4,457 results
-
Plasmid#201296PurposeD3: Promoter of TraesCS3A02G510700 as Golden Gate compatible MoClo PROM partDepositorInsertD3: Promoter of TraesCS3A02G510700
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
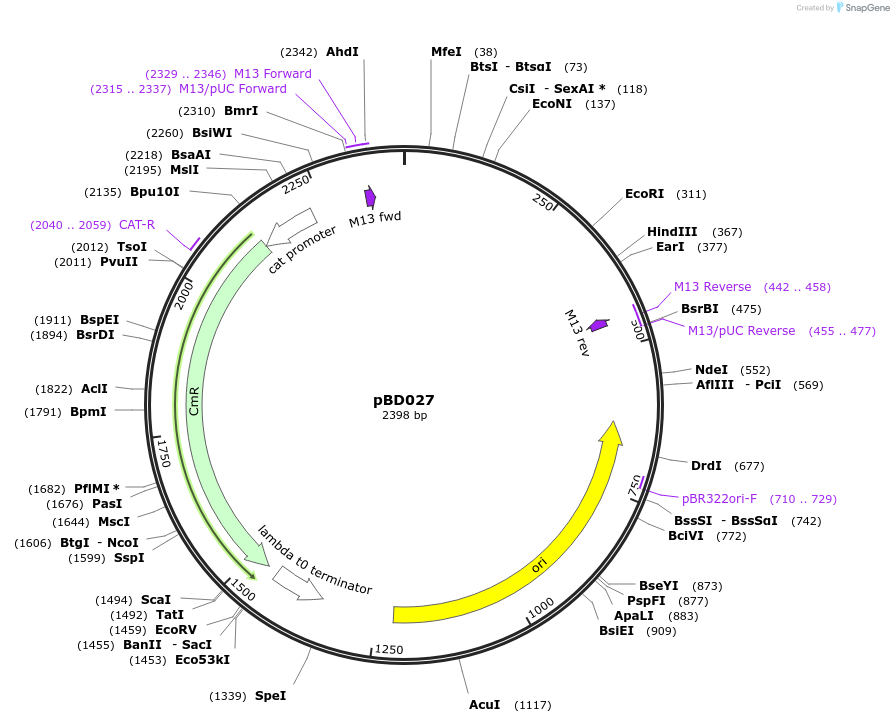
pBD027
Plasmid#201310PurposeavrSr27-3 as Golden Gate compatible MoClo CDS part (no recognition by Sr27)DepositorInsertavrSr27-3
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
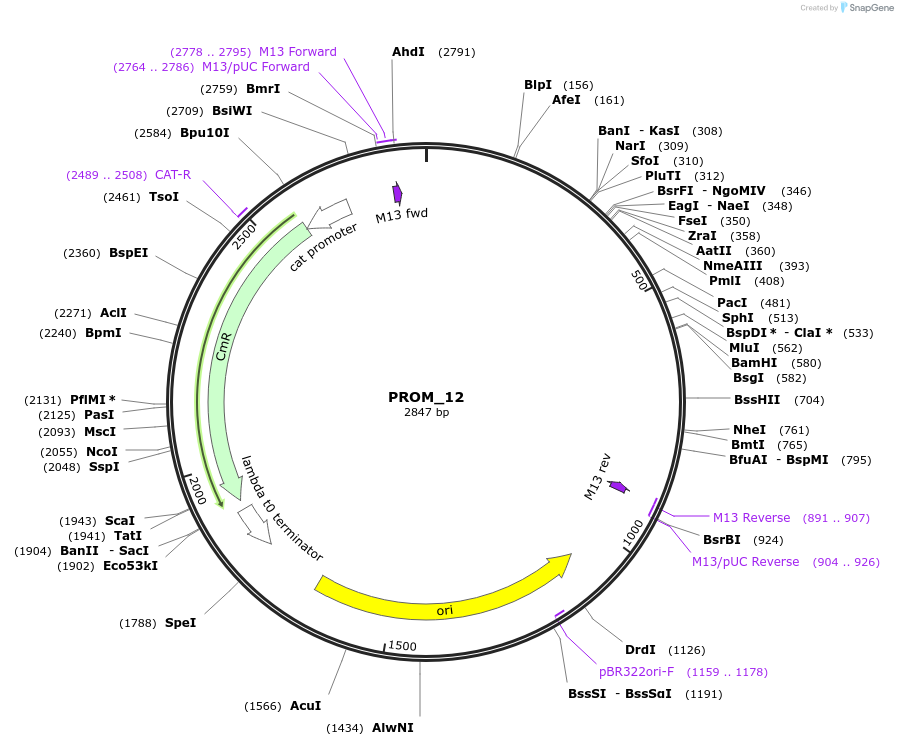
PROM_12
Plasmid#201303PurposeD12: Promoter of TraesCS4D02G131100 as Golden Gate compatible MoClo PROM partDepositorInsertD12: Promoter of TraesCS4D02G131100
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
