We narrowed to 18,764 results for: MUT
-
Plasmid Kit#1000000249PurposeThe T2A Split/Link TurboID Kit uses a sequential Golden Gate assembly method and can be used to assemble mammalian expression vectors for proximity labeling experiments.DepositorAvailable SinceAug. 7, 2024AvailabilityAcademic Institutions and Nonprofits only
-
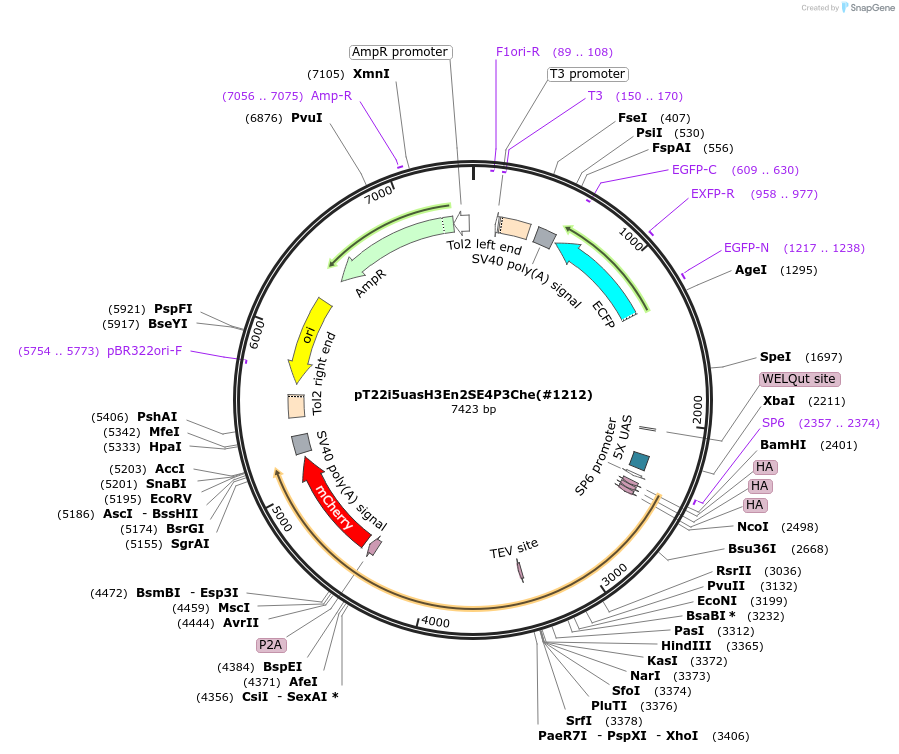
pT22i5uasH3En2SE4P3Che(#1212)
Plasmid#184098Purposecyclofen-inducible chicken EN2 (mut) activation in zebrafish permanent transgenicDepositorInsert3HA-chkEn2(C>S)-ERT2-5'fP2A-mCherry
UseZebrafish transgenesis (blue eyes)Tags3xHAMutationchkEn2: C175S; 5'fP2A: GSG-P2A; ERT2: G400V,…Promoter5xUASAvailable SinceJune 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
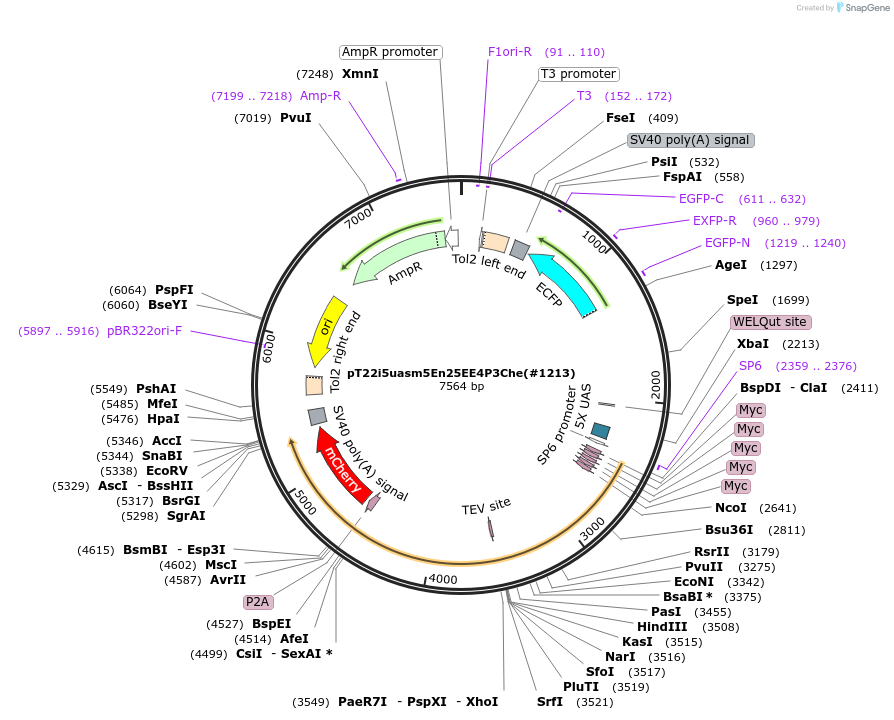
pT22i5uasm5En25EE4P3Che(#1213)
Plasmid#184099Purposecyclofen-inducible chicken EN2 (mut) activation in zebrafish permanent transgenicDepositorInsert5myc-chkEn2(5E)-ERT2-5'fP2A-mCherry
UseZebrafish transgenesis (blue eyes)Tags5xMycMutationchkEn2: S150E, S151E, S153E, S155E, S156E; ERT2: …Promoter5xUASAvailable SinceJune 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
