We narrowed to 2,481 results for: Clu
-
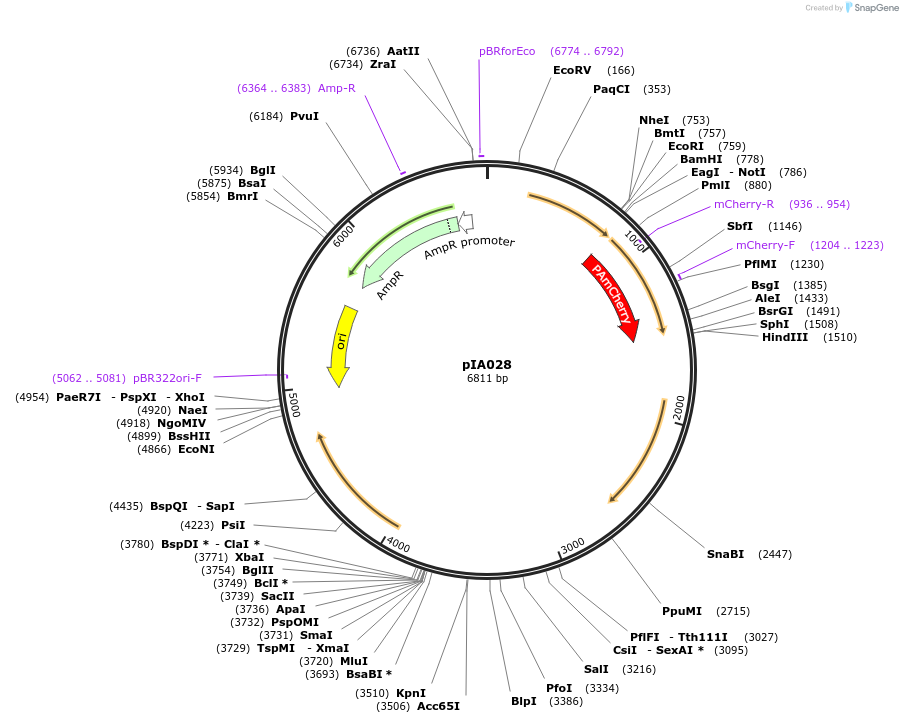
Plasmid#200419PurposethrC::(PAmCherry1 erm2),for PAmCherry1 fusions.PALM integration vector for B. subtilis. Includes PAmCherry1 photoactivatable FP with multi cloning sites (MCS) for fast and interchangeable cloningDepositorArticleTypeEmpty backboneTagsPAmCherry1ExpressionBacterialAvailable SinceAug. 23, 2023AvailabilityAcademic Institutions and Nonprofits only
-
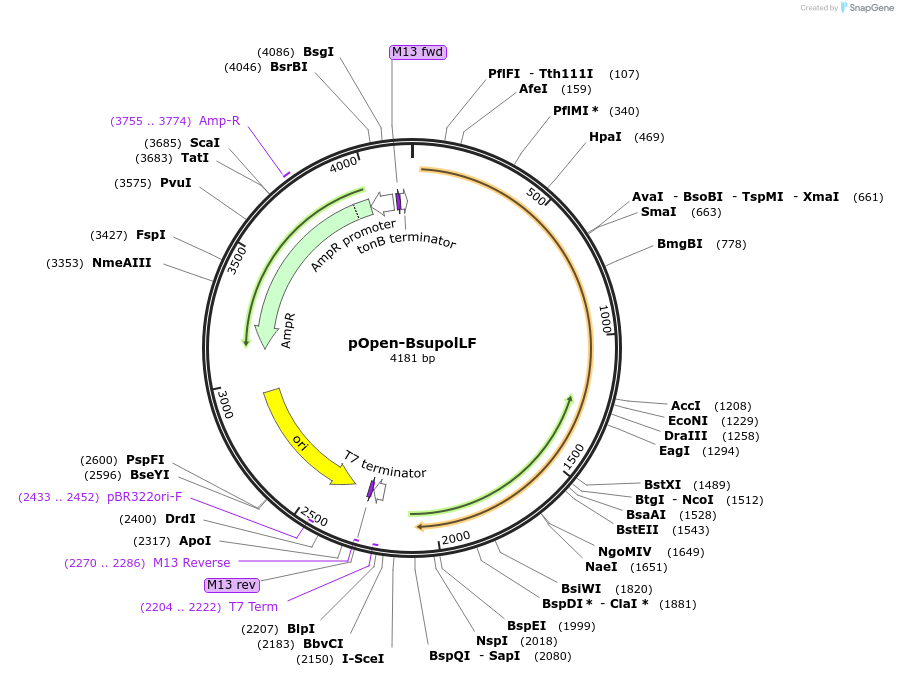
pOpen-BsupolLF
Plasmid#165563PurposeBsu DNA Polymerase I, Large Fragment retains the 5'-3' polymerase activity of the Bacillus subtilis DNA polymerase I, but lacks the 5'-3' exonuclease domain. This large fragment naturally lacks 3'-5' exonuclease activity. Applications include random primer labeling, second strand cDNA synthesis, single dA tailing, and strand displacement DNA synthesis.DepositorInsertBsu DNA Polymerase I, Large Fragment
