We narrowed to 160,365 results for: des
-
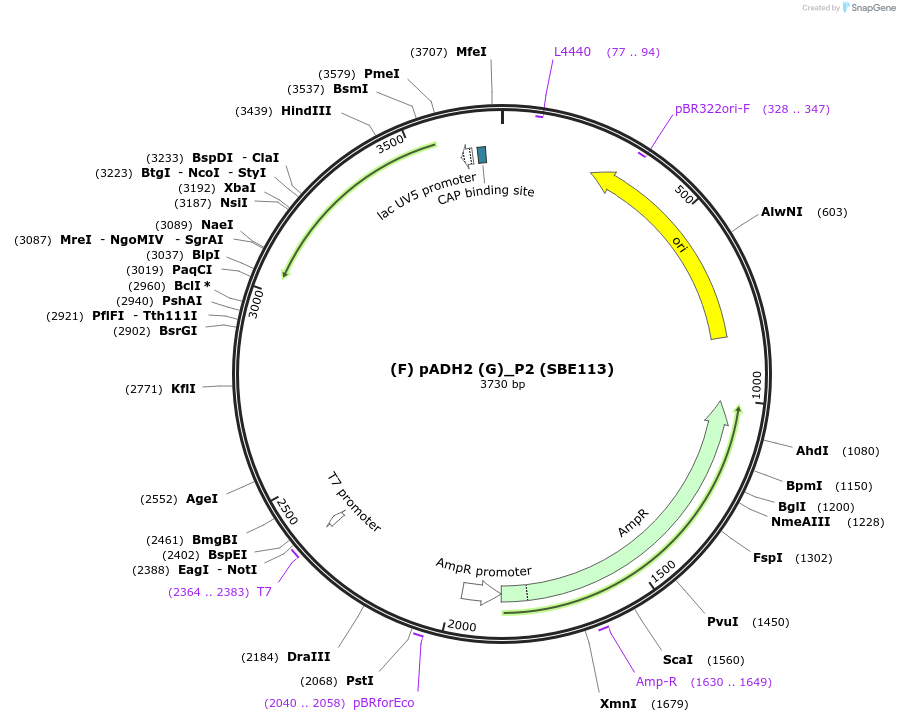
Plasmid#187628Purposepromoter of the gene alcohol dehydrogenase 2 from R. toruloides fro the P2 position (F/G)DepositorInsert(F) pADH2 (G)
ExpressionBacterialAvailable SinceDec. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
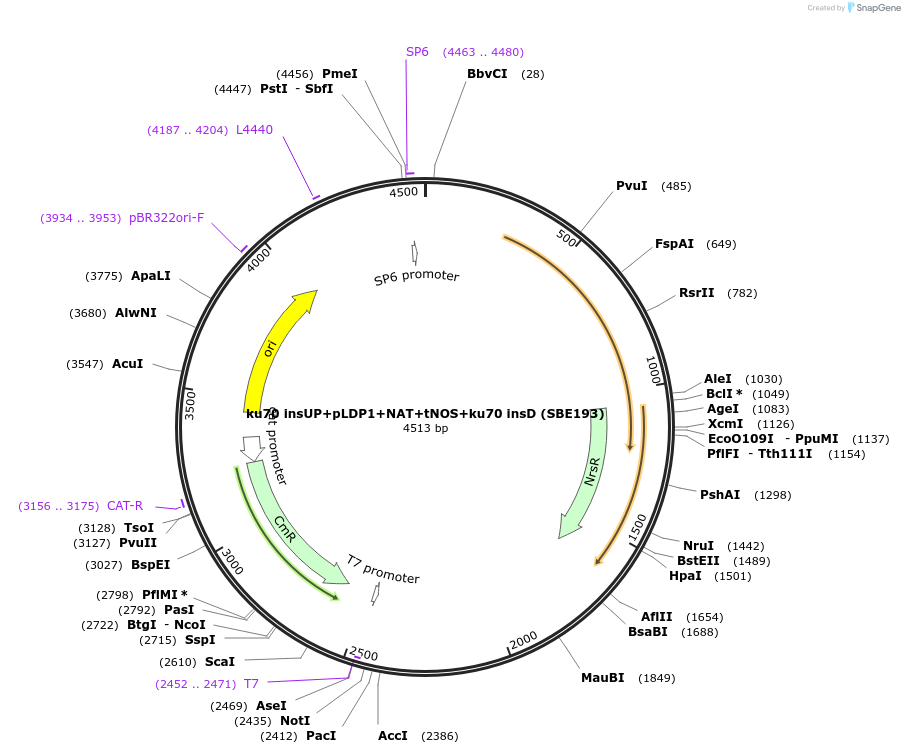
ku70 insUP+pLDP1+NAT+tNOS+ku70 insD (SBE193)
Plasmid#195054Purposeintegration cassette for ku70 deletion and expression of NAT gene under pLDP1 for characterizationDepositorInsertNAT
ExpressionBacterialPromoterpLDP1Available SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
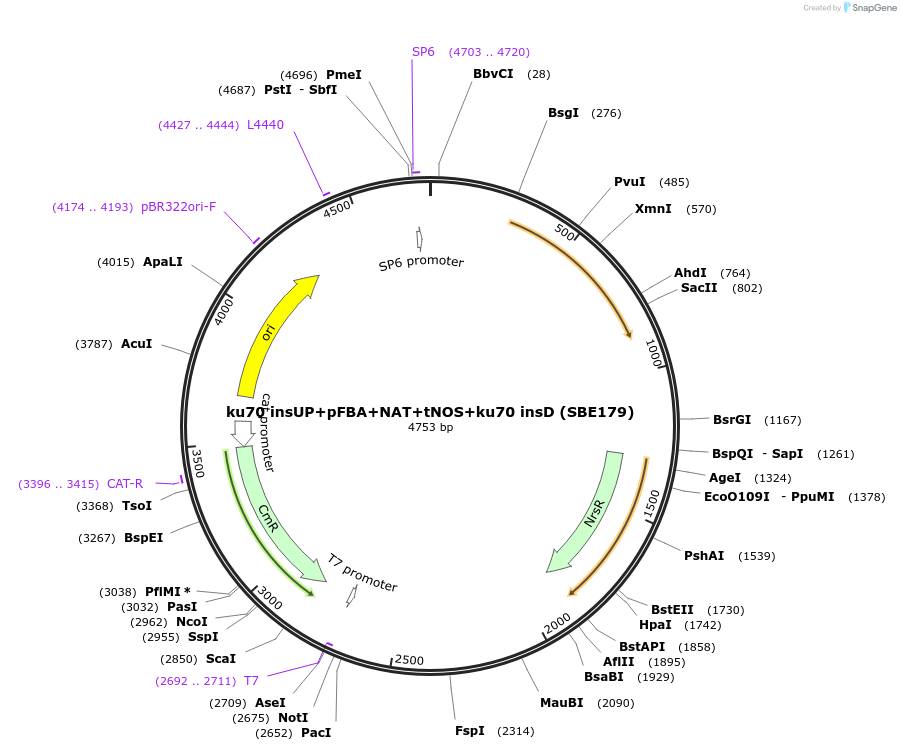
ku70 insUP+pFBA+NAT+tNOS+ku70 insD (SBE179)
Plasmid#195053Purposeintegration cassette for ku70 deletion and expression of NAT gene under pFBA for characterizationDepositorInsertNAT
ExpressionBacterialPromoterpFBAAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
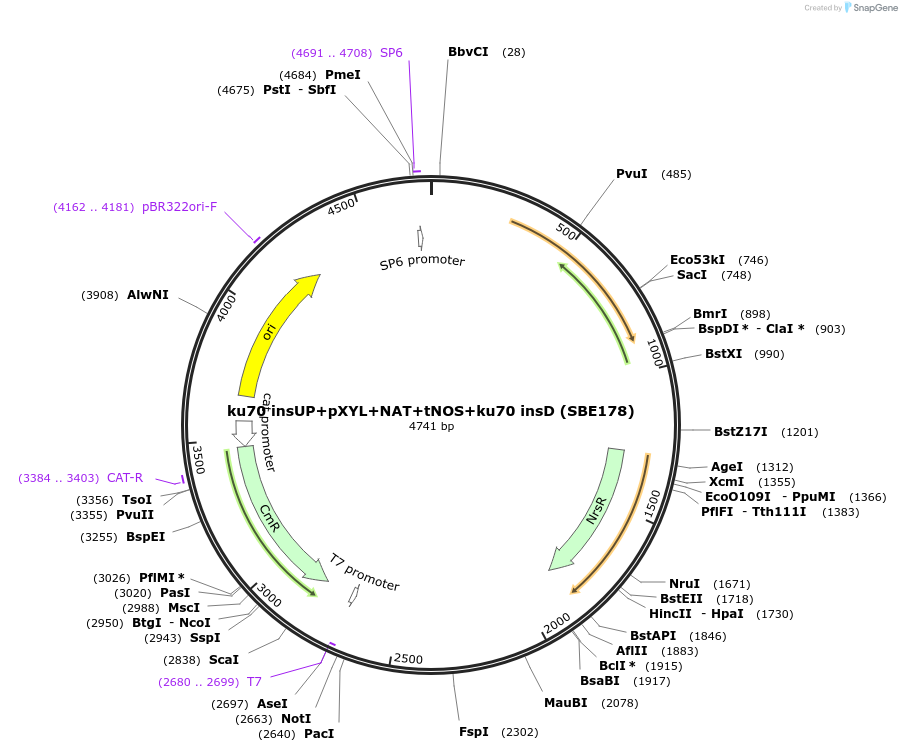
ku70 insUP+pXYL+NAT+tNOS+ku70 insD (SBE178)
Plasmid#195052Purposeintegration cassette for ku70 deletion and expression of NAT gene under pXYL for characterizationDepositorInsertNAT
ExpressionBacterialPromoterpXYLAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
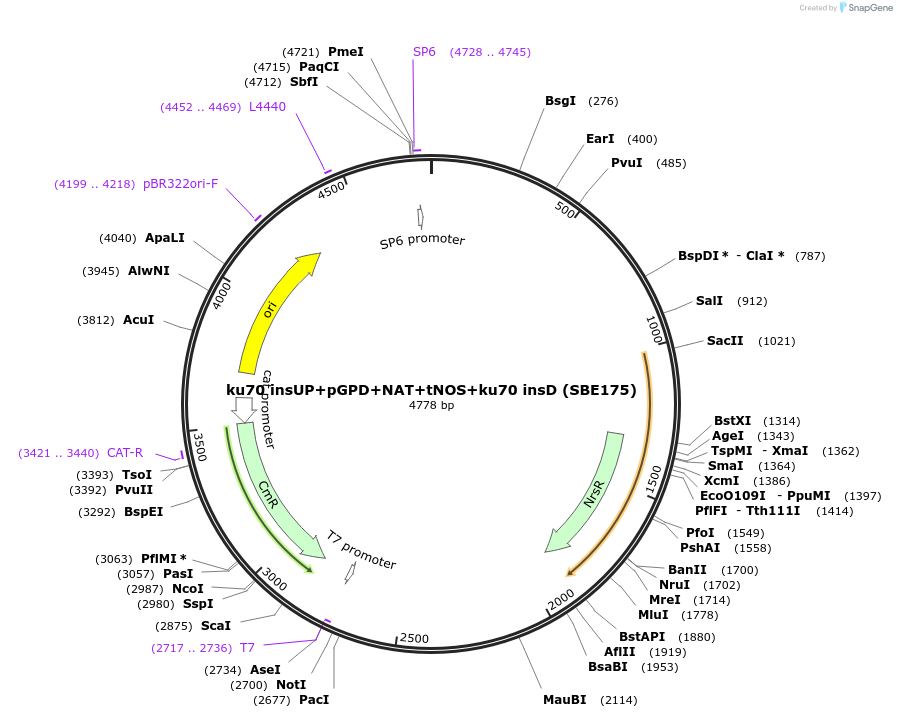
ku70 insUP+pGPD+NAT+tNOS+ku70 insD (SBE175)
Plasmid#195051Purposeintegration cassette for ku70 deletion and expression of NAT gene under pGPD for characterizationDepositorInsertNAT
ExpressionBacterialPromoterpGPDAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
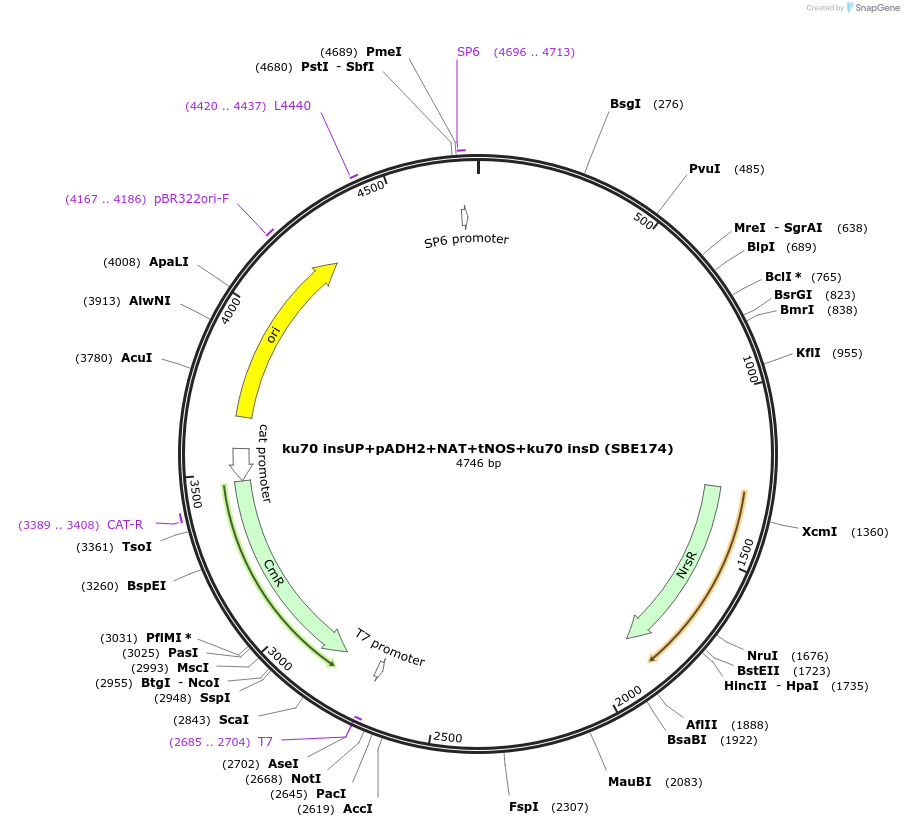
ku70 insUP+pADH2+NAT+tNOS+ku70 insD (SBE174)
Plasmid#195050Purposeintegration cassette for ku70 deletion and expression of NAT gene under pADH2 for characterizationDepositorInsertNAT
ExpressionBacterialPromoterpADH2Available SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
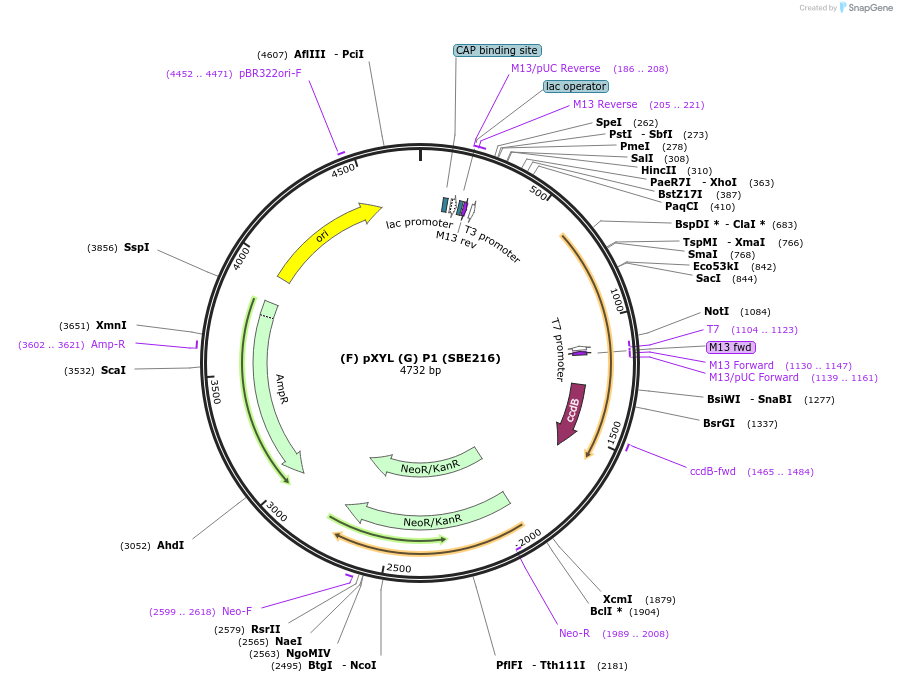
(F) pXYL (G) P1 (SBE216)
Plasmid#195017Purposepromoter of the gene xylose reductase from R. toruloides for the position P2 (F/G)DepositorInsert(F) pXYL (G)
ExpressionBacterialAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
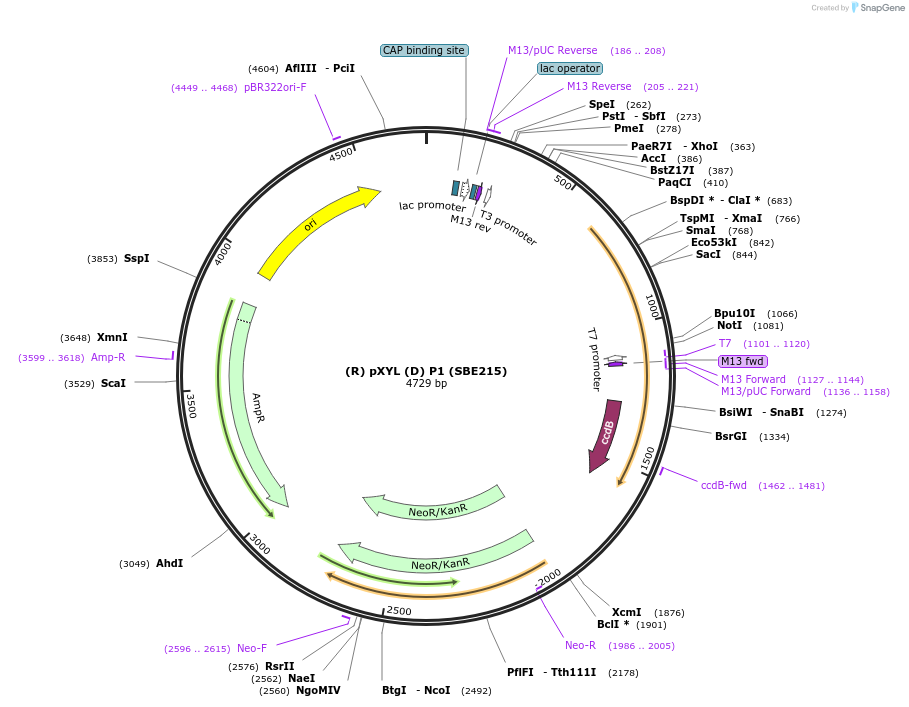
(R) pXYL (D) P1 (SBE215)
Plasmid#195016Purposepromoter of the gene xylose reductase from R. toruloides for the position P1 (R/D)DepositorInsert(R) pXYL (D)
ExpressionBacterialAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
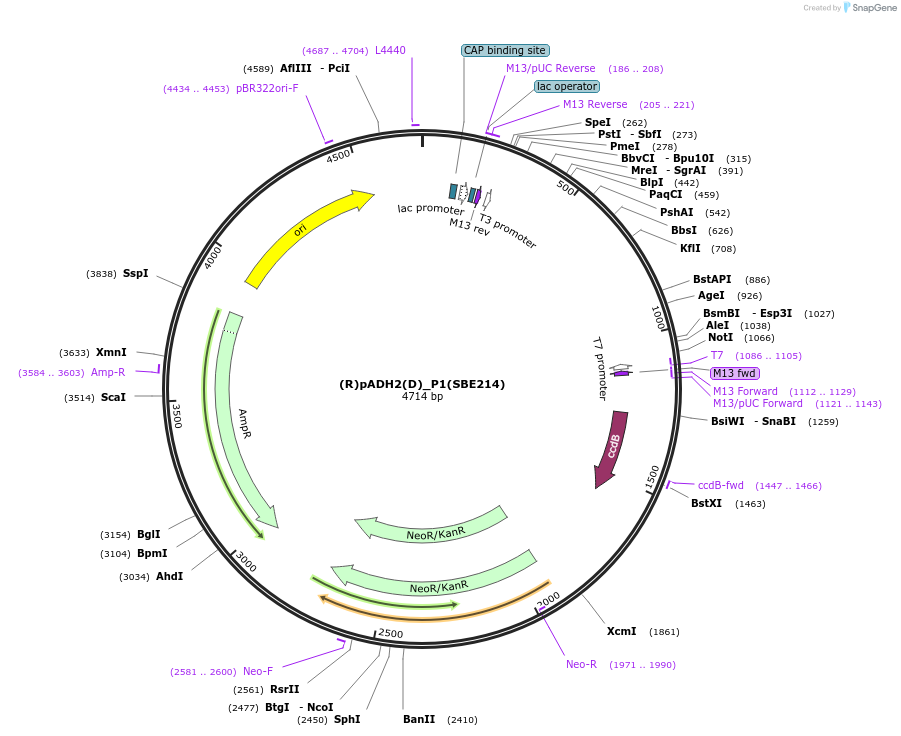
(R)pADH2(D)_P1(SBE214)
Plasmid#195015Purposepromoter of the gene alcohol dehydrogenase 2 from R. toruloides for the position P1DepositorInsert(B) pADH2 (R)
ExpressionBacterialAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
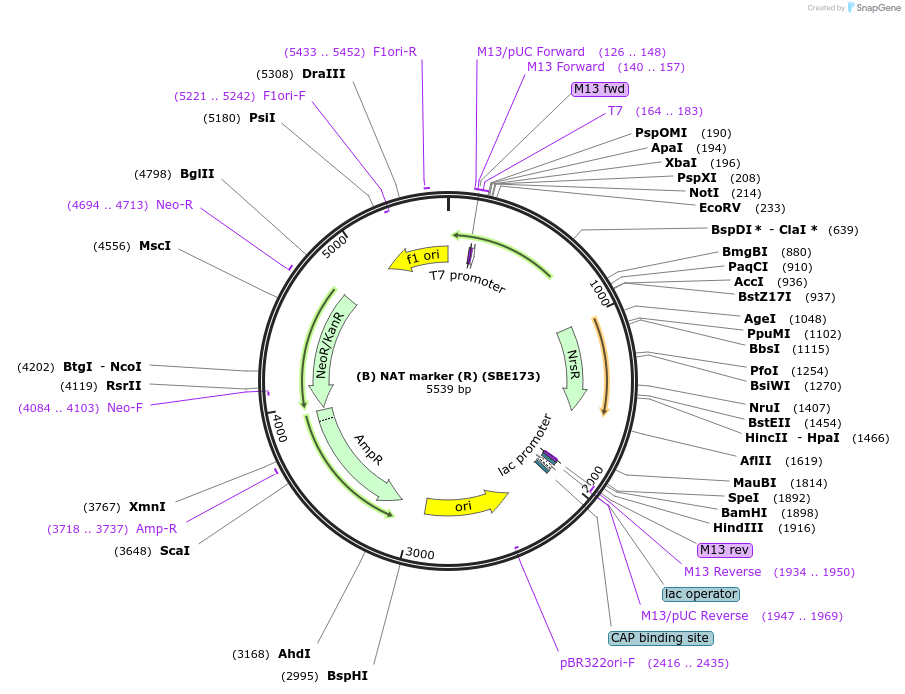
(B) NAT marker (R) (SBE173)
Plasmid#195014Purposenourseothricin (NAT) resistance genes codon-optimized for R. toruloides and expressed under pXYL and tNOSDepositorInsertpXYL+NAT+tNOS
ExpressionBacterialPromoterpXYLAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
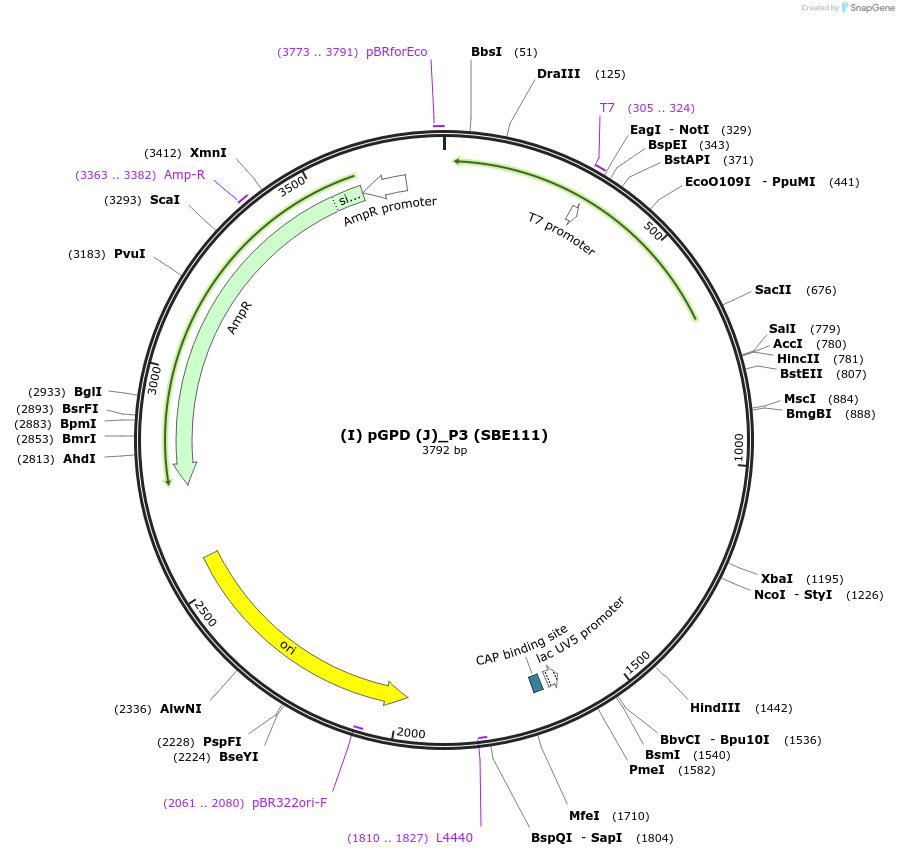
(I) pGPD (J)_P3 (SBE111)
Plasmid#194990Purposepromoter of the gene glyceraldehyde-3-phosphate dehydrogenase from R. toruloides for a P3 position (I/J)DepositorInsert(I) pGPD (J)
ExpressionBacterialAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
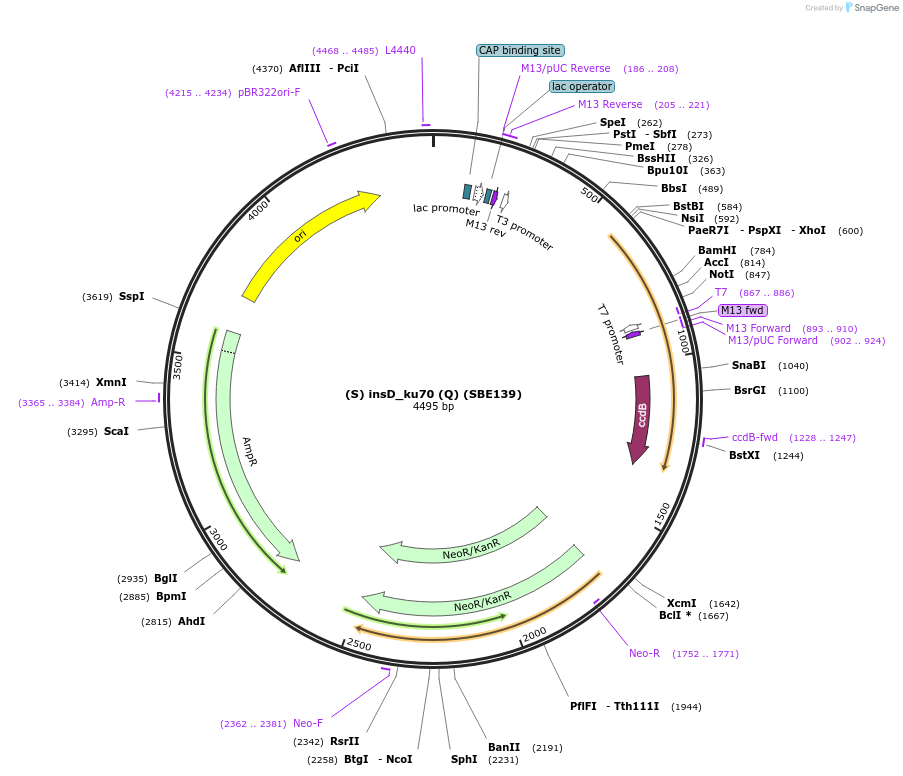
(S) insD_ku70 (Q) (SBE139)
Plasmid#187627Purposethe insertional region downstream for the deletion of the KU70 gene in R. toruloides. Overhangs S/QDepositorInsert(S) insD KU70 (Q)
ExpressionBacterialAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
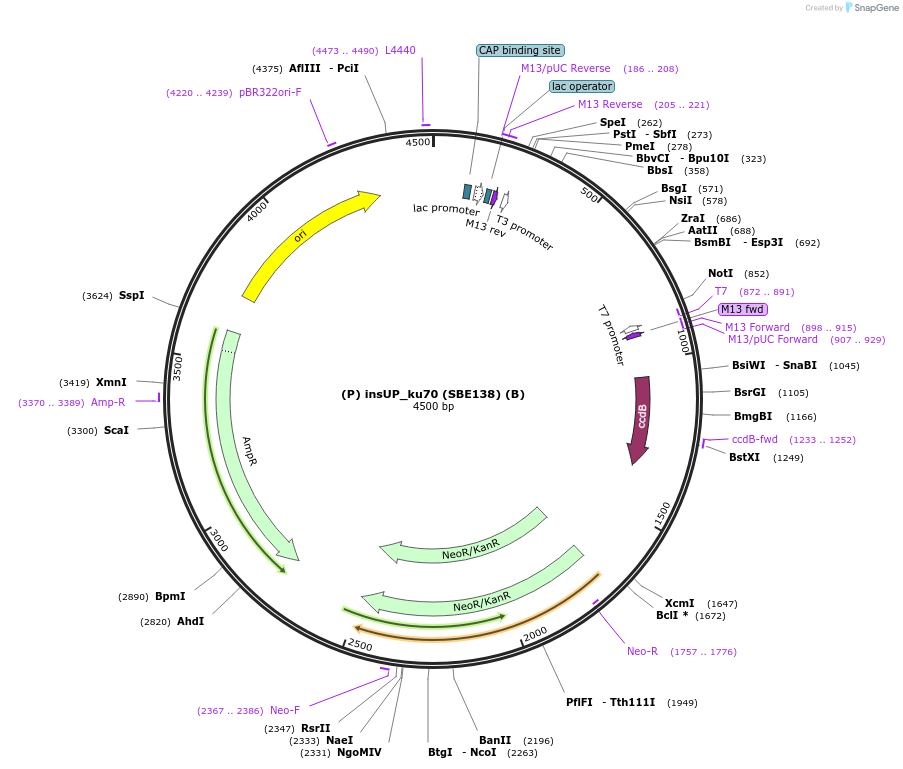
(P) insUP_ku70 (SBE138) (B)
Plasmid#187626Purposethe insertional region upstream for the deletion of the KU70 gene in R. toruloides. Overhangs P/BDepositorInsert(P) insUP KU70 (B)
ExpressionBacterialAvailable SinceNov. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
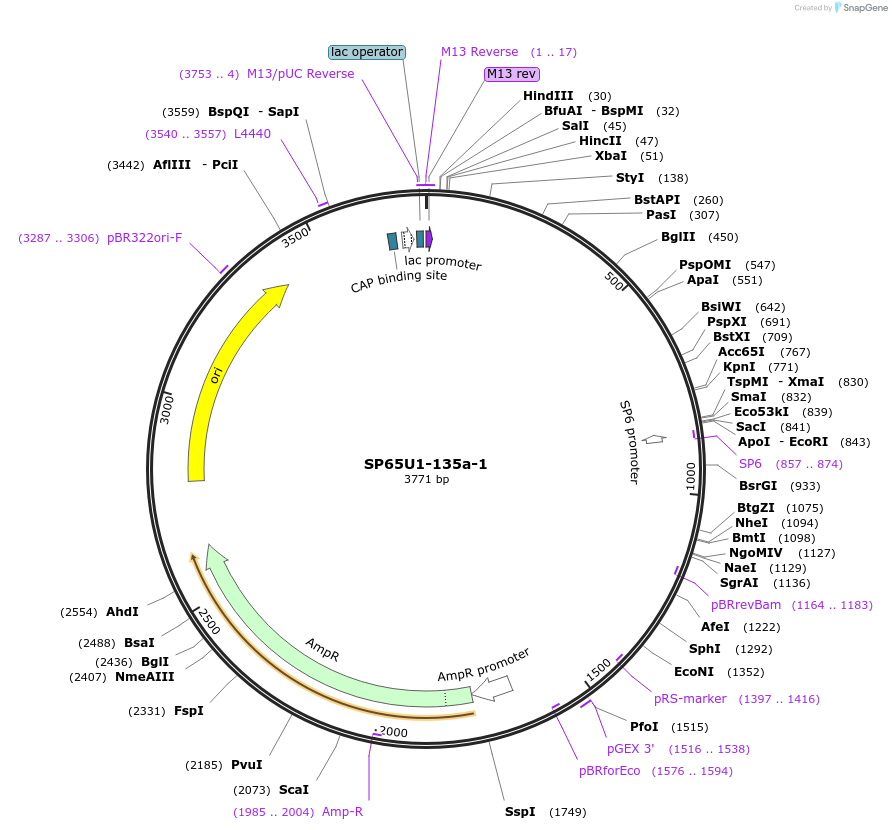
SP65U1-135a-1
Plasmid#105819Purposeplasmid for the overexpression of miR-135a under the U1 expression cassette inserted in the pSP65 plasmidDepositorAvailable SinceJan. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
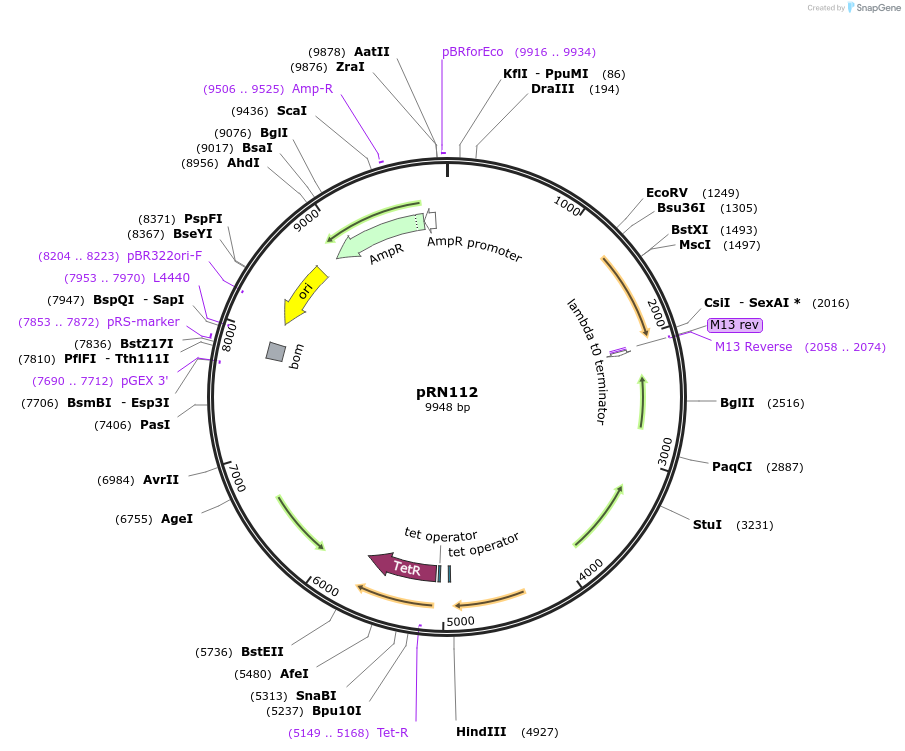
pRN112
Plasmid#84464PurposepJB38-NWMN29-30+ SarA_P1-mAmetrine-TermDepositorInsertSarA_P1-mAmetrine from pRN12
UseSynthetic BiologyAvailable SinceFeb. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
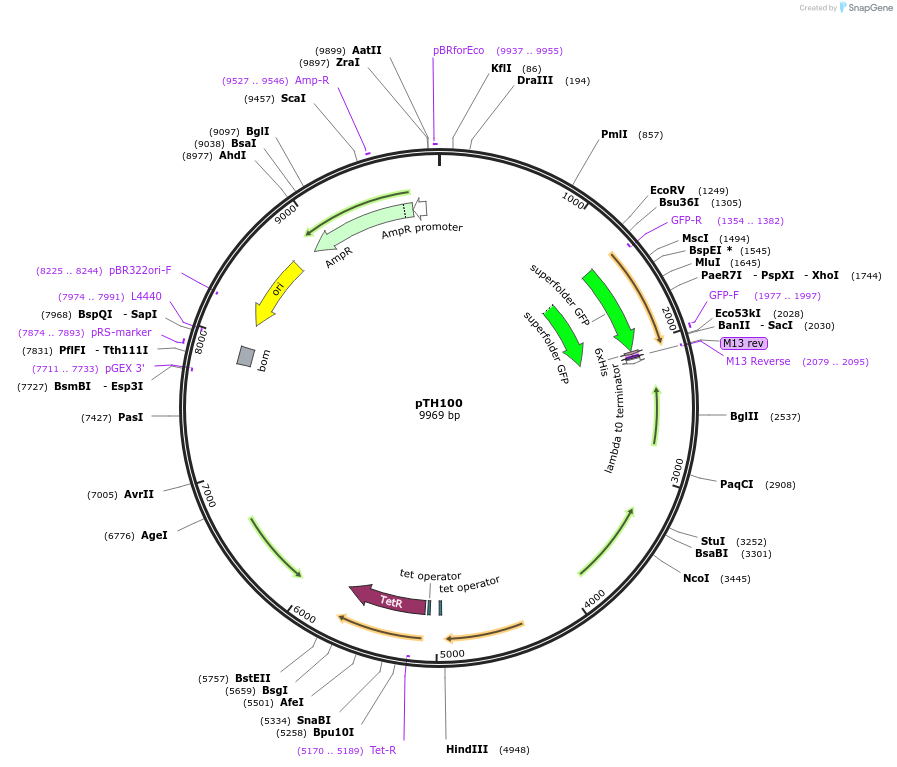
pTH100
Plasmid#84458PurposepJB38-NWMN29-30 + SarA_P1-sGFP-TermDepositorInsertSarA_P1-sGFP from pCM29
Tags6xHis (on sfGFP)Available SinceFeb. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
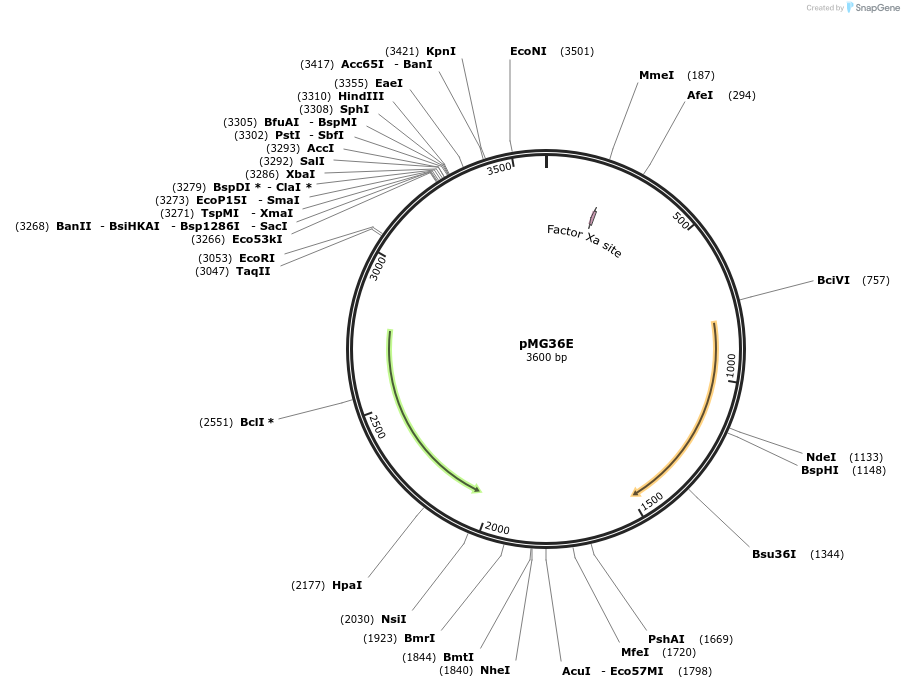
pMG36E
Plasmid#133853PurposeExpression vectorDepositorTypeEmpty backboneExpressionBacterialPromoterP32Available SinceAug. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
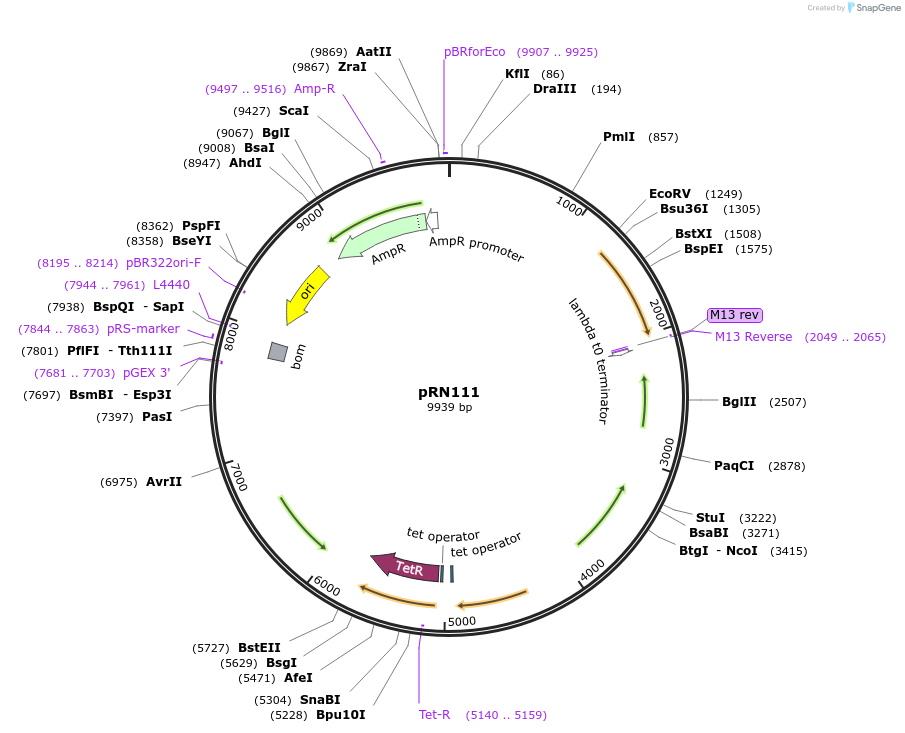
pRN111
Plasmid#84463PurposepJB38-NWMN29-30 + SarA_P1-mCherry-TermDepositorInsertSarA_P1-mCherry from pRN11
Available SinceFeb. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
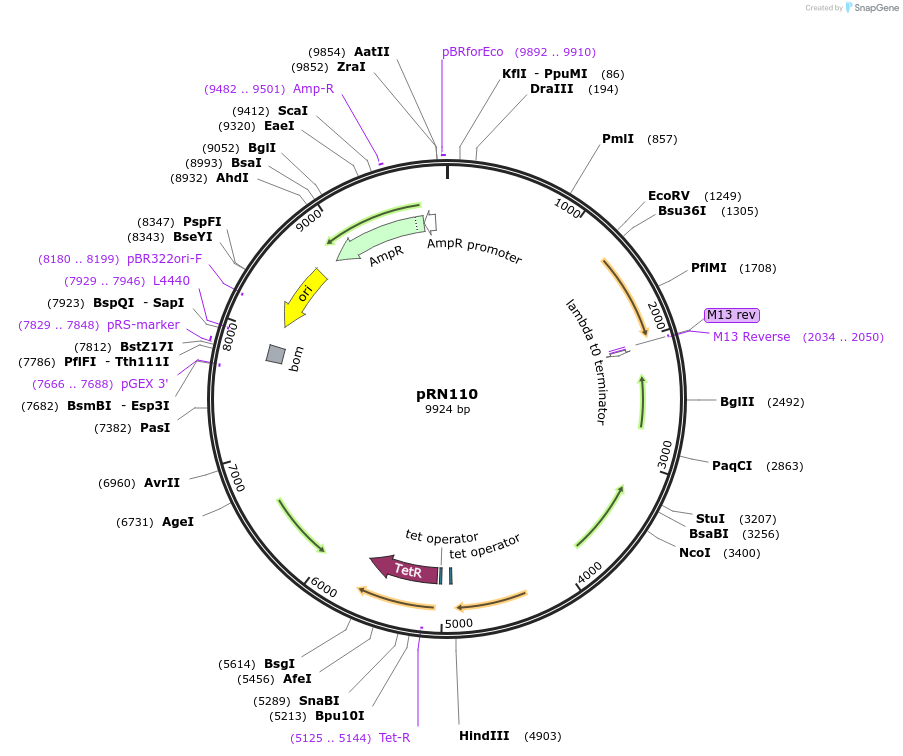
pRN110
Plasmid#84462PurposepJB38-NWMN29-30 + SarA_P1-mKate2-TermDepositorInsertSarA_P1-mKate2 from pRN11
Available SinceFeb. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
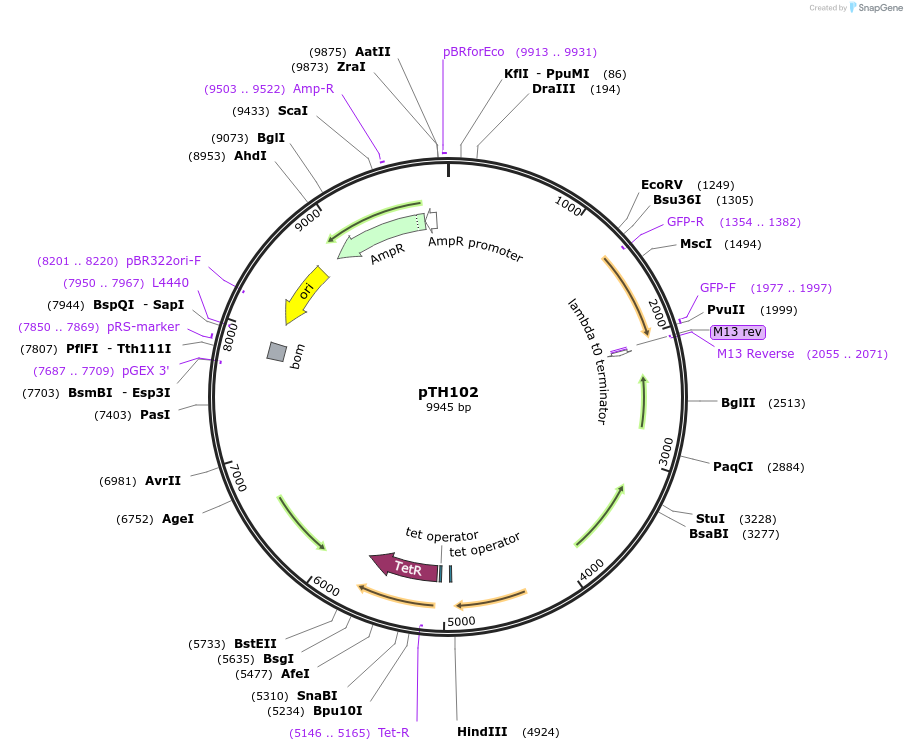
pTH102
Plasmid#84460PurposepJB38-NWMN29-30 + SarA_P1-CFP-TermDepositorInsertSarA_P1-CFP from pTH2
Available SinceFeb. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
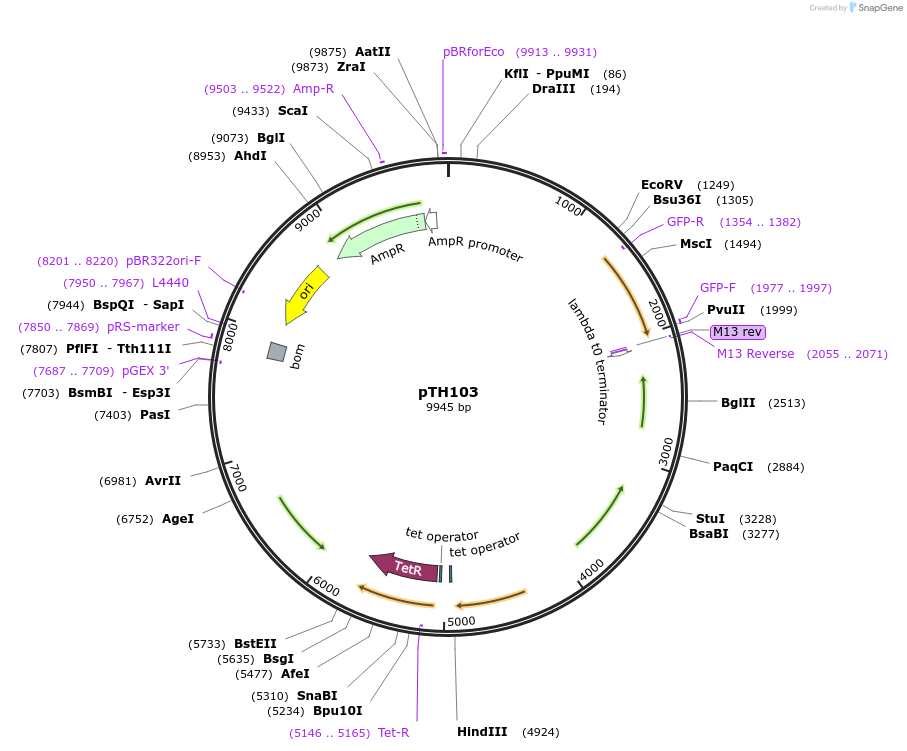
pTH103
Plasmid#84461PurposepJB38-NWMN29-30 + SarA_P1-YFP-TermDepositorInsertSarA_P1-YFP from pTH3
Available SinceFeb. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
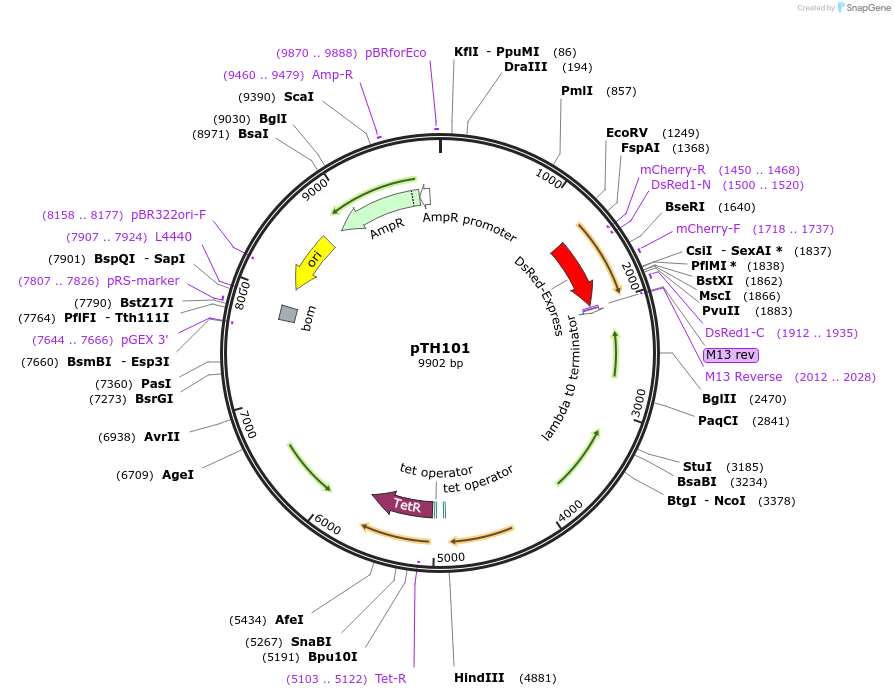
pTH101
Plasmid#84459PurposepJB38-NWMN29-30 + SarA_P1-DsRed-TermDepositorInsertSarA_P1-DsRED from pTH1
Available SinceFeb. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
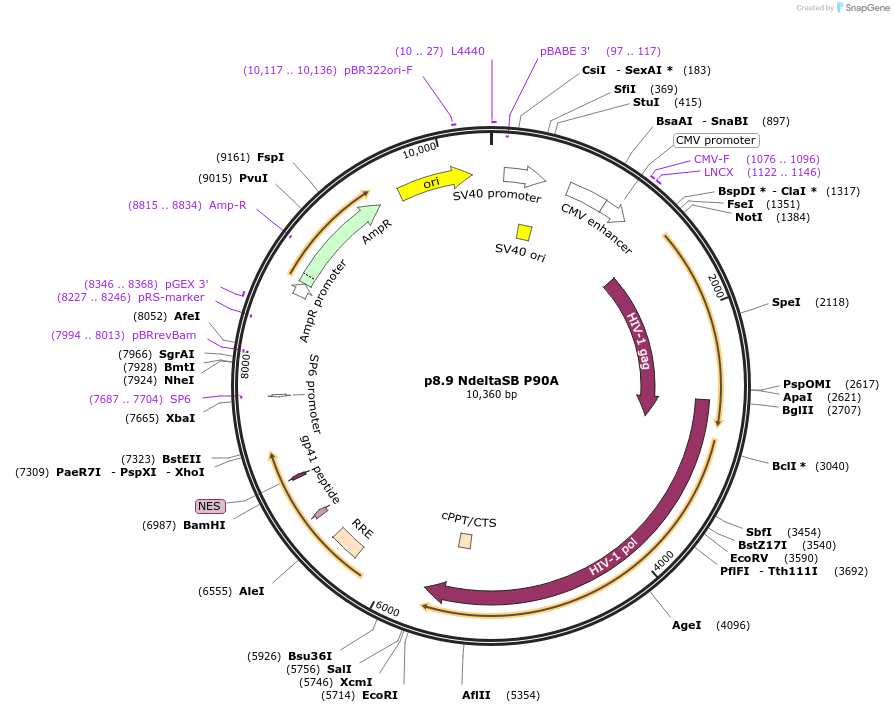
p8.9 NdeltaSB P90A
Plasmid#132930Purposep8.9 NdeltaSB bearing P90A mutant capsid sequenceDepositorInsertgag P90A and pol
UseLentiviralMutationP90AAvailable SinceOct. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
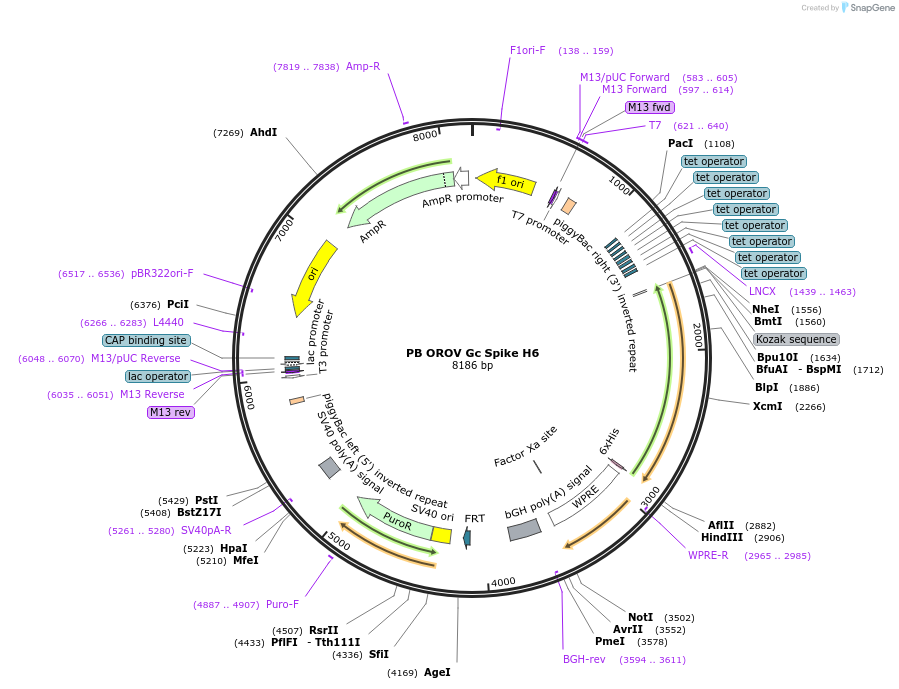
PB OROV Gc Spike H6
Plasmid#229662PurposeMake PiggyBac stable cell line expressing Oropouche virus BeAn 19991 Gc spike region (residues 482–894 of M polyprotein) with C-terminal His6 tagDepositorInsertOROV Gc spike
UsePiggybacTags6xHisExpressionMammalianMutationcontains residues 482-894 onlyPromoterTREAvailable SinceAug. 19, 2025AvailabilityAcademic Institutions and Nonprofits only -
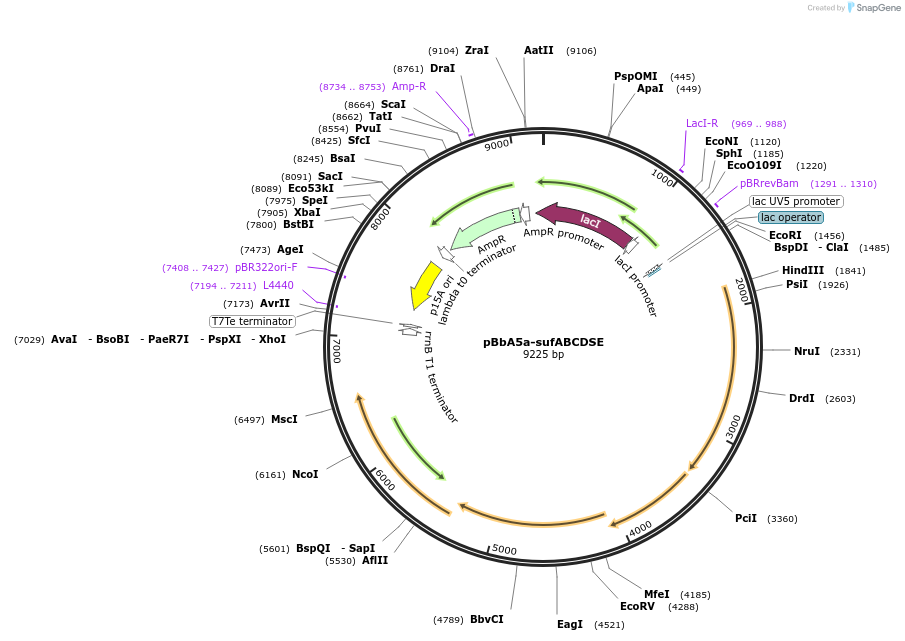
pBbA5a-sufABCDSE
Plasmid#195056PurposeExpresses SUF operon from E. coli from IPTG-inducible promoterDepositorInsertsufABCDSE
ExpressionBacterialAvailable SinceFeb. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
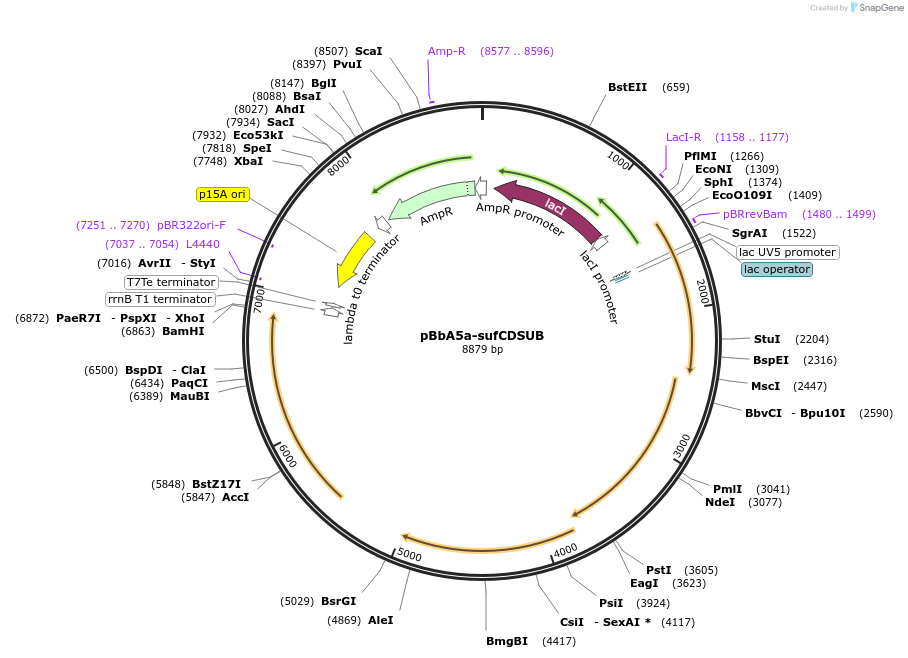
pBbA5a-sufCDSUB
Plasmid#195055PurposeExpresses SUF operon from B. subtilis from IPTG-inducible promoterDepositorInsertsufCDSUB
ExpressionBacterialAvailable SinceFeb. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
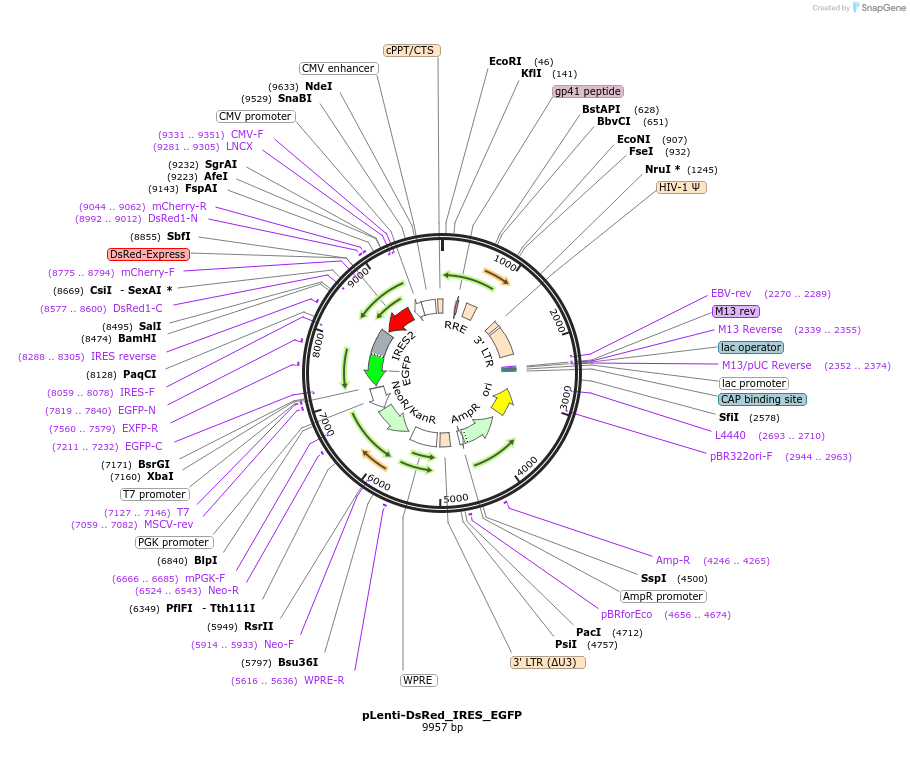
pLenti-DsRed_IRES_EGFP
Plasmid#92194PurposeLentiviral overexpression vector to make stable bicistronic cell line for control (EGFP) screenDepositorInsertDsRed-IRES-EGFP
UseLentiviralExpressionMammalianAvailable SinceJune 26, 2017AvailabilityAcademic Institutions and Nonprofits only -
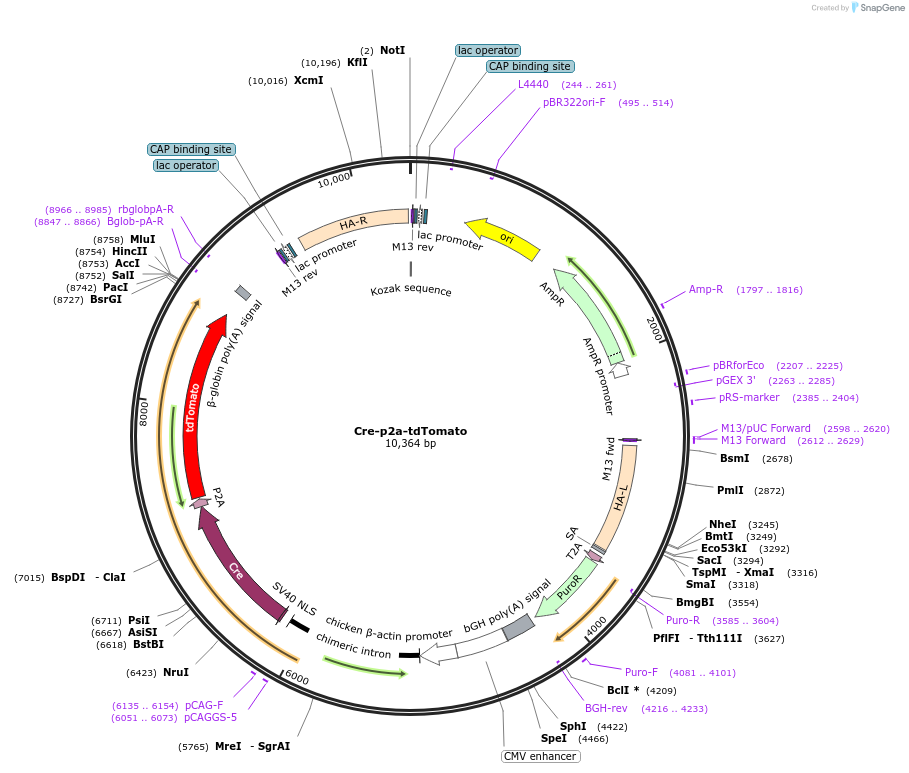
Cre-p2a-tdTomato
Plasmid#176837PurposeExpress Cre in mammalian cells and enable FACS sorting Cre expressing cells by tdTomato expressionDepositorInsertCre-P2A-tdTomato
ExpressionMammalianPromoterCAGGSAvailable SinceNov. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
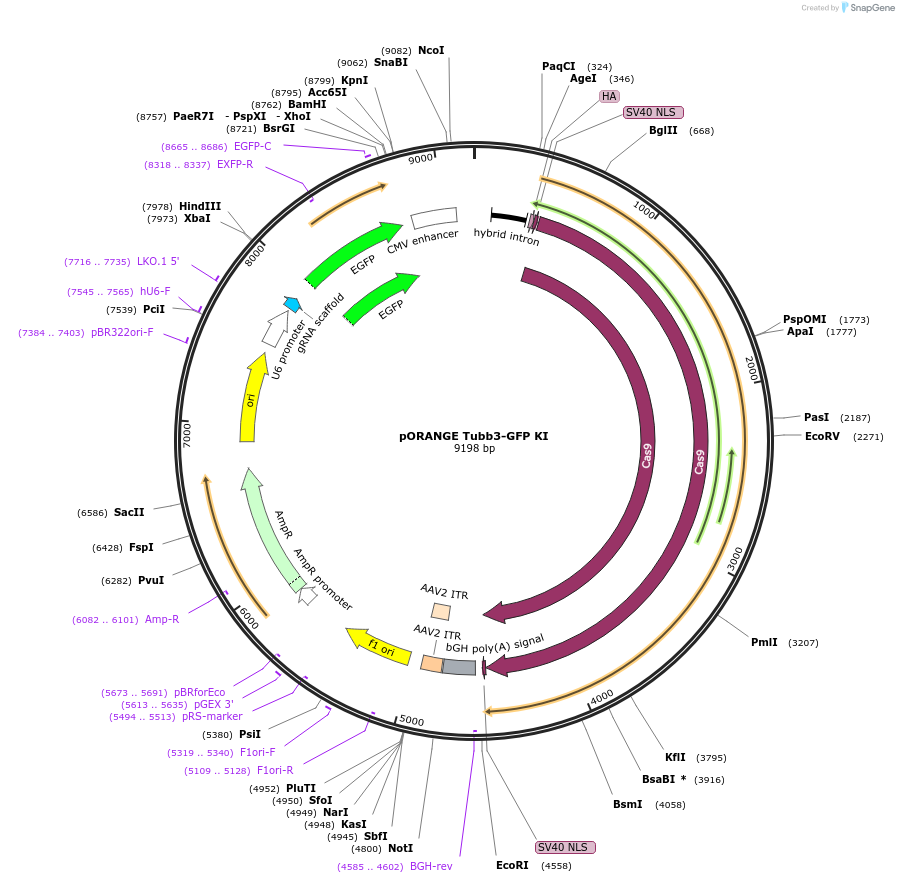
pORANGE Tubb3-GFP KI
Plasmid#131497PurposeEndogenous tagging of β3 Tubulin: C-terminal (amino acid position: STOP codon)DepositorAvailable SinceSept. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
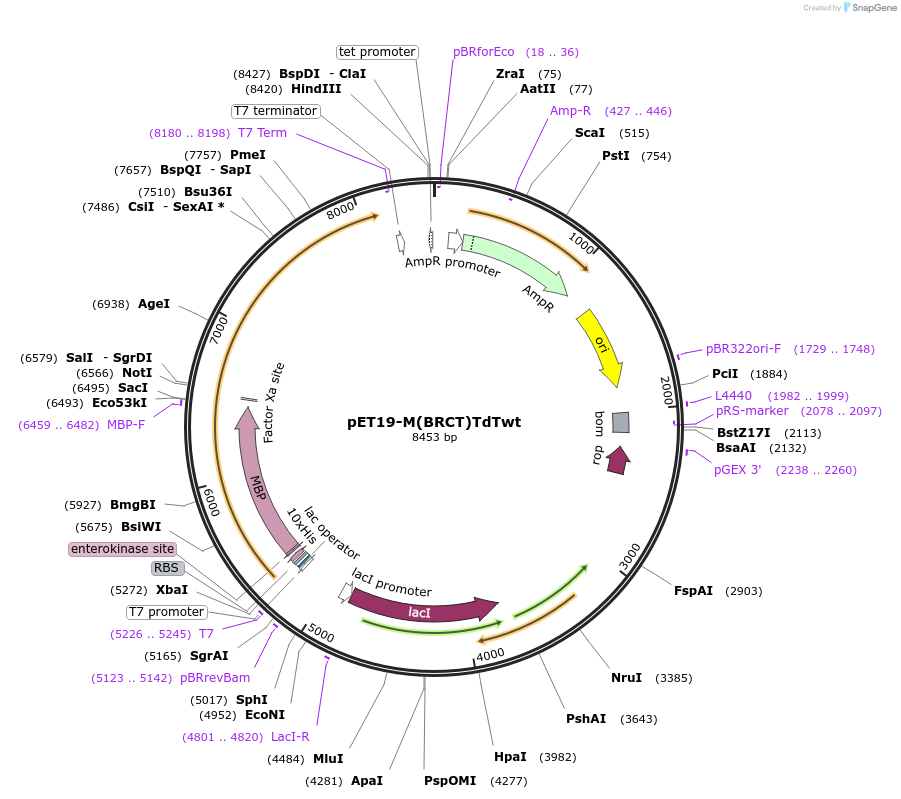
pET19-M(BRCT)TdTwt
Plasmid#207459PurposeBacterial expression of Terminal deoxynucleotidyl transferase (TdT) variant, including the N-terminal BRCT domainDepositorAvailable SinceSept. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
pLenti-DsRed_IRES_SNCA:EGFP
Plasmid#92195PurposeLentiviral overexpression vector to make stable bicistronic cell line for α-Syn screenDepositorInsertDsRed-IRES-SNCA-EGFP
UseLentiviralExpressionMammalianAvailable SinceAug. 7, 2017AvailabilityAcademic Institutions and Nonprofits only -
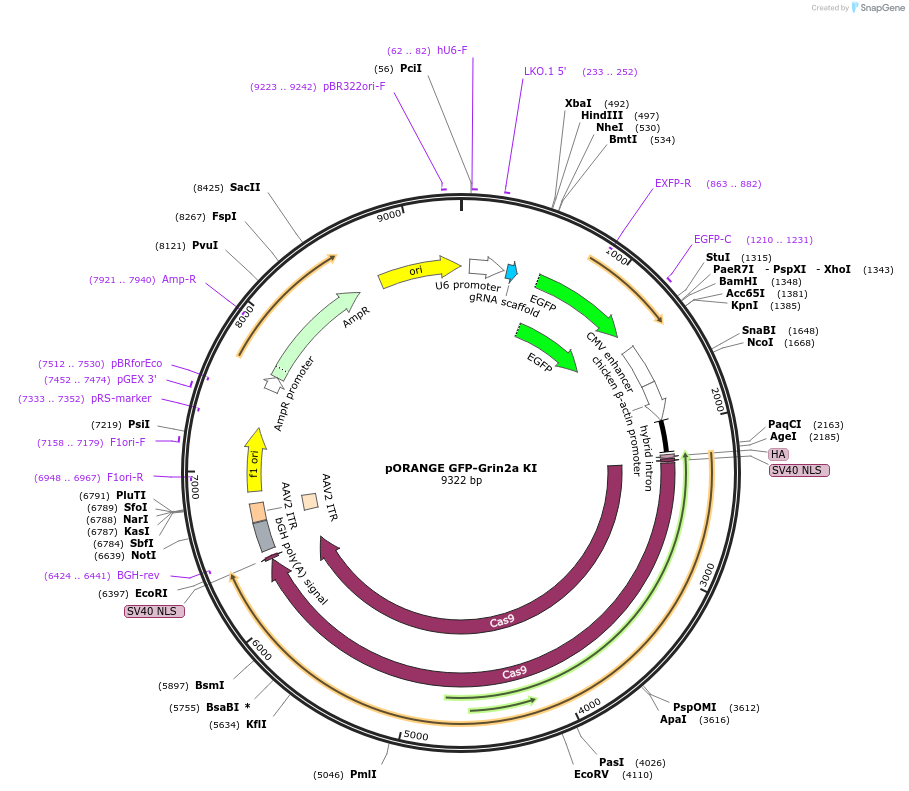
pORANGE GFP-Grin2a KI
Plasmid#131486PurposeEndogenous tagging of GluN2a: N-terminal (amino acid position: A25)DepositorAvailable SinceSept. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
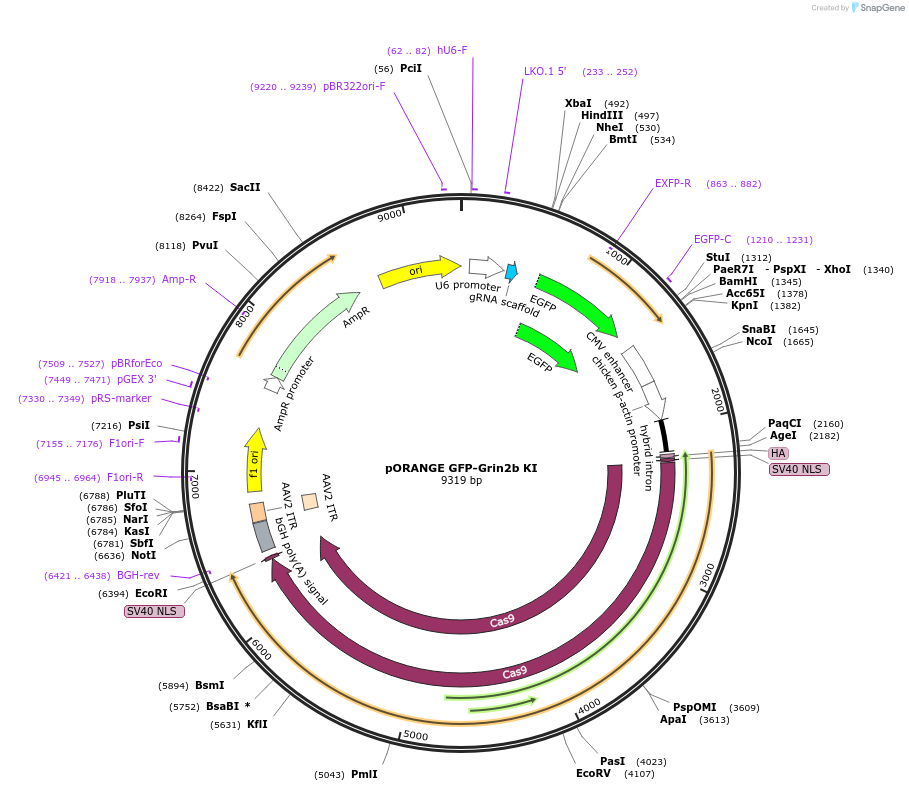
pORANGE GFP-Grin2b KI
Plasmid#131487PurposeEndogenous tagging of GluN2b: N-terminal (amino acid position: S34)DepositorAvailable SinceSept. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
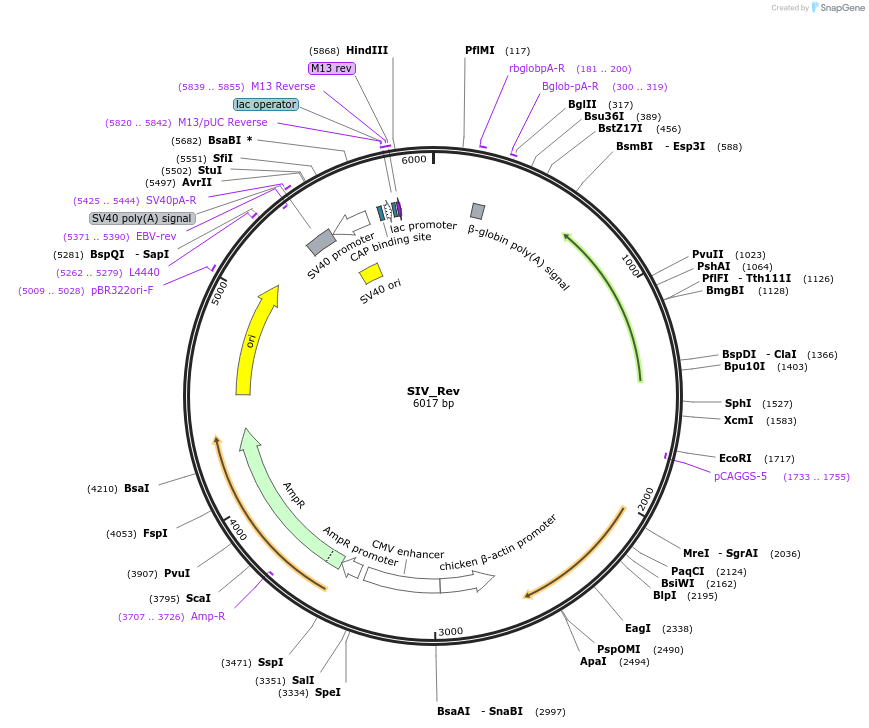
SIV_Rev
Plasmid#236244Purpose3rd generation SIV-based lentiviral packaging plasmid; Contains SIV RevDepositorInsertSIV Rev
UseLentiviralAvailable SinceMay 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
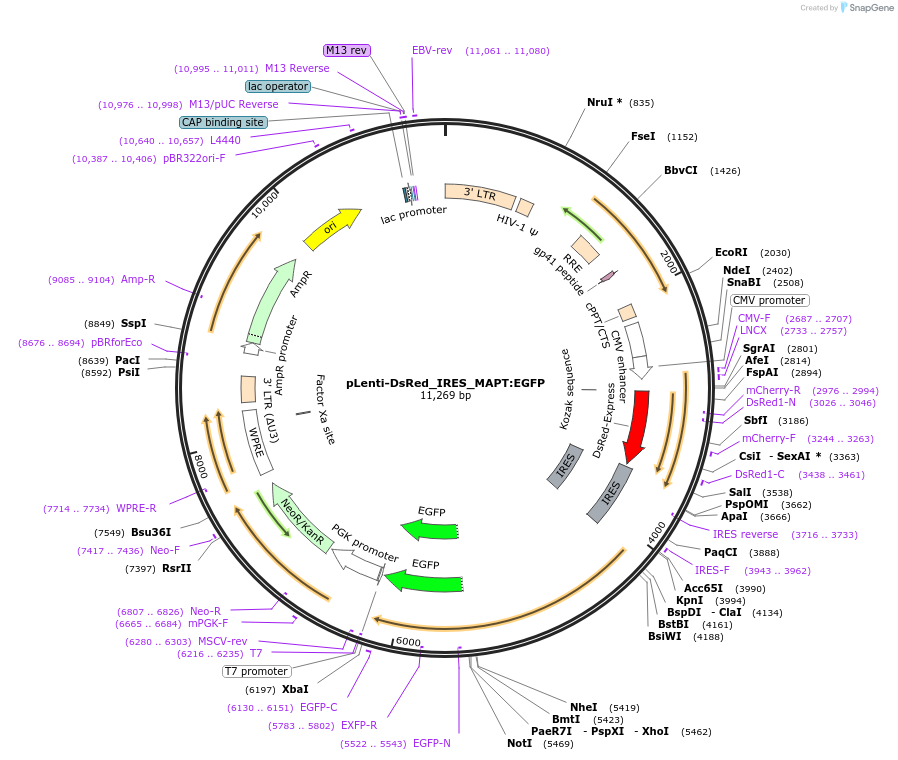
pLenti-DsRed_IRES_MAPT:EGFP
Plasmid#92196PurposeLentiviral overexpression vector to make stable bicistronic cell line for Tau screenDepositorInsertDsRed-IRES-MAPT-EGFP
UseLentiviralExpressionMammalianAvailable SinceAug. 7, 2017AvailabilityAcademic Institutions and Nonprofits only -
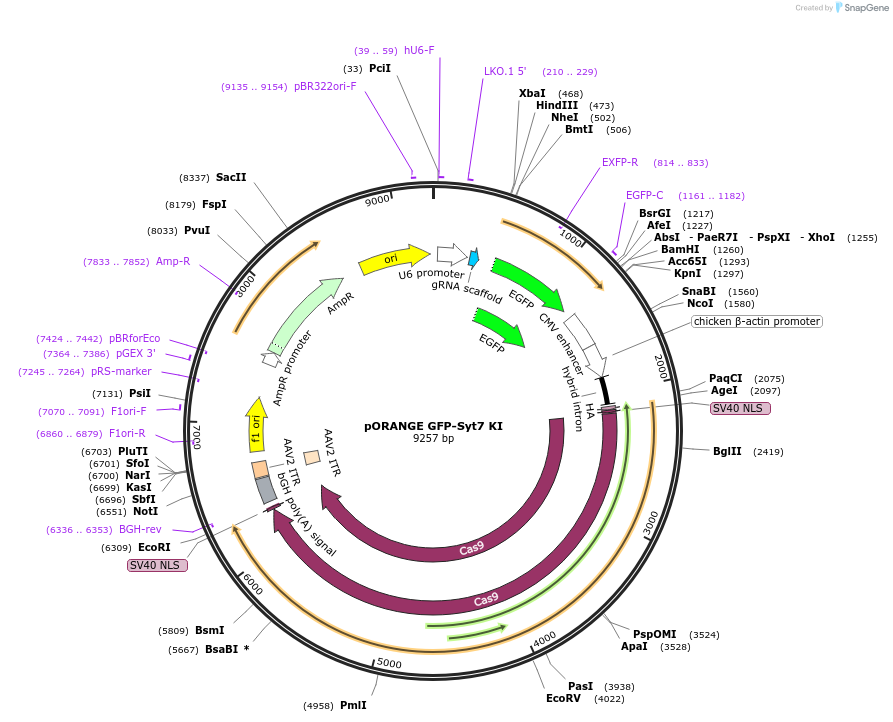
pORANGE GFP-Syt7 KI
Plasmid#131488PurposeEndogenous tagging of Synaptotagmin-7: N-terminal (amino acid position: P5)DepositorAvailable SinceSept. 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
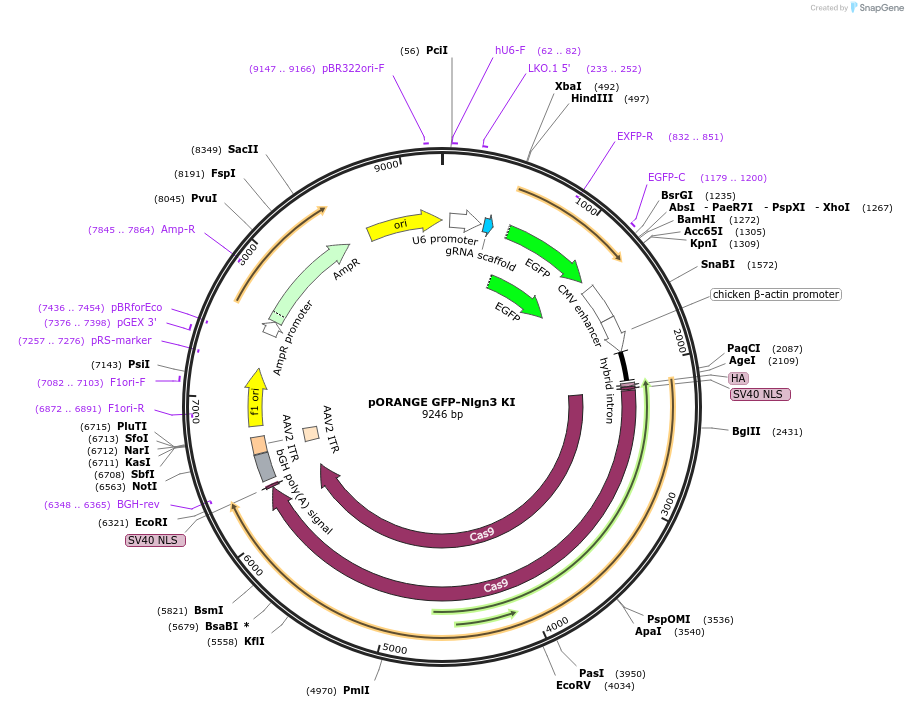
pORANGE GFP-Nlgn3 KI
Plasmid#131501PurposeEndogenous tagging of Neuroligin-3: N-terminal (amino acid position: T37)DepositorUseCRISPRExpressionMammalianPromoterU6 and CbhAvailable SinceSept. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
pENTRY-GFP
Plasmid#15301DepositorInserthrGFP
ExpressionMammalianAvailable SinceJuly 12, 2007AvailabilityAcademic Institutions and Nonprofits only -
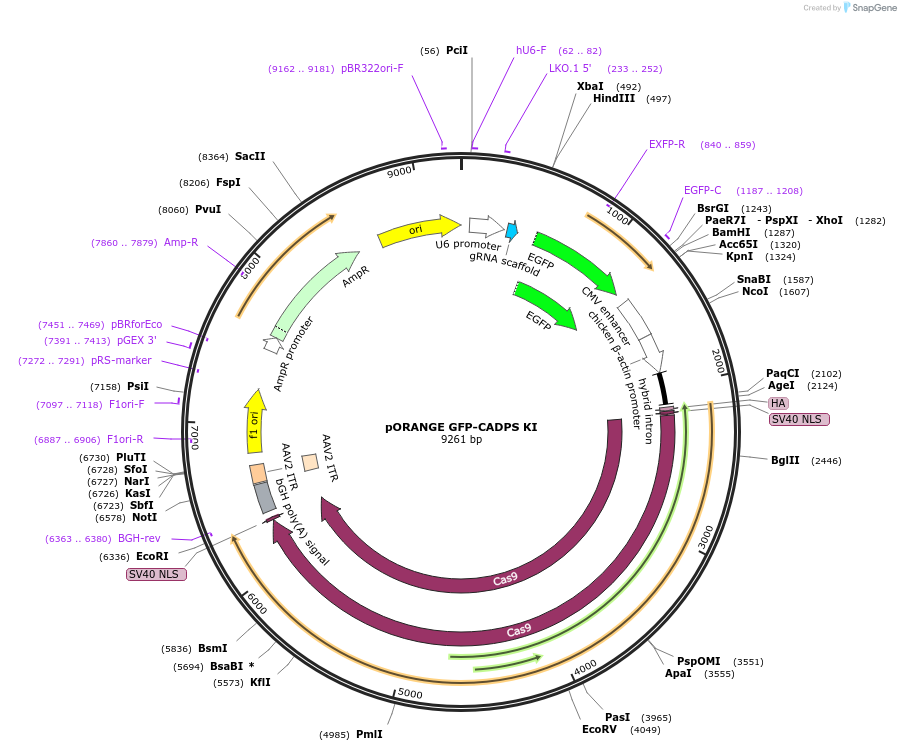
pORANGE GFP-CADPS KI
Plasmid#131482PurposeEndogenous tagging of CAPS1: N-terminal (amino acid position: S39)DepositorAvailable SinceSept. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
pFUGW ORANGE Tubb3-GFP KI _ mCherry-KASH
Plasmid#131508PurposeEndogenous tagging of β3 Tubulin: C-terminal (amino acid position: STOP codon)DepositorUseLentiviralExpressionMammalianPromoterhUBCAvailable SinceSept. 20, 2019AvailabilityAcademic Institutions and Nonprofits only