We narrowed to 9,859 results for: Coli
-
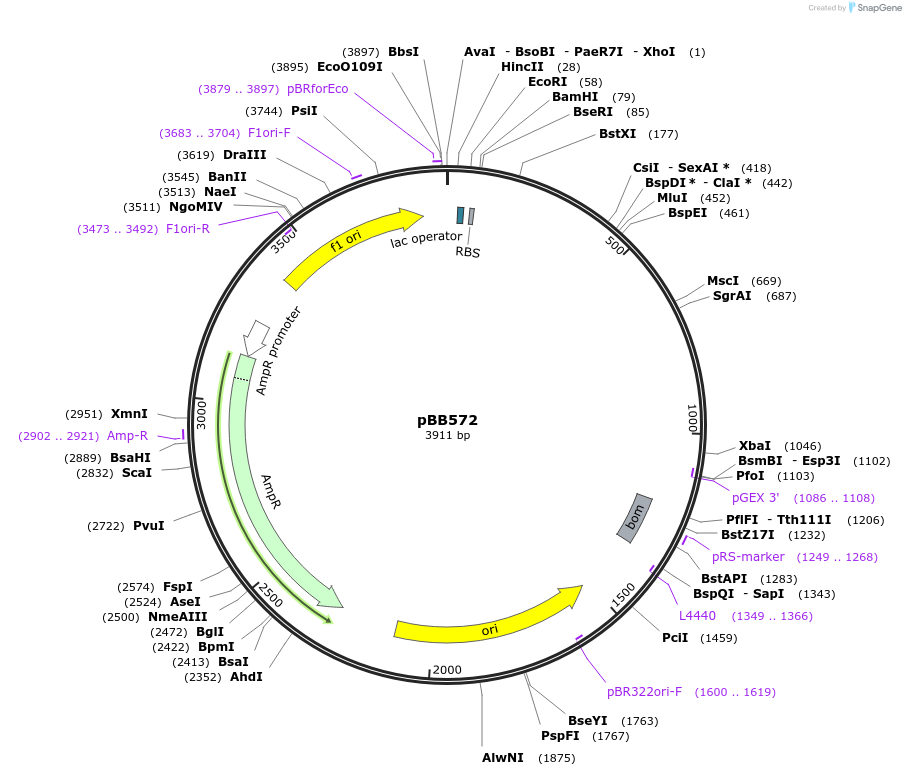
Plasmid#27397DepositorAvailable SinceMarch 28, 2011AvailabilityAcademic Institutions and Nonprofits only
-
pRM006
Plasmid#98922PurposeExpress Histag-SUMO-FtsZ for purification in E coliDepositorInsertFtsZ
TagsHistag-SUMOExpressionBacterialPromoterT7Available SinceAug. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
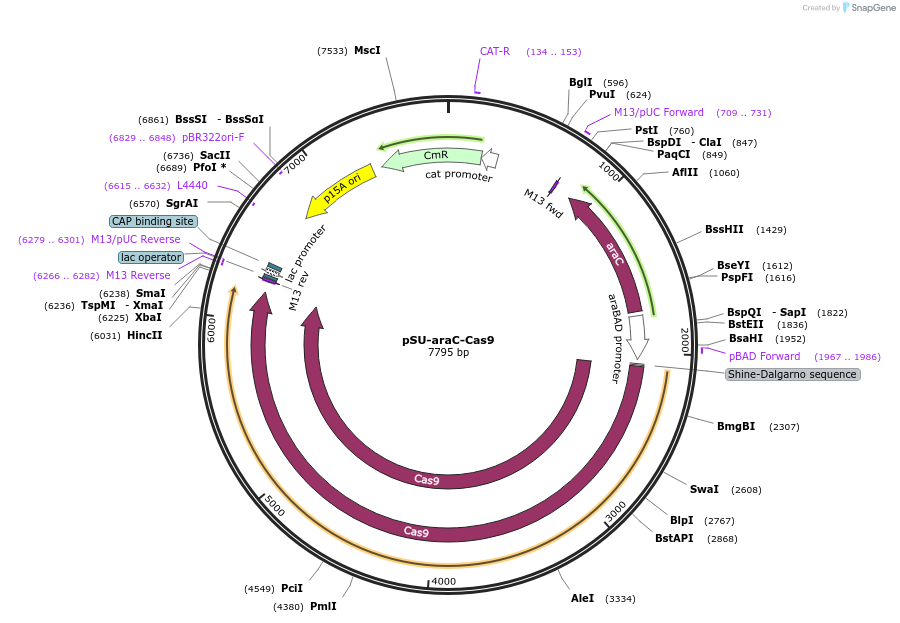
pSU-araC-Cas9
Plasmid#120423PurposeMultigene editing in the Escherichia coli genome using the CRISPR-Cas9 systemDepositorInsertCas9
UseCRISPRExpressionBacterialPromoteraraBADAvailable SinceFeb. 15, 2019AvailabilityAcademic Institutions and Nonprofits only -