UseSynthetic BiologyAvailable SinceAug. 17, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
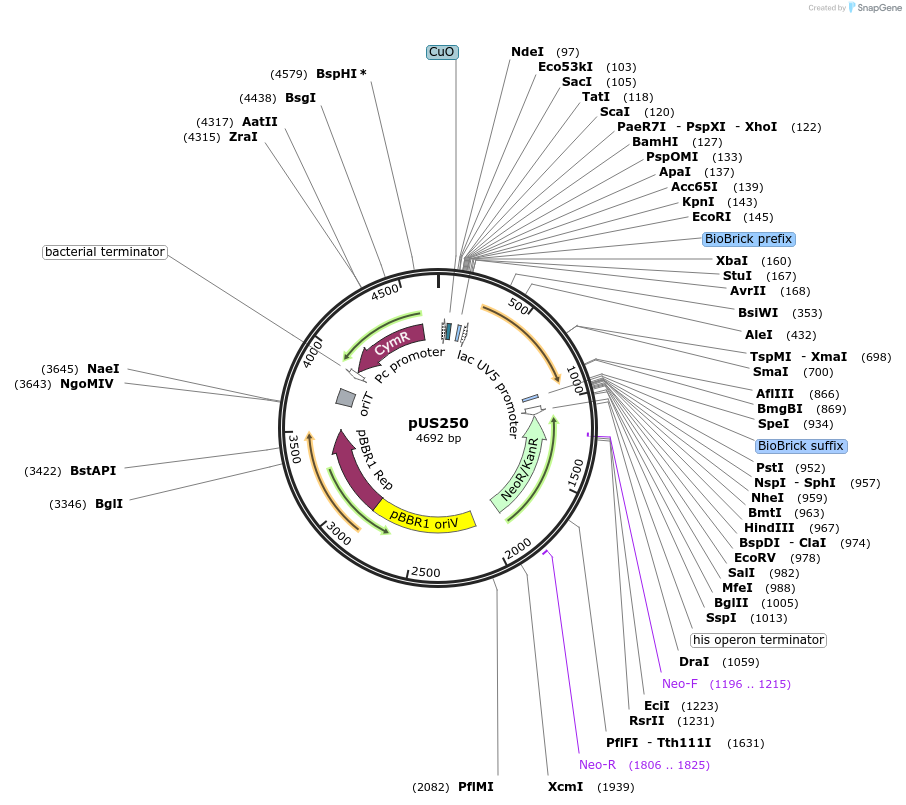
pUS250
Plasmid#198322PurposeCumate-inducible gene expression in diverse gram negative bacteria. Features also include mobilisation via oriT, a gigantic multiple cloning site, and blue/white screening via amilCP chromoproteinDepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterialPromotercumate-inducibleAvailable SinceApril 12, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
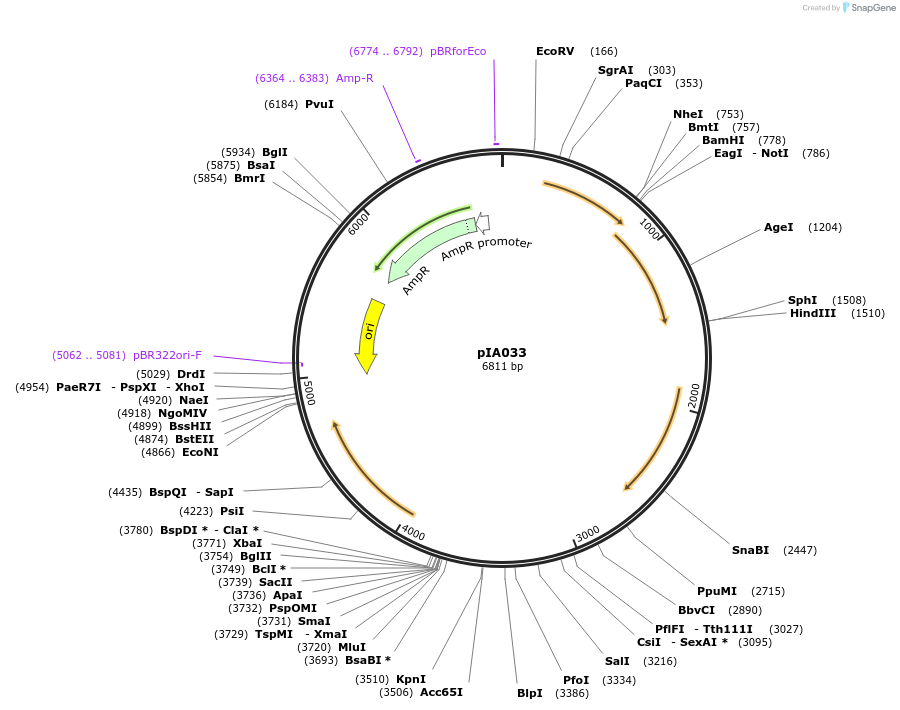
pIA033
Plasmid#200420PurposethrC::(bsPamCherry1 erm2), for bsPAmCherry1 fusions. PALM integration vector includes codon optimized PAmCherry1 FP for B. subtilis. MCS for fast and interchangeable cloning.DepositorArticleTypeEmpty backboneTagsbsPAmCherryExpressionBacterialAvailable SinceMarch 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
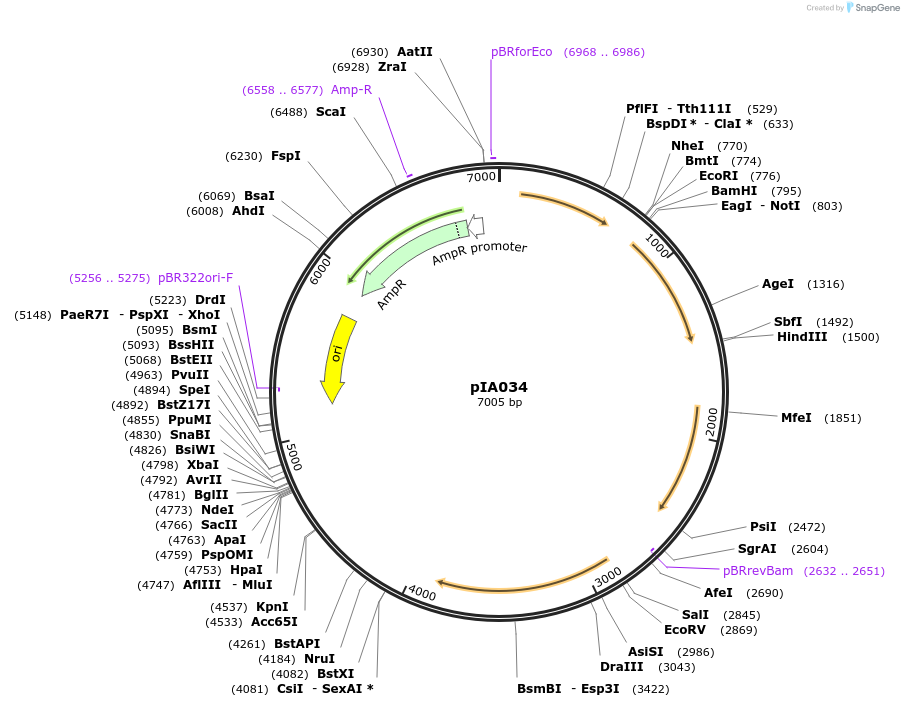