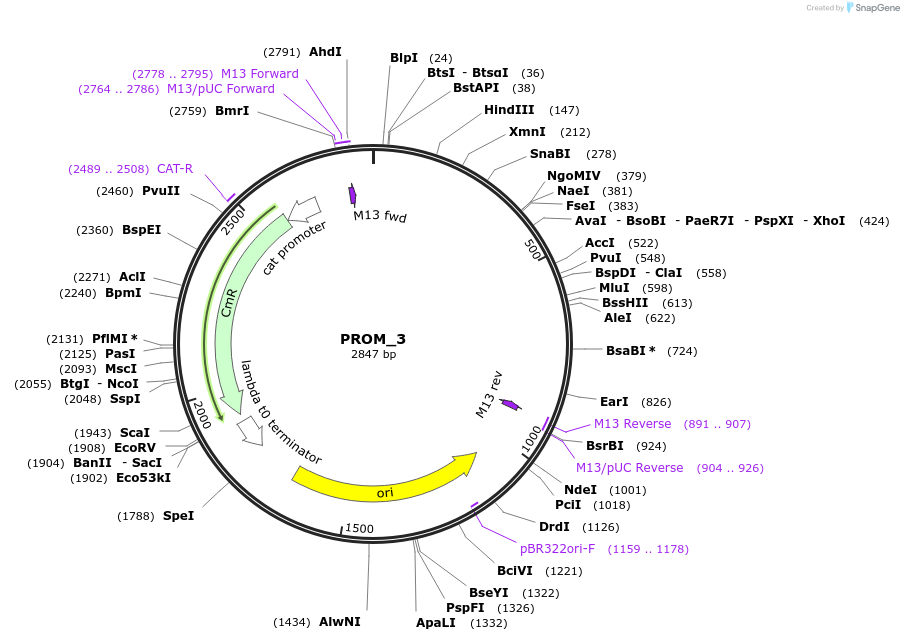
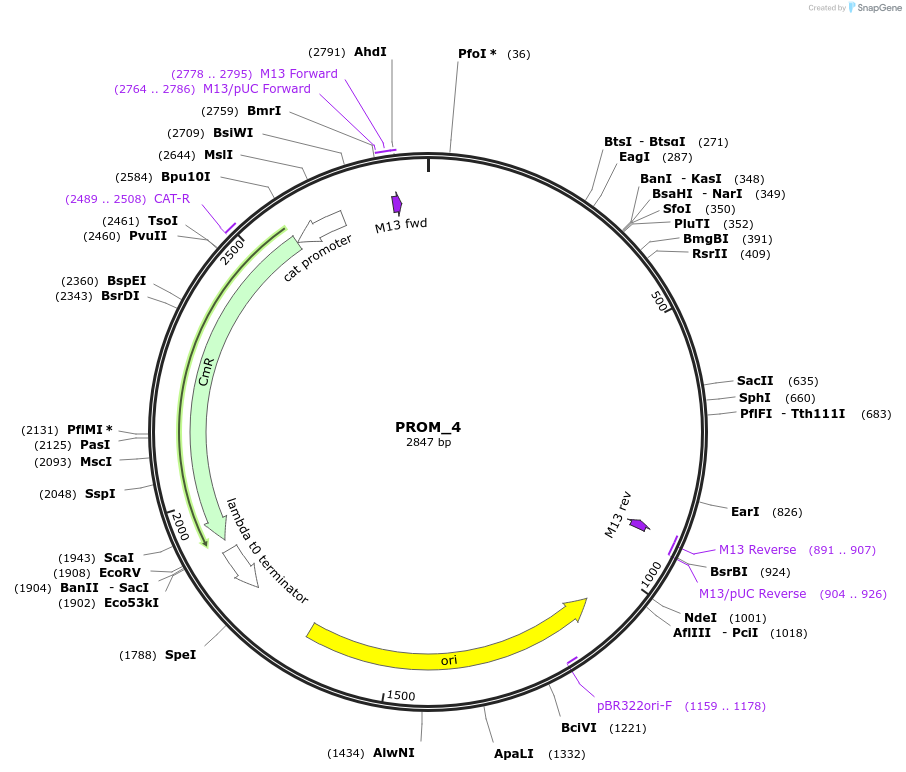
PROM_4
Plasmid#201297PurposeD4: Promoter of TraesCS3B02G552700 as Golden Gate compatible MoClo PROM partDepositorInsertD4: Promoter of TraesCS3B02G552700
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
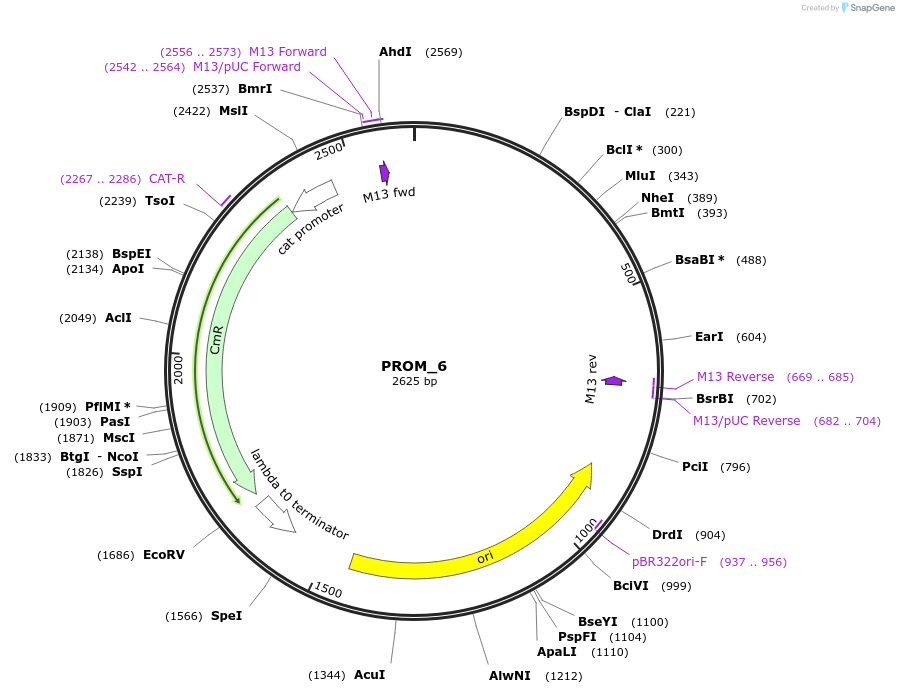
PROM_6
Plasmid#201299PurposeD6: Promoter of TraesCS3B02G578800 as Golden Gate compatible MoClo PROM partDepositorInsertD6: Promoter of TraesCS3B02G578800
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
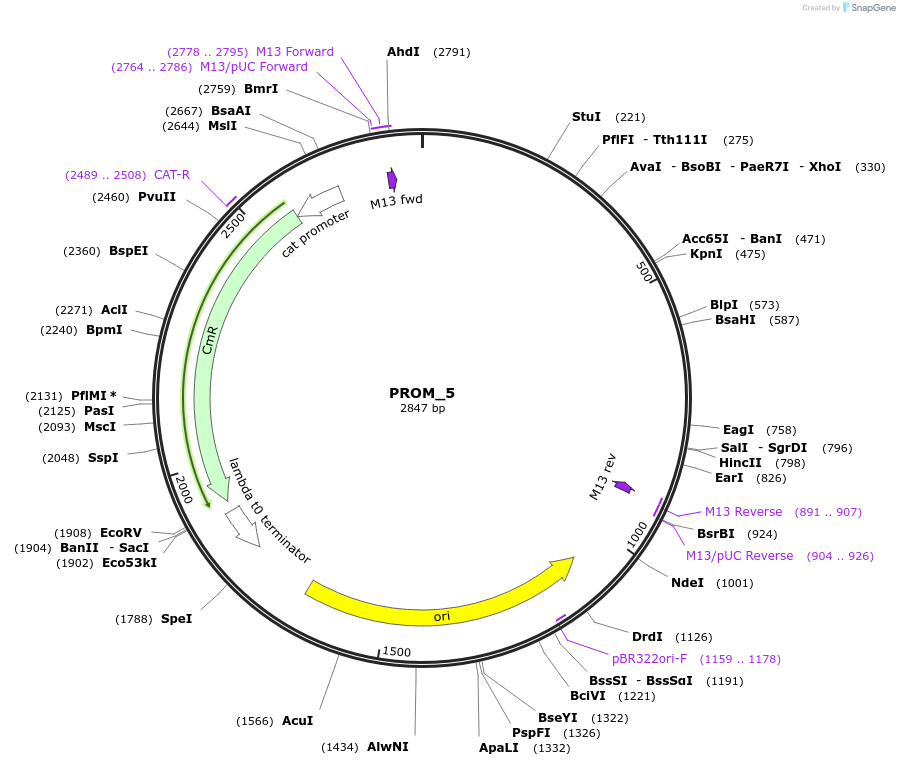
PROM_5
Plasmid#201298PurposeD5: Promoter of TraesCS3B02G553900 as Golden Gate compatible MoClo PROM partDepositorInsertD5: Promoter of TraesCS3B02G553900
