We narrowed to 4,248 results for: fas
-
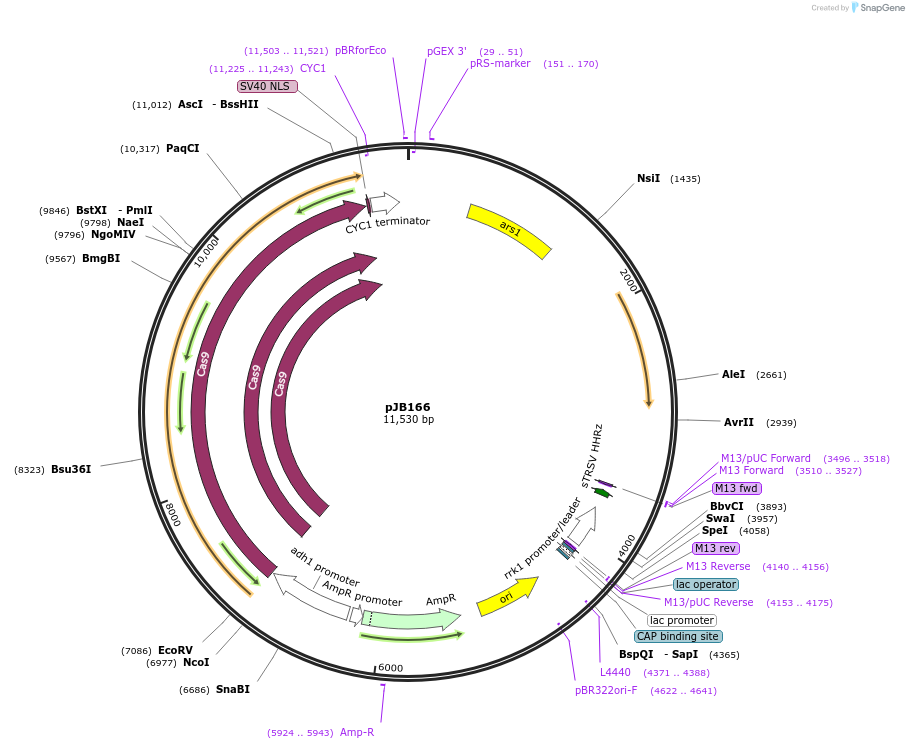
Plasmid#86998PurposeCRISPR/Cas9 in fission yeast using fluoride selectionDepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceFeb. 22, 2017AvailabilityAcademic Institutions and Nonprofits only
-
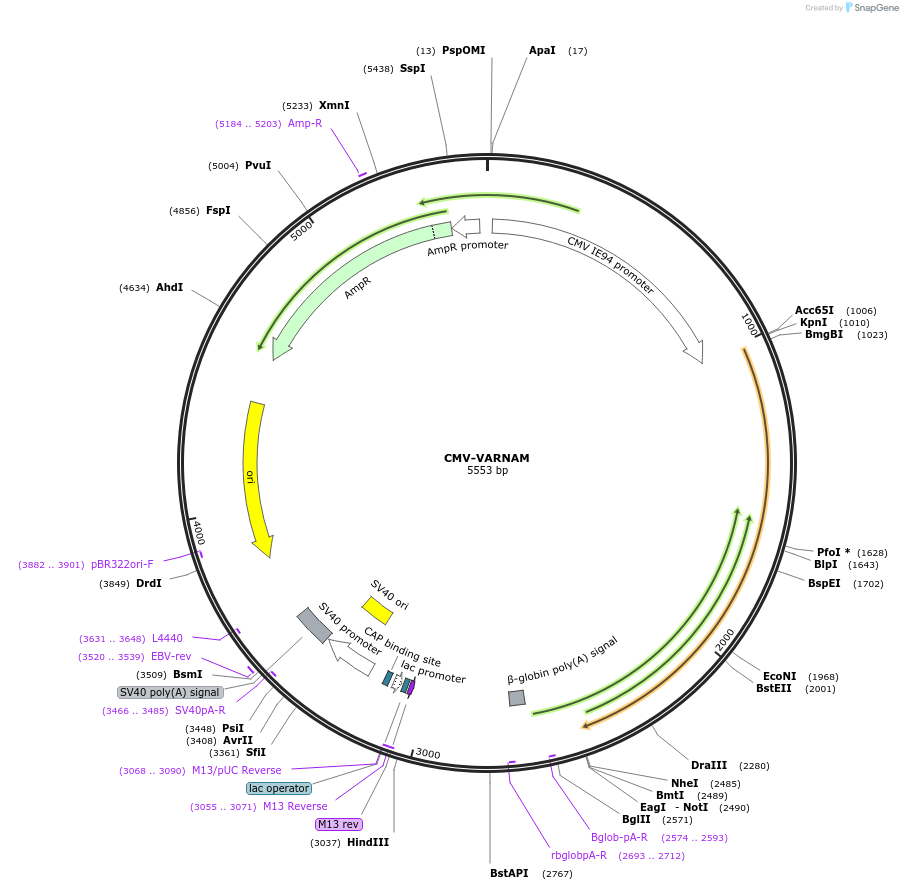
CMV-VARNAM
Plasmid#115552PurposeRed fluorescent voltage indicator for mammalian expressionDepositorInsertVARNAM
TagsGolgi and ER export signalExpressionMammalianPromoterCMV IE94Available SinceNov. 12, 2018AvailabilityAcademic Institutions and Nonprofits only -
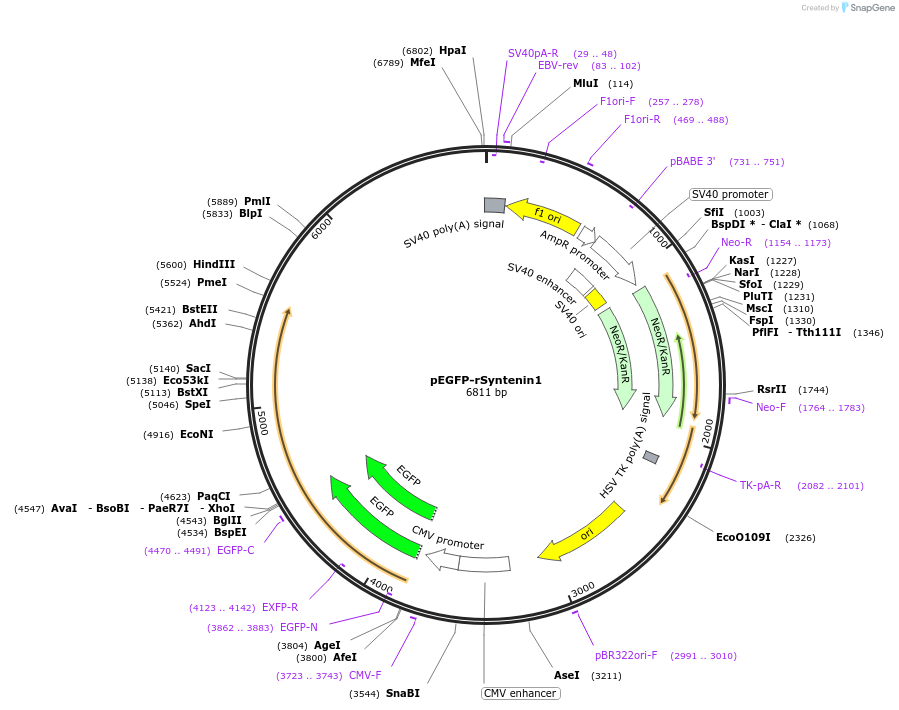
pEGFP-rSyntenin1
Plasmid#89431Purposemammalian expressio of EGFP tagged rat Syntenin1DepositorInsertSDCBP