pIA034
Plasmid#200421PurposeamyE::(bsdronPA cm), for bsdronPA fusions. PALM integration vector includes codon optimized dronPA FP for B. subtilis. MCS for fast and interchangeable cloning.DepositorArticleTypeEmpty backboneTagsbsdronPAExpressionBacterialAvailable SinceAug. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
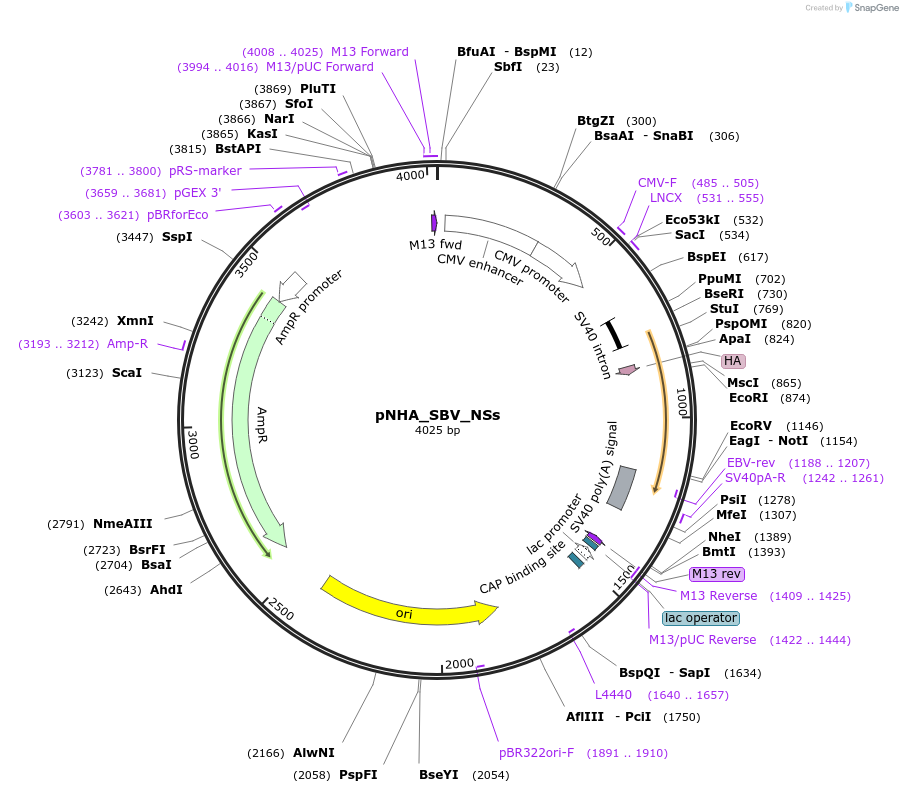
pNHA_SBV_NSs
Plasmid#208932PurposeExpresses N-terminally HA-tagged Schmallenberg orthobunyavirus NSs protein from CMV promoter in mammalian cellsDepositorInsertNSs
TagsHAExpressionMammalianMutationDeleted 50 bp from 5' end (including start c…PromoterCMVAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
pJT122
Plasmid#31396DepositorInsertpJT122
UseSynthetic BiologyExpressionBacterialAvailable SinceOct. 4, 2011AvailabilityAcademic Institutions and Nonprofits only -
pLV-Enh eGFP Reporter-huKC2-IGR21