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
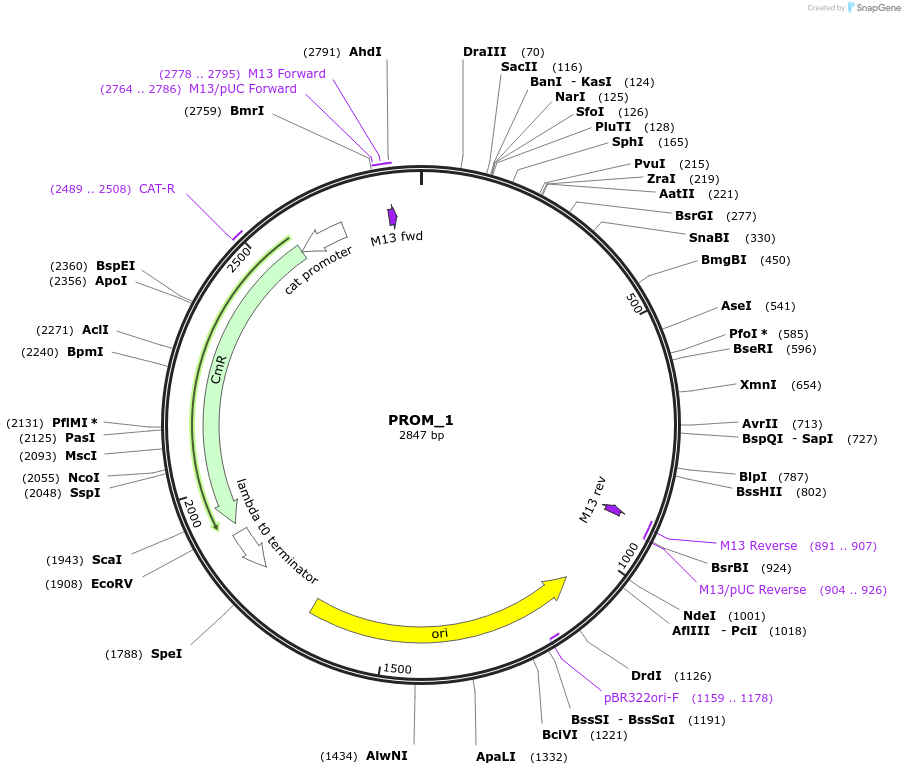
PROM_1
Plasmid#201294PurposeD1: Promoter of TraesCS1A02G332800 as Golden Gate compatible MoClo PROM partDepositorInsertD1: Promoter of TraesCS1A02G332800
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
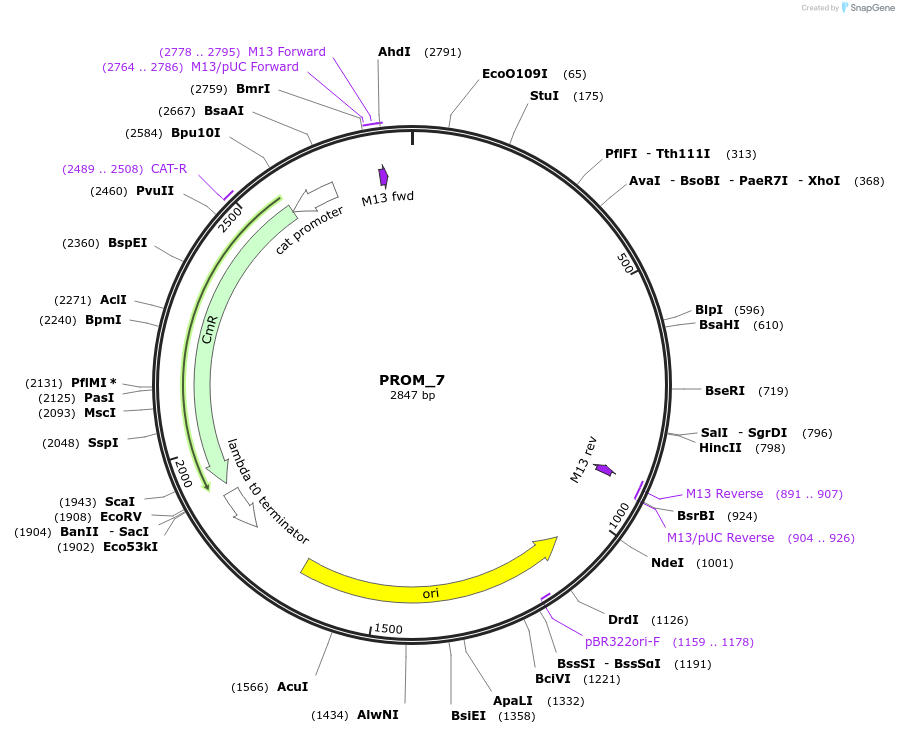
PROM_7
Plasmid#201300PurposeD7: Promoter of TraesCS3D02G316100 as Golden Gate compatible MoClo PROM partDepositorInsertD7: Promoter of TraesCS3D02G316100
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
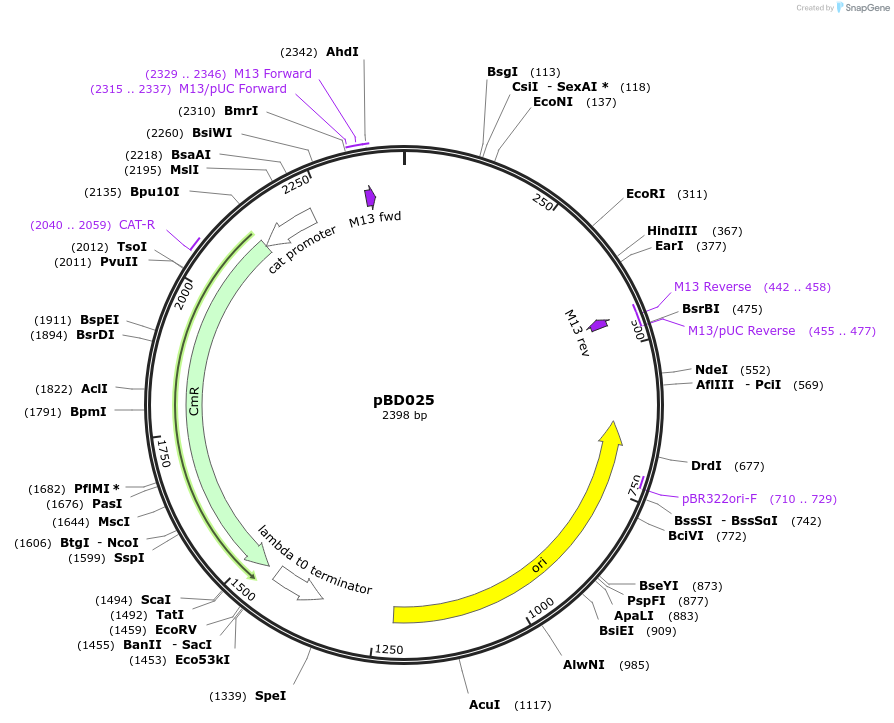
pBD025
Plasmid#201308PurposeAvrSr27-1 as Golden Gate compatible MoClo CDS partDepositorInsertAvrSr27-1