TagsEGFPExpressionMammalianPromoterCMVAvailable SinceMay 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
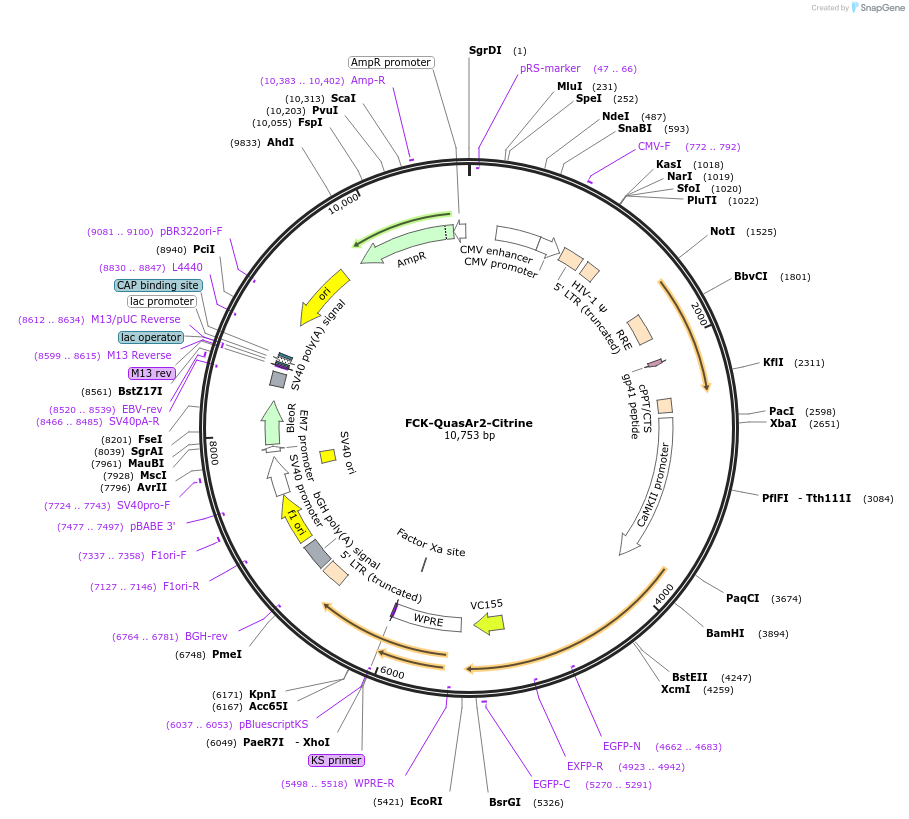
FCK-QuasAr2-Citrine
Plasmid#59172PurposeFluorescent membrane voltage sensorDepositorInsertQuasAr2-Citrine eFRET
UseLentiviralExpressionMammalianAvailable SinceJan. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
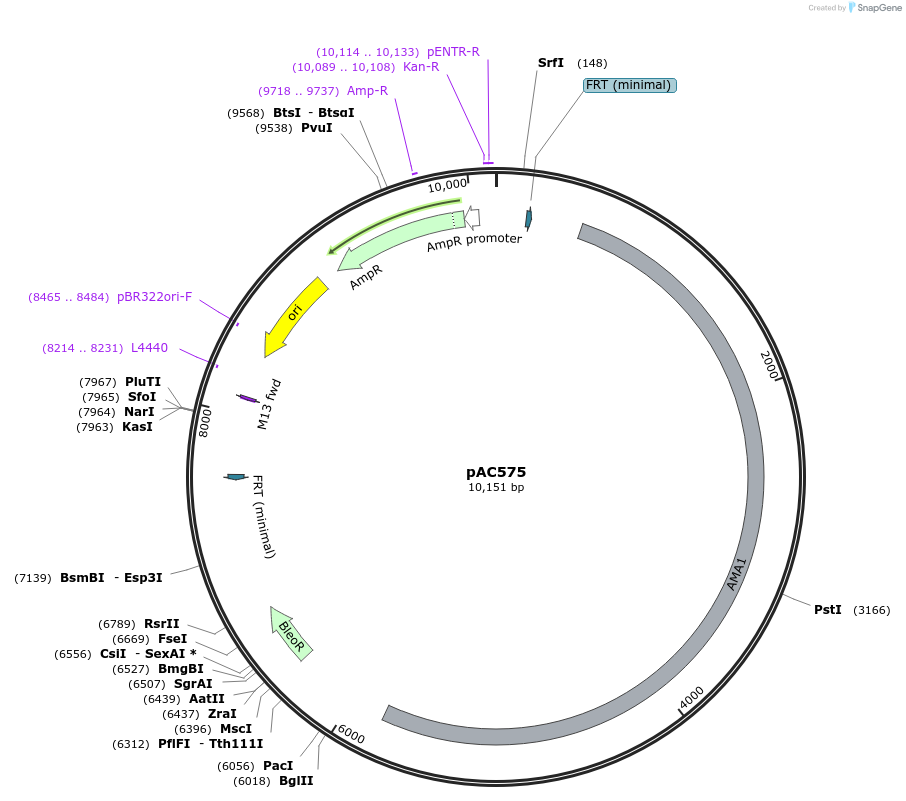
pAC575
Plasmid#127976PurposeAMA1 plasmid with ble selection marker, and USER cassette (PacI/Nt.BbvCI)DepositorInsertble (bleomycin resistance marker)
UseFungal expressionExpressionBacterialPromoterAspergillus nidulans trpC promoterAvailable SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