Plasmid#59456PurposeLentiviral GFP reporter for testing enhancer activity in different cell types. Insert is an enhancer identified in the intergenic region of the human keratin type II cluster.DepositorInserthuKrt1 3’intergenic region
UseLentiviralPromoterMu Hsp68 minimal promoterAvailable SinceOct. 2, 2014AvailabilityAcademic Institutions and Nonprofits only -
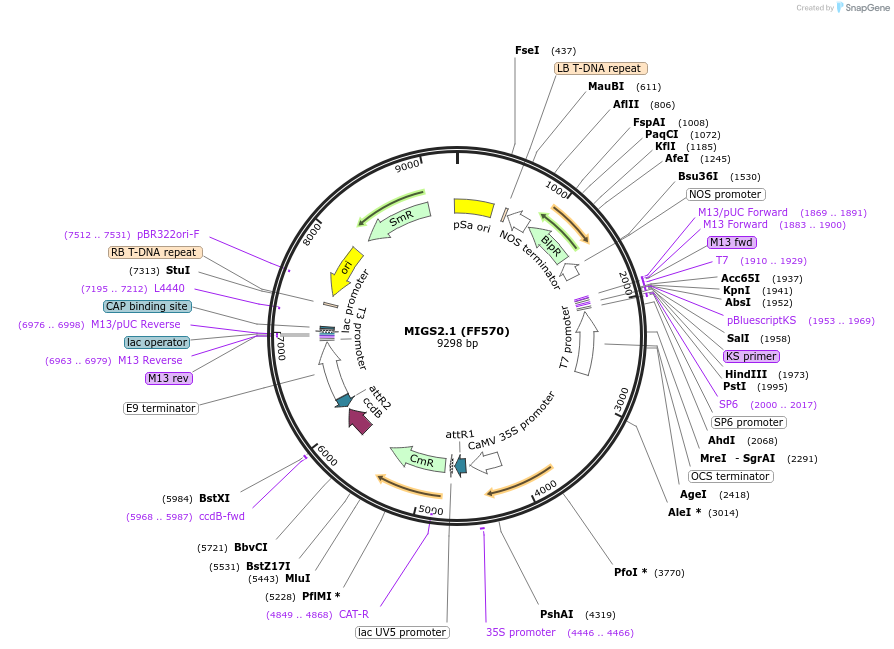
MIGS2.1 (FF570)
Plasmid#35247PurposeExpression of miR173 precursor under UBQ10 promoter; includes miR173 target site after 35S promoter (Basta selection marker)DepositorTypeEmpty backboneExpressionPlantPromoterCaMV 35SAvailable SinceMarch 23, 2012AvailabilityAcademic Institutions and Nonprofits only -
pSC301b
Plasmid#31852DepositorInsertGFP (including ATG start codon)
ExpressionBacterialPromoterSOD and noneAvailable SinceDec. 1, 2011AvailabilityAcademic Institutions and Nonprofits only -
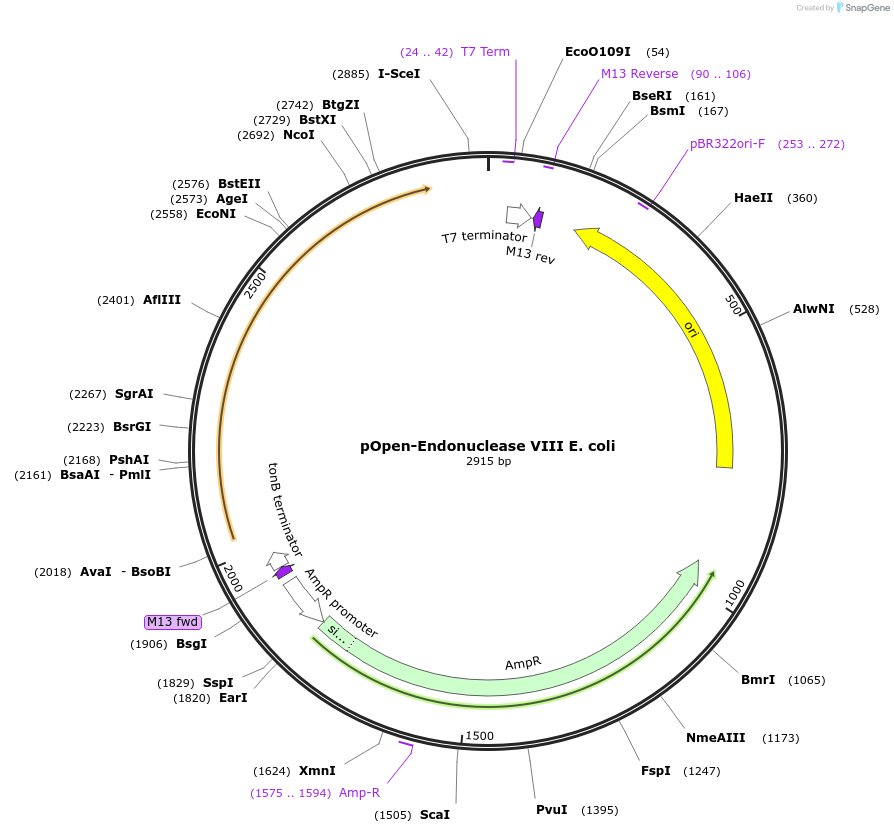
pOpen-Endonuclease VIII E. coli