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
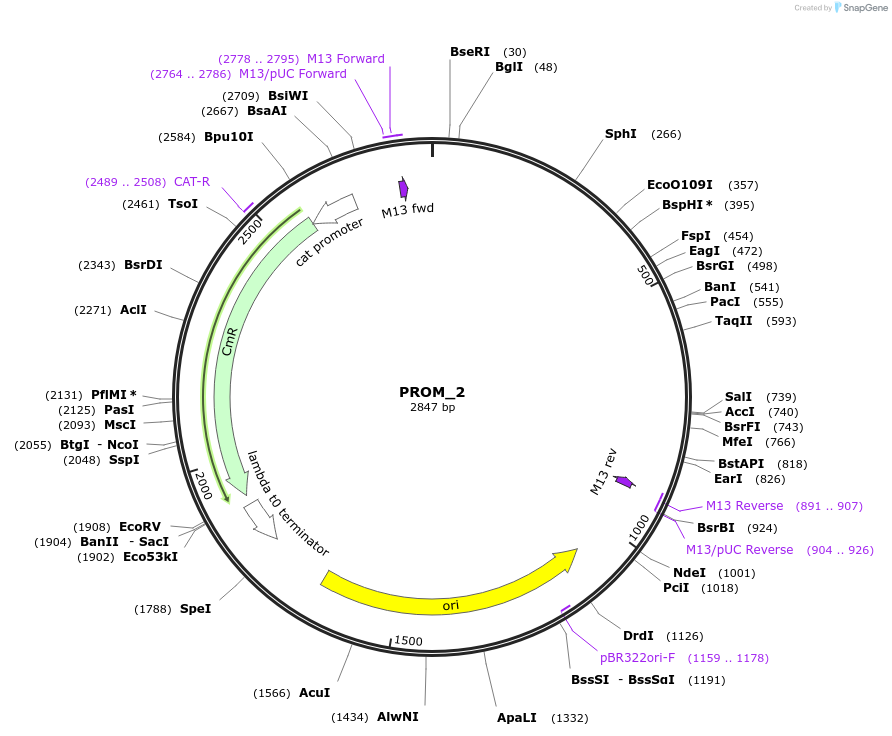
PROM_2
Plasmid#201295PurposeD2: Promoter of TraesCS2B02G533000 as Golden Gate compatible MoClo PROM partDepositorInsertD2: Promoter of TraesCS2B02G533000
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
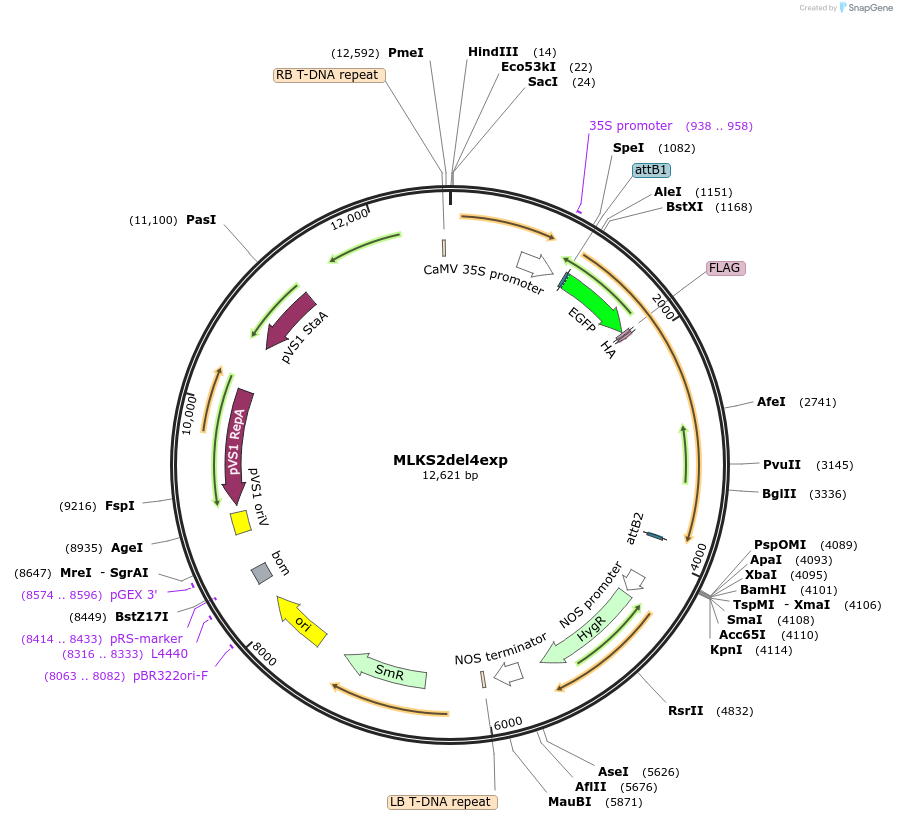
MLKS2del4exp
Plasmid#130861PurposeExpression of plant nuclear envelope LINC protein fused to fluorescent protein and affinity tags for cell biologyDepositorInsertMLKS2
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of terminal four amino acidsAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
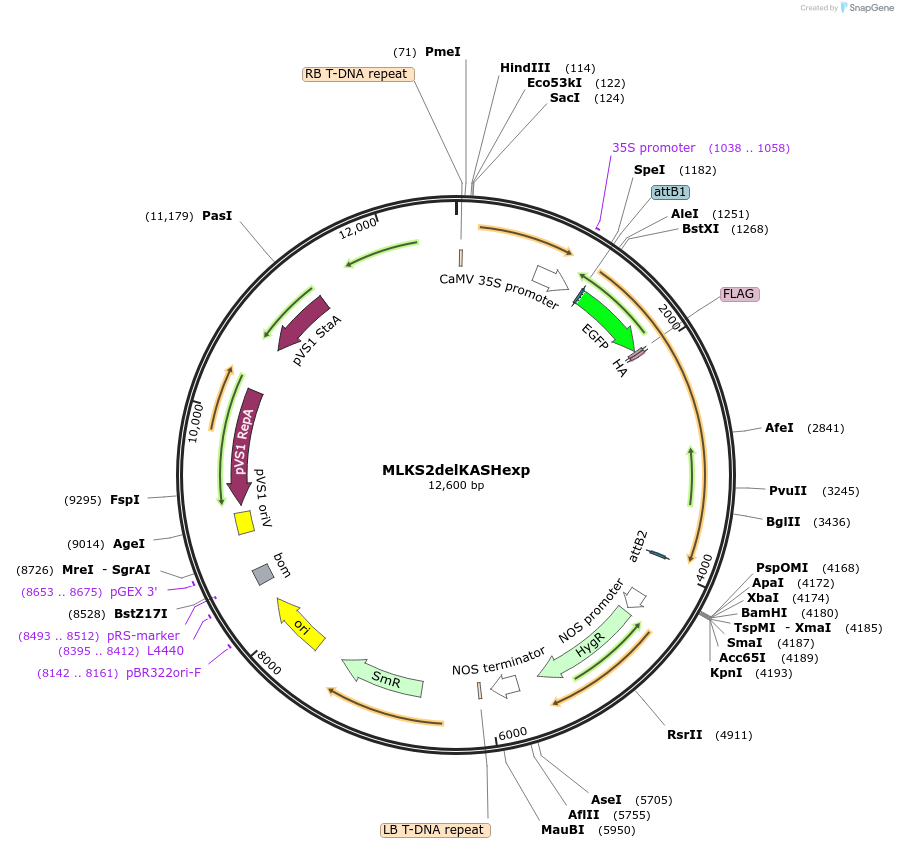