pJAT13araE
Plasmid#18987DepositorAvailable SinceOct. 27, 2008AvailabilityAcademic Institutions and Nonprofits only -
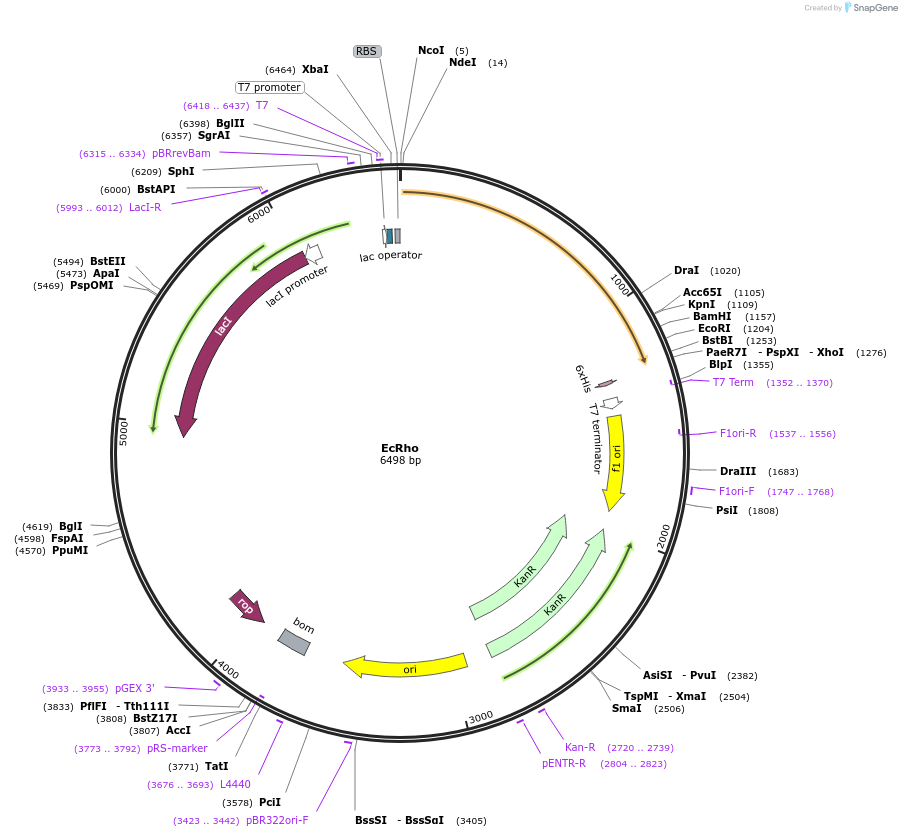
EcRho
Plasmid#113121PurposeExpresses E. coli RhoDepositorInsertRho
TagsMGHExpressionBacterialPromoterT7Available SinceOct. 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
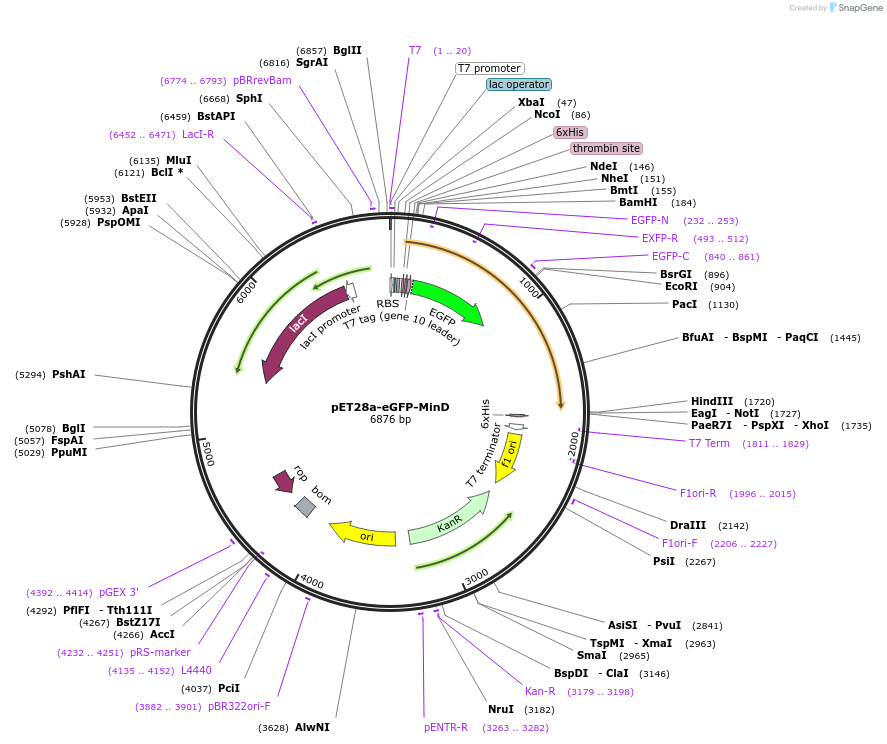
pET28a-eGFP-MinD
Plasmid#133622PurposeExpression vector for the Escherichia coli MinD with an N-terminal fusion of a His-tag for purification and an EGFP for visualization.DepositorAvailable SinceFeb. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
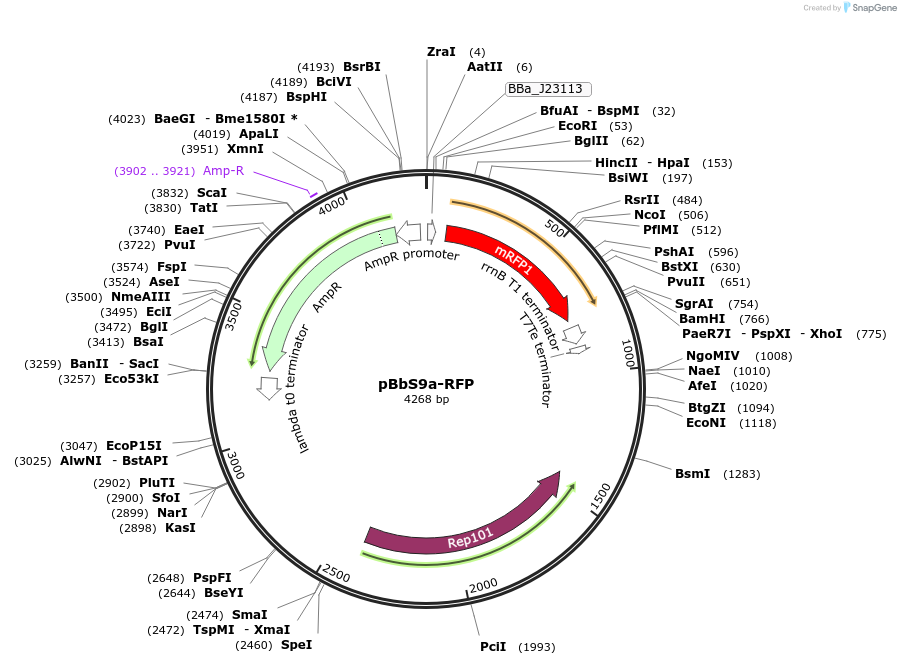
pBbS9a-RFP