Plasmid#165561PurposeBifunctional DNA glycosylase with DNA N-glycosylase and AP lyase activities; The N-glycosylase activity releases damaged pyrimidines, including thymine glycol and uracil glycol. The AP lyase activity cleaves DNA phosphodiester backbone at AP sites via beta and delta-elimination, creating a 1 nucleotide DNA gap with 5' and 3' phosphate termini.DepositorInsertEndonuclease VIII E. coli
UseSynthetic BiologyAvailable SinceAug. 16, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
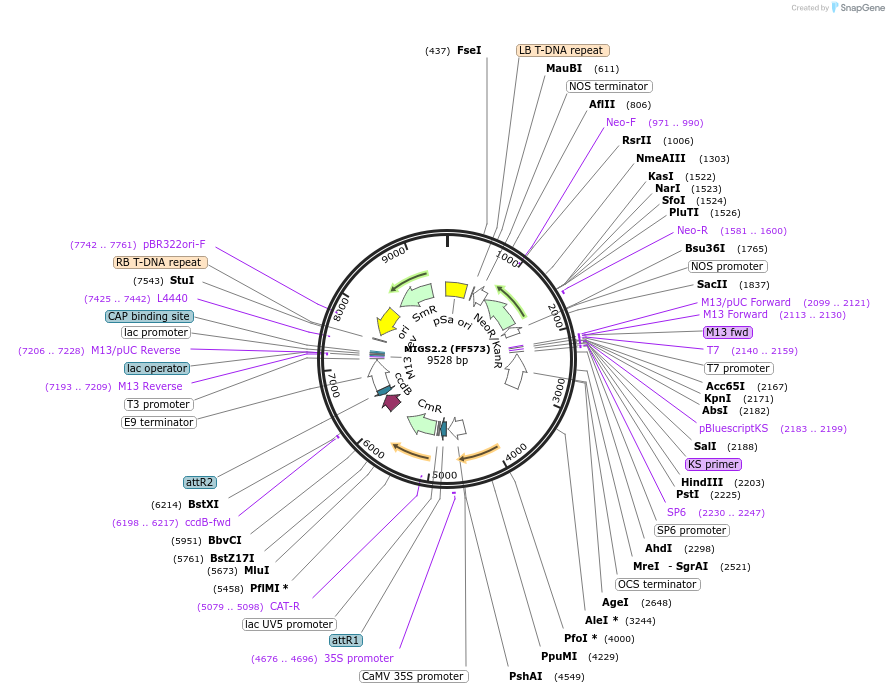
MIGS2.2 (FF573)
Plasmid#35248PurposeExpression of miR173 precursor under UBQ10 promoter; includes miR173 target site after 35S promoter (Kanamycin selection marker)DepositorTypeEmpty backboneExpressionPlantPromoterCaMV 35SAvailable SinceMarch 23, 2012AvailabilityAcademic Institutions and Nonprofits only -
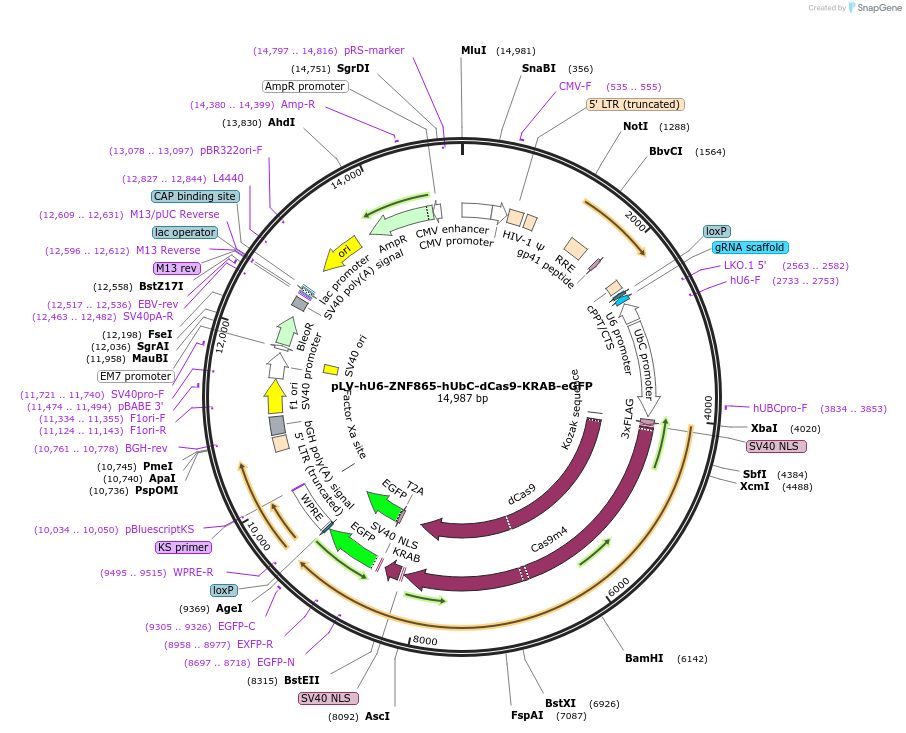
pLV-hU6-ZNF865-hUbC-dCas9-KRAB-eGFP