MLKS2delKASHexp
Plasmid#130862PurposeExpression of plant nuclear envelope LINC protein fused to fluorescent protein and affinity tags for cell biologyDepositorInsertMLKS2
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of KASH domainAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
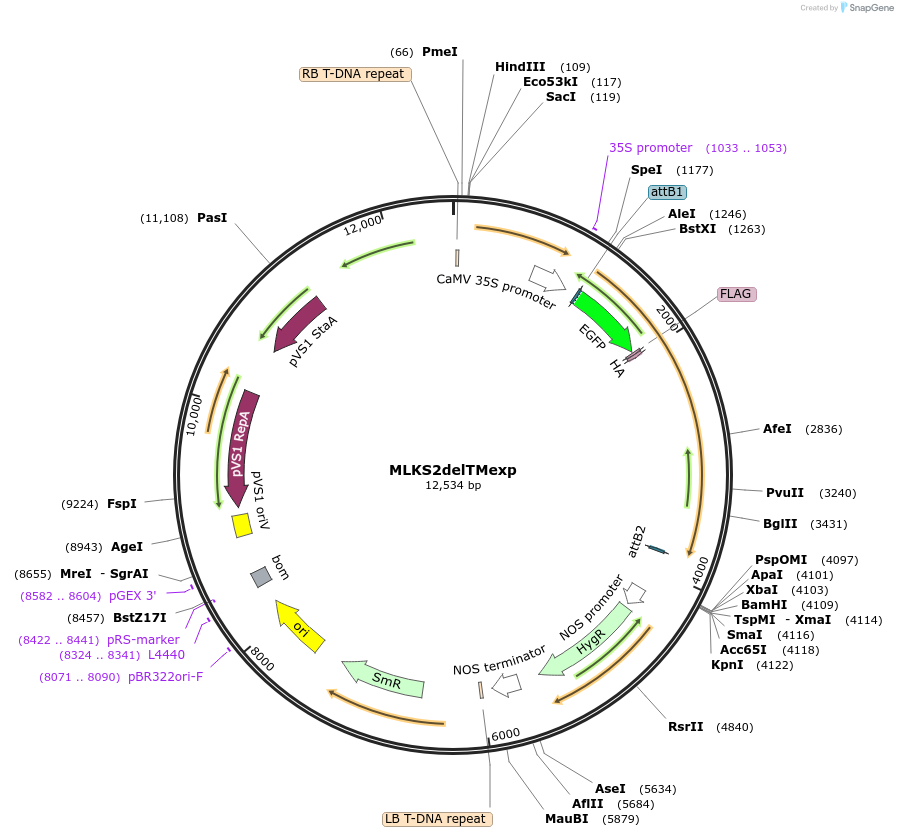
MLKS2delTMexp
Plasmid#130863PurposeExpression of plant nuclear envelope LINC protein fused to fluorescent protein and affinity tags for cell biologyDepositorInsertMLKS2
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of transmembrane domainAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
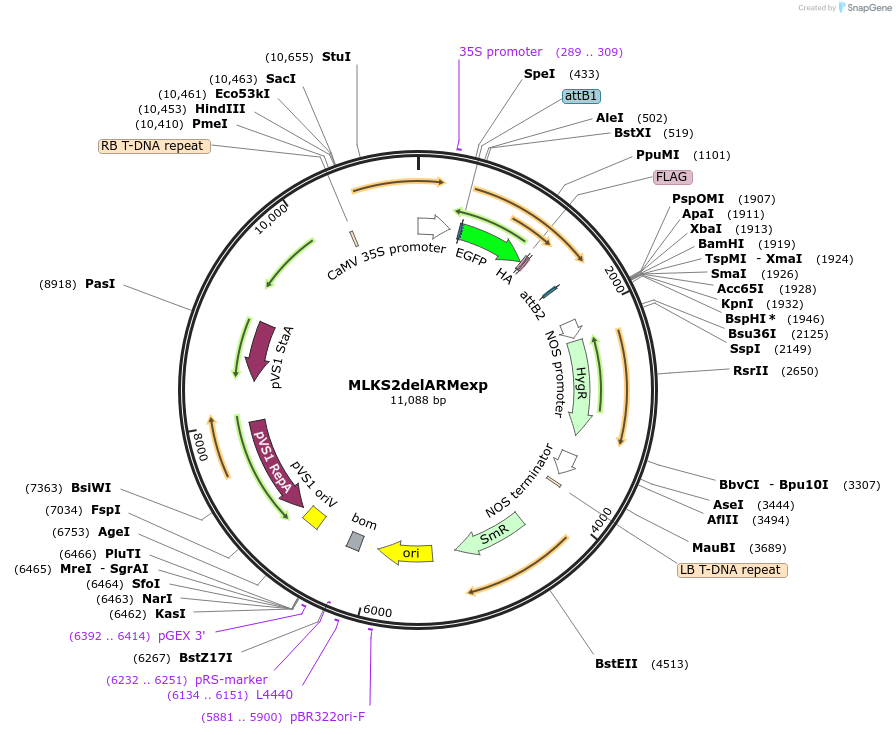
MLKS2delARMexp