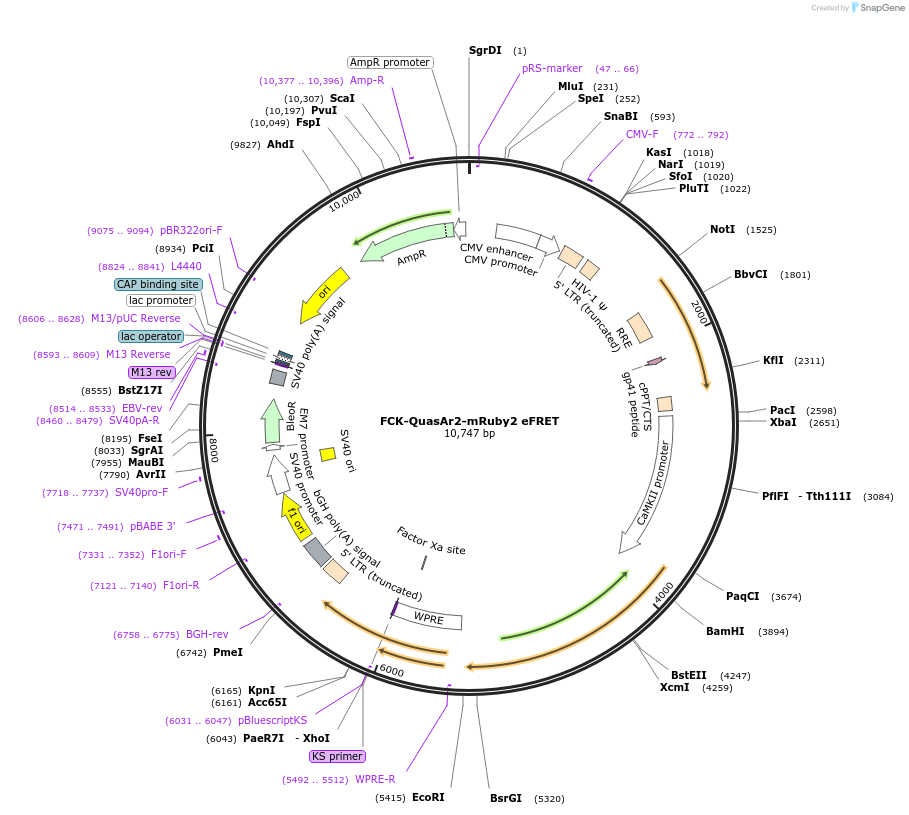
FCK-QuasAr2-mRuby2 eFRET
Plasmid#59174PurposeFluorescent membrane voltage sensorDepositorInsertQuasAr2-mRuby2 eFRET
UseLentiviralExpressionMammalianAvailable SinceJan. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
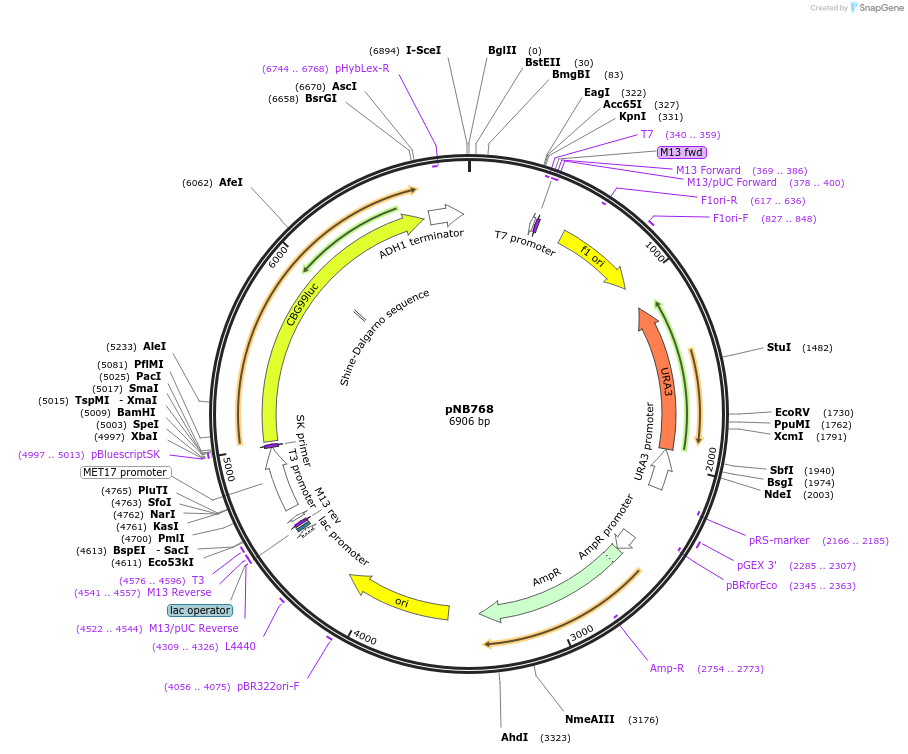
pNB768
Plasmid#60139PurposeGreen click beetle luciferase from Promega driven by MET17 promoter.DepositorInsertCBG99
ExpressionYeastPromoterMET17prAvailable SinceNov. 13, 2014AvailabilityAcademic Institutions and Nonprofits only -
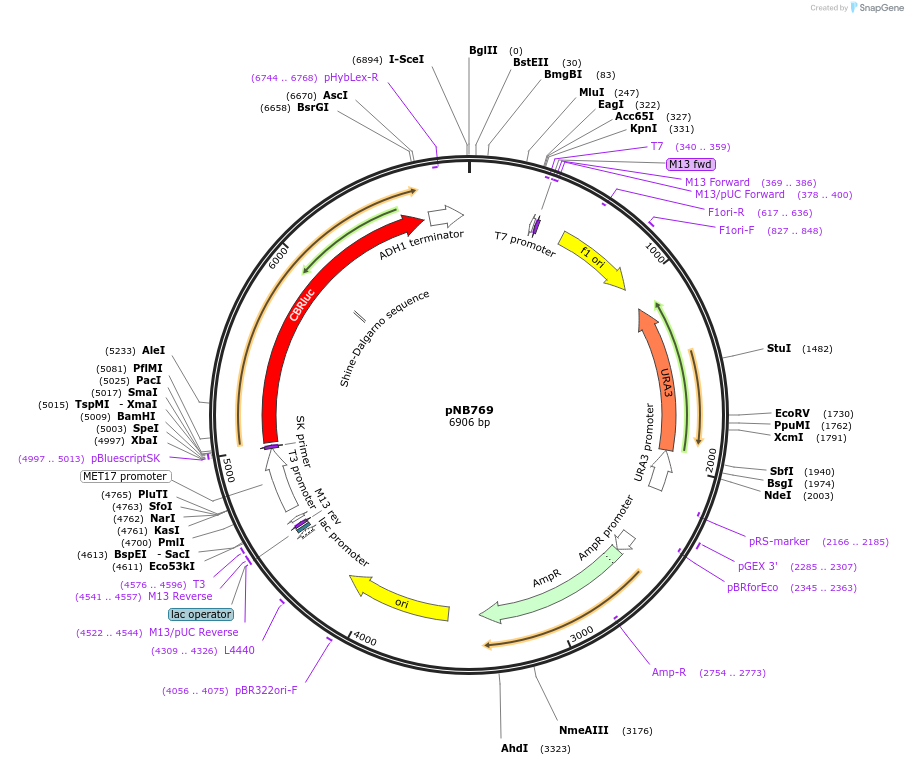
pNB769
Plasmid#60138PurposeRed click beetle luciferase from Promega driven by MET17 promoter.DepositorInsertCBR