Plasmid#126172PurposeConstitutive expression vector for Escherichia coli. BglBrick compatible.DepositorInsertRFP
UseSynthetic BiologyExpressionBacterialPromoterJ23113Available SinceJune 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
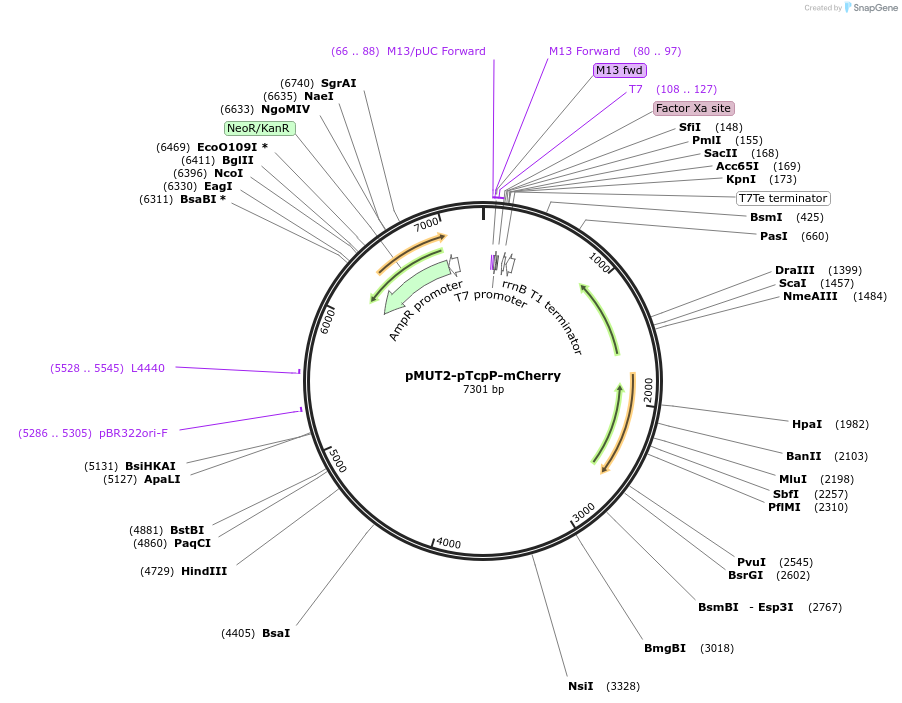
pMUT2-pTcpP-mCherry
Plasmid#192861PurposepMUT2 based vector for sensing human primary bile acids in E. coli Nissle 1917DepositorInsertTcpP-CadC fusion protein and cognate promoter inducible by human primary bile acids
ExpressionBacterialAvailable SinceApril 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
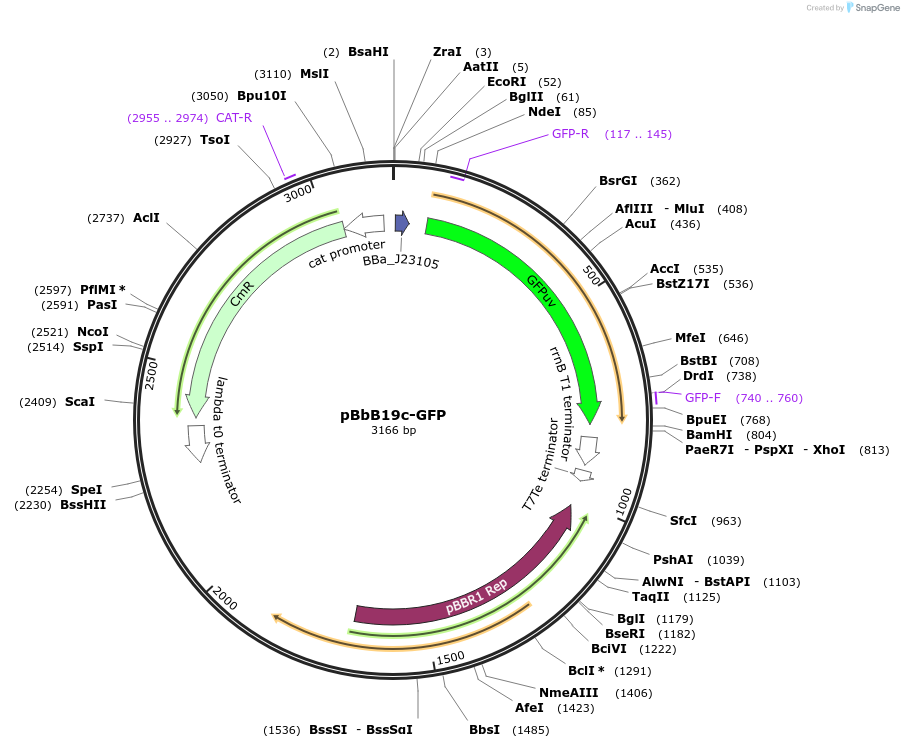
pBbB19c-GFP