Plasmid#130864PurposeExpression of plant nuclear envelope LINC protein fused to fluorescent protein and affinity tags for cell biologyDepositorInsertMLKS2
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of armadillo domainAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
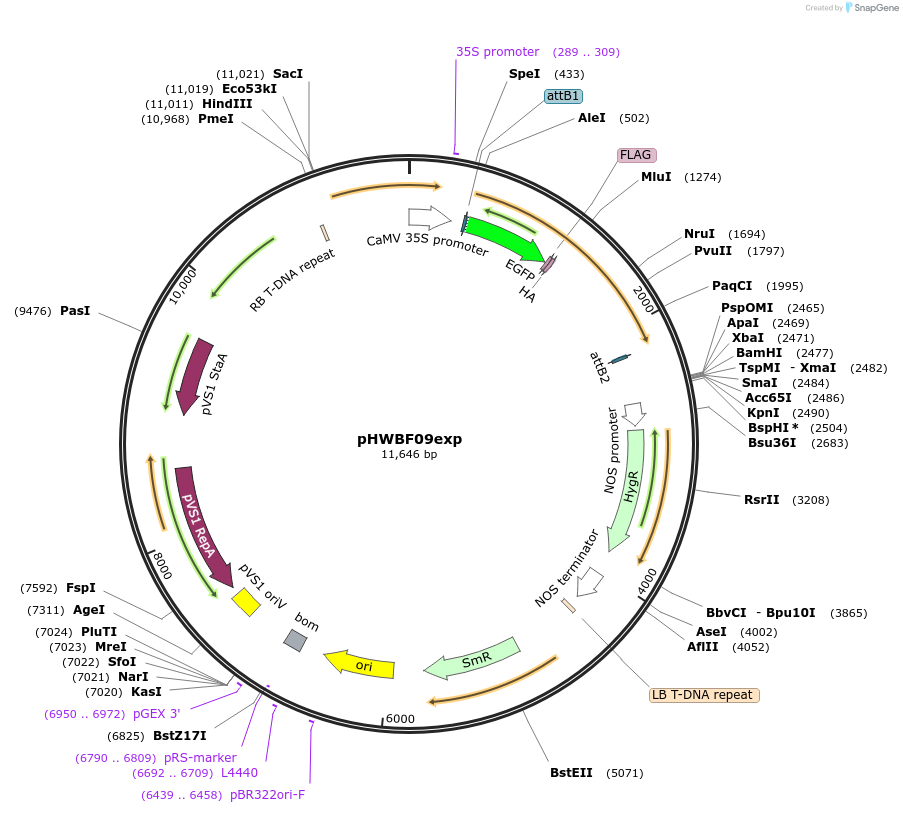
pHWBF09exp
Plasmid#130833PurposeExpression of plant LINC fluorescent fusion protein for cell biologyDepositorInsertMLKP1
TagseGFP-FLAG-HAExpressionPlantPromoterCaMV 35SAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
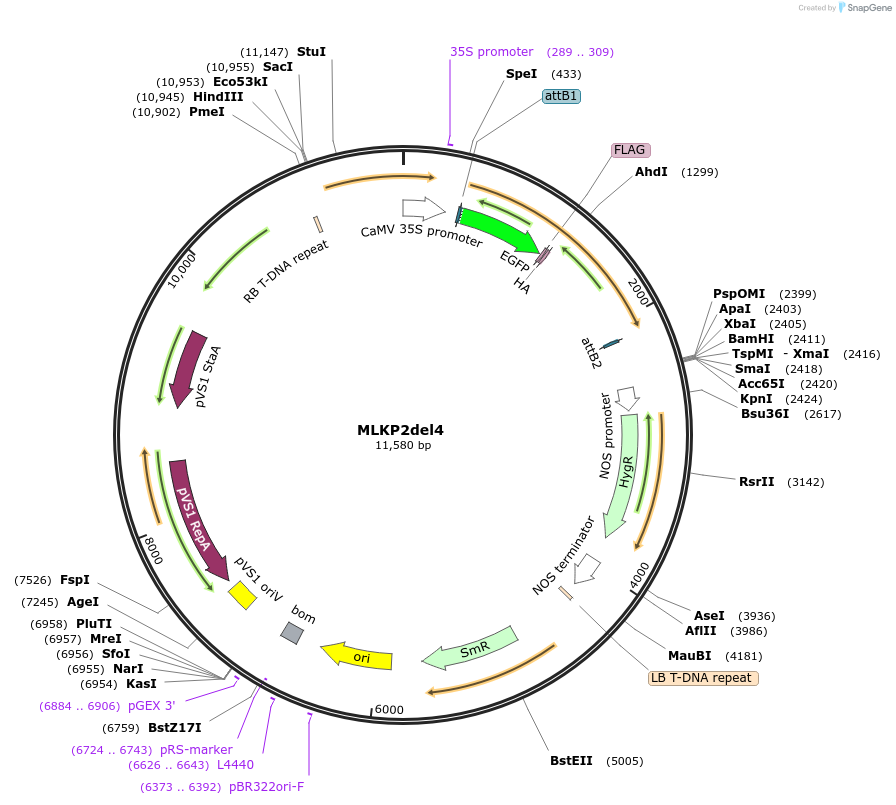
MLKP2del4
Plasmid#130837PurposeExpression of plant LINC fluorescent fusion protein for cell biologyDepositorInsertMLKP2