Plasmid#241308Purpose3rd gen lenti vector encoding dCas9 (s. Pyogenes) fused with the KRAB and a 2A eGFP fluorescence marker. Includes guide for suppression of the ZNF865 geneDepositorInsertZNF865 sgRNA (ZNF865 Human)
UseLentiviralAvailable SinceDec. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
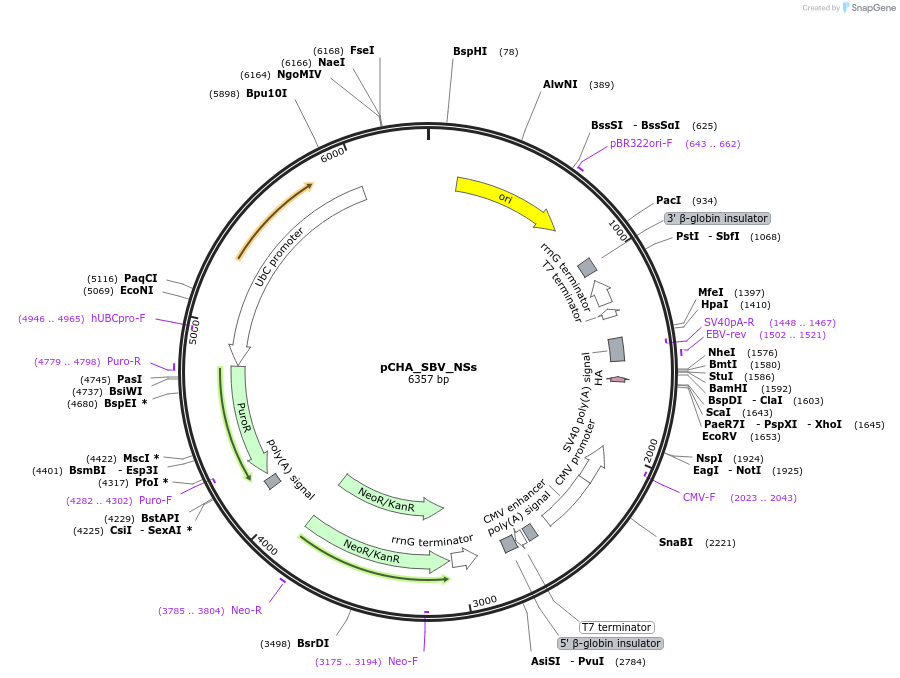
pCHA_SBV_NSs
Plasmid#208938PurposeExpresses C-terminally HA-tagged Schmallenberg orthobunyavirus NSs protein from CMV promoter in mammalian cellsDepositorInsertNSs
TagsHAExpressionMammalianMutationDeleted 47 bp at 5' end and 510 bp from 3&#x…PromoterCMVAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
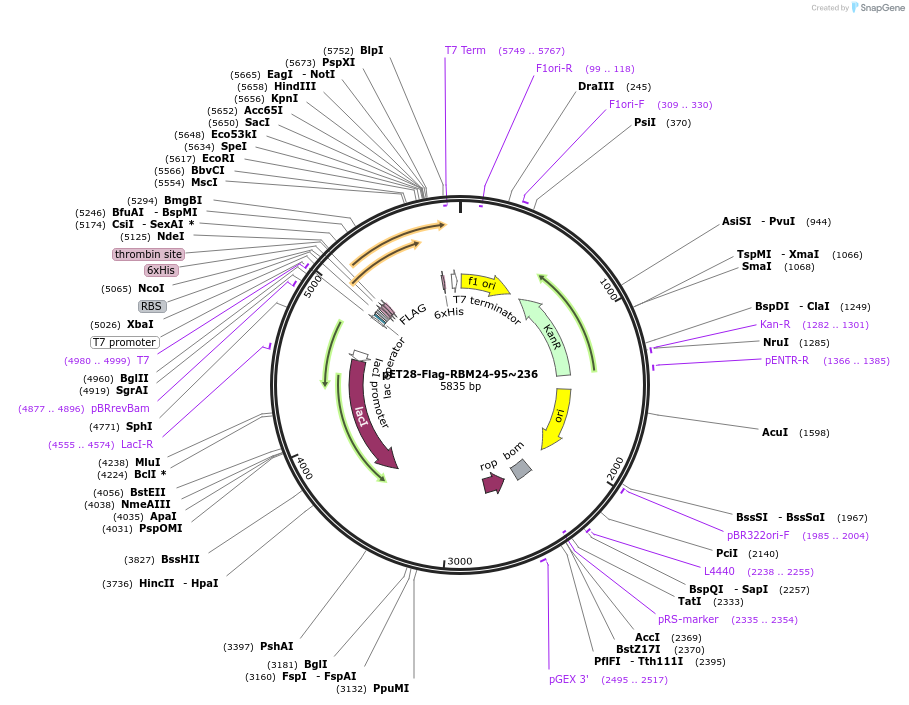
pET28-Flag-RBM24-95~236
Plasmid#208634Purposefor E. coli expression of truncated Flag tagged human RBM24 (ENST00000379052), aa95~236DepositorInserthuman RBM24 (ENST00000216146) aa 95~236 (RBM24 Human)
Tags6xHis and FlagExpressionBacterialMutationonly aa 95~236, other residues not includedPromoterT7Available SinceJan. 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