Plasmid#126139PurposeConstitutive expression vector for Escherichia coli. BglBrick compatible.DepositorInsertGFPuv
UseSynthetic BiologyExpressionBacterialPromoterJ23105Available SinceJune 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
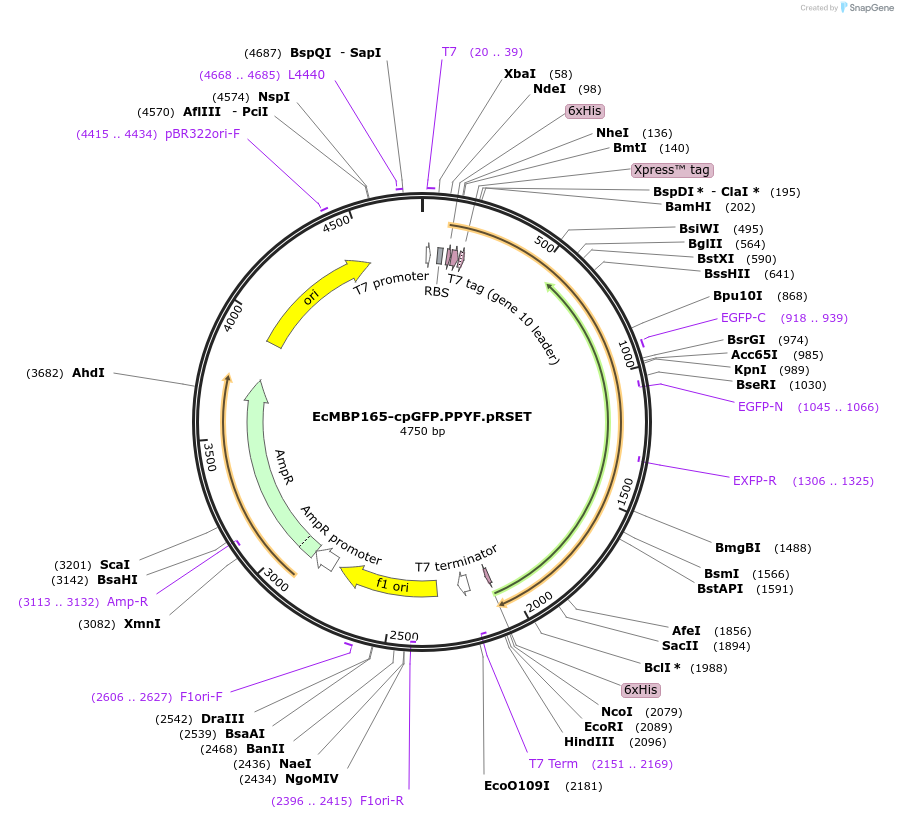
EcMBP165-cpGFP.PPYF.pRSET
Plasmid#33372Purposea single-wavelength maltose sensor constructed from E. coli MalE and cpGFPDepositorInsertMBP based Maltose Biosensor
Tags6xHISExpressionBacterialMutationcpGFP146 variant from GCaMP2 inserted between AA1…PromoterT7Available SinceJan. 13, 2012AvailabilityAcademic Institutions and Nonprofits only -
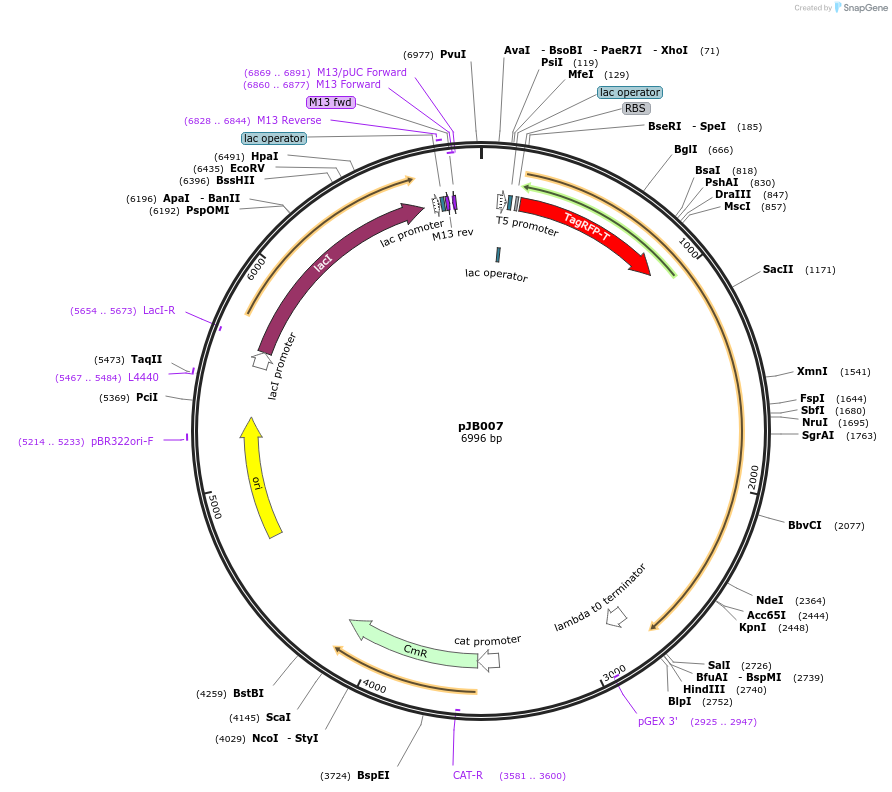
pJB007
Plasmid#98995PurposeExpress FtsI tagged with tagRFP-t in E coliDepositorInsertFtsI