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of terminal four amino acid residuesPromoterCaMV 35SAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
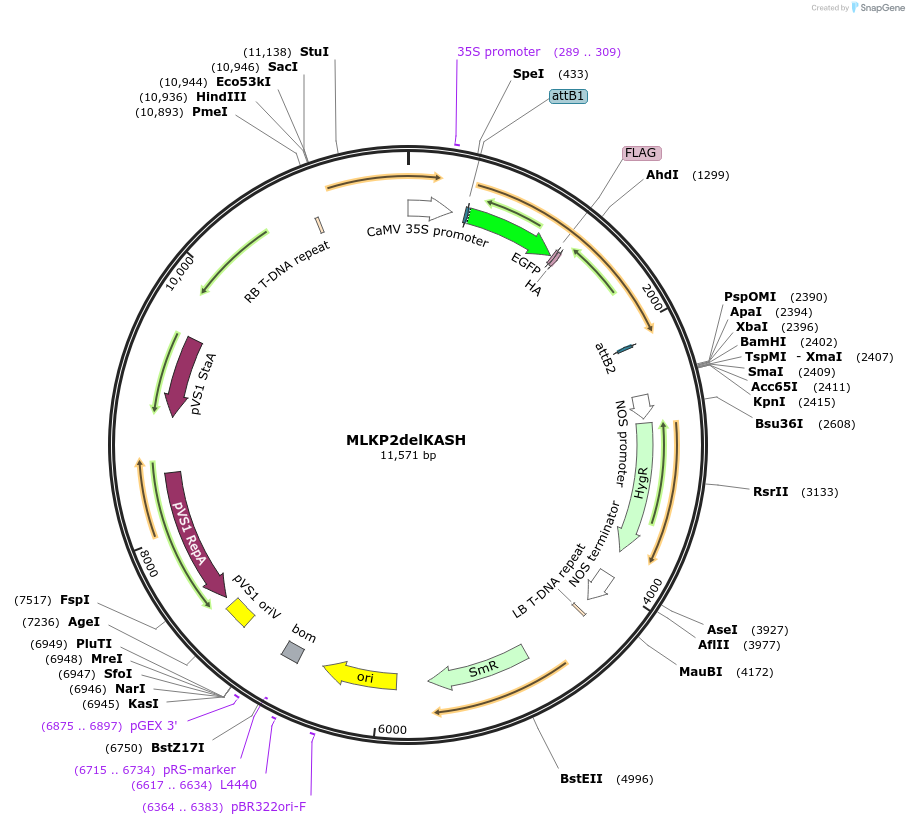
MLKP2delKASH
Plasmid#130838PurposeExpression of plant LINC fluorescent fusion protein for cell biologyDepositorInsertMLKP2
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of KASH domain, addition of three alani…PromoterCaMV 35SAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
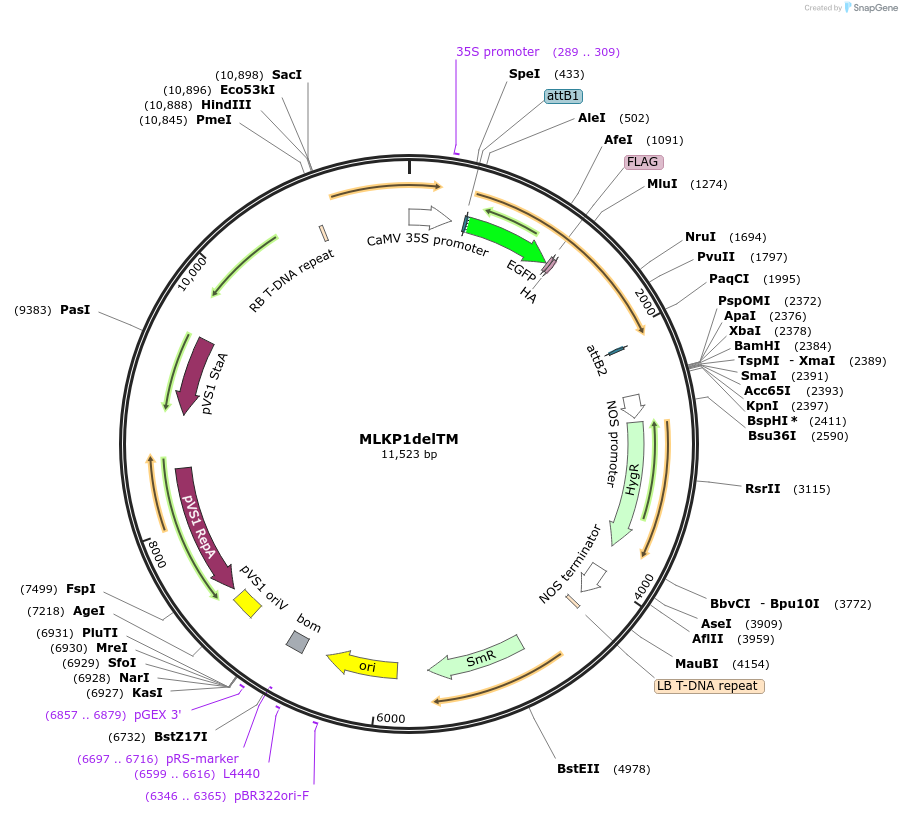
MLKP1delTM
Plasmid#130841PurposeExpression of plant LINC fluorescent fusion protein for cell biologyDepositorInsertMLKP1