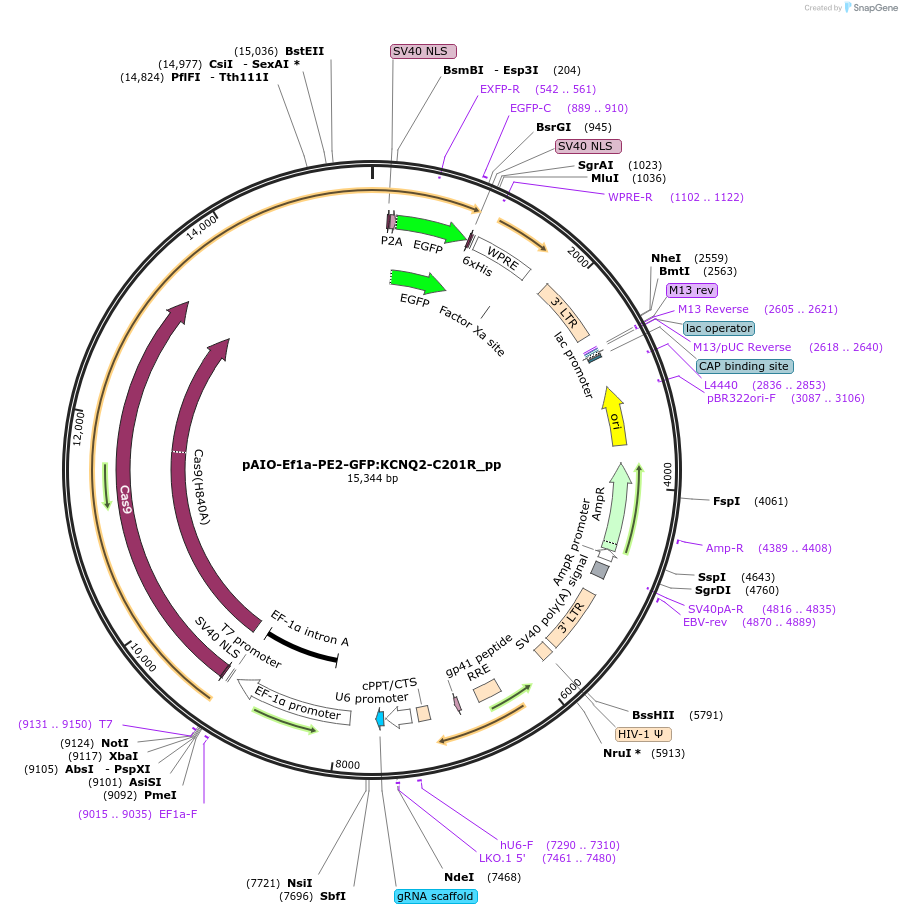
pAIO-Ef1a-PE2-GFP:KCNQ2-C201R_pp
Plasmid#185061PurposeEf1a driven PE2 with pegRNA for editing C201R mutation (including silent PAM mutation) in KCNQ2. See Addgene plasmid #184445DepositorInsertU6:pegRNA:scaffold:PBS+RT template
UseCRISPR and LentiviralExpressionMammalianPromoterU6Available SinceNov. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
TJP3
Plasmid#25310PurposeBacterial expression for structure determination; may not be full ORFDepositorInsertTJP3:G508-L808:cH6 (TJP3 Human)
TagsHisExpressionBacterialMutationContains amino acids G508-L808Available SinceApril 4, 2016AvailabilityIndustry, Academic Institutions, and Nonprofits -
pLV-Enh eGFP Reporter-muKC2-IGR4
Plasmid#59273PurposeLentiviral GFP reporter for testing enhancer activity in different cell types. Insert is an enhancer identified in the intergenic region of the keratin type II cluster.DepositorInsertIntergenic 3kb downstream Krt82
UseLentiviralPromoterMu Hsp68 minimal promoterAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
pLV-Enh eGFP Reporter-muKC2-IGR7
Plasmid#59276PurposeLentiviral GFP reporter for testing enhancer activity in different cell types. Insert is an enhancer identified in the intergenic region of the keratin type II cluster.DepositorInsertintergenic 4kb upstream krt1
UseLentiviralPromoterMu Hsp68 minimal promoterAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only -
pLV-Enh eGFP Reporter-muKC2-IGR8
Plasmid#59277PurposeLentiviral GFP reporter for testing enhancer activity in different cell types. Insert is an enhancer identified in the intergenic region of the keratin type II cluster.DepositorInsertIntergenic 2kb downstream krt79
UseLentiviralPromoterMu Hsp68 minimal promoterAvailable SinceDec. 3, 2014AvailabilityAcademic Institutions and Nonprofits only