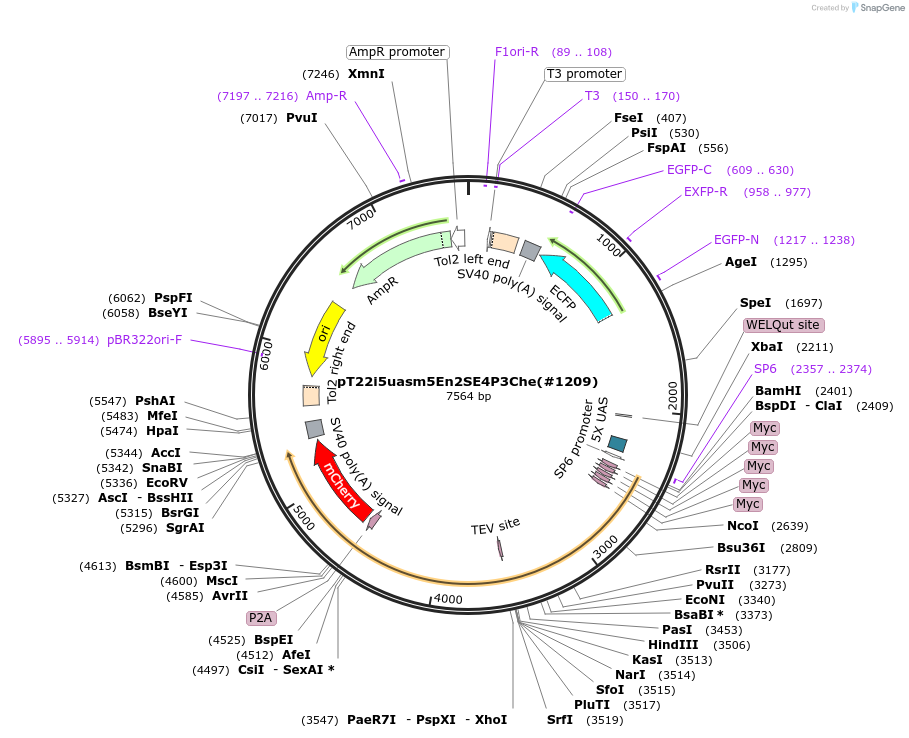
pT22i5uasm5En2SE4P3Che(#1209)
Plasmid#184097Purposecyclofen-inducible chicken EN2 (mut) activation in zebrafish permanent transgenicDepositorInsert5myc-chkEn2(C>S)-ERT2-5'fP2A-mCherry
UseZebrafish transgenesis (blue eyes)Tags5xMycMutationchkEn2: C175S; 5'fP2A: GSG-P2A; ERT2: G400V,…Promoter5xUASAvailable SinceJune 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
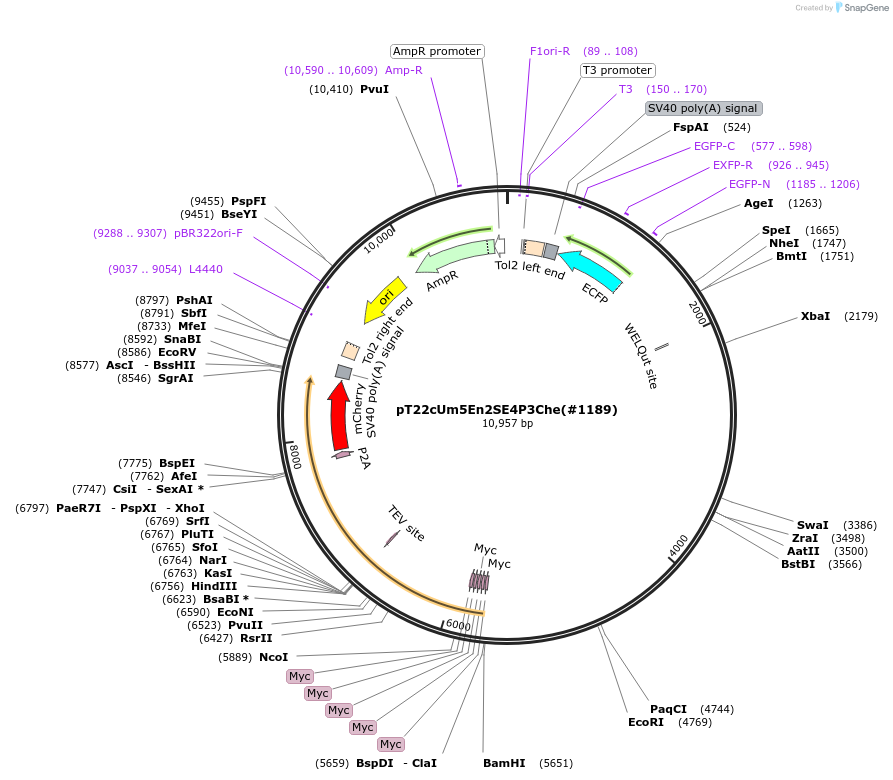
pT22cUm5En2SE4P3Che(#1189)
Plasmid#184095Purposecyclofen-inducible chicken EN2 (mut) activation in zebrafish permanent transgenicDepositorInsert5myc-chkEn2(C>S)-ERT2-5'fP2A-mCherry
UseZebrafish transgenesis (blue eyes)Tags5xMycMutationchkEn2: C175S; 5'fP2A: GSG-P2A; ERT2: G400V,…PromoterZf-UbiAvailable SinceJune 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
p077 pCS xTRPS1 (M)
Plasmid#11047DepositorInsertTRPS1 mut (trps1 Frog)
UseMammalian, avian, and zebrafishMutationTwo Cys residues in the GATA-type zinc finger wer…Available SinceDec. 6, 2005AvailabilityAcademic Institutions and Nonprofits only -
Cancer Pathways ORFs Kit