ExpressionYeastPromoterMET17prAvailable SinceNov. 13, 2014AvailabilityAcademic Institutions and Nonprofits only -
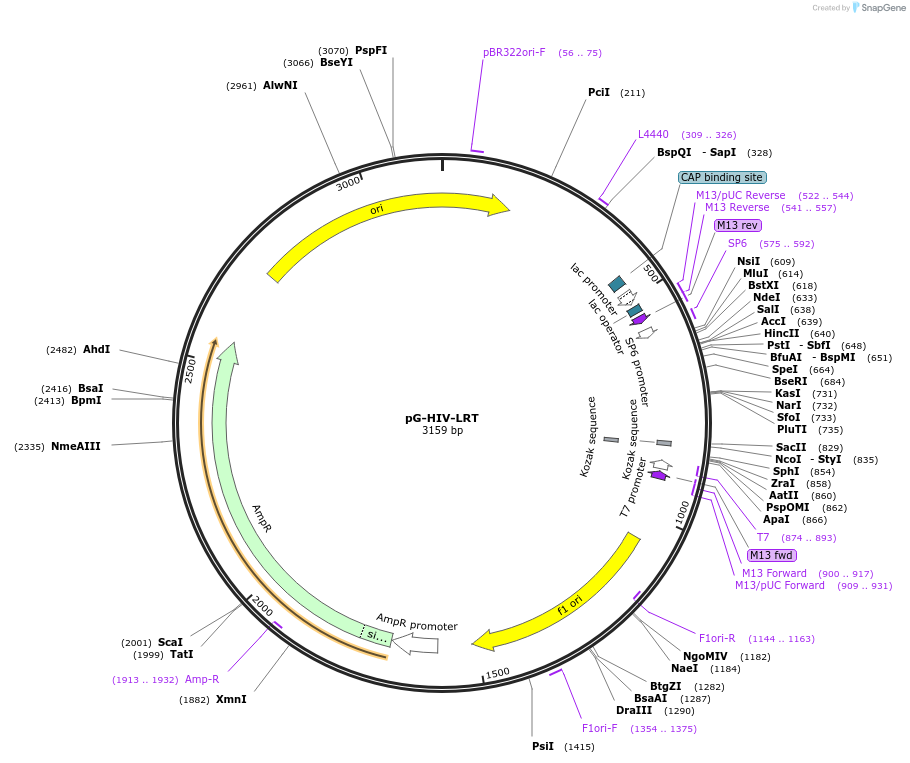
pG-HIV-LRT
Plasmid#104592PurposeStandard for qPCRDepositorInsertHIV-1 late reverse transcripts
UseCloning vectorAvailable SinceJan. 3, 2018AvailabilityAcademic Institutions and Nonprofits only -
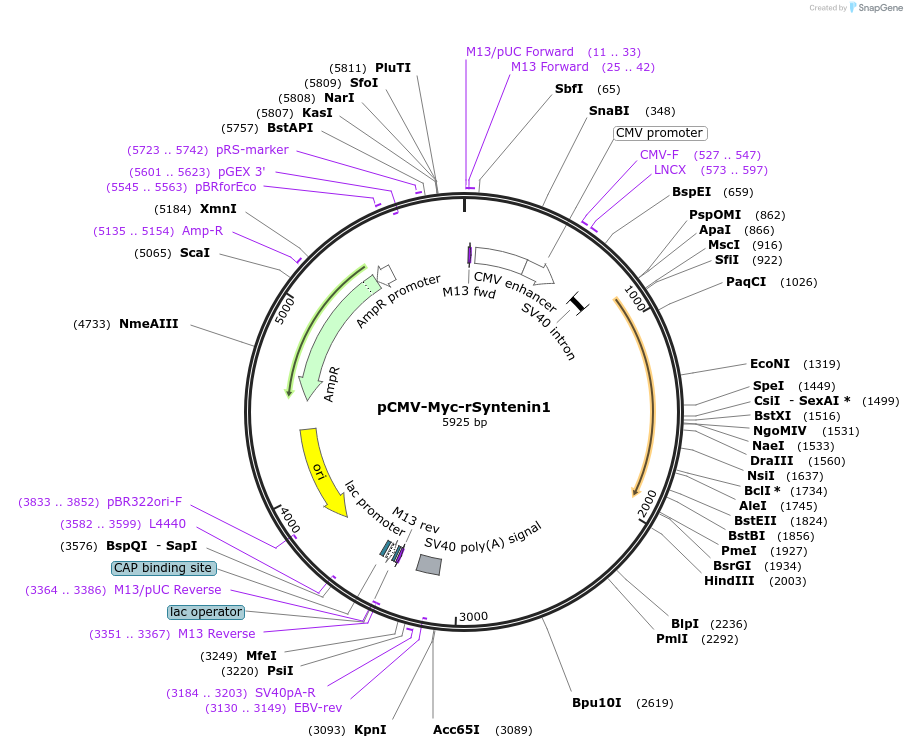
pCMV-Myc-rSyntenin1
Plasmid#89435Purposemammalian expression of rat Syntenin1DepositorInsertSDCBP
TagsMycExpressionMammalianPromoterCMVAvailable SinceMay 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
pFBD-ESCRT 0
Plasmid#21499DepositorInsertpFastBac Dual-His/MBP/Hrs-GST/STAM1 (STAM Human, Mouse)