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of transmembrane domainPromoterCaMV 35SAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
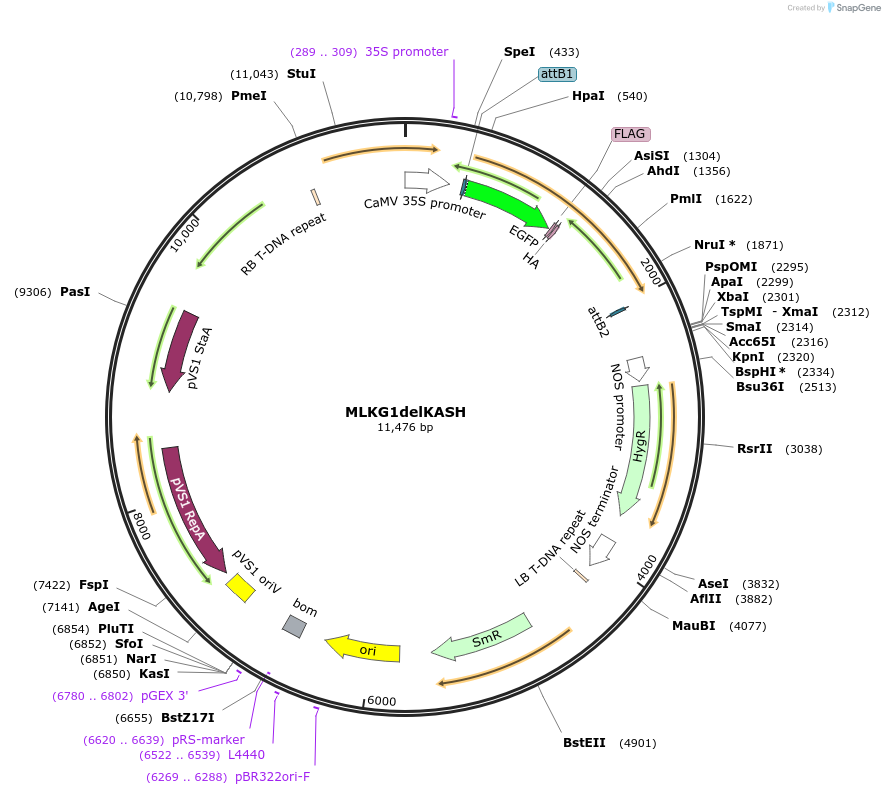
MLKG1delKASH
Plasmid#130842PurposeExpression of plant LINC fluorescent fusion protein for cell biologyDepositorInsertMLKG1
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of KASH domainPromoterCaMV 35SAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
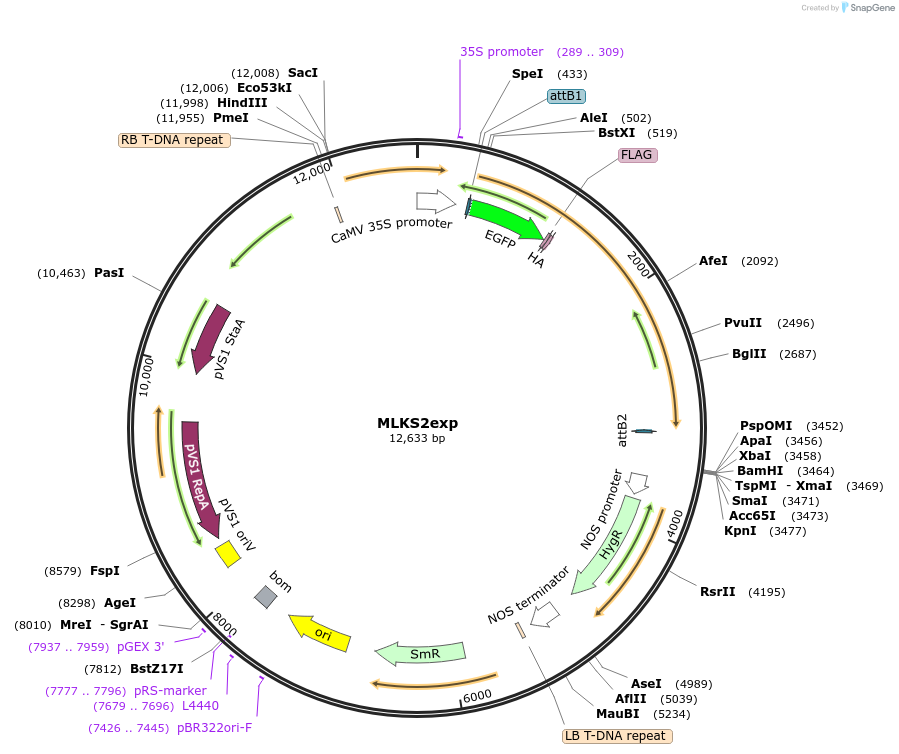
MLKS2exp
Plasmid#130856PurposeExpression of plant nuclear envelope LINC protein fused to fluorescent protein and affinity tags for cell biologyDepositorInsertMLKS2
TagseGFP-FLAG-HAExpressionPlantAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits