4,457 results
-
Plasmid#246300PurposeEvaluation of HbU6.2m3 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertHbU6.2m3 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-T (-29 from TSS), G-to-A (-24)PromoterHbU6.2m3 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
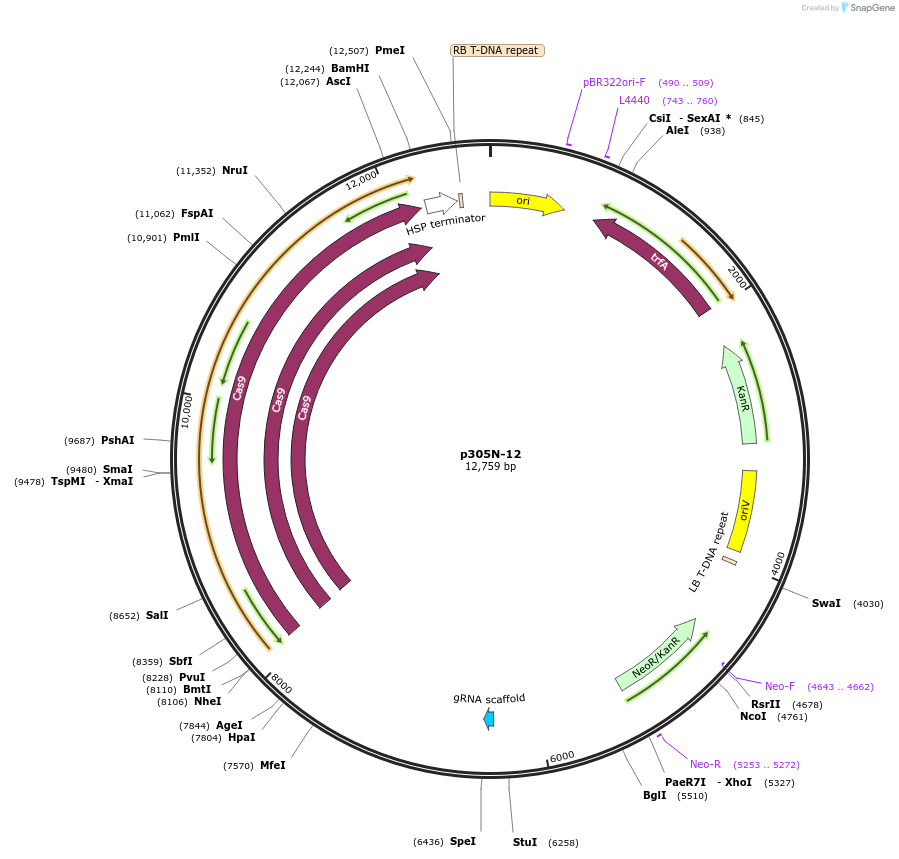
p305N-12
Plasmid#246295PurposeEvaluation of CiU6.4 promoter (Pol III promoter) for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertCiU6.4 promoter
UseCRISPRExpressionPlantPromoterCiU6.4 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
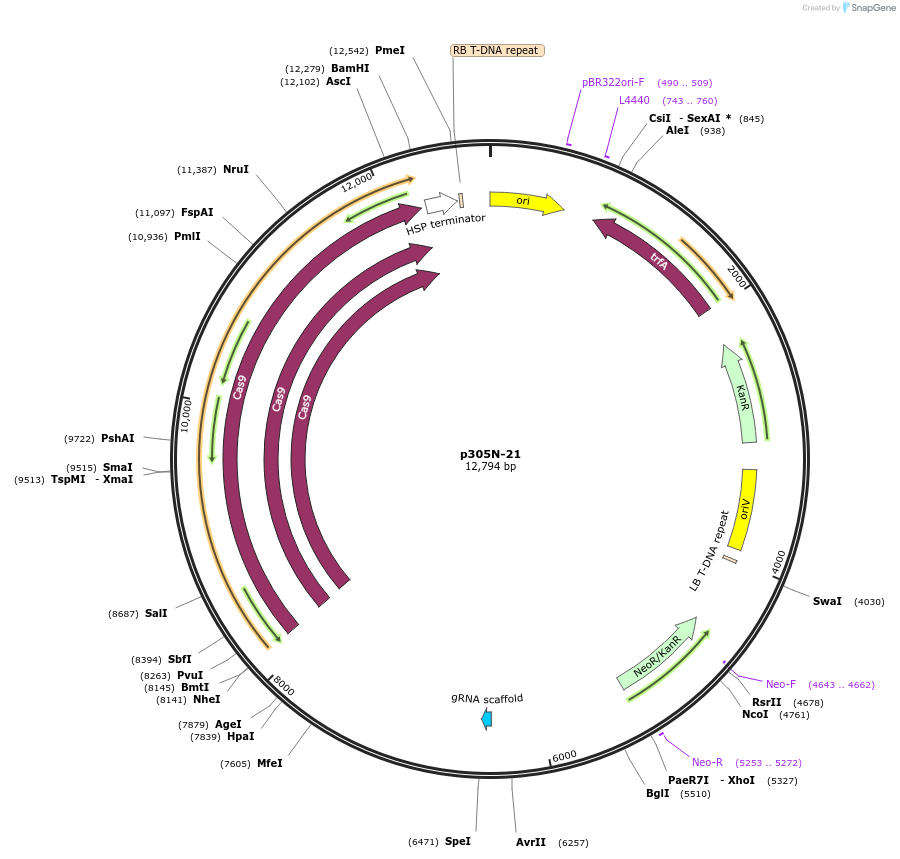
p305N-21
Plasmid#246304PurposeEvaluation of MtU6.6-104 promoter (Pol III promoter) deletion (104 bp) for CRISPR-Cas9 editing of mEGFP in Nicotiana benthamiana and poplar reporter linesDepositorInsertMtU6.6-104 promoter
UseCRISPRExpressionPlantPromoterMtU6.6-104 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
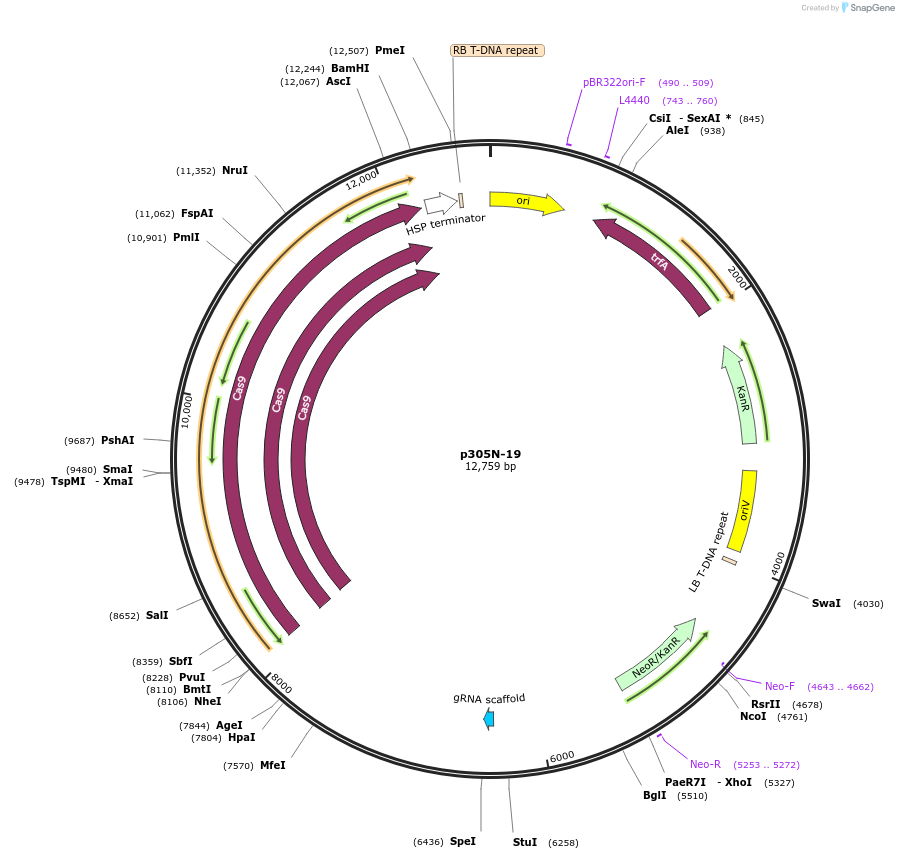
p305N-19
Plasmid#246302PurposeEvaluation of MdU6.1 promoter (Pol III promoter) for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertMdU6.1 promoter
UseCRISPRExpressionPlantPromoterMdU6.1 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
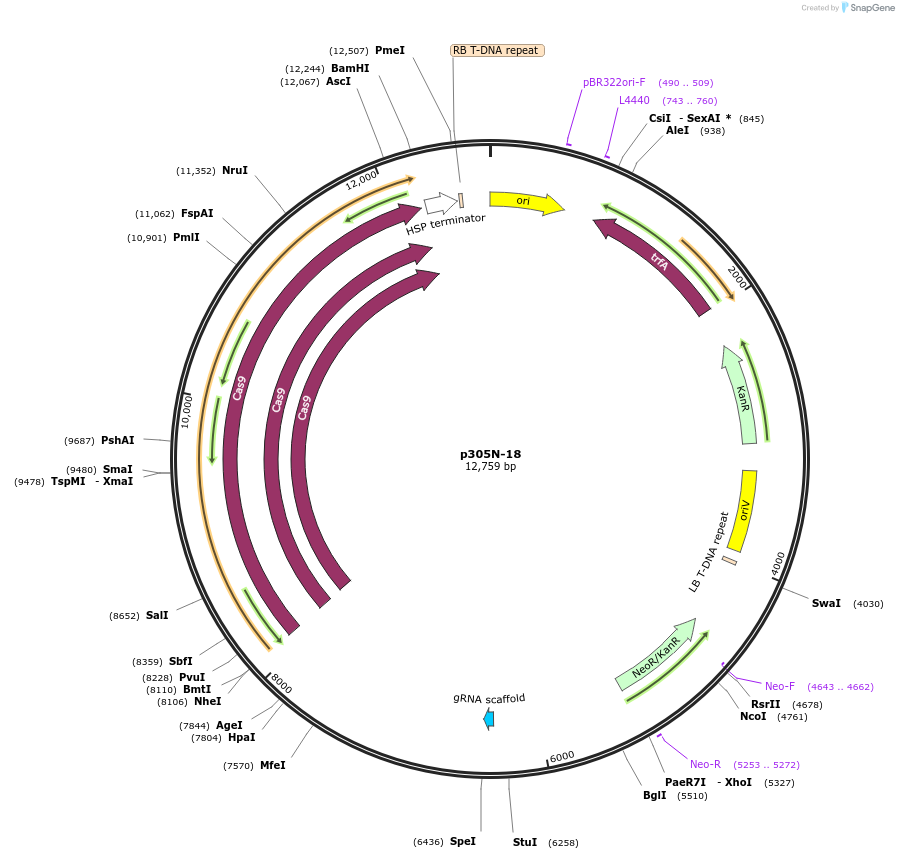
p305N-18
Plasmid#246301PurposeEvaluation of MdU3.1 promoter (Pol III promoter) for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertMdU3.1 promoter
UseCRISPRExpressionPlantPromoterMdU3.1 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
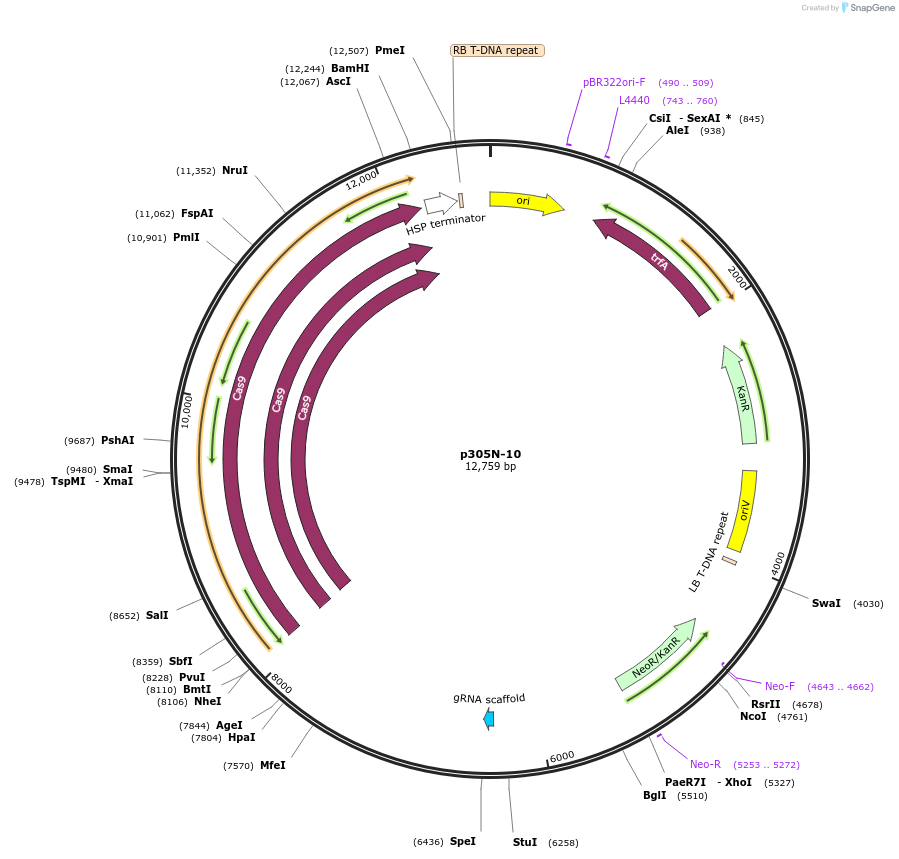
p305N-10
Plasmid#246293PurposeEvaluation of CiU6.1 promoter (Pol III promoter) for CRISPR-Cas9 editing of mEGFP in Nicotiana benthamiana and poplar reporter linesDepositorInsertCiU6.1 promoter
UseCRISPRExpressionPlantPromoterCiU6.1 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
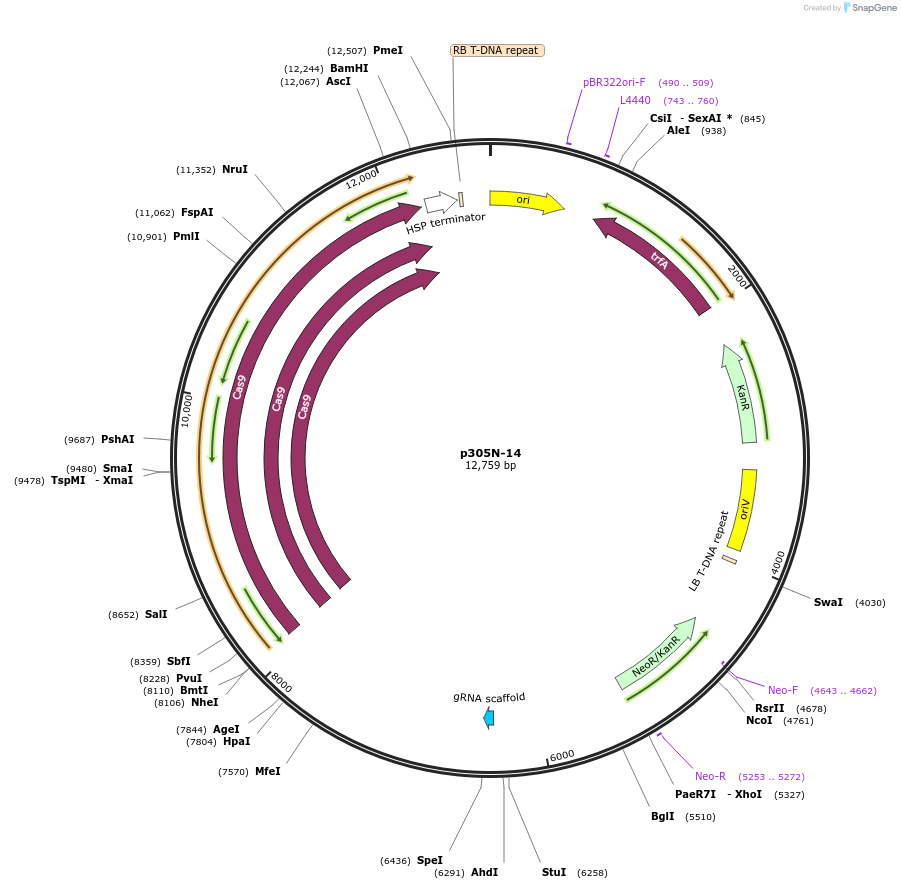
p305N-14
Plasmid#246297PurposeEvaluation of CiU6.6c16 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertCiU6.6c16 promoter
UseCRISPRExpressionPlantMutationT-to-C (-2 from TSS)PromoterCiU6.6c16 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