TagsGST and His-MBPExpressionInsectMutationnoneAvailable SinceSept. 9, 2009AvailabilityAcademic Institutions and Nonprofits only -
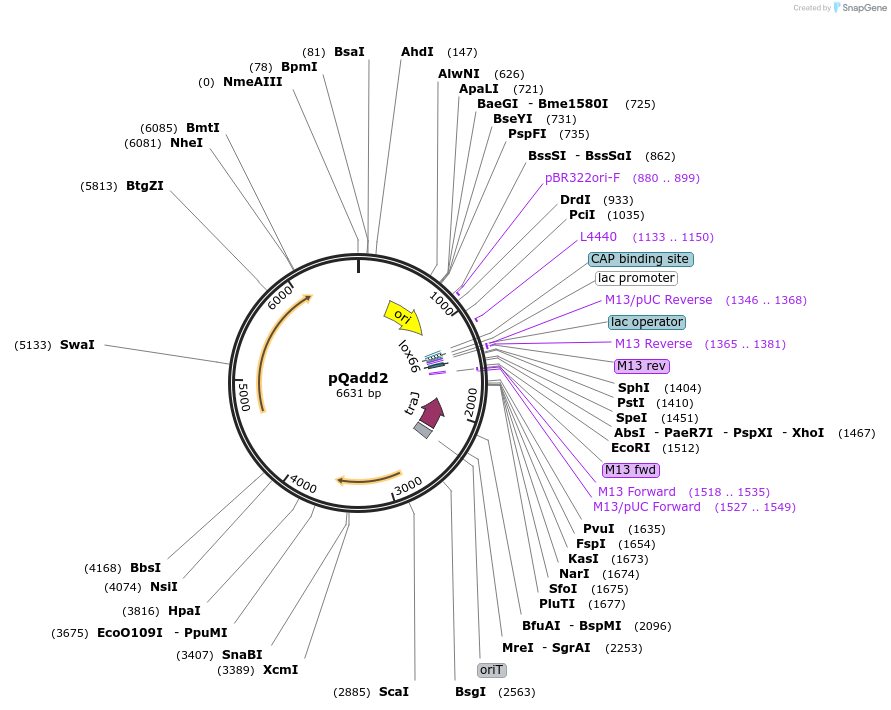
pQadd2
Plasmid#135662PurposeClostridial vector, carrying lox511/66 and loxFAS/71 sites separated by restriction sites in order to add cargo. Use in combination with pQcre1 to insert cargo into lox sites, inserted with pQadd1F/R.DepositorInsertLox511/66 and LoxFas/71
UseClostridial vector, pamβ1 originExpressionBacterialAvailable SinceJuly 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
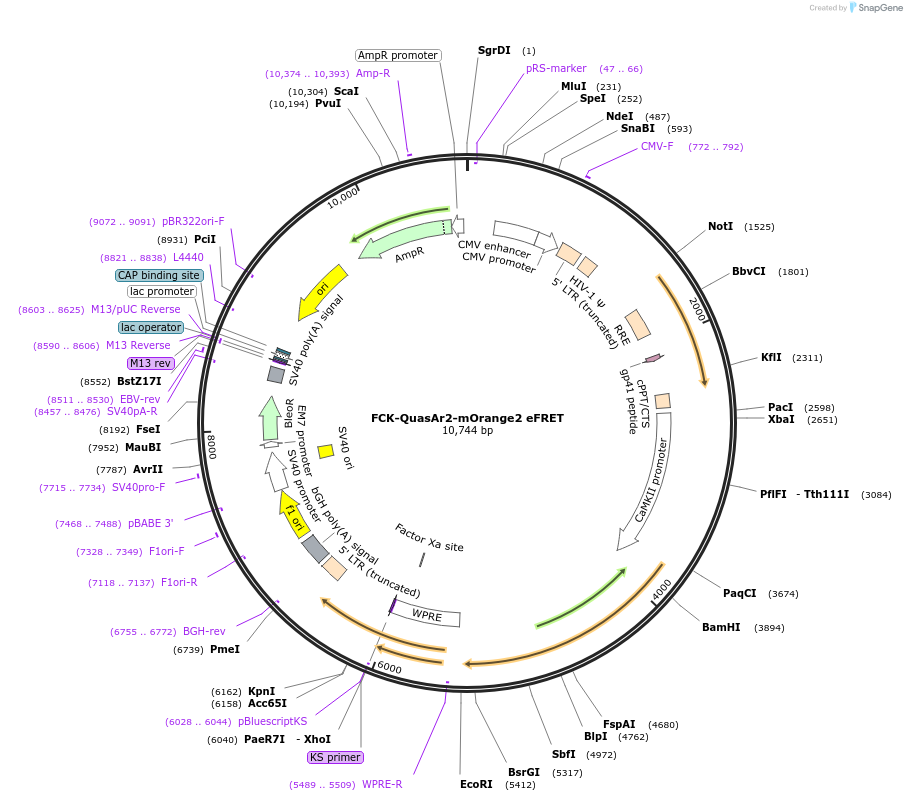
FCK-QuasAr2-mOrange2 eFRET
Plasmid#59173PurposeFluorescent membrane voltage sensorDepositorInsertQuasAr2-mOrange2 eFRET