TagsTagRFP-TExpressionBacterialPromoterT5-lacAvailable SinceAug. 9, 2017AvailabilityAcademic Institutions and Nonprofits only -
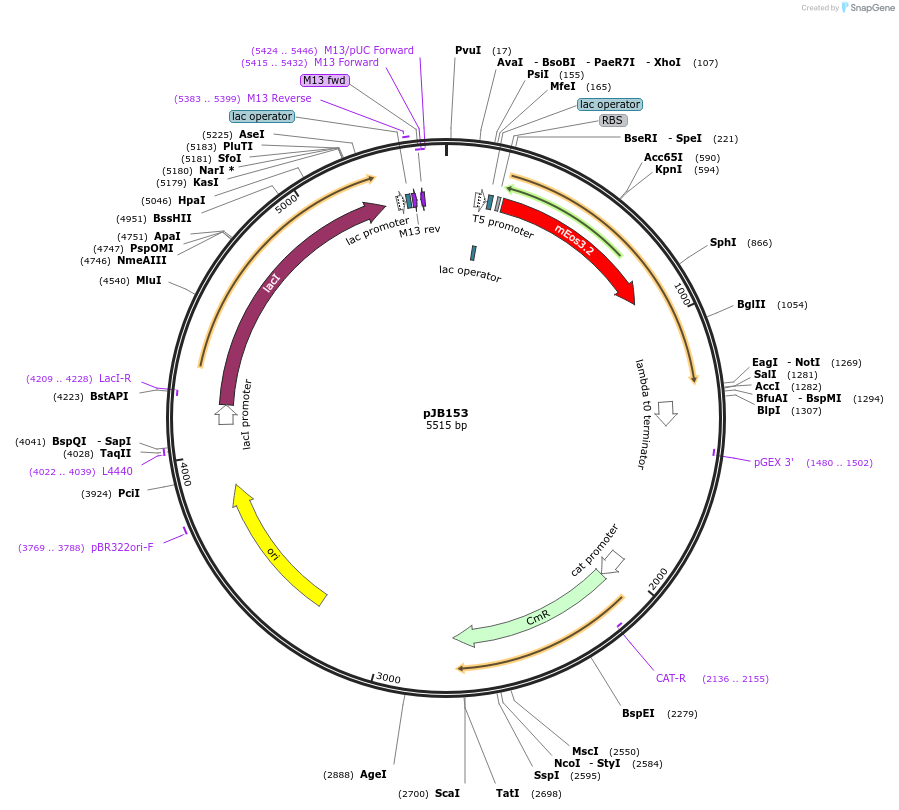
pJB153
Plasmid#92328PurposeExpress mEos3.2 tagged ZapA in E coliDepositorInsertZapA
TagsmEos3.2ExpressionBacterialPromoterT5-lacAvailable SinceAug. 9, 2017AvailabilityAcademic Institutions and Nonprofits only -
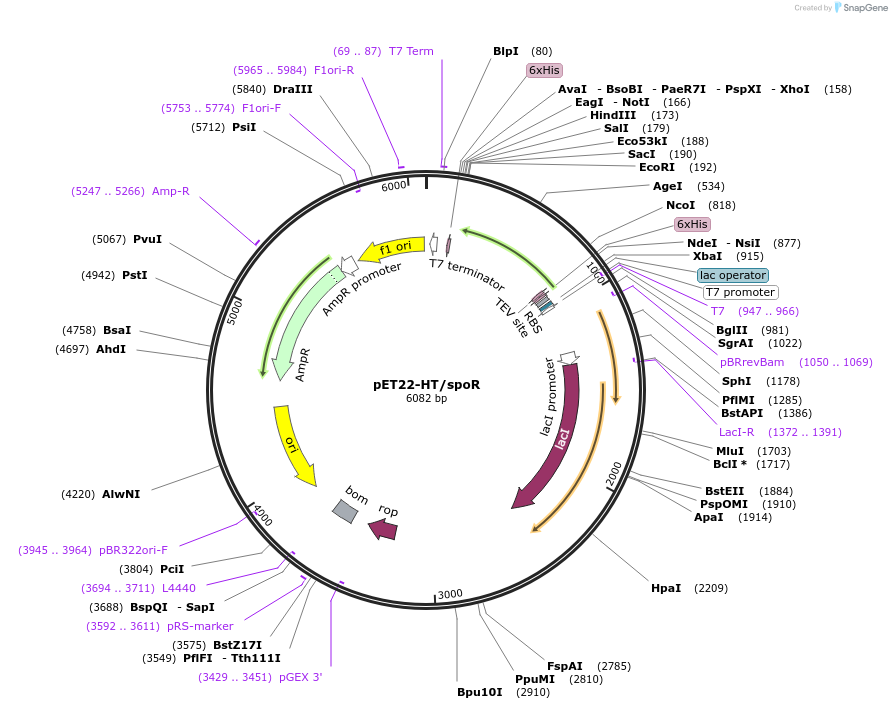
pET22-HT/spoR
Plasmid#73631PurposeProduces Escherichia coli guanylate kinase (SpoR), N-terminal His6 tagDepositorInsertguanylate kinase
TagsHis6ExpressionBacterialPromoterT7Available SinceFeb. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
