We narrowed to 21,231 results for: omp
-
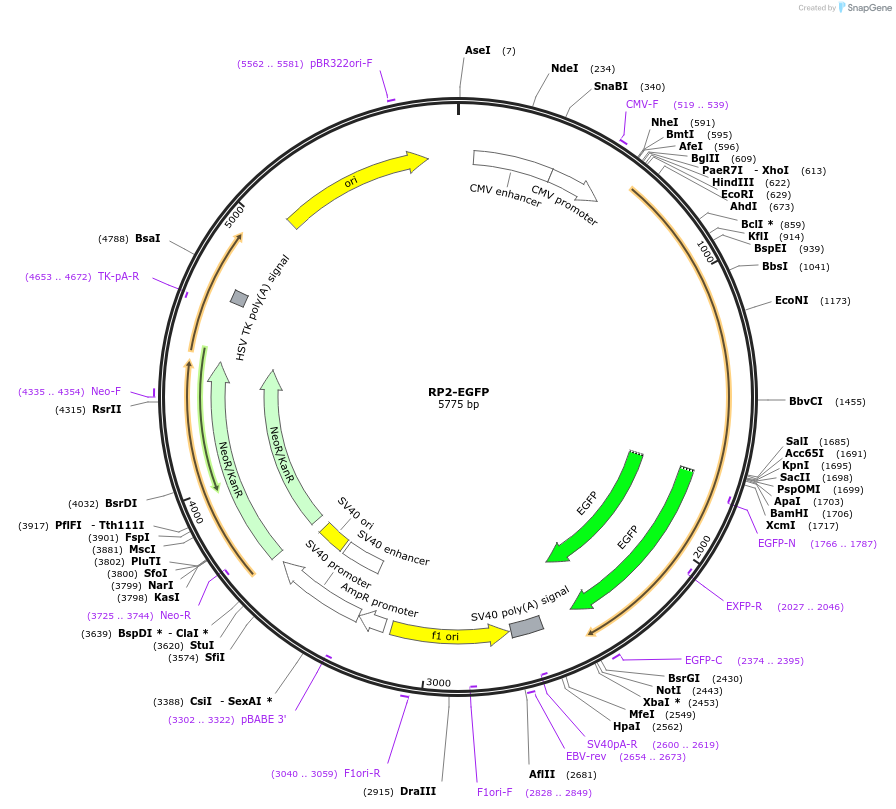
Plasmid#86073PurposeA mammalian expression plasmid encoding retinitis pigmentosa 2 (RP2) with C-terminus EGFP.DepositorAvailable SinceMay 9, 2017AvailabilityAcademic Institutions and Nonprofits only
-
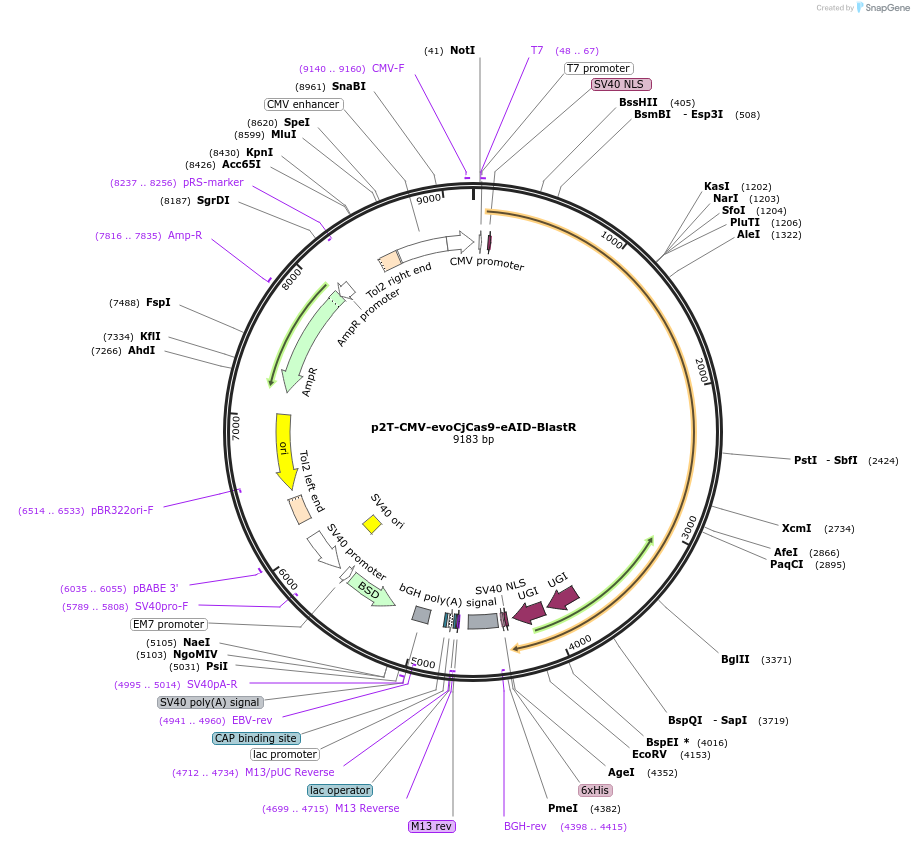
p2T-CMV-evoCjCas9-eAID-BlastR
Plasmid#194062PurposeMammalian expression of evoCjCas9 eAIDDepositorInsertevoCjCas9-eAID
UseCRISPRExpressionMammalianAvailable SinceSept. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
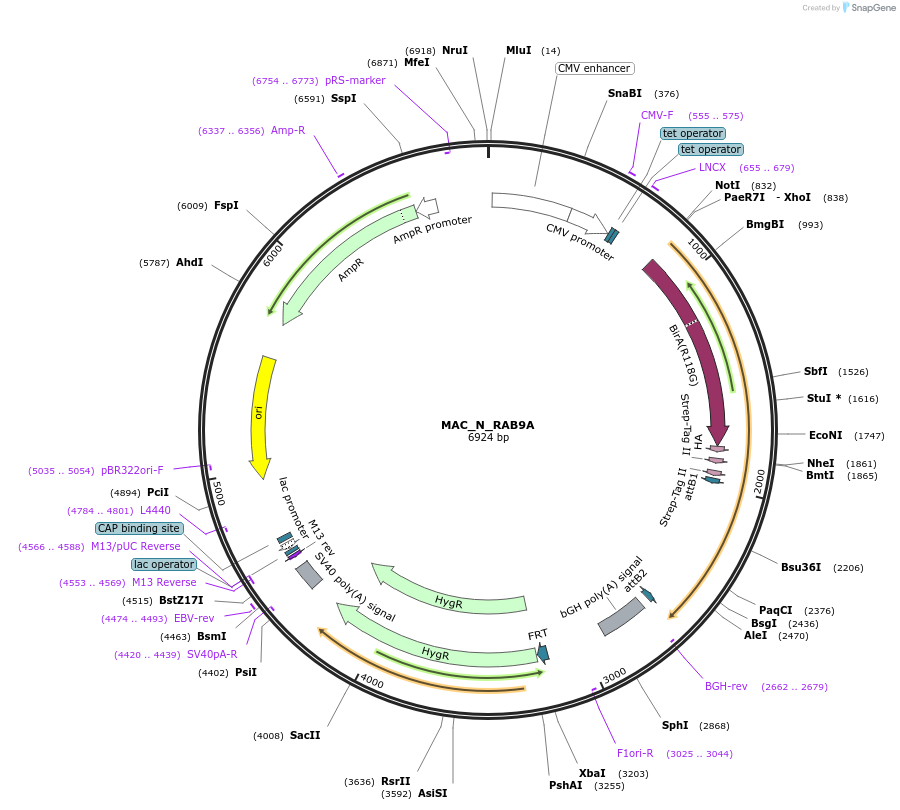