UseLentiviralExpressionMammalianAvailable SinceJan. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
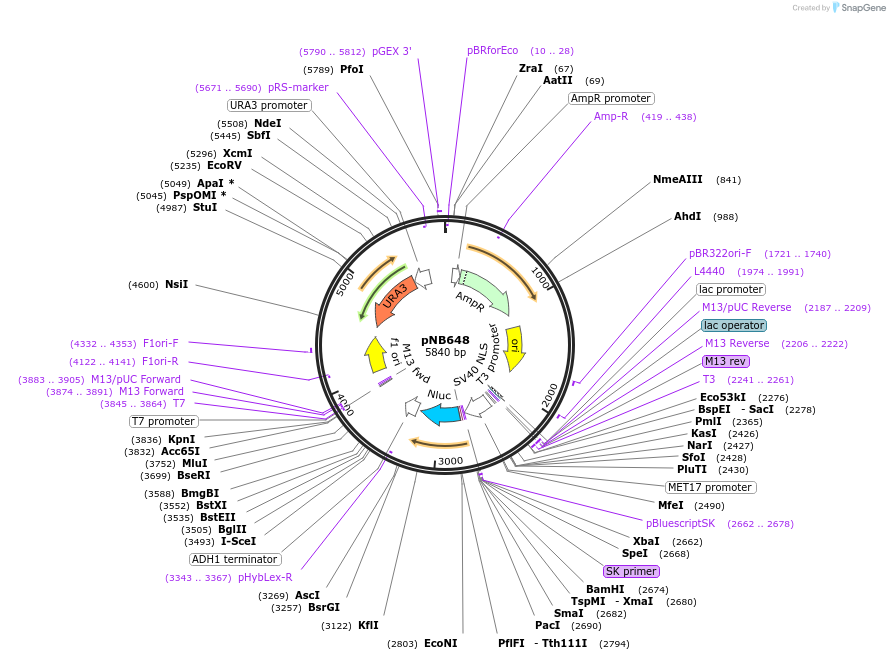
pNB648
Plasmid#60140PurposeNanoLuc luciferase with a nuclear localization signal (NLS) driven by MET17 promoter.DepositorInsertNLuc
TagsNuclear localization signal (NLS)ExpressionYeastPromoterMET17prAvailable SinceMarch 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
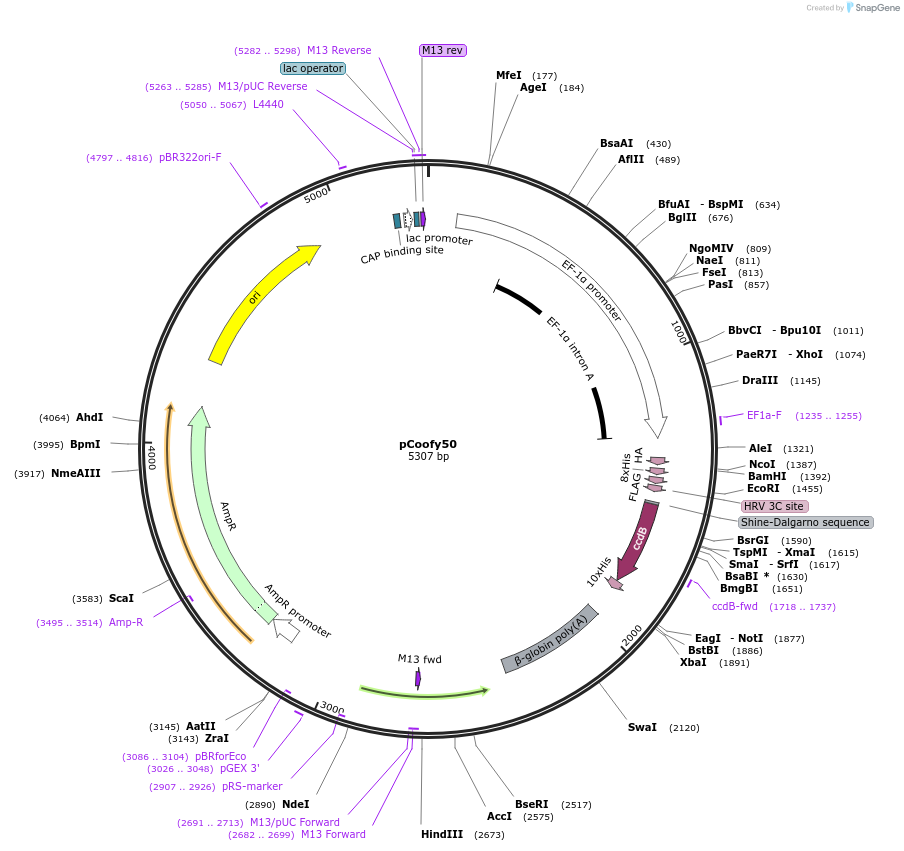
pCoofy50
Plasmid#55189PurposeMammalian expression vector for parallel SLIC cloning containing N-terminal HA, 8x His and FLAG tags. C-terminal tags can be added during SLIC.DepositorTypeEmpty backboneTags8x His, FLAG, HA, PreScission site (3C protease s…ExpressionMammalianPromoterEF-1aAvailable SinceAug. 