Plasmid Kit#1000000072PurposeA set of vectors containing cDNAs for studying oncogenic signaling pathways. cDNAs are either wild-type, constitutively active mutants, or dominant negative mutants.DepositorAvailable SinceMarch 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 2 (positions 437-527)
Pooled Library#174296PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library N501Y Tile 1 (positions 333-436)
Pooled Library#174295PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 2 (positions 437-527)
Pooled Library#174294PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 1 (positions 333-436)
Pooled Library#174293PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 1, positions 333-436) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
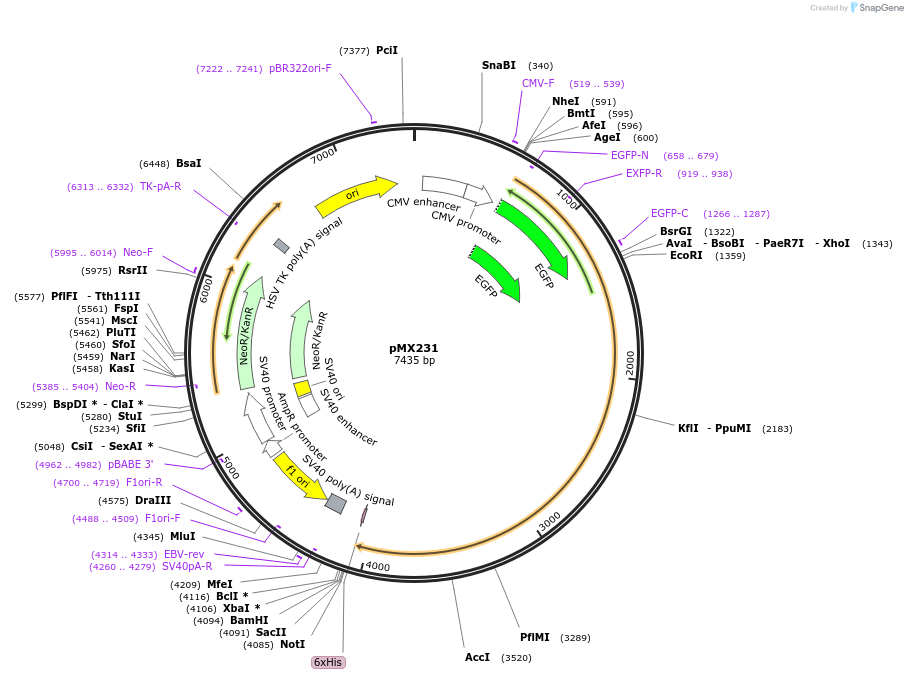
pMX231
Plasmid#23003DepositorAvailable SinceFeb. 8, 2010AvailabilityAcademic Institutions and Nonprofits only -
p068 pCS mTRPS1 (M)
Plasmid#11040DepositorInsertTRPS1 mut (Trps1 Mouse)
UseXenopus, mammalian, avian, and zebrafishMutationTwo Cys residues at positions 917 and 920 in the …Available SinceDec. 2, 2005AvailabilityAcademic Institutions and Nonprofits only -
p075 pCS xTRPS1 delN delC119 (M)
Plasmid#11045DepositorInsertTRPS1 delN delC119 mut (trps1 Frog)
UseXenopus, mammalian, avian, and zebrafishMutationaa806-1153 of xTRPS1. Two Cys residues in the GA…Available SinceDec. 6, 2005AvailabilityAcademic Institutions and Nonprofits only -
p063 pCS xTRPS1 delta N (M)
Plasmid#11036DepositorInsertTRPS1 deltaN mut (trps1 Frog)
UseMammalian, avian, and zebrafishMutationaa806-1153 of xTRPS1. Two Cys residues in the GA…Available SinceDec. 2, 2005AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library C
Pooled Library#157973PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library B
Pooled Library#157972PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library A
Pooled Library#157971PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library 1
Pooled Library#168776PurposePooled library expressing barcoded SARS-CoV-2 RNA Binding Domain mutationsDepositorExpressionYeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library 2
Pooled Library#168777PurposePooled library expressing barcoded SARS-CoV-2 RNA Binding Domain mutationsDepositorExpressionYeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only