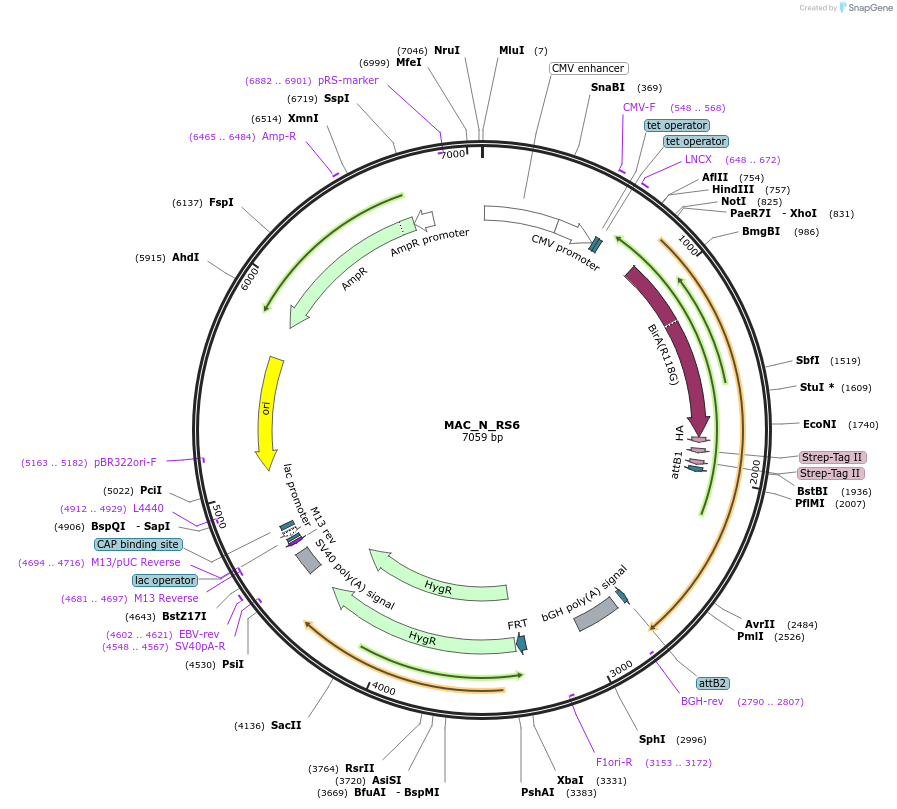
MAC_N_RAB9A
Plasmid#111693PurposeMAC-tagged gene expressionDepositorInsertRAB9A (RAB9A Human)
ExpressionMammalianAvailable SinceJune 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
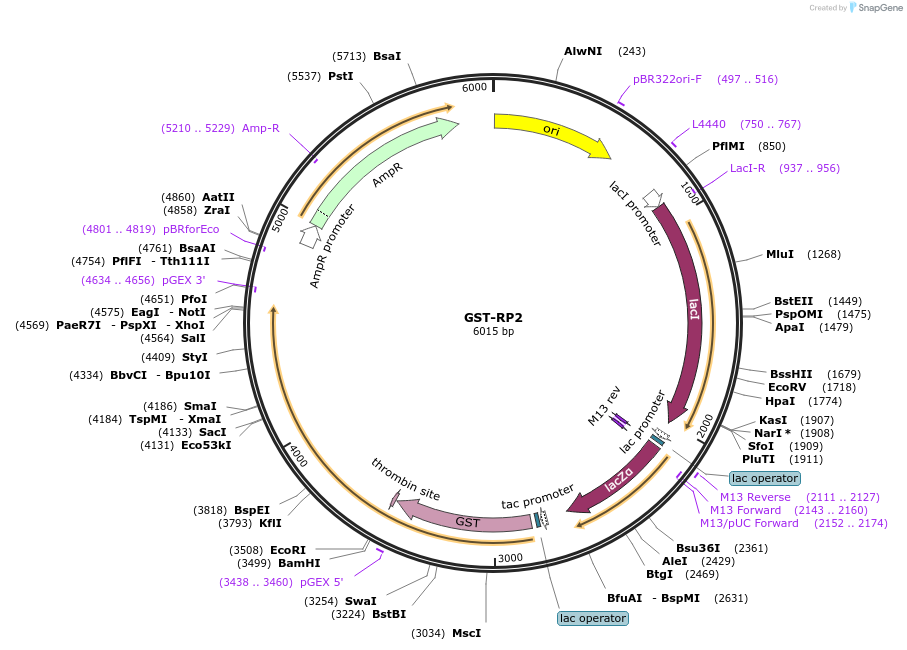
GST-RP2
Plasmid#86072PurposeA bacterial expression plasmid encoding retinitis pigmentosa 2 (RP2) with N-terminus GST-tag.DepositorAvailable SinceMay 9, 2017AvailabilityAcademic Institutions and Nonprofits only -
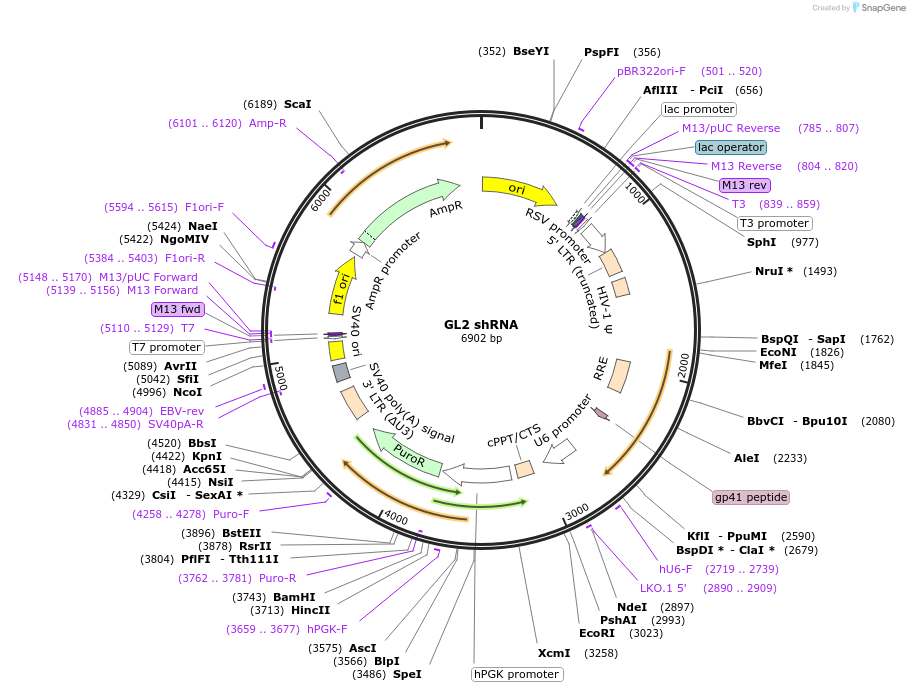
GL2 shRNA
Plasmid#86083PurposeA lentiviral construct for building stable cell lines expressing GL2 shRNA via puromycin selection.DepositorInsertcontrol shRNA