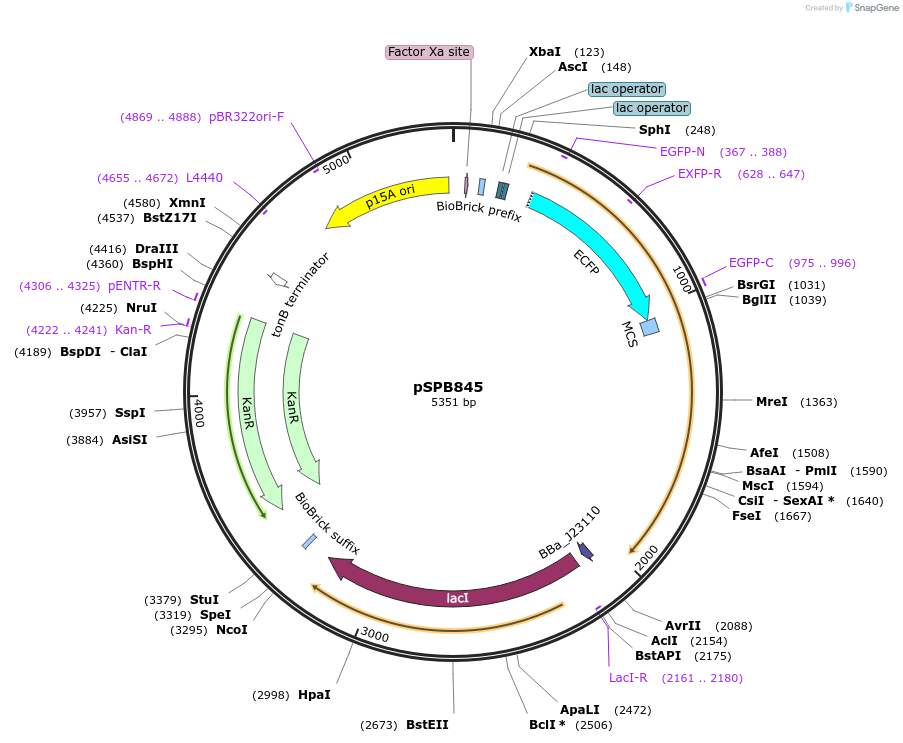
pSPB845
Plasmid#128296Purposeasymmetric plasmid partitioning E. coli - regulatory plasmidDepositorInsertPLlacO1 - lp-cfp-parB, BBa_J23110 - lacI
ExpressionBacterialAvailable SinceJuly 10, 2019AvailabilityAcademic Institutions and Nonprofits only -
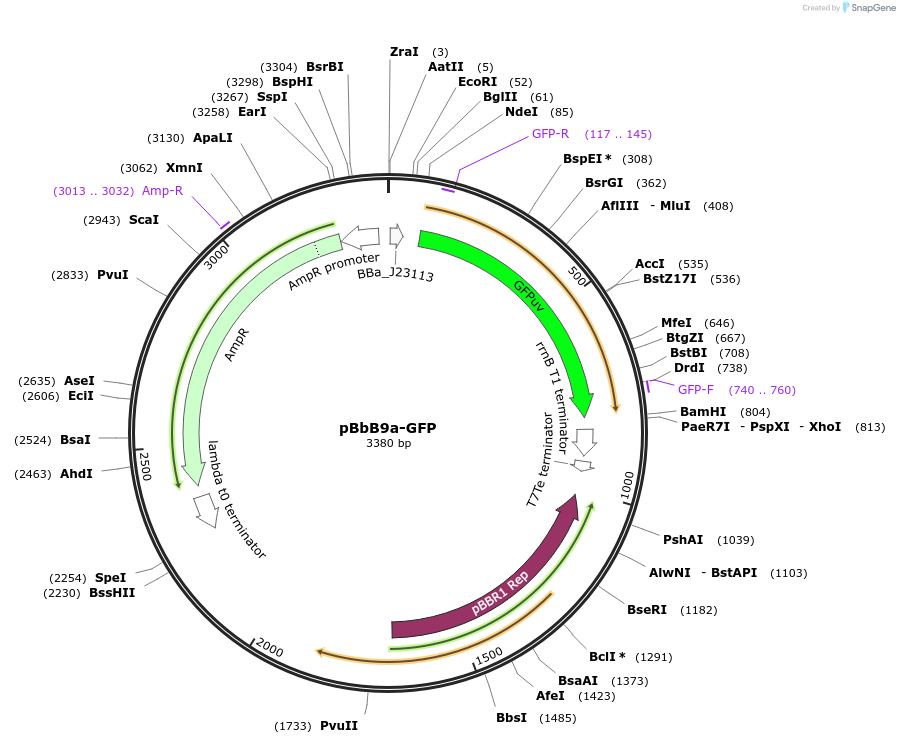
pBbB9a-GFP
Plasmid#126112PurposeConstitutive expression vector for Escherichia coli. BglBrick compatible.DepositorInsertGFPuv
UseSynthetic BiologyExpressionBacterialPromoterJ23113Available SinceJune 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
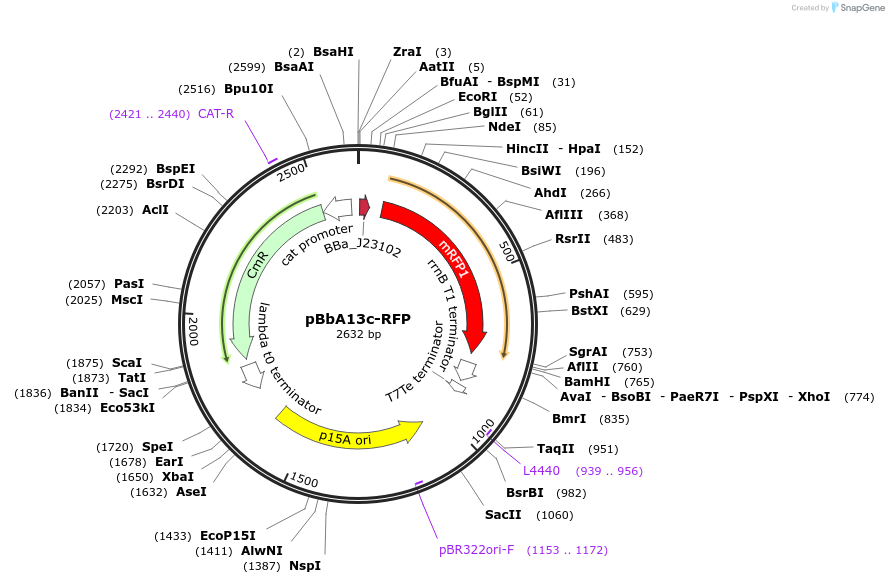
pBbA13c-RFP
