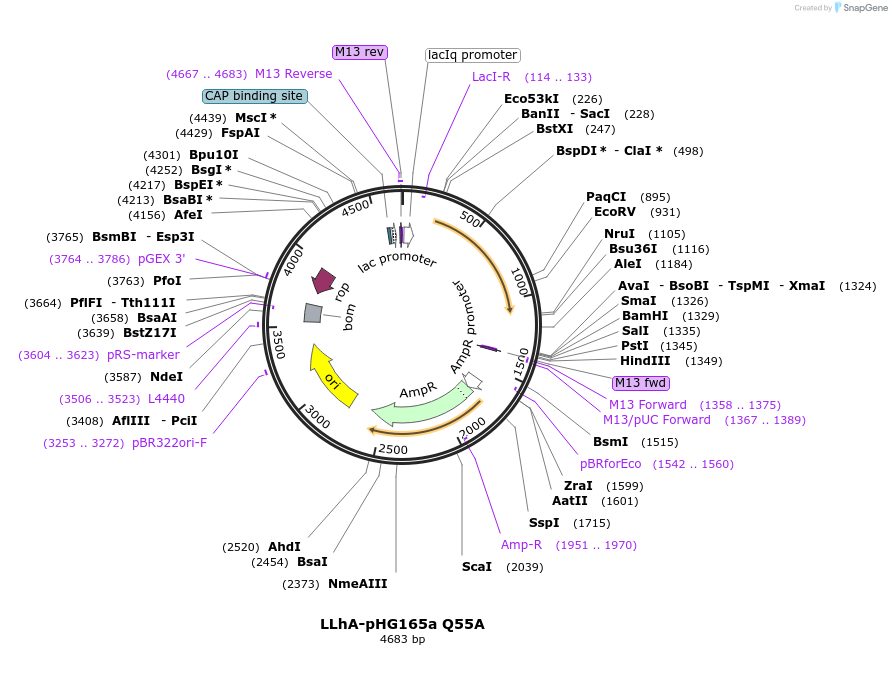
LLhA-pHG165a Q55A
Plasmid#90052PurposeConstitutive expression of a chimeric LacI:AscG transcription repressorDepositorInsertLLhA/Q55A
ExpressionBacterialMutationQ55APromoterIqAvailable SinceJune 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
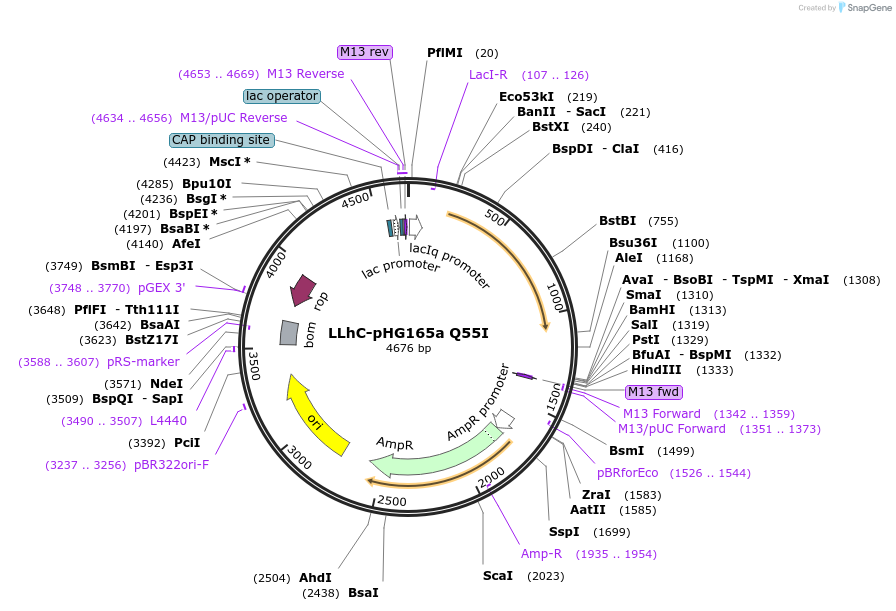
LLhC-pHG165a Q55I
Plasmid#90053PurposeConstitutive expression of a chimeric LacI:CytR transcription repressorDepositorInsertLLhC/Q55I
ExpressionBacterialMutationQ55IPromoterIqAvailable SinceJune 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
pGL3-PIM2mut3
Plasmid#53742Purposepromoter region of PIM2 lacking the 2 forkhead binding sitesDepositorInsertPIM2 (PIM2 Human)
UseLuciferase; Reporter vectorMutationdeletion of 2 forkhead binding sites (bp# 395-497…Available SinceAug. 21, 2014AvailabilityAcademic Institutions and Nonprofits only