PromoterU6Available SinceMay 26, 2017AvailabilityAcademic Institutions and Nonprofits only -
pBABE flag M33
Plasmid#1951DepositorAvailable SinceMarch 23, 2006AvailabilityAcademic Institutions and Nonprofits only -
FLAG-Amida
Plasmid#15362DepositorAvailable SinceAug. 17, 2007AvailabilityAcademic Institutions and Nonprofits only -
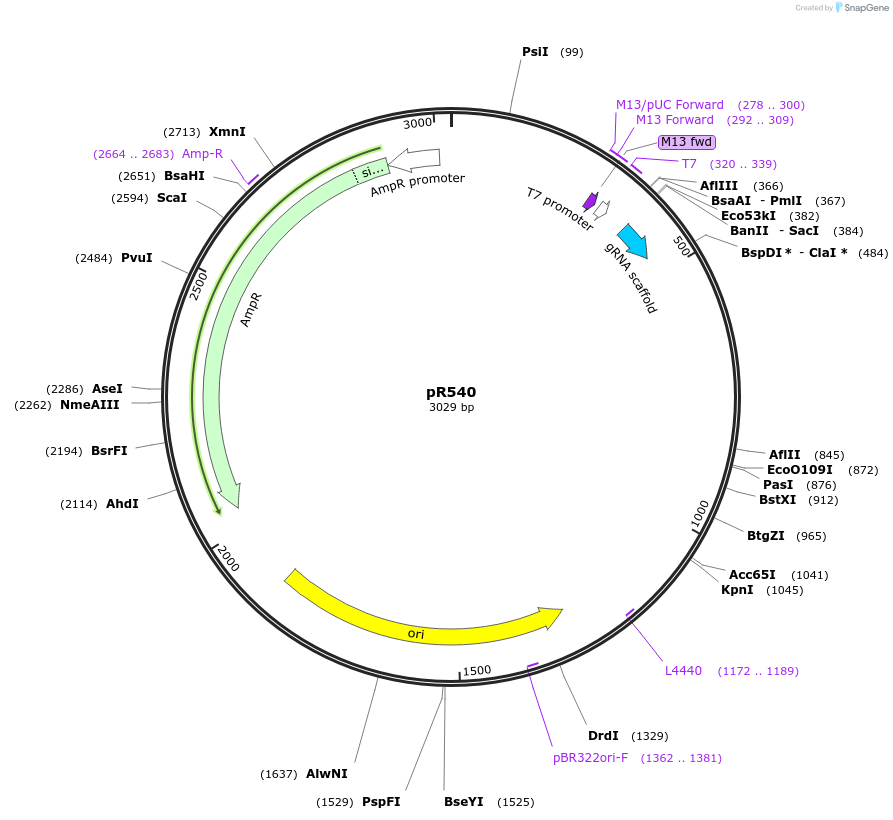
pR540
Plasmid#207885Purposetemplate for 2 gRNA amplificationDepositorInsertscaffold-pU6
ExpressionBacterialAvailable SinceNov. 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