Plasmid#126098PurposeConstitutive expression vector for Escherichia coli. BglBrick compatible.DepositorInsertRFP
UseSynthetic BiologyExpressionBacterialPromoterJ23102Available SinceJune 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
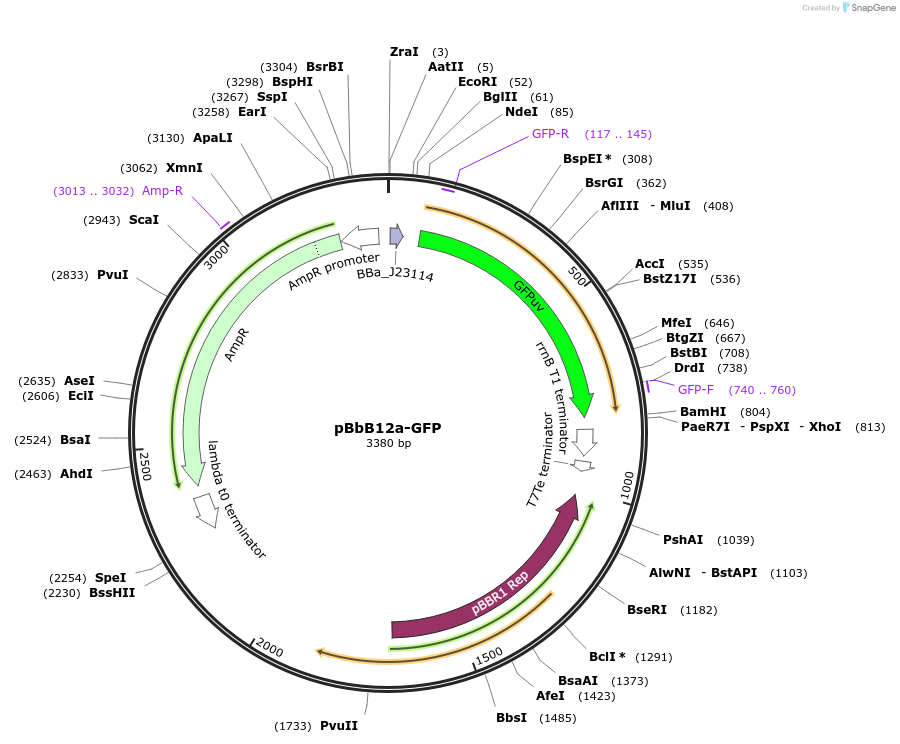
pBbB12a-GFP
Plasmid#126115PurposeConstitutive expression vector for Escherichia coli. BglBrick compatible.DepositorInsertGFPuv
UseSynthetic BiologyExpressionBacterialPromoterJ23151Available SinceJune 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
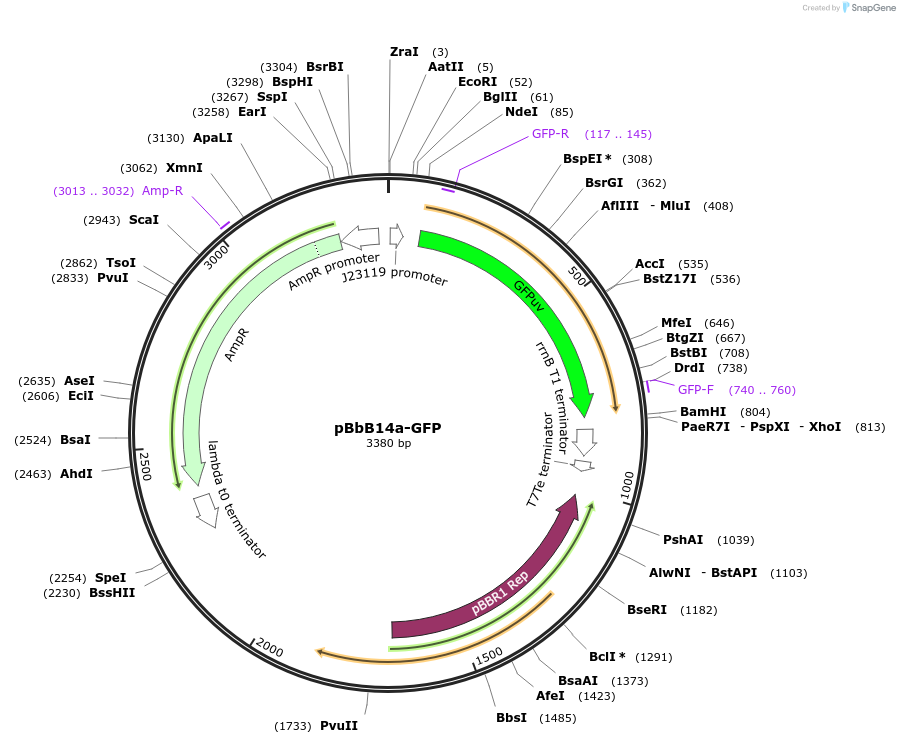
pBbB14a-GFP
Plasmid#126117PurposeConstitutive expression vector for Escherichia coli. BglBrick compatible.DepositorInsertGFPuv
UseSynthetic BiologyExpressionBacterialPromoterJ23119Available SinceJune 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
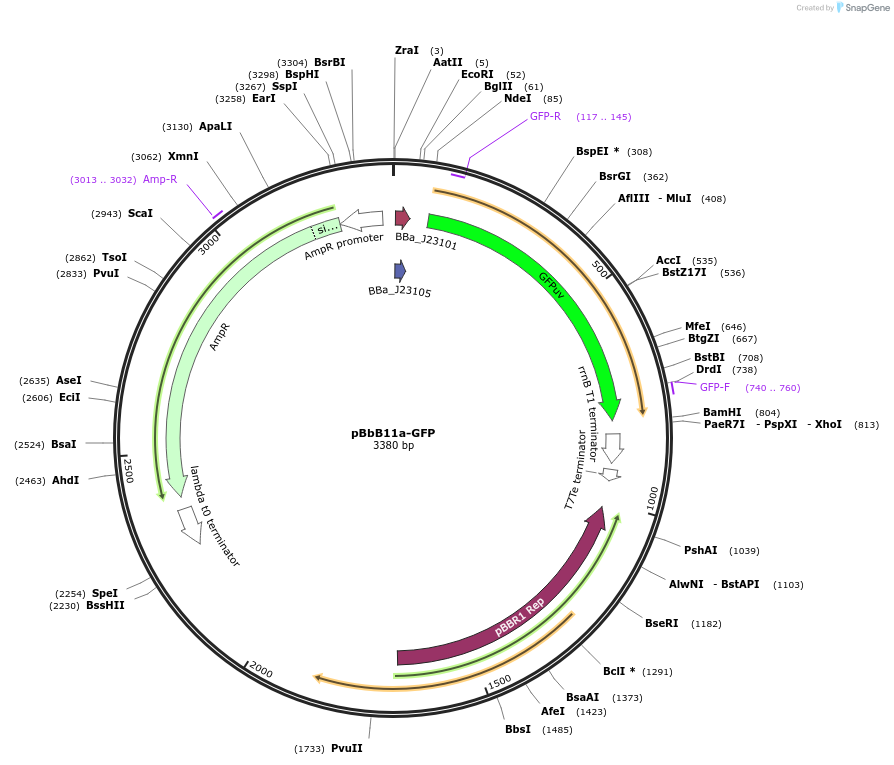
pBbB11a-GFP
Plasmid#126114PurposeConstitutive expression vector for Escherichia coli. BglBrick compatible.DepositorInsertGFPuv
UseSynthetic BiologyExpressionBacterialPromoterJ23150Available SinceJune 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
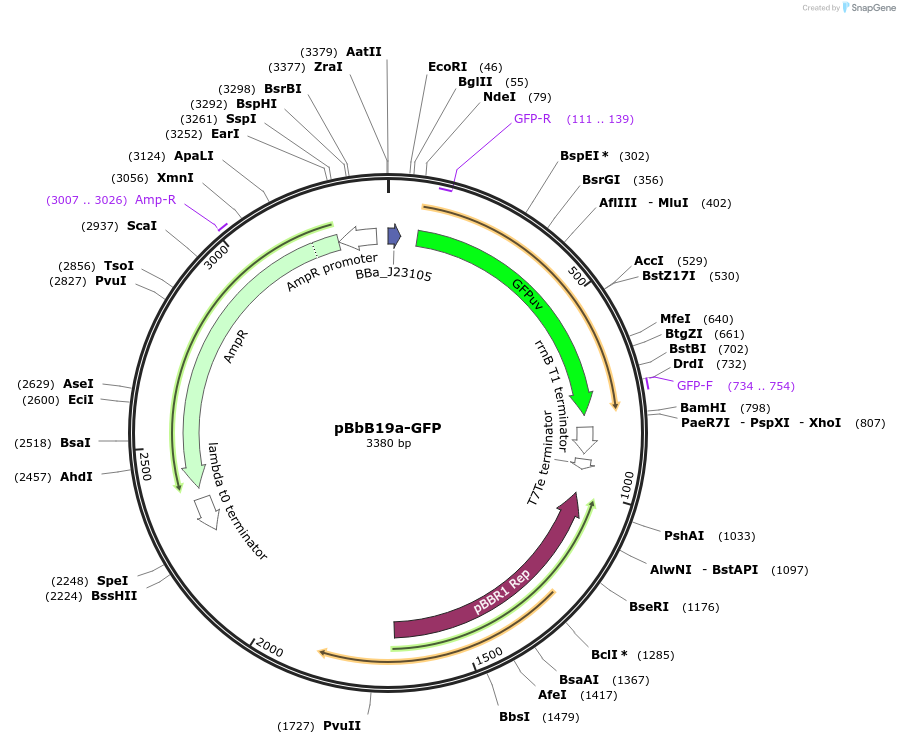
pBbB19a-GFP
Plasmid#126131PurposeConstitutive expression vector for Escherichia coli. BglBrick compatible.DepositorInsertGFPuv
UseSynthetic BiologyExpressionBacterialPromoterJ23105Available SinceApril 15, 2021AvailabilityAcademic Institutions and Nonprofits only