26, 2014AvailabilityAcademic Institutions and Nonprofits only -
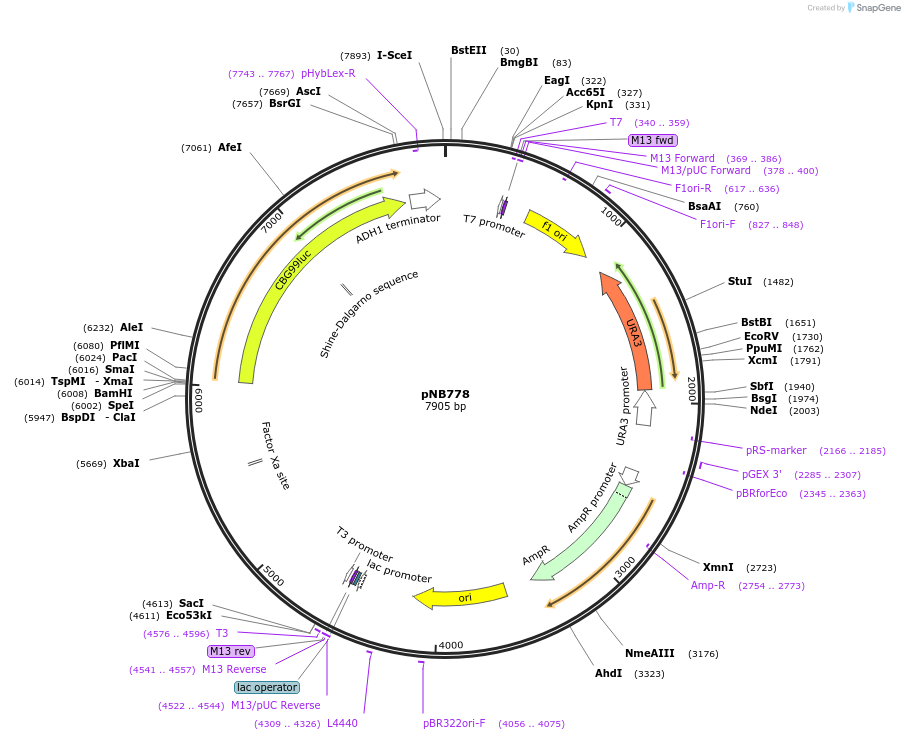
pNB778
Plasmid#60154PurposeGreen click beetle luciferase (Promega) driven by LEU1 promoter.DepositorInsertCBG99
ExpressionYeastPromoterLEU1prAvailable SinceNov. 13, 2014AvailabilityAcademic Institutions and Nonprofits only -
Kohinoor/pcDNA3
Plasmid#67771PurposeKohinoor for mammalian cell expression in cytosolDepositorInsertKohinoor
ExpressionMammalianPromoterCMVAvailable SinceSept. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
Kohinoor-Actin/pcDNA3
Plasmid#67776PurposeActin-targetted Kohinoor for mammalian cell expressionDepositorInsertKohinoor