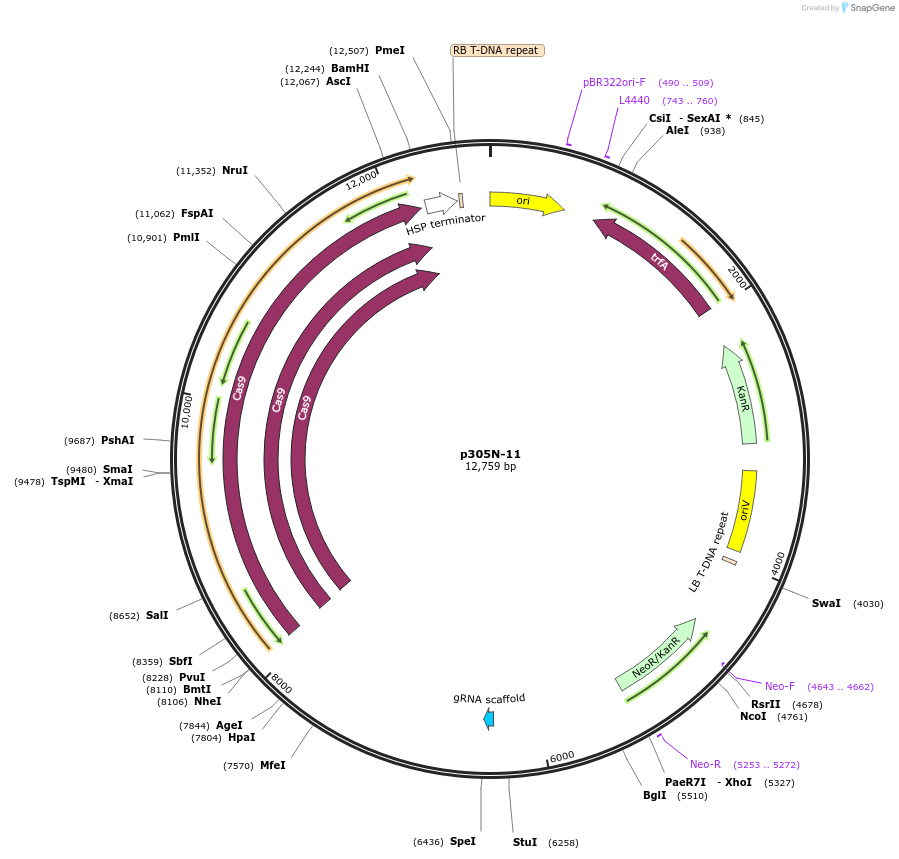
p305N-11
Plasmid#246294PurposeEvaluation of CiU6.3c8 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertCiU6.3c8 promoter
UseCRISPRExpressionPlantMutationA-to-C (-23 from TSS)PromoterCiU6.3c8 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
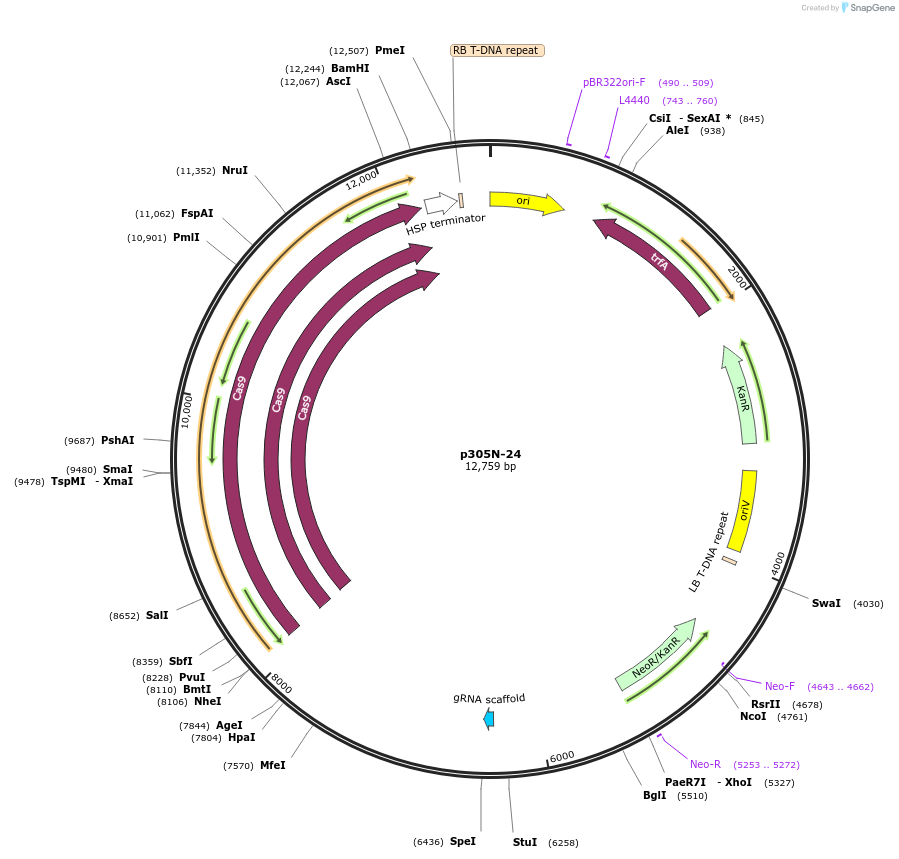
p305N-24
Plasmid#246307PurposeEvaluation of a synthetic Pol III promoter based on the MtU6.6 promoter for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertMtU6.6m4 promoter
UseCRISPRExpressionPlantPromoterMtU6.6m4 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
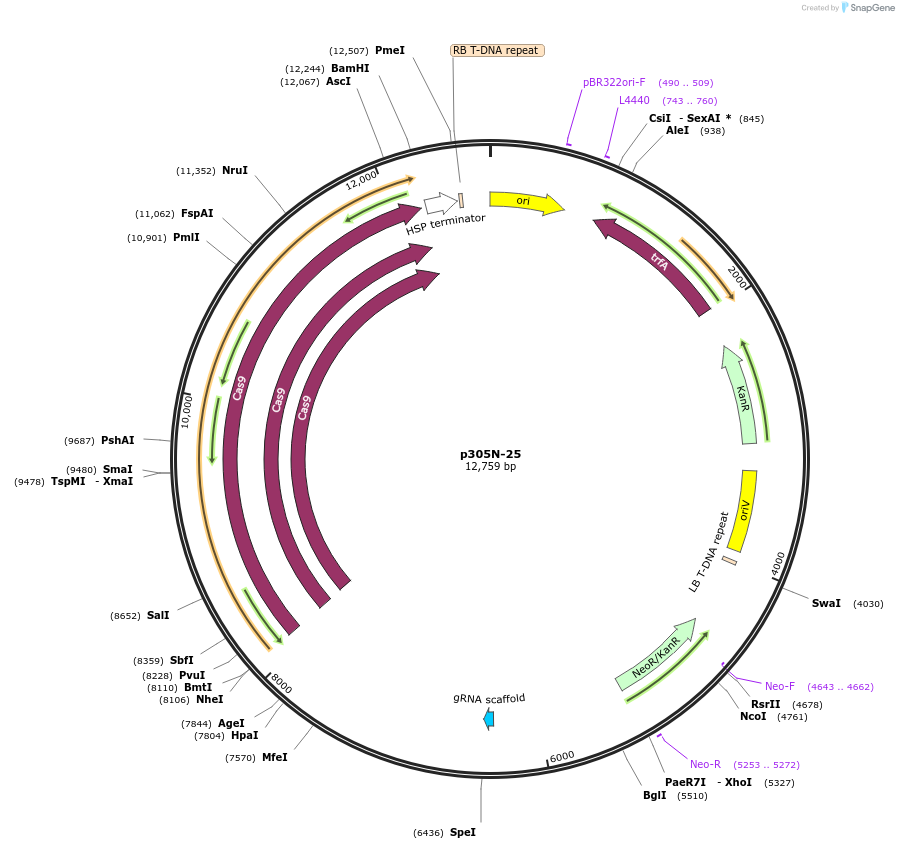
p305N-25
Plasmid#246308PurposeEvaluation of a synthetic Pol III promoter based on the MtU6.6 promoter for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertMtU6.6m5 promoter
UseCRISPRExpressionPlantPromoterMtU6.6m5 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
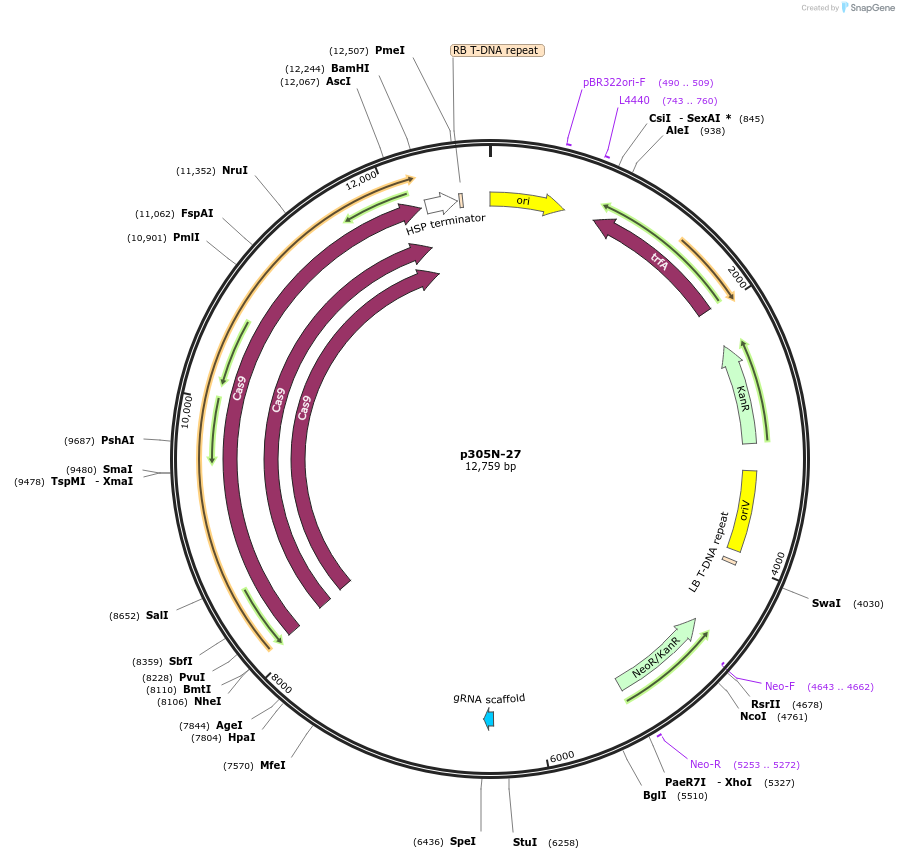
p305N-27
Plasmid#246310PurposeEvaluation of PtU6.1 promoter (Pol III promoter) for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.1 promoter
UseCRISPRExpressionPlantPromoterPtU6.1 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
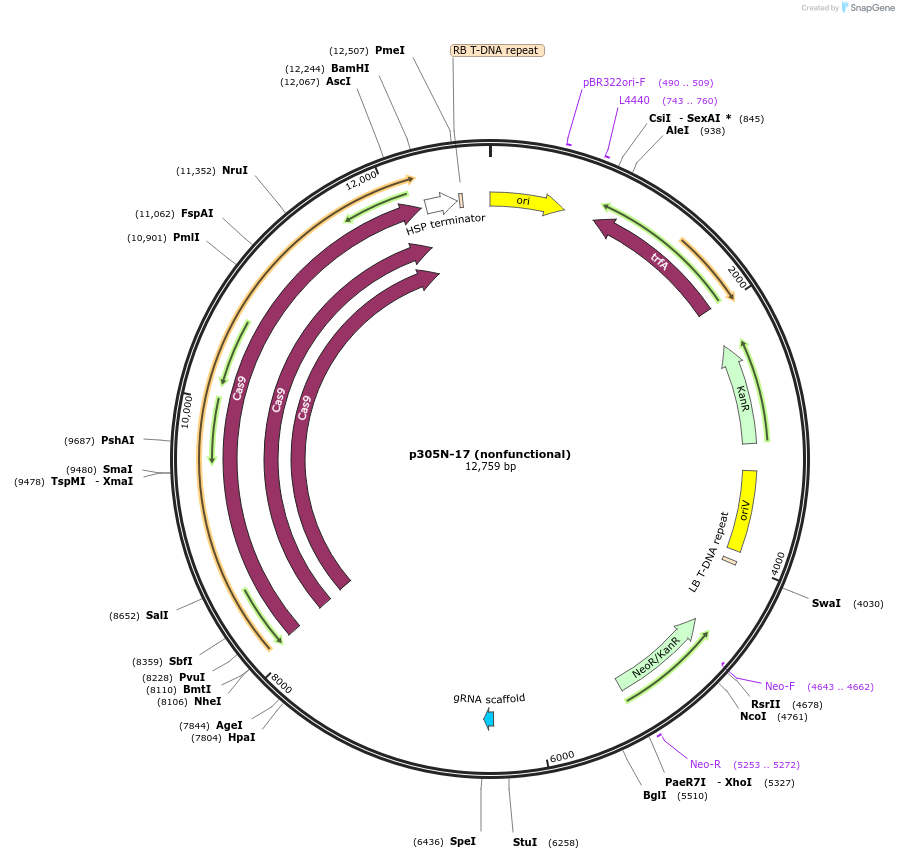
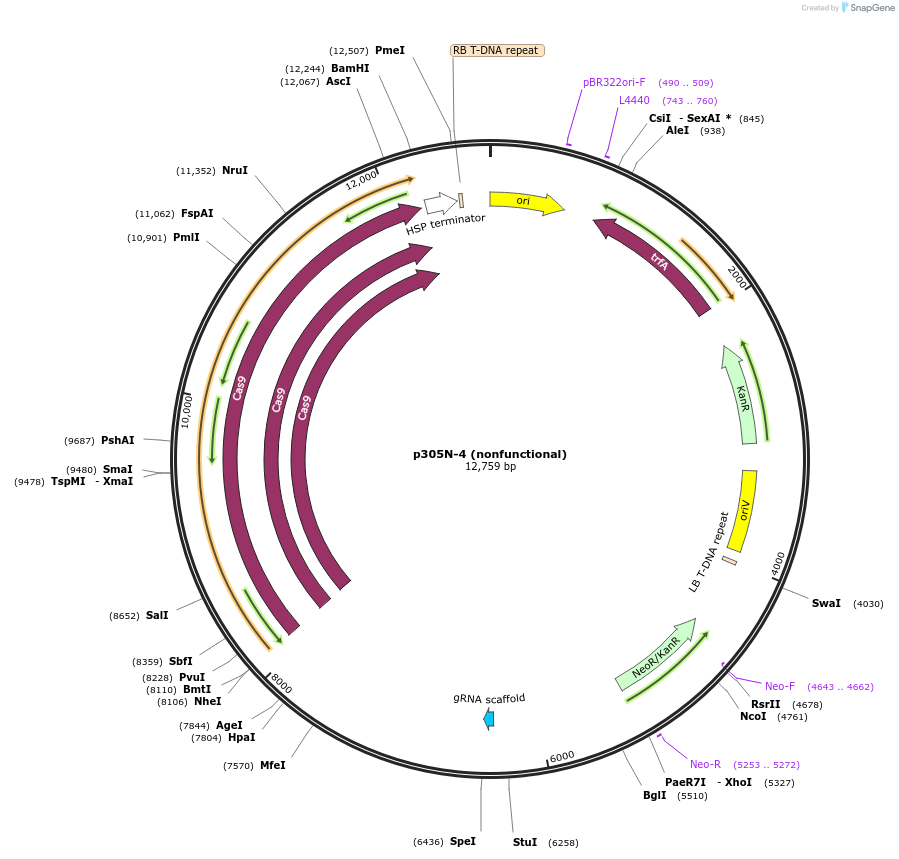
p305N-4 (nonfunctional)
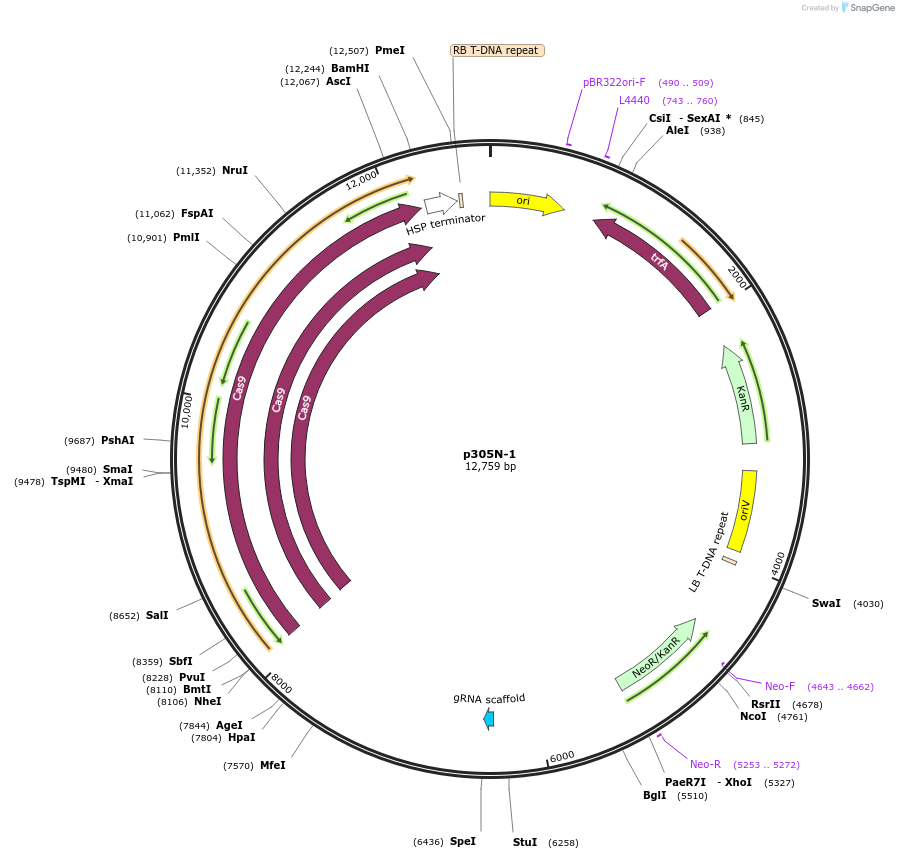
Plasmid#246287PurposeEvaluation of AtU6.1c1 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.1c1 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationG-to-C at -15 from TSSPromoterAtU6.1c1 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
p305N-1
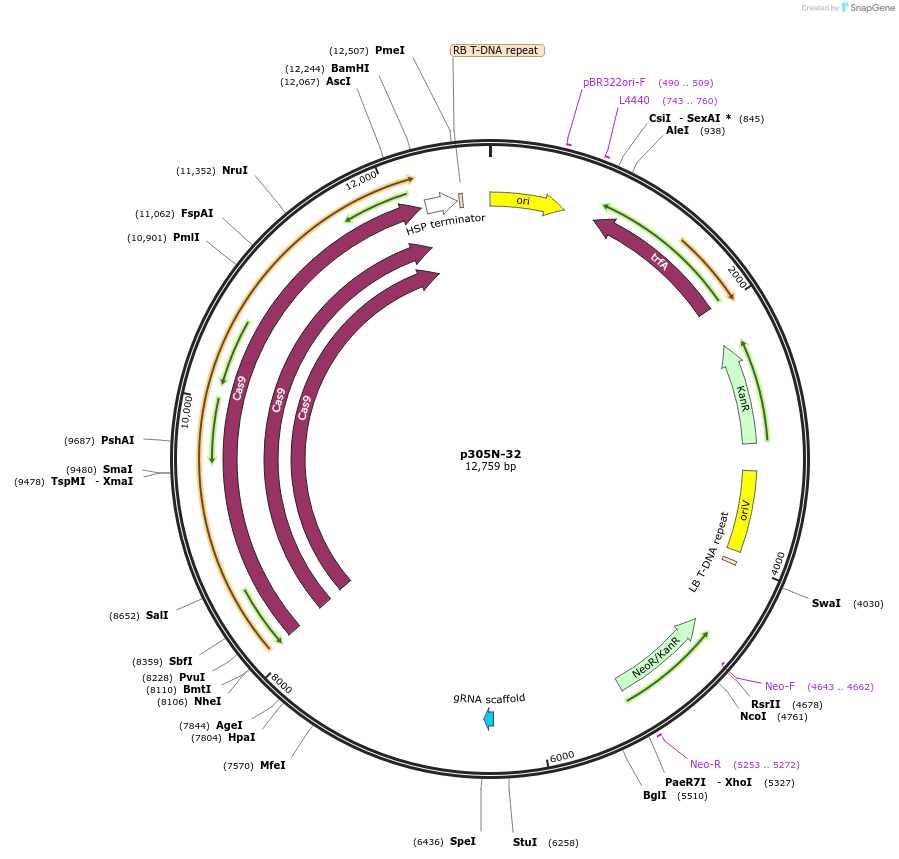
Plasmid#246284PurposeEvaluation of AtU3b promoter (Pol III promoter) for CRISPR-Cas9 editing of mEGFP in Nicotiana benthamiana and poplar reporter linesDepositorInsertAtU3b promoter
UseCRISPRExpressionPlantPromoterAtU3b promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
p305N-32
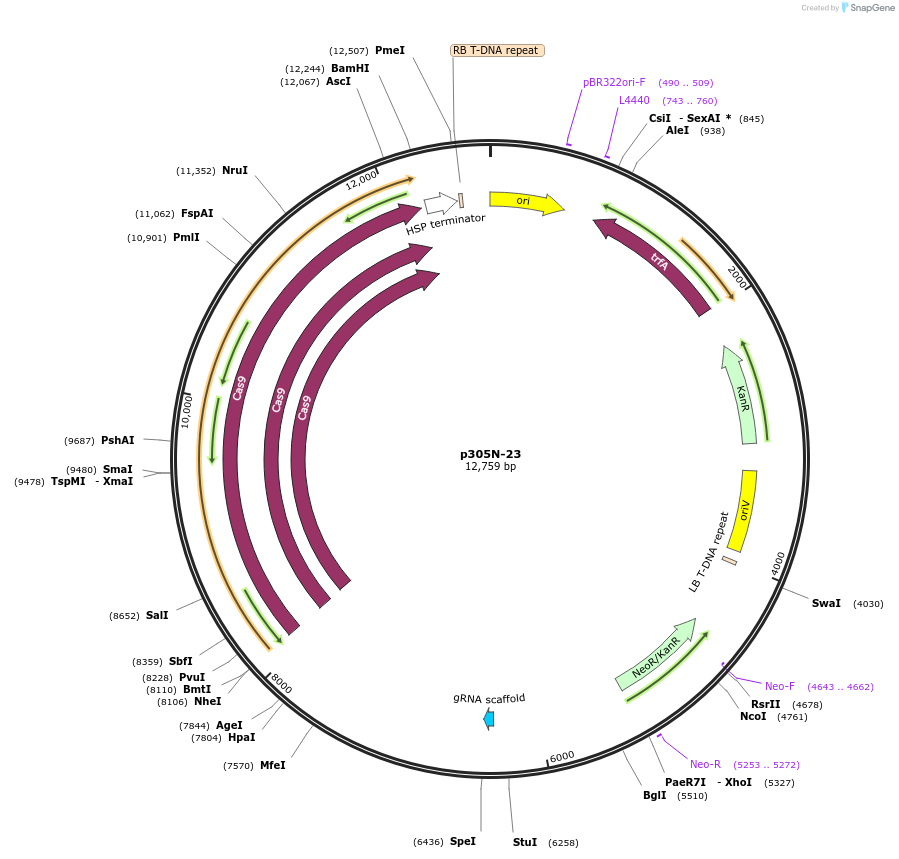
Plasmid#246315PurposeEvaluation of VvU6.1 promoter (Pol III promoter) for CRISPR-Cas9 editing of mEGFP in Nicotiana benthamiana and poplar reporter linesDepositorInsertVvU6.1 promoter
UseCRISPRExpressionPlantPromoterVvU6.1 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
p305N-23
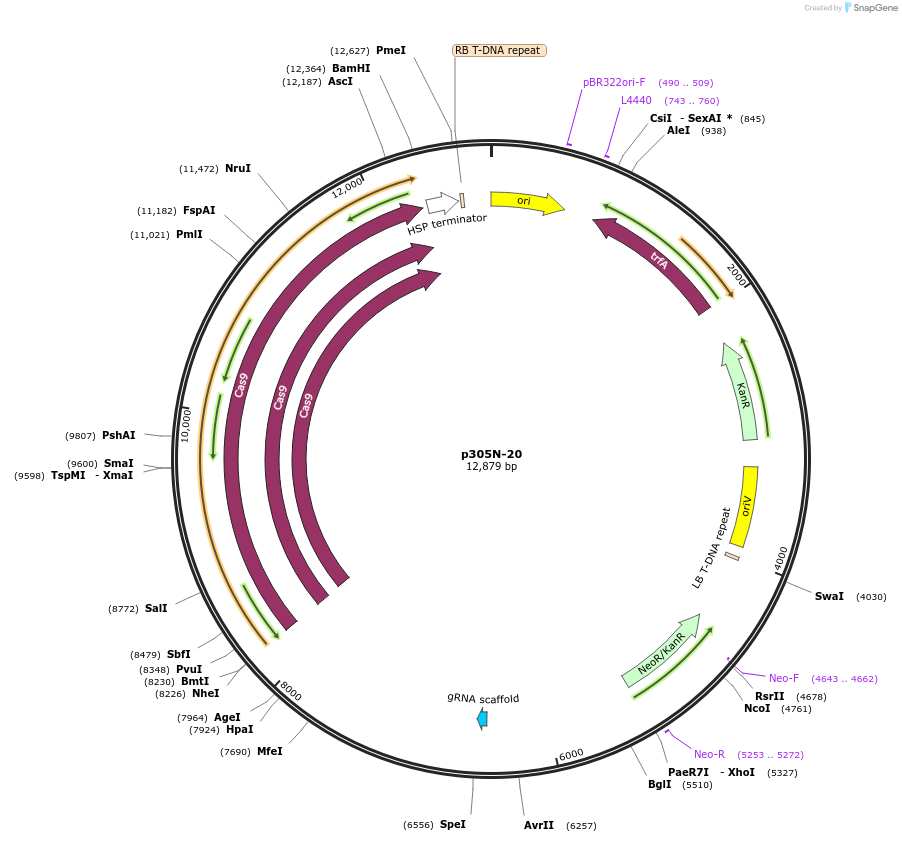
Plasmid#246306PurposeEvaluation of a synthetic Pol III promoter based on the MtU6.6 promoter for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertMtU6.6m1 promoter
UseCRISPRExpressionPlantPromoterMtU6.6m1 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
p305N-20
Plasmid#246303PurposeEvaluation of MtU6.6-189 promoter (Pol III promoter) deletion (189 bp) for CRISPR-Cas9 editing of mEGFP in Nicotiana benthamiana and poplar reporter linesDepositorInsertMtU6.6-189 promoter
UseCRISPRExpressionPlantPromoterMtU6.6-189 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
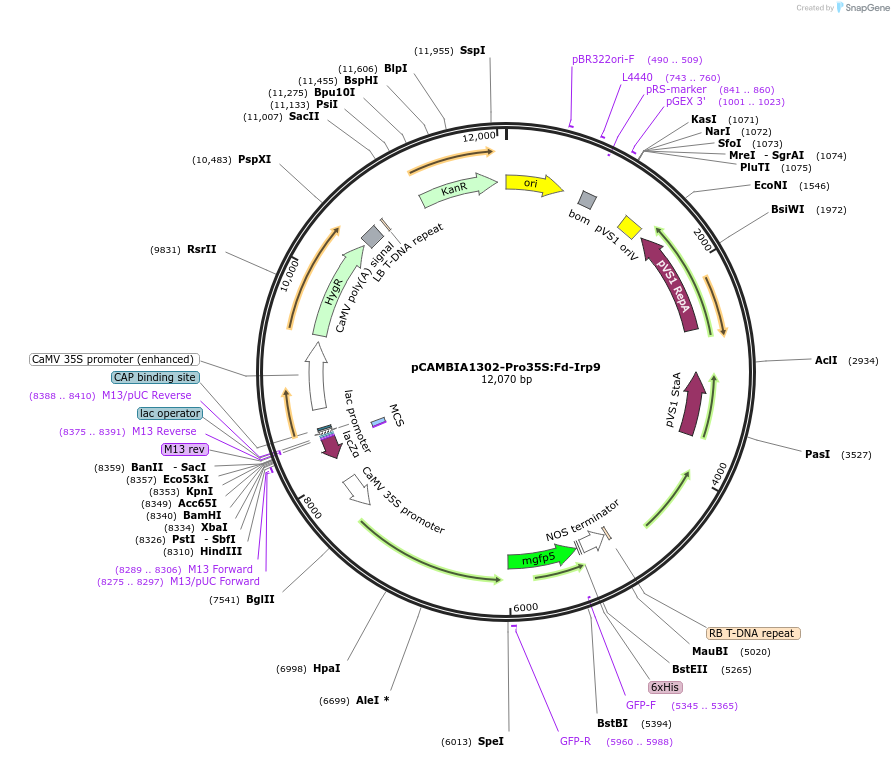
pCAMBIA1302-Pro35S:Fd-Irp9
Plasmid#246318PurposeOverexpression of a bacterial bifunctional salicylate synthase, Irp9, targeted to the chloroplastDepositorInsertFd-Irp9
ExpressionPlantPromotercauliflower mosaic virus 35SAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
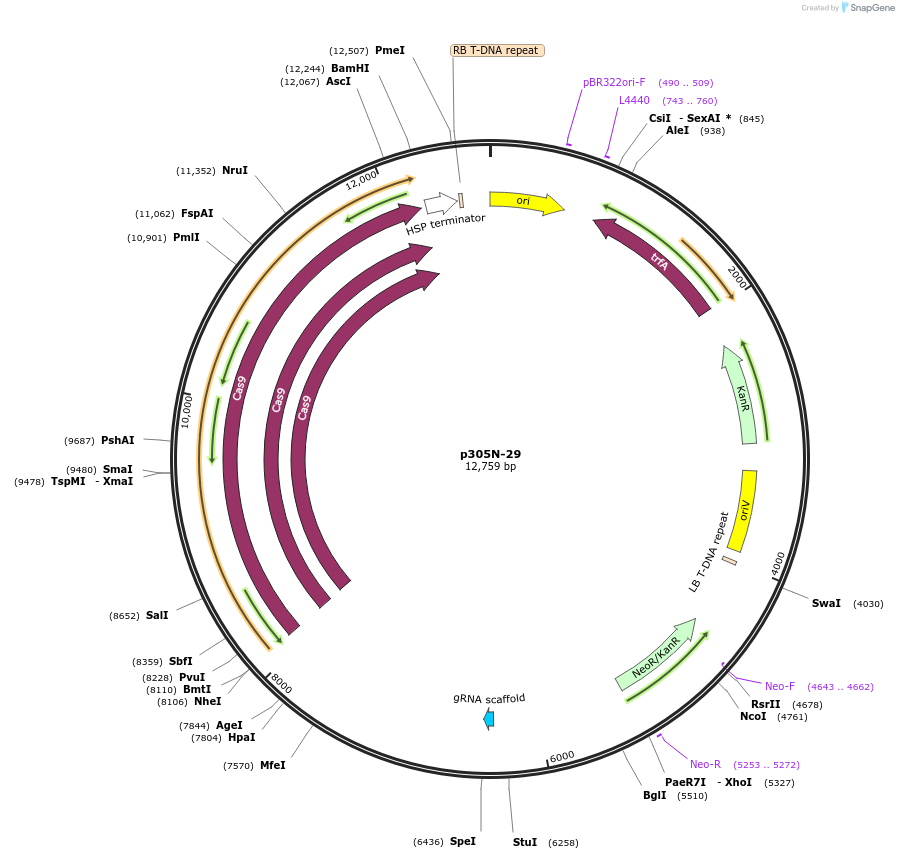
p305N-29
Plasmid#246312PurposeEvaluation of PtU6.2 promoter (Pol III promoter) for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2 promoter
UseCRISPRExpressionPlantPromoterPtU6.2 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
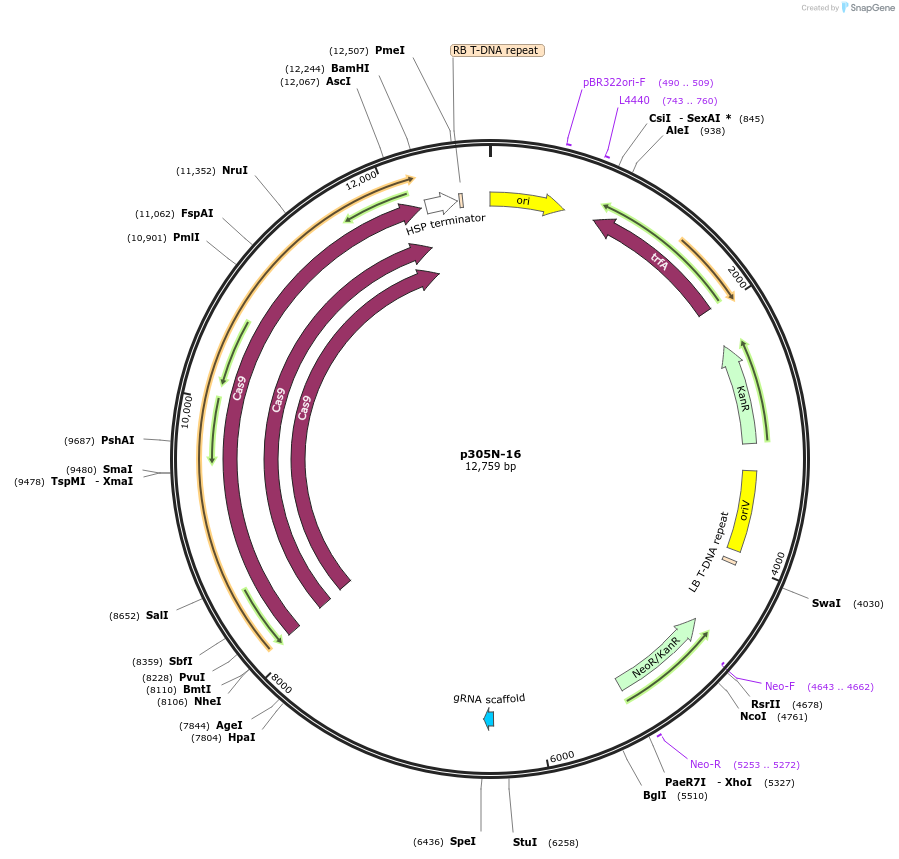
p305N-16
Plasmid#246299PurposeEvaluation of HbU6.2m1 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertHbU6.2m1 promoter
UseCRISPRExpressionPlantMutationA-to-G (-56 from TSS), C-to-T (-29), G-to-A (-24)PromoterHbU6.2m1 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
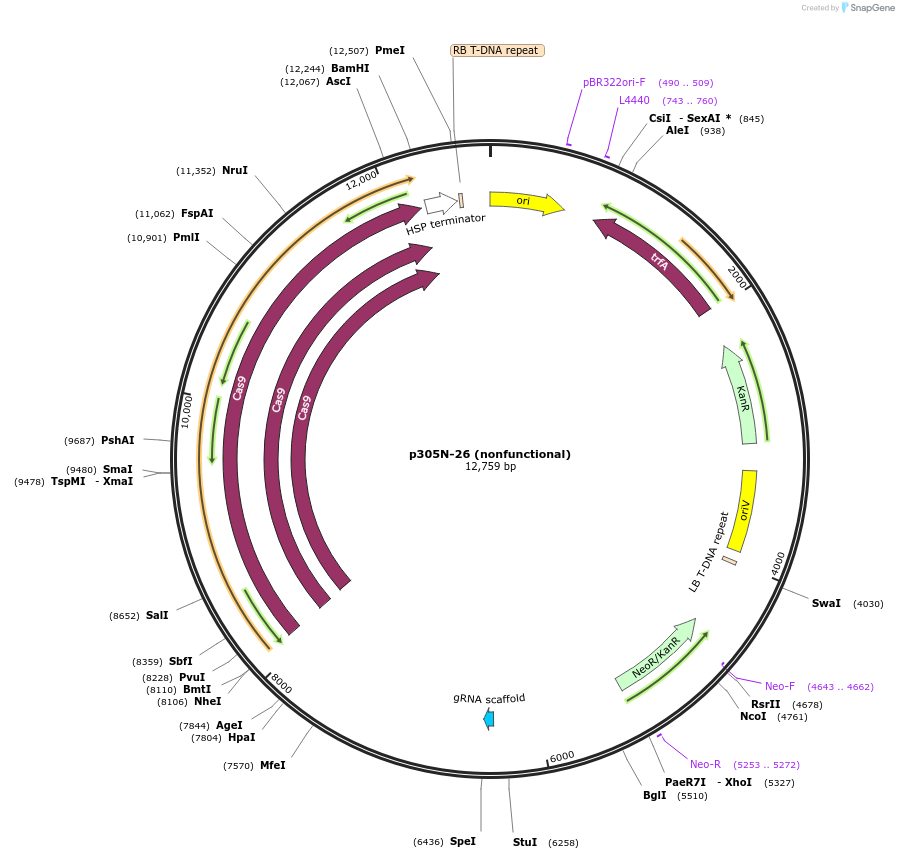
p305N-26 (nonfunctional)
Plasmid#246309PurposeEvaluation of MtU6.6m7 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertMtU6.6m7 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-A (-63 from TSS), T-to-G (-57)PromoterMtU6.6m7 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
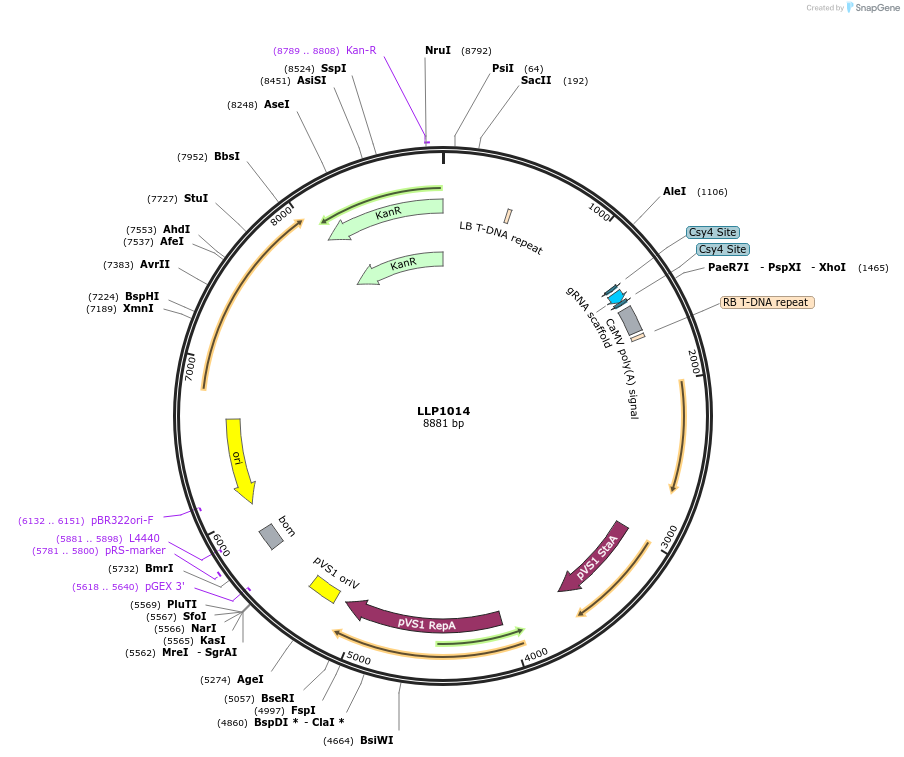
LLP1014
Plasmid#239936PurposeHsp18.2 (heat inducible) promoter driving the expression of sgRNA-BDepositorInsertHsp18.2::sgRNA-B
UseSynthetic BiologyExpressionPlantMutationN/AAvailable SinceOct. 29, 2025AvailabilityAcademic Institutions and Nonprofits only -
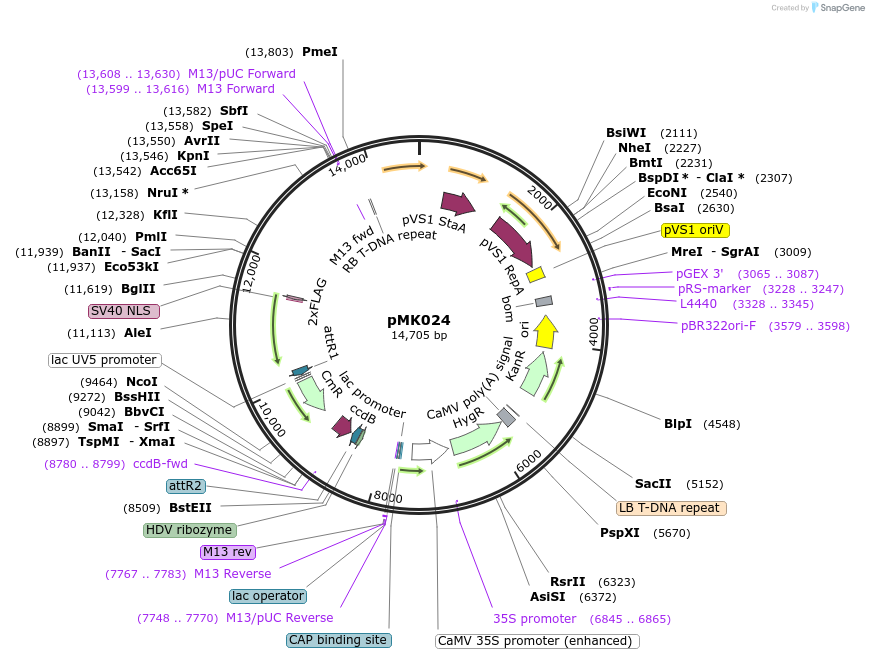
pMK024
Plasmid#236571PurposeCloning vector for PaqCI Golden Gate assembly of ISAam1 guides into ISAam1 plant expression vectorDepositorTypeEmpty backboneExpressionPlantAvailable SinceOct. 28, 2025AvailabilityAcademic Institutions and Nonprofits only -
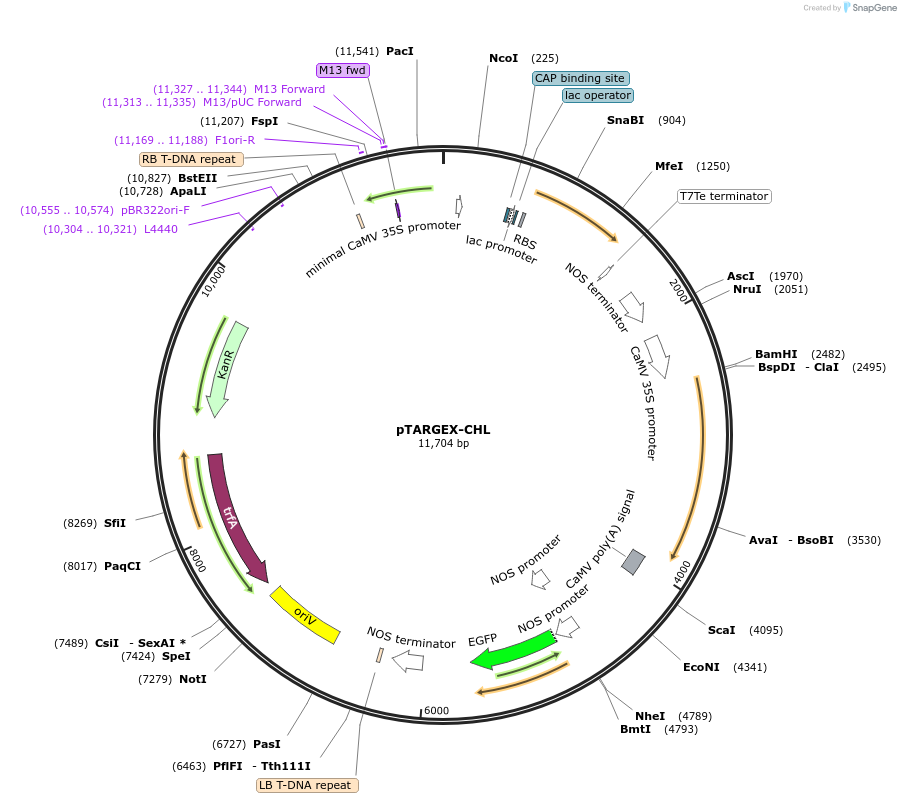
pTARGEX-CHL
Plasmid#242447PurposeTargets high levels of recombinant protein to chloroplast. Empty vector. Clone via SapI restriction site. mCherry dropout cassette will enable Red/White screening in E.coli.DepositorTypeEmpty backboneTagsTransit peptide from N. tabacum RbcSExpressionPlantPromoterdouble 35SAvailable SinceOct. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
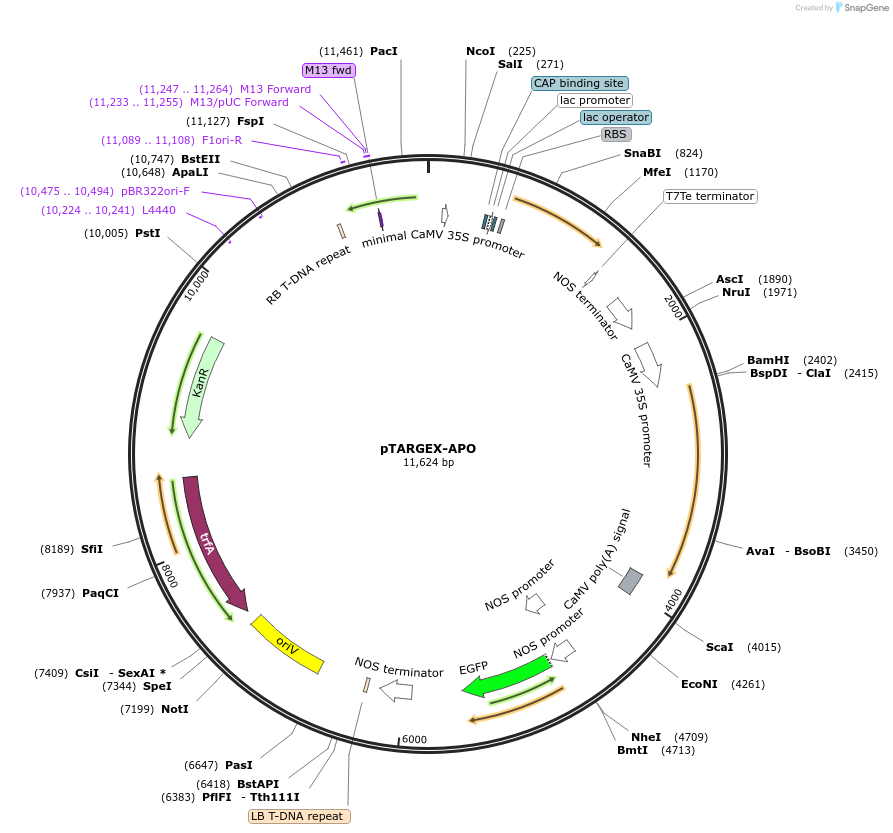
pTARGEX-APO
Plasmid#242446PurposeTargets high levels of recombinant protein to apoplast. Empty vector. Clone via SapI restriction site. mCherry dropout cassette will enable Red/White screening in E.coli.DepositorTypeEmpty backboneTagsN-terminal fusion with PR1b signal peptideExpressionPlantPromoterdouble 35SAvailable SinceOct. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
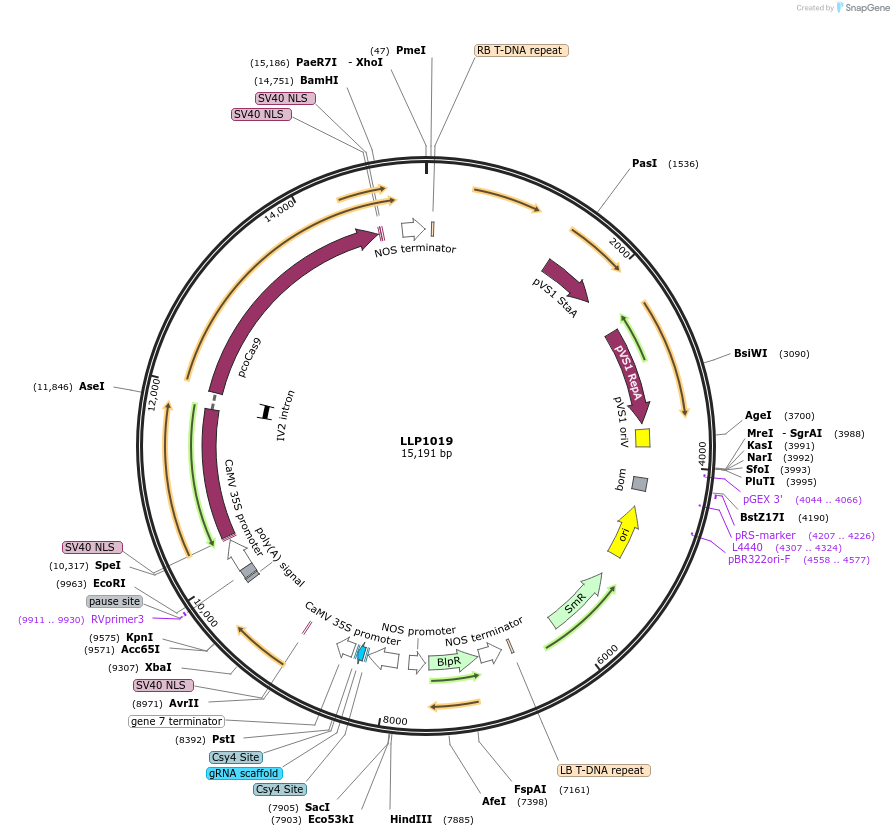
LLP1019
Plasmid#239941PurposeCaMV 35S driving the expression of dCas9-ZAT10(1x)DepositorInsertCaMV 35S::dCas9-ZAT10(1x)
UseCRISPR and Synthetic BiologyExpressionPlantMutationN/AAvailable SinceOct. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
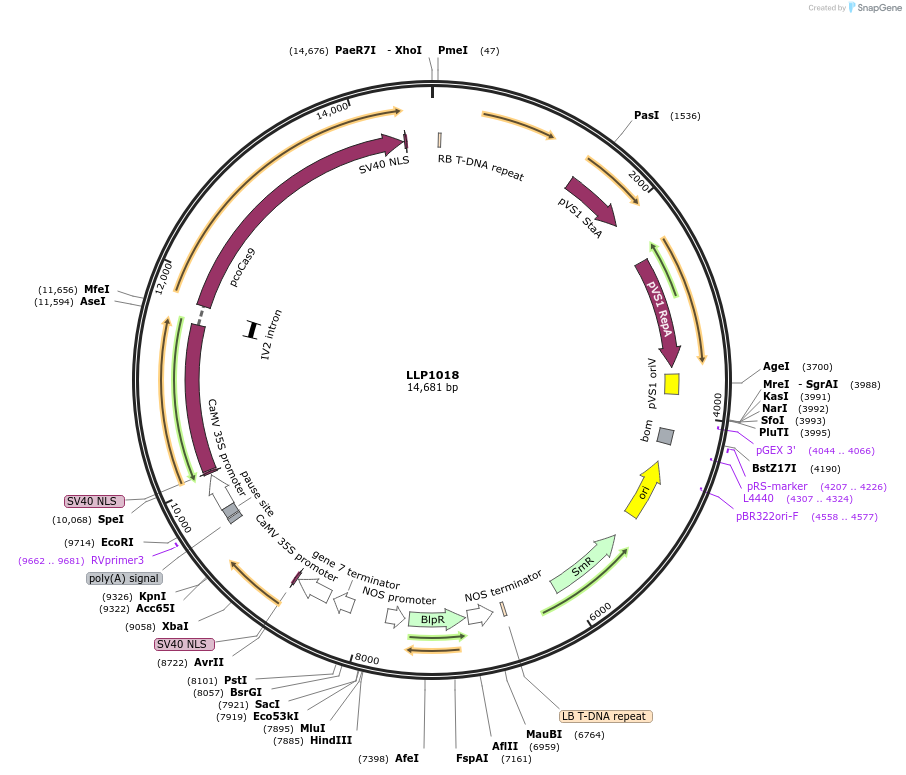
LLP1018
Plasmid#239940PurposeCaMV 35S driving the expression of dCas9DepositorInsertCaMV 35S::dCas9
UseCRISPR and Synthetic BiologyExpressionPlantMutationN/AAvailable SinceOct. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
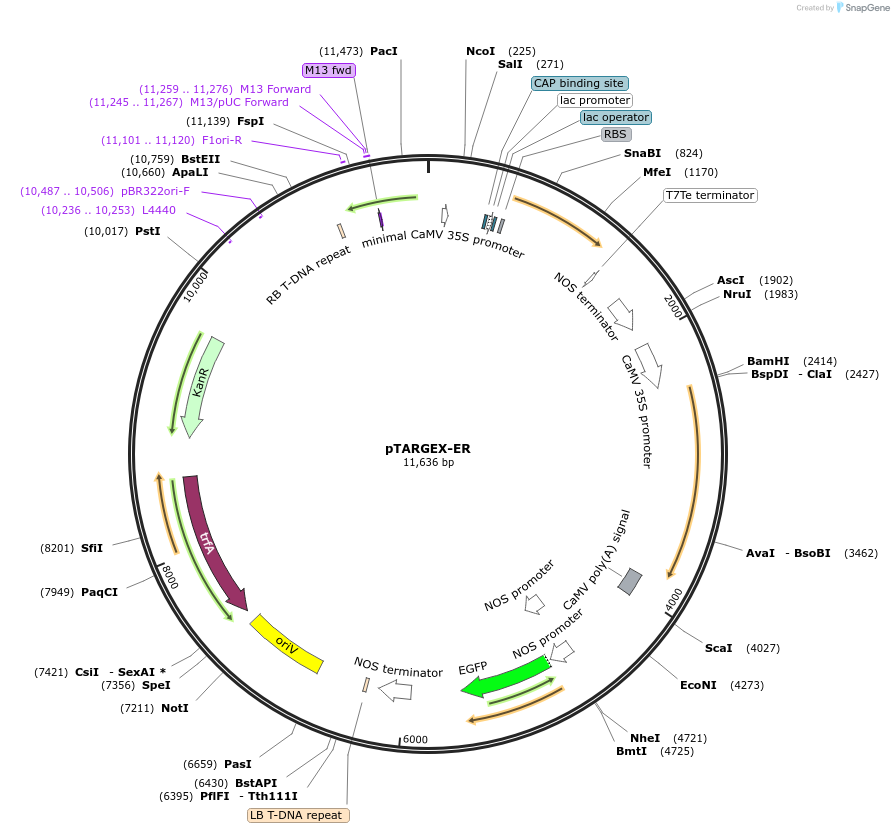
pTARGEX-ER
Plasmid#242449PurposeTargets high levels of recombinant protein for retention in ER. Empty vector. Clone via SapI restriction site. mCherry dropout cassette will enable Red/White screening in E.coli.DepositorTypeEmpty backboneTagsER-retrieval motif (KDEL) and Signal peptide deri…ExpressionPlantPromoterdouble 35SAvailable SinceOct. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
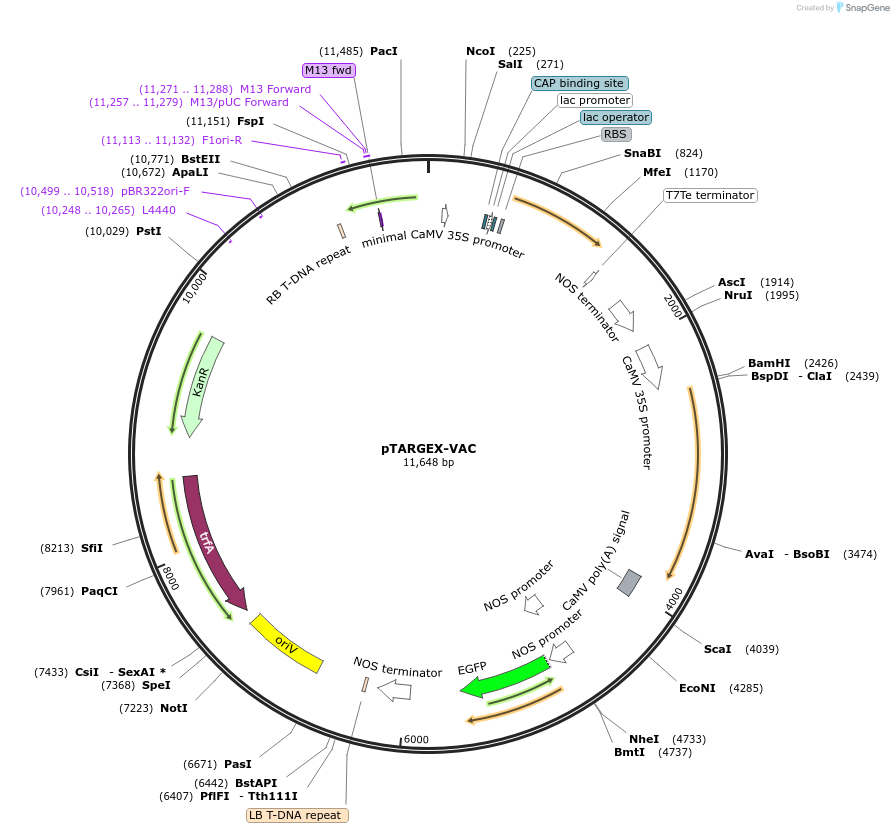
pTARGEX-VAC
Plasmid#242450PurposeTargets high levels of recombinant protein to vacuole. Empty vector. Clone via SapI restriction site. mCherry dropout cassette will enable Red/White screening in E.coli.DepositorTypeEmpty backboneTagsC-terminal propeptide (CTPP) derived from N. taba…ExpressionPlantPromoterdouble 35SAvailable SinceOct. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
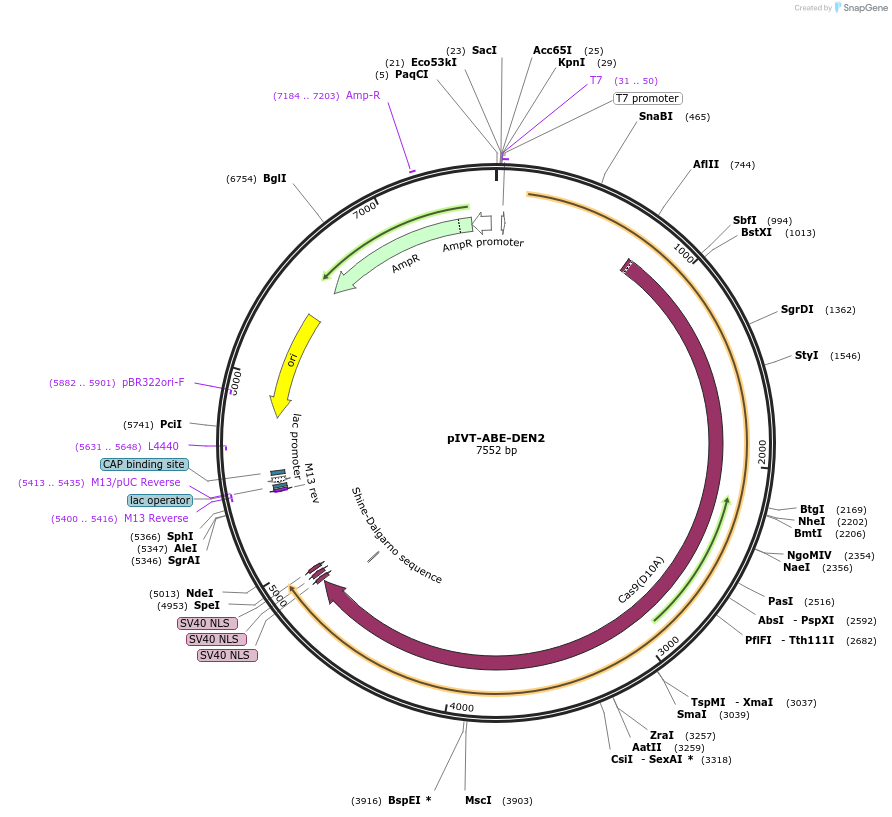
pIVT-ABE-DEN2
Plasmid#238020Purposefor DNA-free adenine base editing in rice and wheat or other plantsDepositorInsertTadA8e-nSpCas9(D10A)
ExpressionPlantMutationD10A for SpCas9PromoterT7Available SinceOct. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
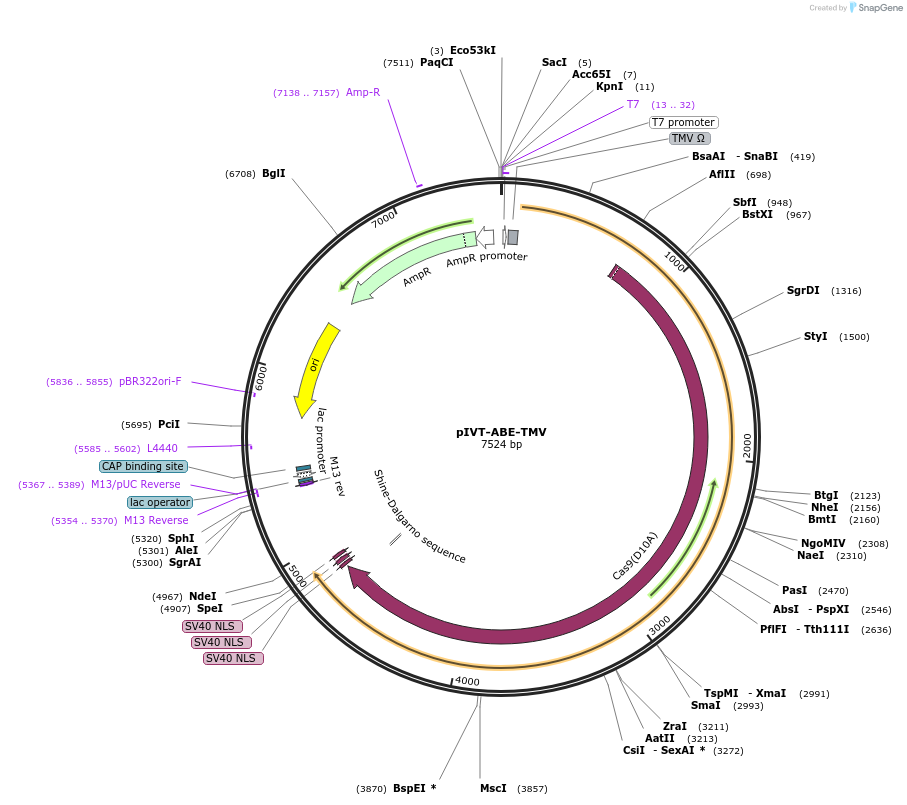
pIVT-ABE-TMV
Plasmid#238021Purposefor DNA-free adenine base editing in rice and wheat or other plantsDepositorInsertTadA8e-nSpCas9(D10A)
ExpressionPlantMutationD10A for SpCas9PromoterT7Available SinceOct. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
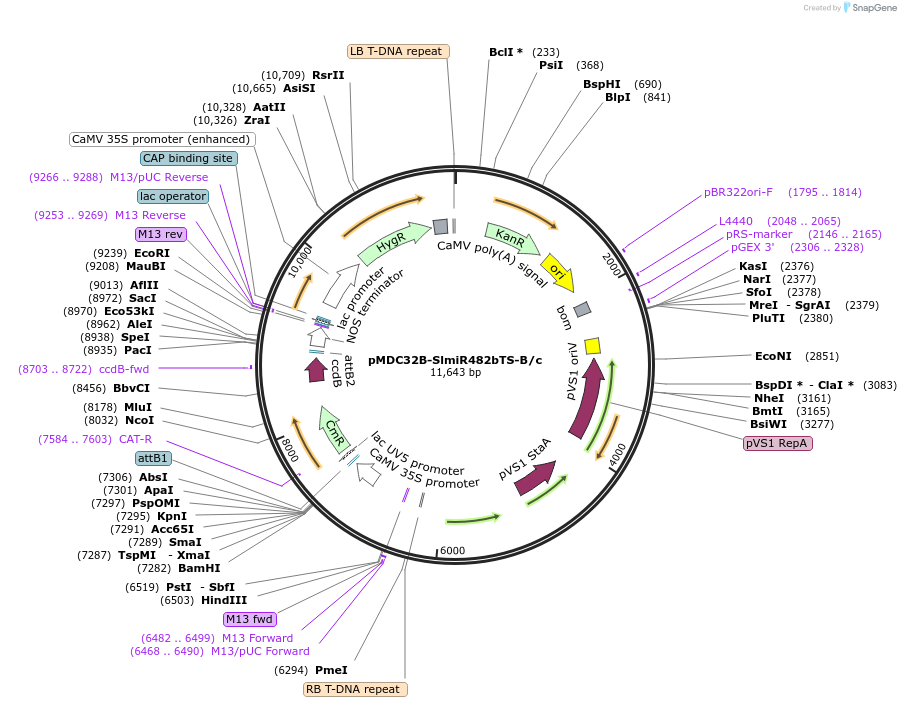
pMDC32B-SlmiR482bTS-B/c
Plasmid#234369PurposeCloning syn-tasiRNA sequences in minimal SlmiR482bTS precursors for expression in tomatoDepositorInsertsB/c
SlmiR482bTS
ExpressionPlantAvailable SinceOct. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
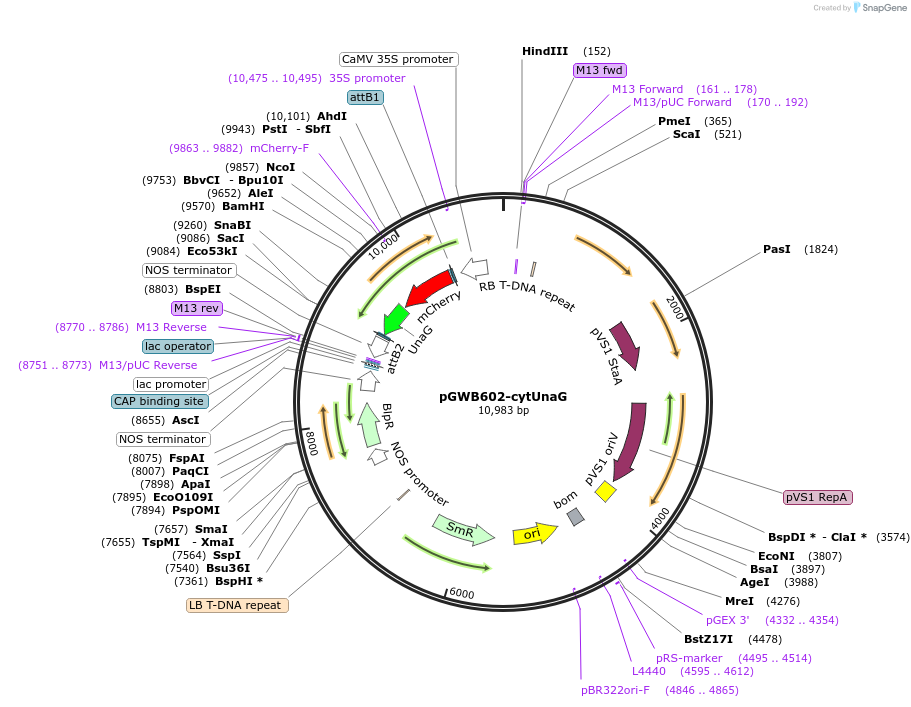
pGWB602-cytUnaG
Plasmid#220101PurposeFor Agrobacterium-mediated transformation. Cytosol-localized mCherry-UnaG.DepositorInsertmCherry-UnaG
ExpressionPlantAvailable SinceOct. 15, 2025AvailabilityAcademic Institutions and Nonprofits only -
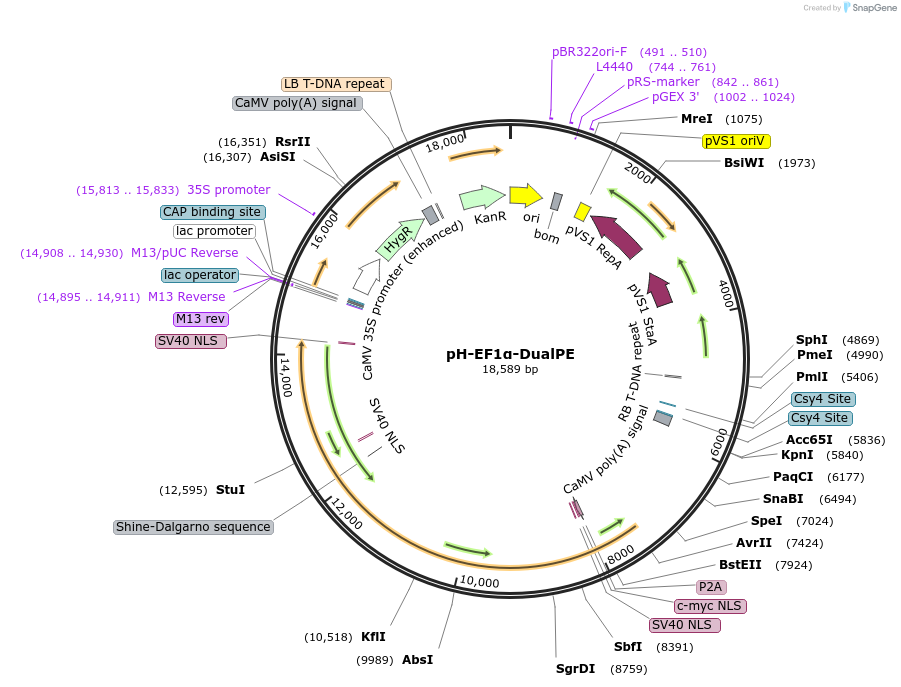
pH-EF1α-DualPE
Plasmid#236417PurposeFor plant prime editing including large DNA fragment editing in Solanum lycopersicum or other dicotyledonsDepositorInsertEF1α promoter
UseCRISPRExpressionPlantAvailable SinceOct. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
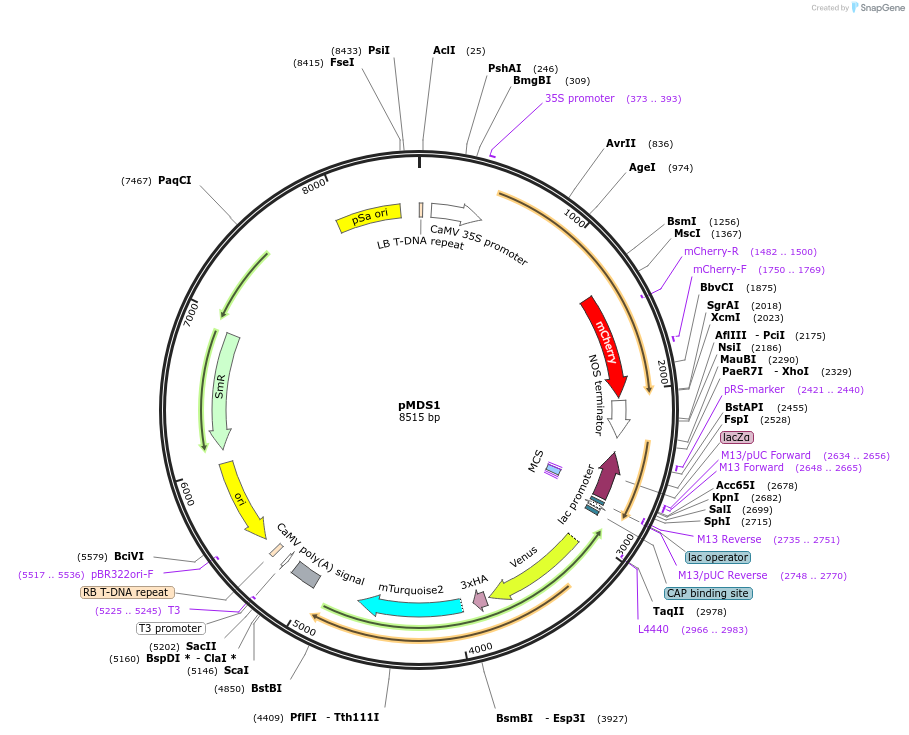
pMDS1
Plasmid#239460PurposeFor simultaneous monitoring of both transcription and translation of genes in plant tissues: C-terminus fusionDepositorTypeEmpty backboneTagsVENUS:3xHA:2A:mTurqN7ExpressionBacterial and PlantAvailable SinceOct. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
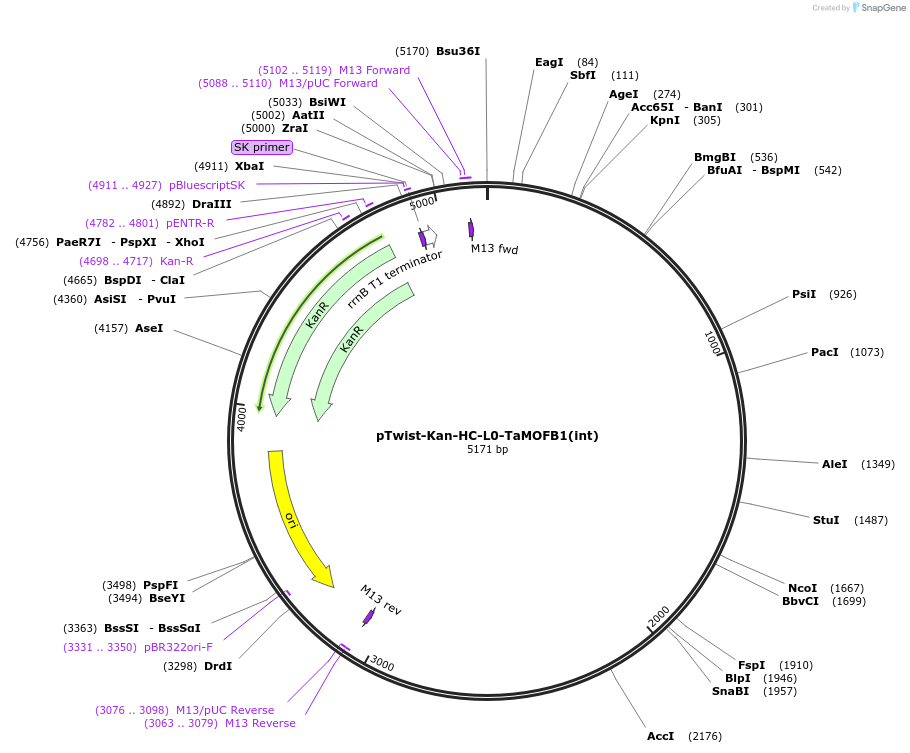
pTwist-Kan-HC-L0-TaMOFB1(int)
Plasmid#242005PurposeL0 CDS1 Golden Gate part of the pre-mRNA (excluding UTRs) of Triticum aestivum MORE FLORET 1 (MOF-1) / MULTI-FLORET SPIKELET 2 (MFS-2) B-subgenome homoeolog (TraesCS2B02G420900)DepositorInsertTaMOF-B1_with_introns
UseSynthetic BiologyExpressionPlantAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
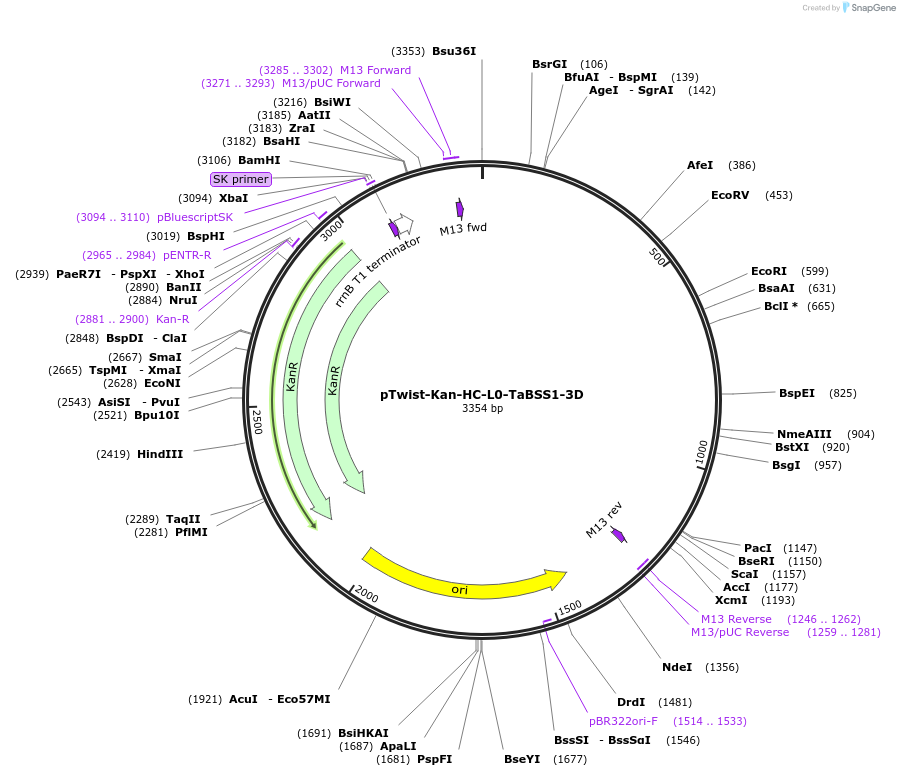
pTwist-Kan-HC-L0-TaBSS1-3D
Plasmid#242008PurposeL0 3U* Golden Gate part of 3' UTR and downstream regulatory regions of Triticum aestivum MANY-NODED DWARF 1 (MND1) B-subgenome homoeolog (TraesCS7B02G413900)DepositorInsertTaBSS1-3D
UseSynthetic BiologyExpressionPlantAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
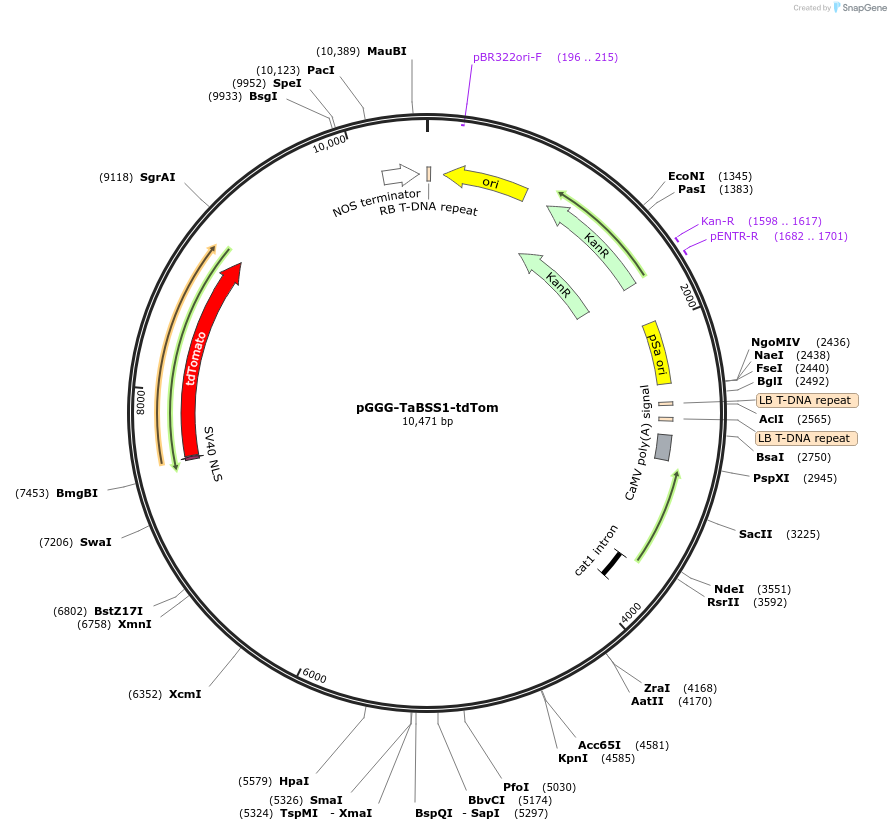
pGGG-TaBSS1-tdTom
Plasmid#242014PurposeL2 Golden Gate construct for in planta (wheat) expression of transgenic tdTomato driven by the Basal Spikelet Specific 1 regulatory environment.DepositorInserttdTomato
TagsNLSExpressionPlantPromoterBSS1Available SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
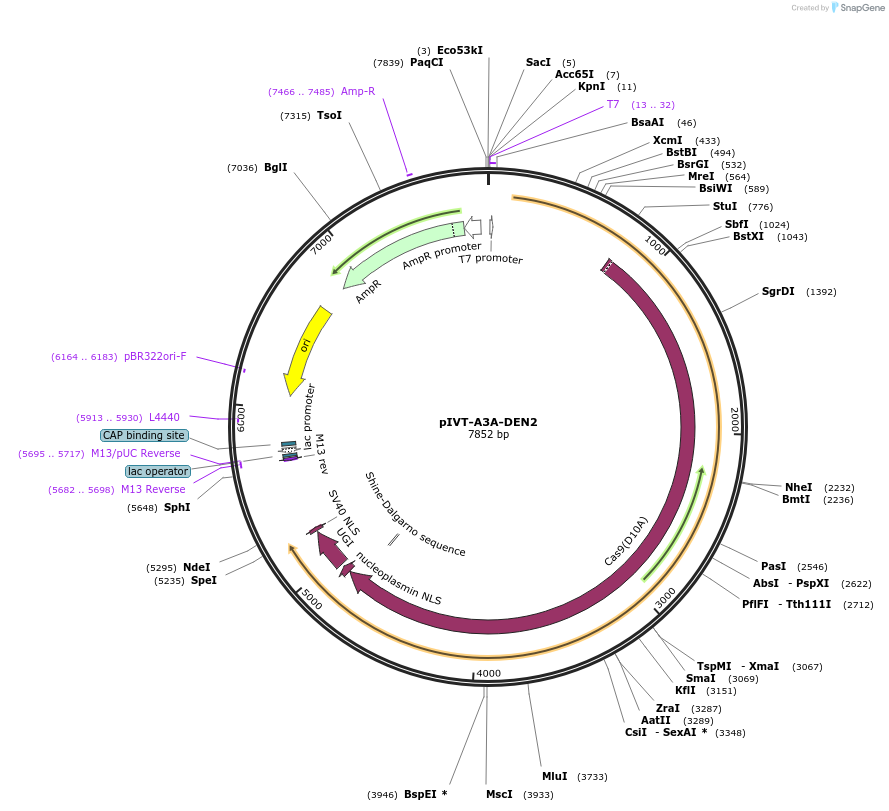
pIVT-A3A-DEN2
Plasmid#238019Purposefor DNA-free cytosine base editing in rice and wheat or other plantsDepositorInsertA3A-nSpCas9(D10A)-UGI
ExpressionPlantMutationD10A for SpCas9PromoterT7Available SinceOct. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
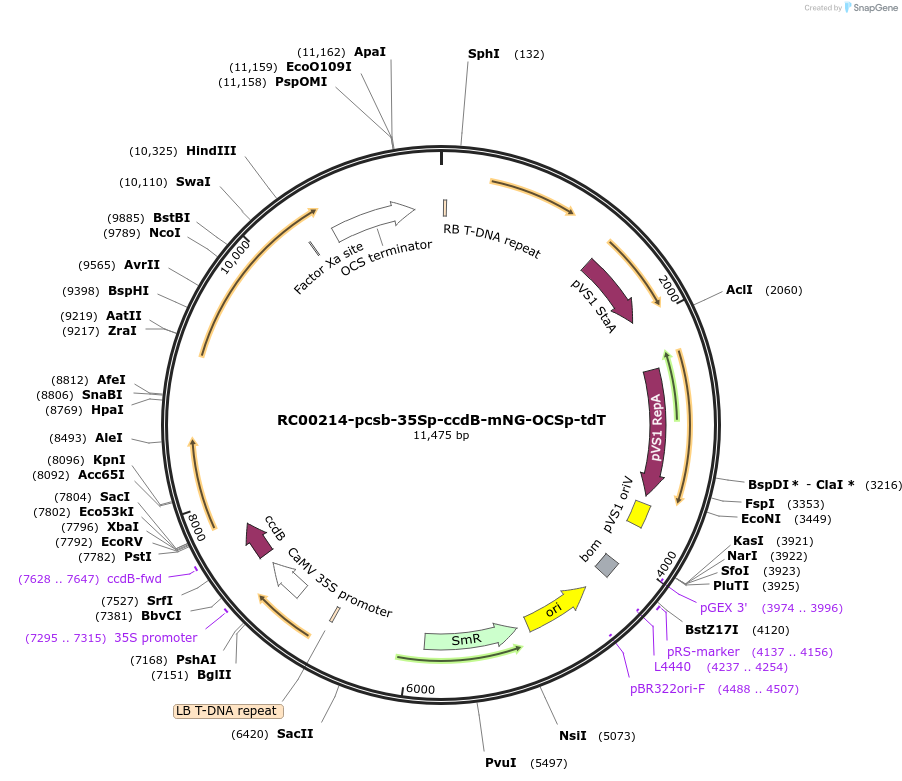
RC00214-pcsb-35Sp-ccdB-mNG-OCSp-tdT
Plasmid#209019PurposeDual-fluorescence acceptor plasmid for one-step insertion of a 5' leader of interest between the CaMV35S promoter and mNeonGreen via BpiI. A tdTomato expression cassette serves as internal control.DepositorInsertsccdB
mNeonGreen
tdTomato
ExpressionPlantMutationcodon optimized for N. benthamianaPromoterCaMV35S and Octopine SynthaseAvailable SinceOct. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
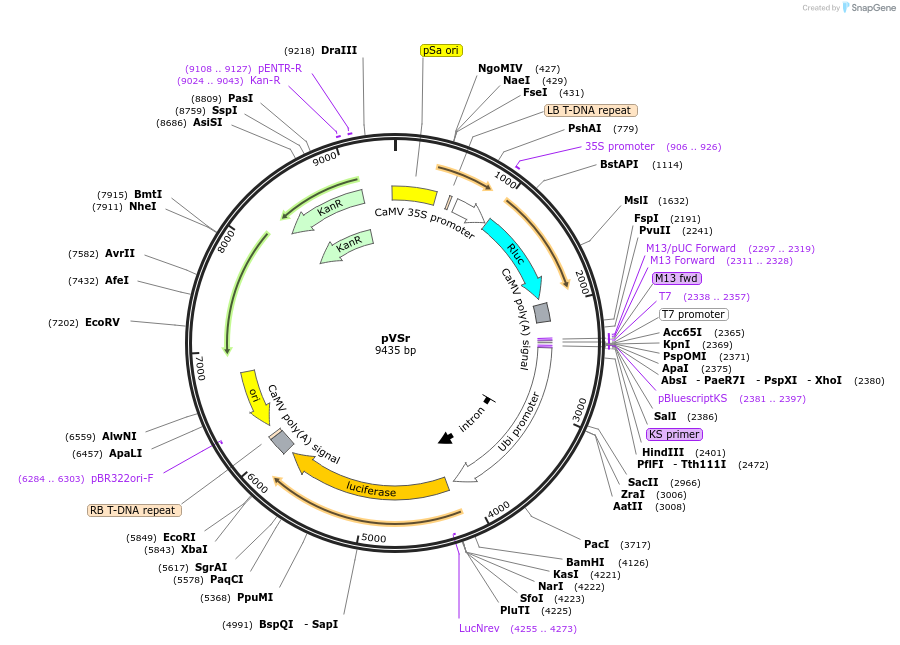
pVSr
Plasmid#234930PurposeVirus-induced gene silencing (VIGS) reporter vectorDepositorTypeEmpty backboneExpressionPlantAvailable SinceSept. 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
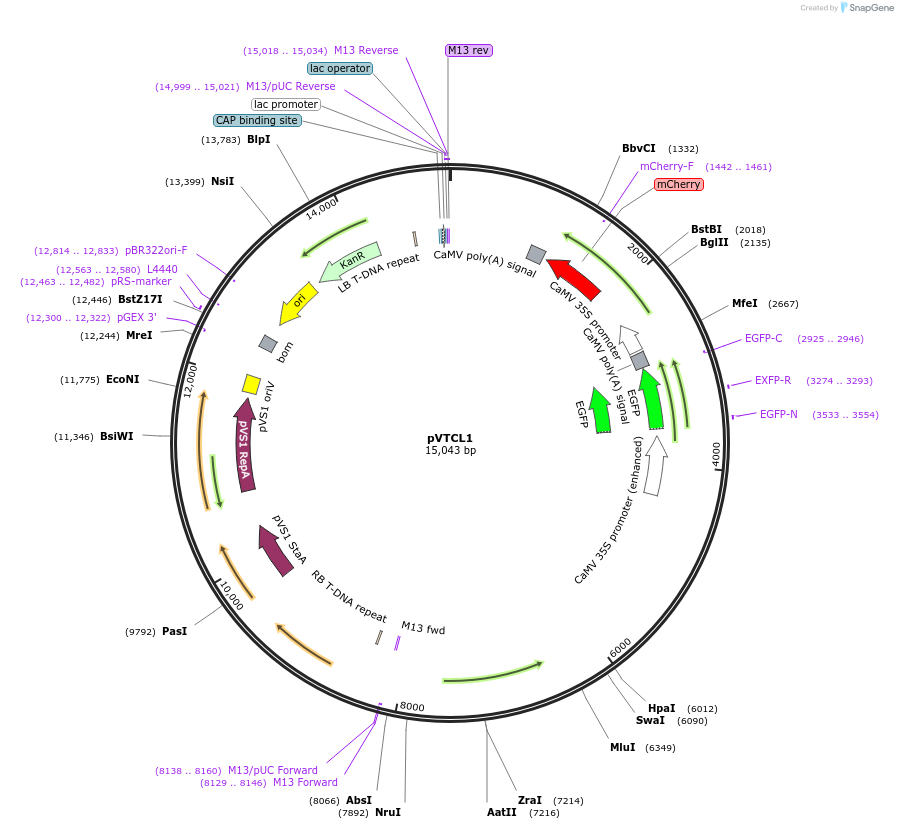
pVTCL1
Plasmid#234919PurposeAll-in-one subcellar localization vector. Construct containing EGFP tag and cell nucleus-targeted mCherry for co-localization study in plants.DepositorTypeEmpty backboneExpressionPlantAvailable SinceSept. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
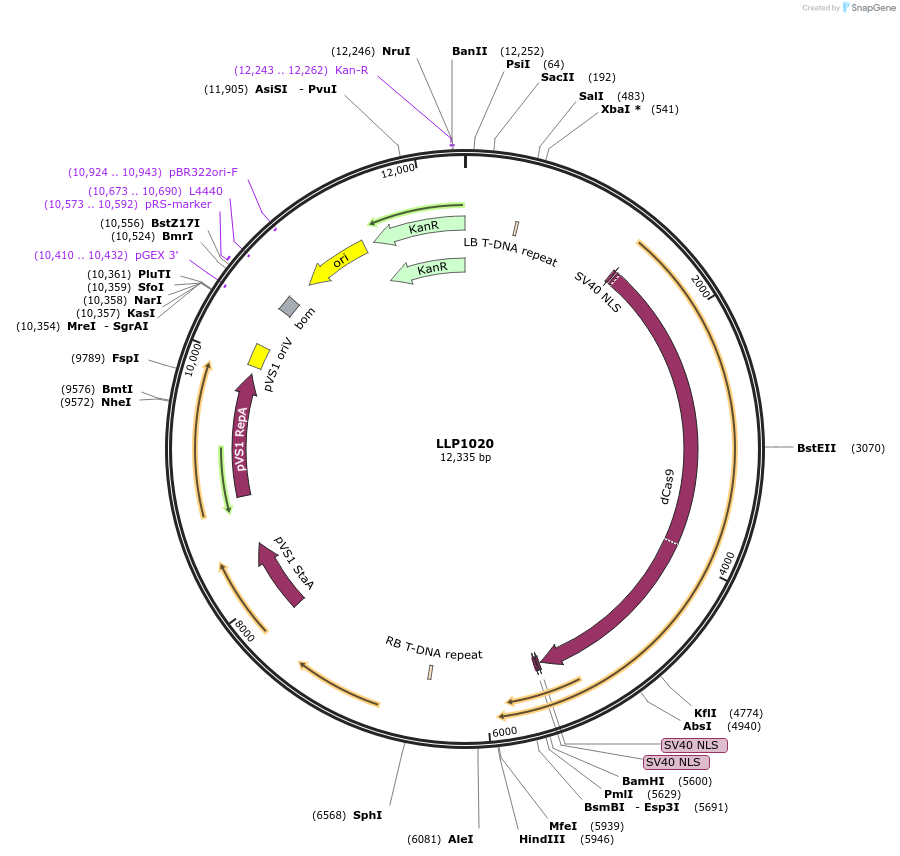
LLP1020
Plasmid#239942PurposeAtACT2 promoter driving the expression of dCas9-ZAT10(2x)DepositorInsertAtACT2::dCas9-ZAT10(2x)
UseCRISPR and Synthetic BiologyExpressionPlantMutationN/AAvailable SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
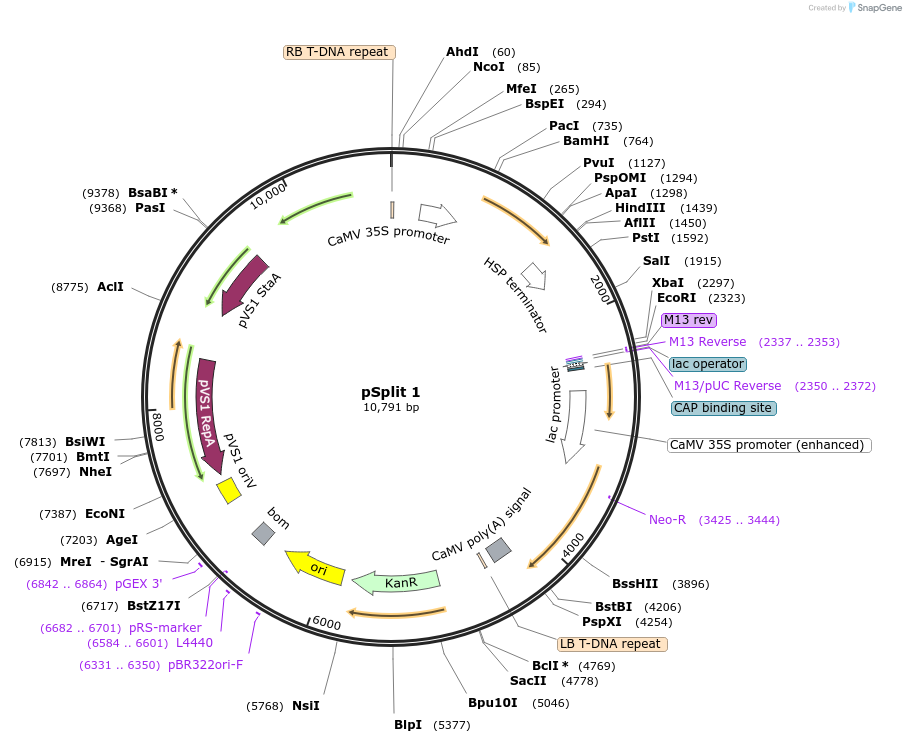
pSplit 1
Plasmid#240576PurposeDestination vector for pSplit system in plantsDepositorInsertNpuDnaE(N)
ExpressionPlantAvailable SinceSept. 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
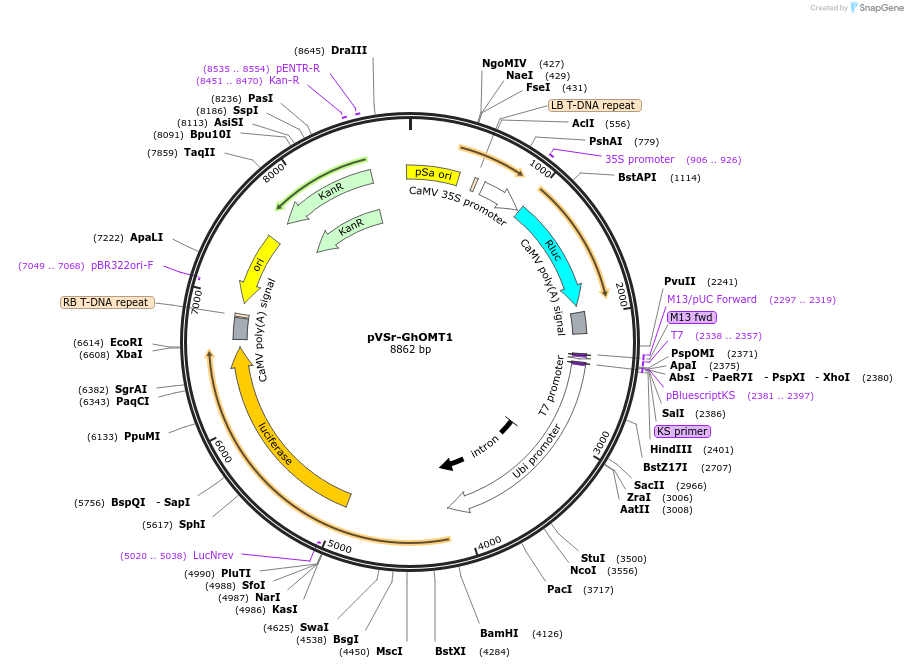
pVSr-GhOMT1
Plasmid#234932PurposeThis virus-induced gene silencing (VIGS) reporter vector is used to evaluate the silencing of GhOMT1 geneDepositorInsertPartial coding region sequence of GhOMT1
ExpressionPlantAvailable SinceSept. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
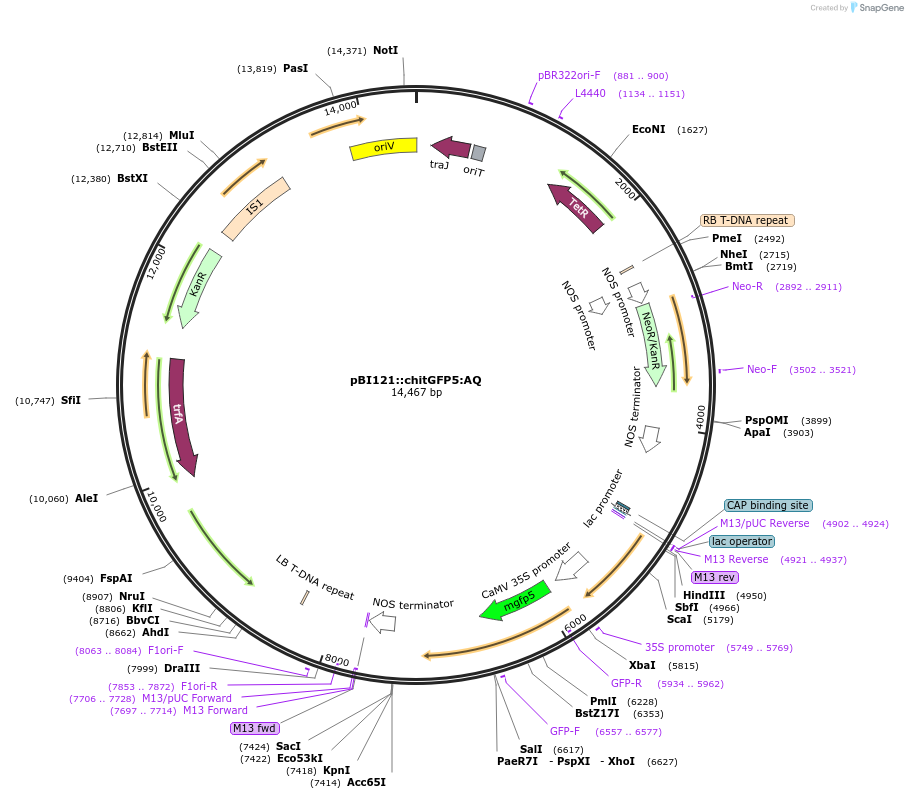
pBI121::chitGFP5:AQ
Plasmid#240450PurposeExpression of indicator protein fusion (soluble modified GFP5 & Aequorin) for monitoring calcium concentrations in cell walls of higher plants. See Resource Information.DepositorInsertFusion of Chitinase signal, smGFP5, and Aequorin
ExpressionBacterial and PlantMutationGFP5 for expression plants (PMID 9122158); Solubl…PromoterCauliflower mosaic virus 35S promoter (GenBank AJ…Available SinceSept. 5, 2025AvailabilityAcademic Institutions and Nonprofits only -
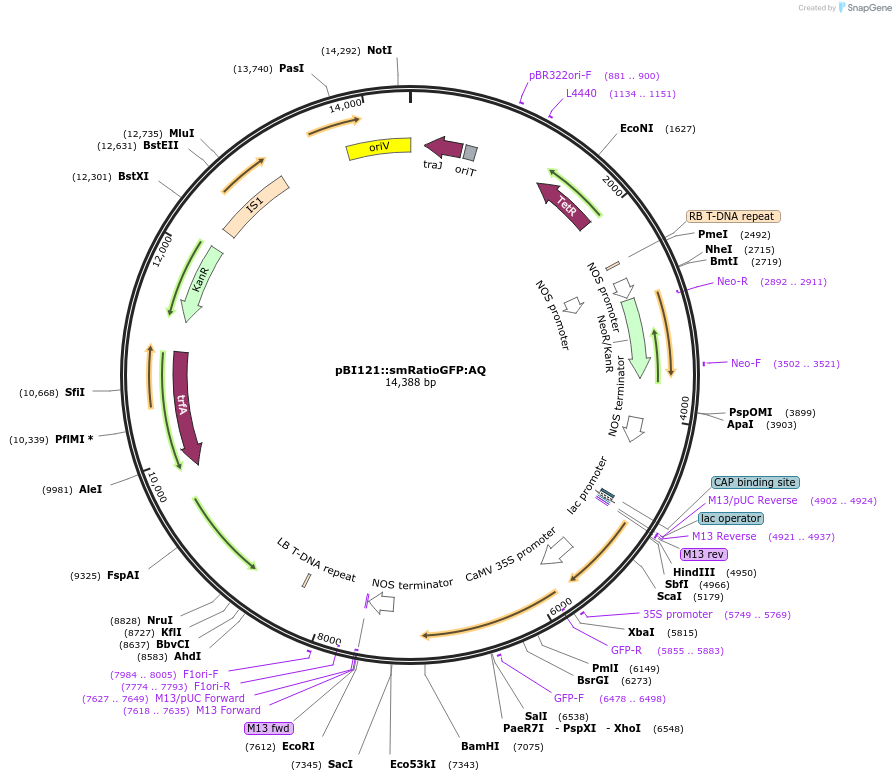
pBI121::smRatioGFP:AQ
Plasmid#240453PurposeExpression of indicator protein fusion (soluble modified ratiometric pHluorin & Aequorin) for monitoring calcium concentrations and pH in the cytoplasm of higher plants. See Resource Information.DepositorInsertFusion of soluble modified ratiometric pHluorin and Aequorin
ExpressionBacterial and PlantMutationRatiometric GFP (ratiometric pHluorin) (PMID 9671…PromoterCauliflower mosaic virus 35S promoter (GenBank AJ…Available SinceSept. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
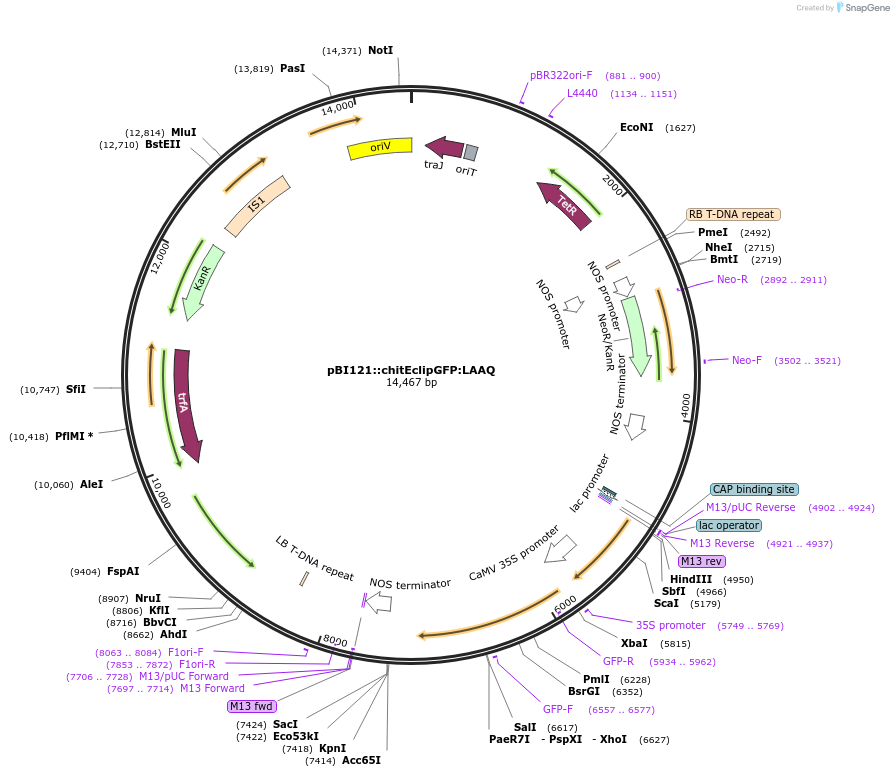
pBI121::chitEclipGFP:LAAQ
Plasmid#240452PurposeExpression of indicator protein fusion (modified ecliptic pHluorin & low affinity Aequorin) for monitoring calcium concentrations and pH in cell walls of higher plants. See Resource Information.DepositorInsertFusion of Chitinase signal, soluble modified ecliptic pHluorin and low affinity Aequorin (D119A)
ExpressionBacterial and PlantMutationEcliptic GFP (ecliptic pHluorin) (PMID 9671304); …PromoterCauliflower mosaic virus 35S promoter (GenBank AJ…Available SinceSept. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
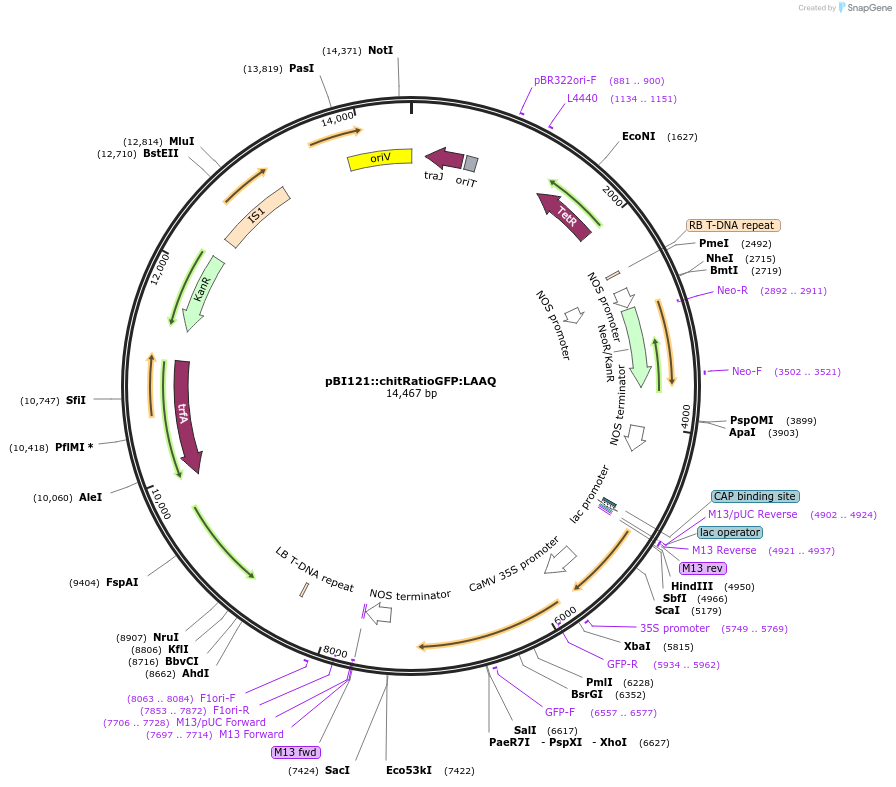
pBI121::chitRatioGFP:LAAQ
Plasmid#240451PurposeExpression of indicator protein fusion (modified ratiometric pHluorin & low affinity Aequorin) for monitoring calcium concentrations and pH in cell walls of higher plants. See Resource Information.DepositorInsertFusion of chitinase signal, soluble modified ratiometric pHluorin and low affinity Aequorin (D119A)
ExpressionBacterial and PlantMutationRatiometric GFP (ratiometric pHluorin) (PMID 9671…PromoterCauliflower mosaic virus 35S promoter (GenBank AJ…Available SinceSept. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
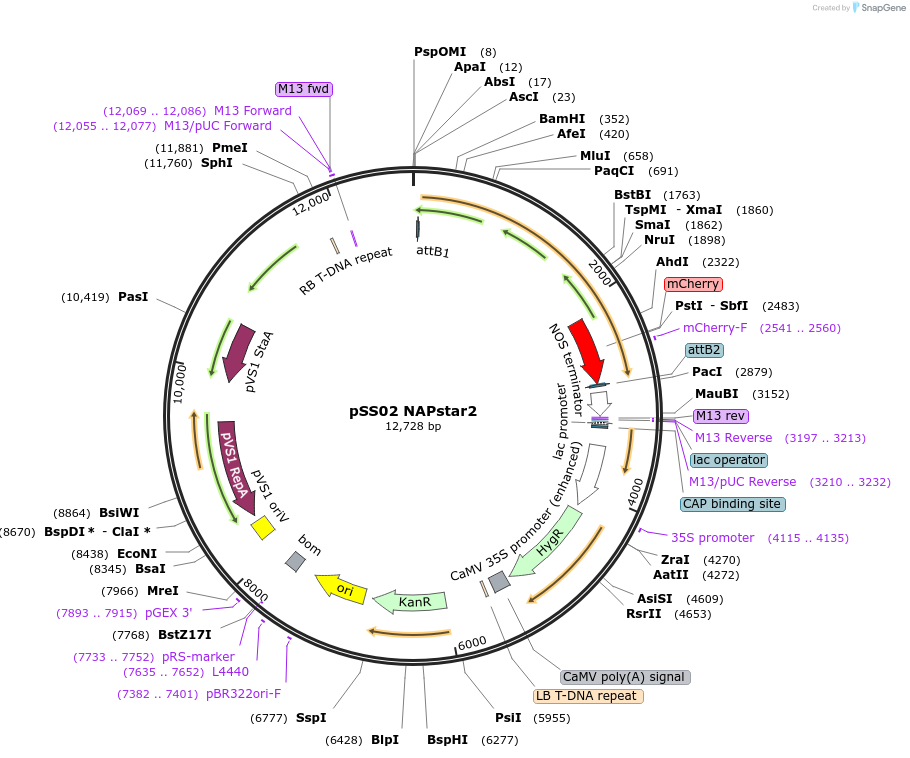
pSS02 NAPstar2
Plasmid#241272PurposeExpresses NADPH/NADP+ biosensor NAPstar2 in Arabidopsis thalianaDepositorInsertcyt-NAPstar2
ExpressionPlantAvailable SinceSept. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
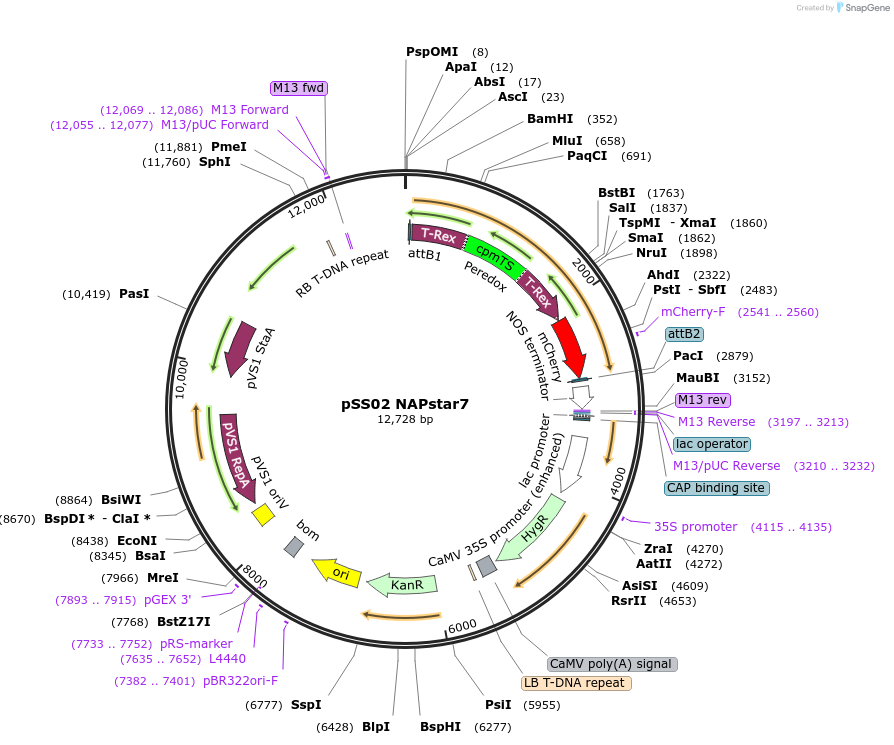
pSS02 NAPstar7
Plasmid#241224PurposeExpresses NADPH/NADP+ biosensor NAPstar7 in Arabidopsis thalianaDepositorInsertcyt-NAPstar7
ExpressionPlantAvailable SinceSept. 3, 2025AvailabilityAcademic Institutions and Nonprofits only