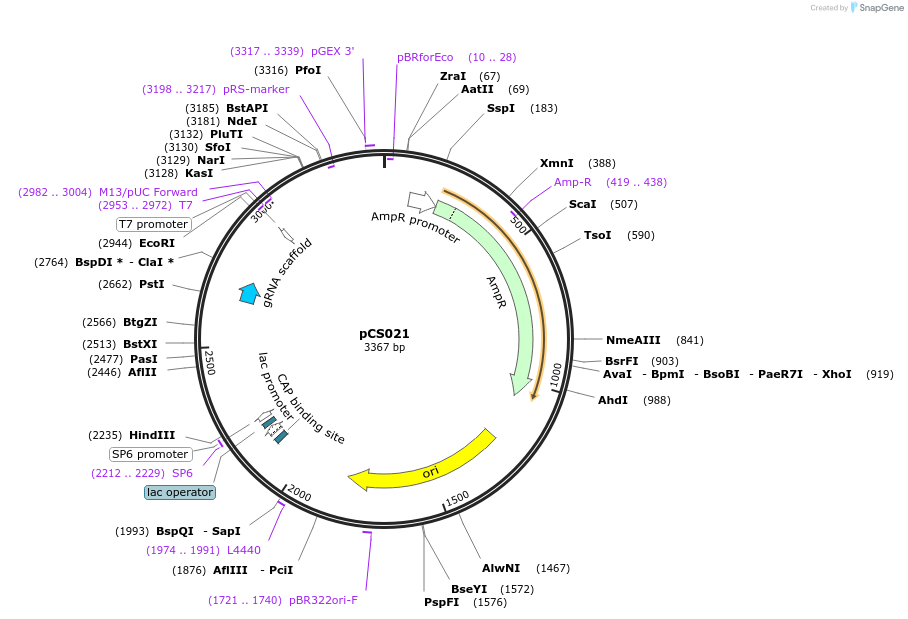
pCS021
Plasmid#207917PurposeLevel 0 for gRNA in E2 module (5' CCAT-3' ACTA)DepositorInsertpU6-AarI-scaffold-term
UseCRISPRAvailable SinceNov. 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
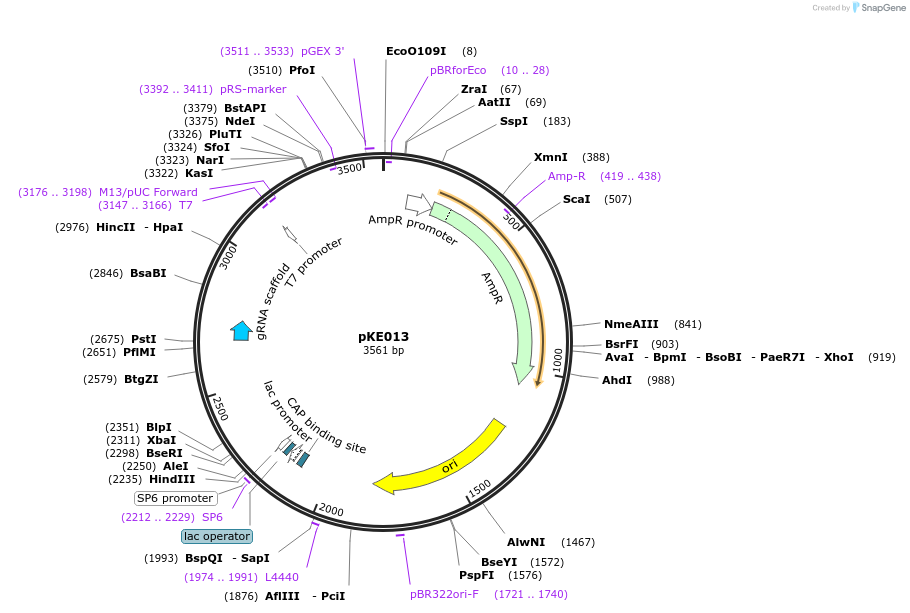
pKE013
Plasmid#207916PurposeLevel 0 for gRNA in E1 module (5' CTGC-3' CCAT)DepositorInsertpU3-AarI-scaffold-term
UseCRISPRAvailable SinceNov. 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
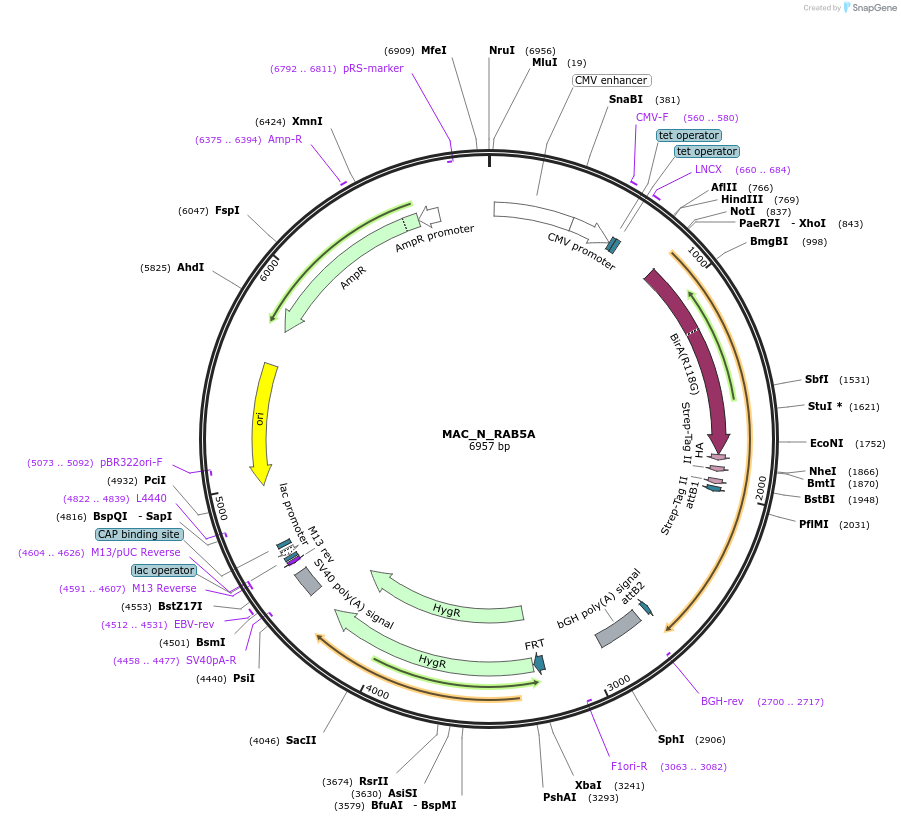
MAC_N_RAB5A
Plasmid#111692PurposeMAC-tagged gene expressionDepositorInsertRAB5A (RAB5A Human)
ExpressionMammalianAvailable SinceJune 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
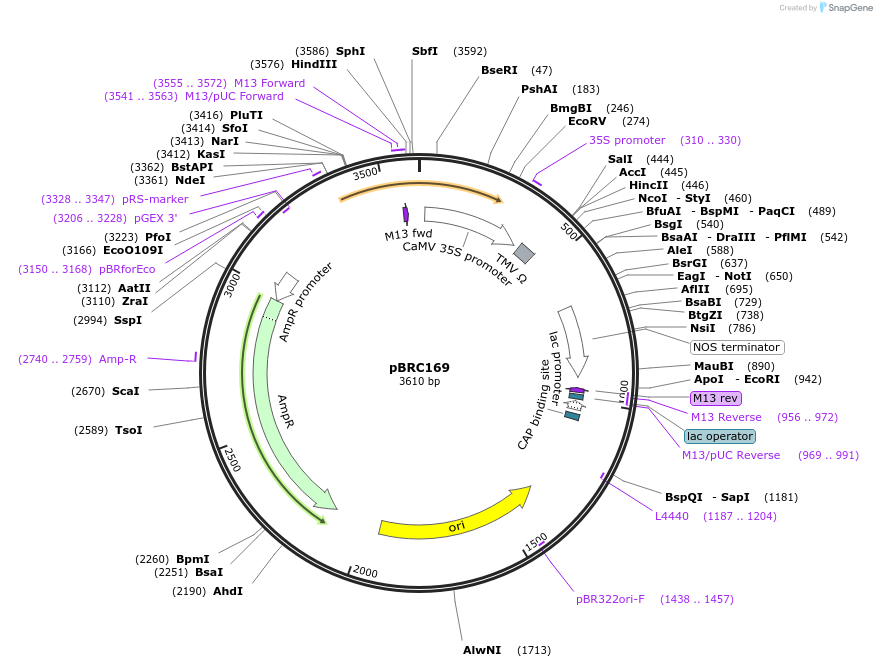
pBRC169
Plasmid#79670PurposeDsREDmonomer-based BiFC vector for transient expression of RC (169-225) in plants.DepositorTypeEmpty backboneUseReporter plasmidTagsDsRED-monomer (169–225)/RC169ExpressionPlantPromoterCaMV35SAvailable SinceAug. 18, 2016AvailabilityAcademic Institutions and Nonprofits only -
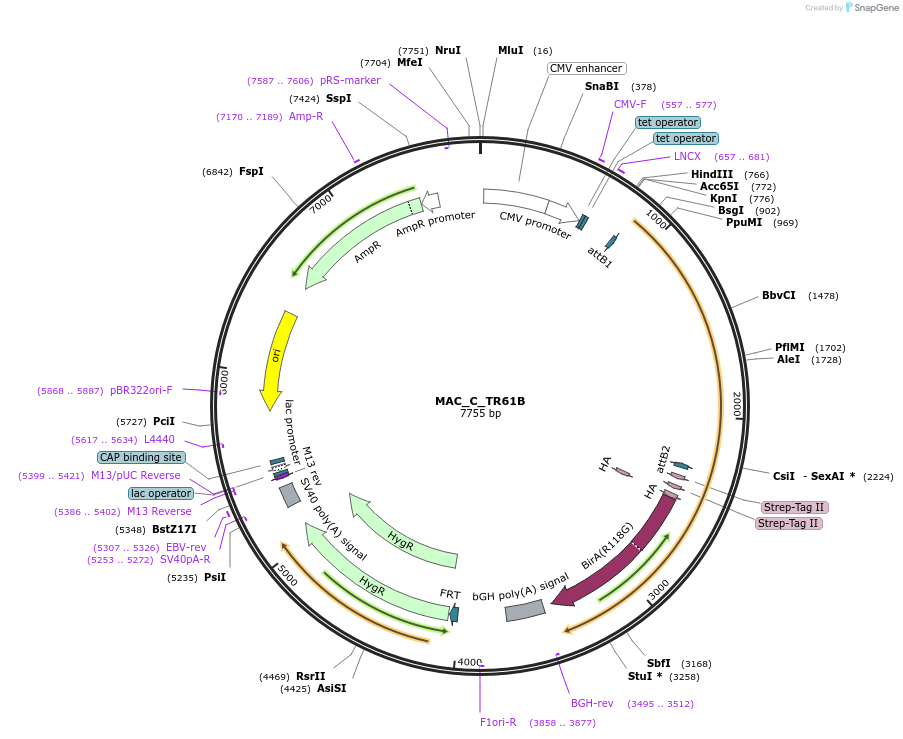
MAC_C_TR61B
Plasmid#111679PurposeMAC-tagged gene expressionDepositorInsertTR61B (TRMT61B Human)
ExpressionMammalianAvailable SinceJune 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
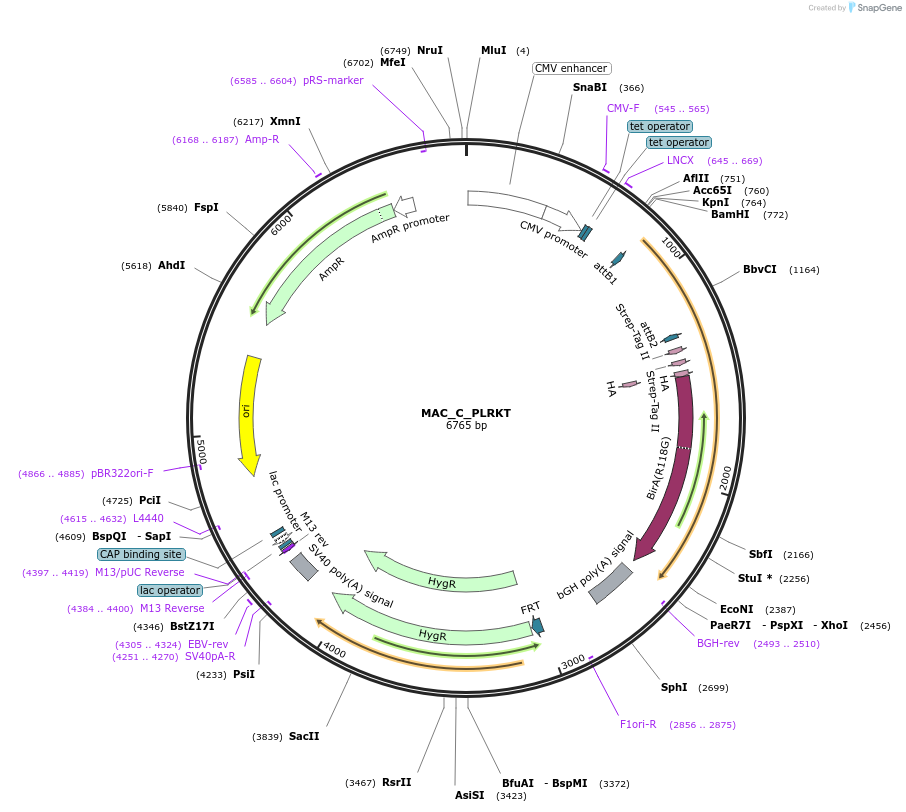
MAC_C_PLRKT
Plasmid#111673PurposeMAC-tagged gene expressionDepositorInsertPLRKT (PLGRKT Human)
ExpressionMammalianAvailable SinceJune 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
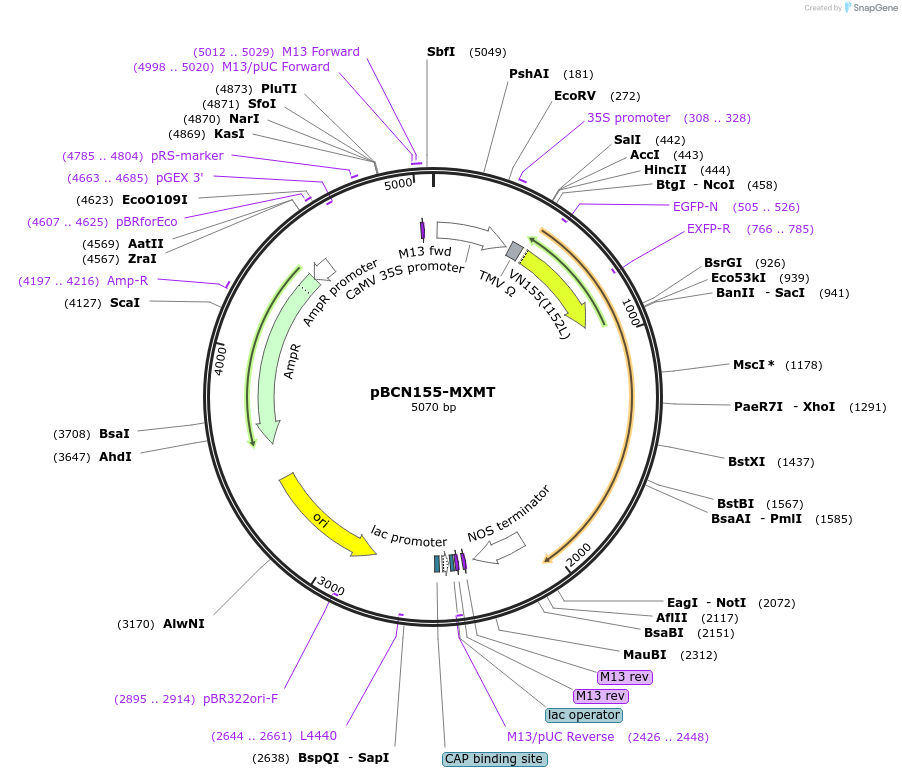
pBCN155-MXMT
Plasmid#79815PurposeCFP-based BiFC vector for plants (Positive control: MXMT)DepositorInsertCaMXMT1
UseReporter plasmidTagsCN155/CFP (1–155)ExpressionPlantPromoterCaMV 35S promoter and CaMV35SAvailable SinceAug. 18, 2016AvailabilityAcademic Institutions and Nonprofits only -
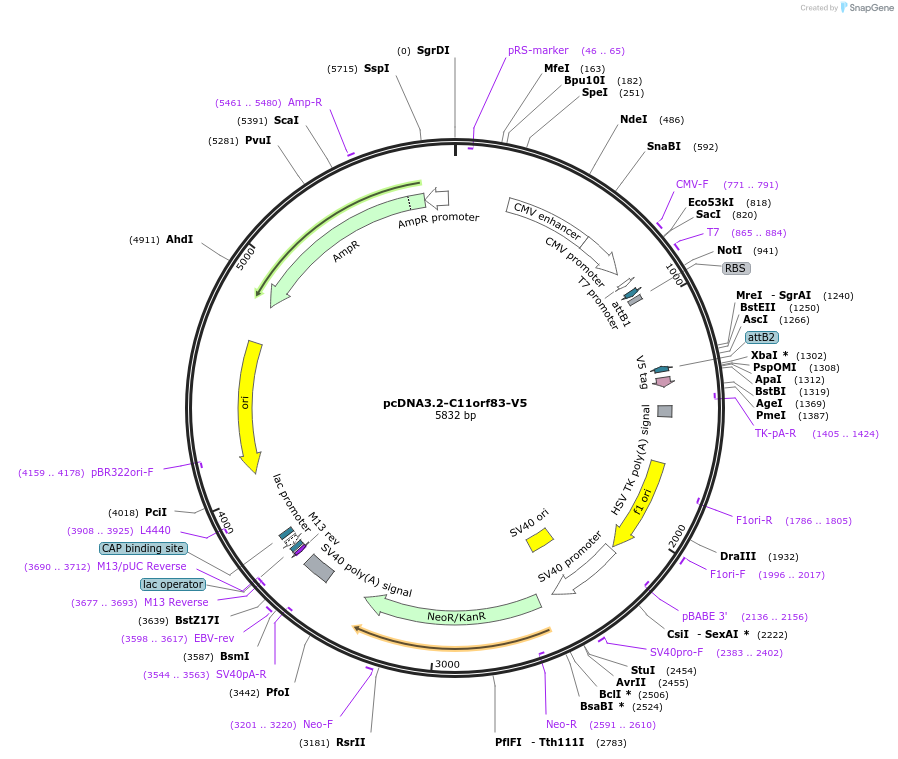
pcDNA3.2-C11orf83-V5
Plasmid#65843PurposeMammalian expression of C terminally V5-tagged C11orf83/UQCC3DepositorAvailable SinceJuly 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
-
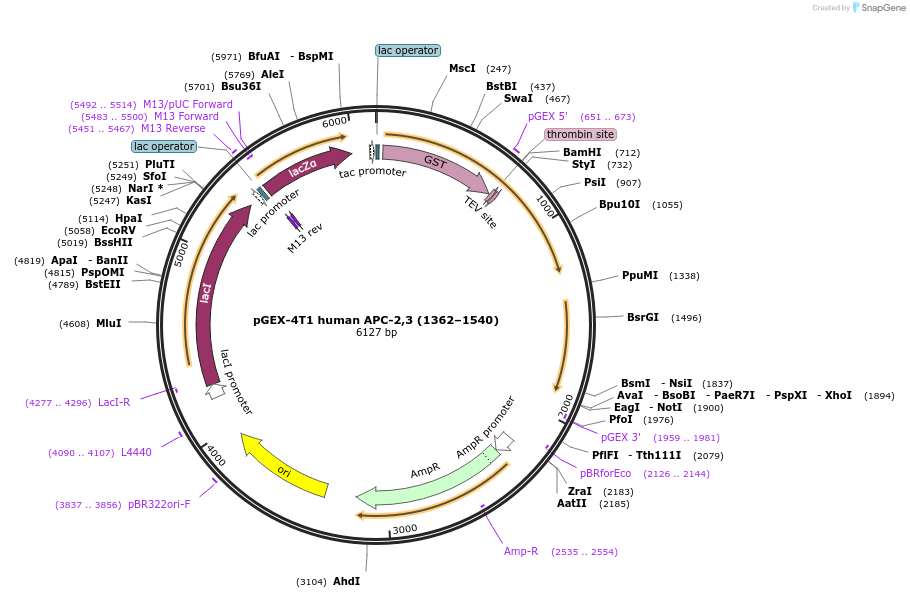
pGEX-4T1 human APC-2,3 (1362–1540)
Plasmid#132631Purposeexpresses human APC-2,3 (1362–1540) in E. coliDepositorInsertAPC-2,3 (1362–1540)
ExpressionBacterialAvailable SinceOct. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
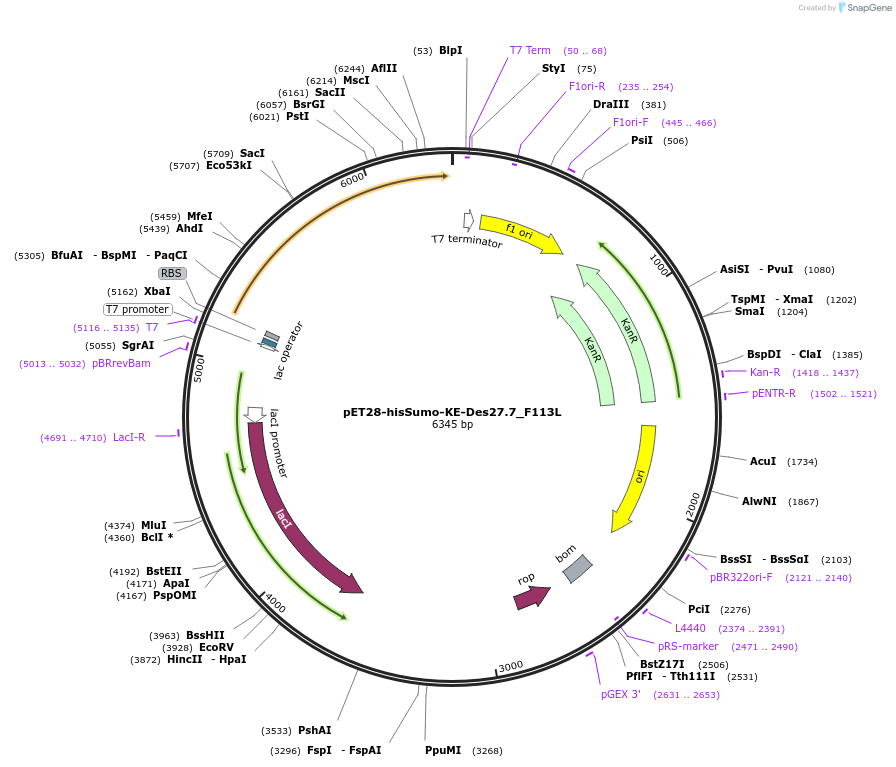
pET28-hisSumo-KE-Des27.7_F113L
Plasmid#248016PurposeExpression of synthetic Kemp eliminase (Des27.7 F113L)DepositorInsertKE-Des27.7_F113L
Tags12xHis-bdSumoExpressionBacterialMutationF113LPromoterT7Available SinceDec. 11, 2025AvailabilityAcademic Institutions and Nonprofits only -
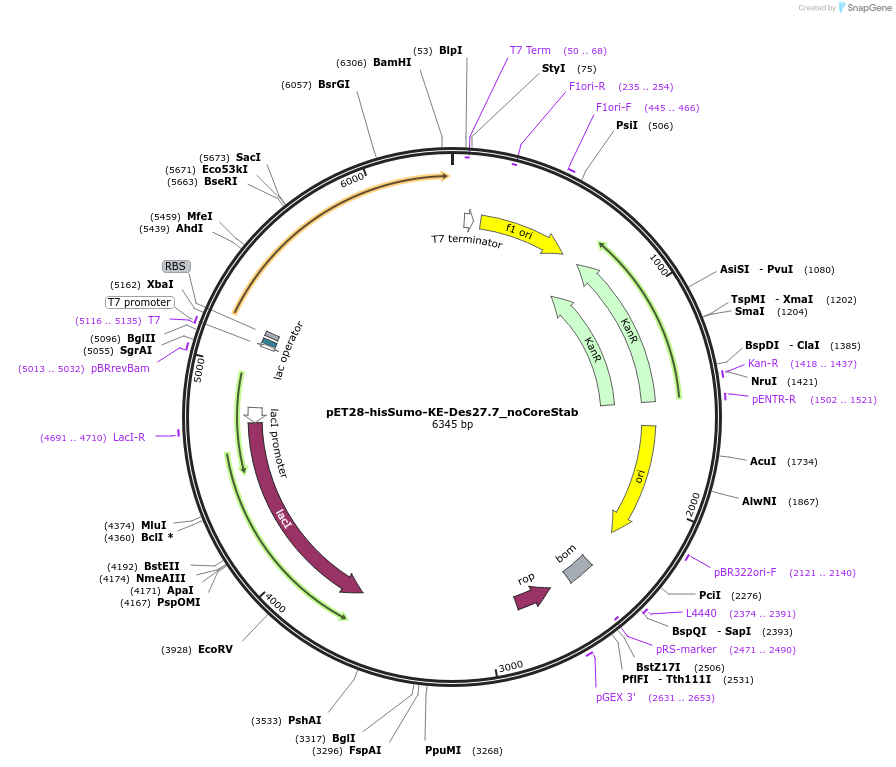
pET28-hisSumo-KE-Des27.7_noCoreStab
Plasmid#248014PurposeExpression of synthetic Kemp eliminase (Ablation of Des27.7 ModularAssembly&PROSS&ActiveSite stability)DepositorInsertKE-Des27.7_noCoreStab
Tags12xHis-bdSumoExpressionBacterialPromoterT7Available SinceDec. 11, 2025AvailabilityAcademic Institutions and Nonprofits only