TagsActinExpressionMammalianPromoterCMVAvailable SinceSept. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
Kohinoor-Paxillin/pcDNA3
Plasmid#67775PurposePaxilin-targetted Kohinoor for mammalian cell expressionDepositorInsertKohinoor
TagsPaxillinExpressionMammalianPromoterCMVAvailable SinceSept. 4, 2015AvailabilityAcademic Institutions and Nonprofits only -
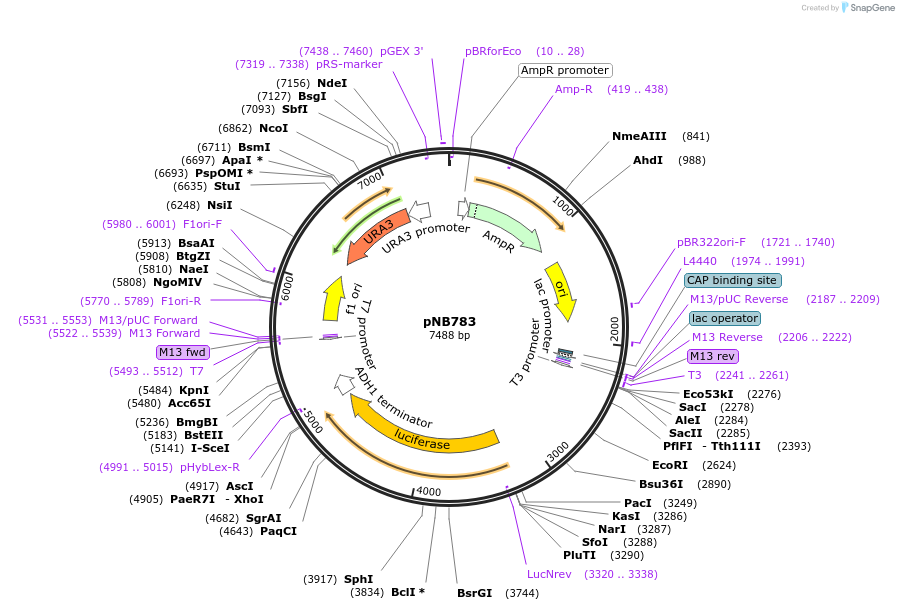
pNB783
Plasmid#60162PurposeFirefly luciferase (Promega) driven by RNR1 promoter.DepositorInsertFLuc
ExpressionYeastMutationA4V & S504G (no functional effect)PromoterRNR1prAvailable SinceMarch 25, 2015AvailabilityAcademic Institutions and Nonprofits only