4,458 results
-
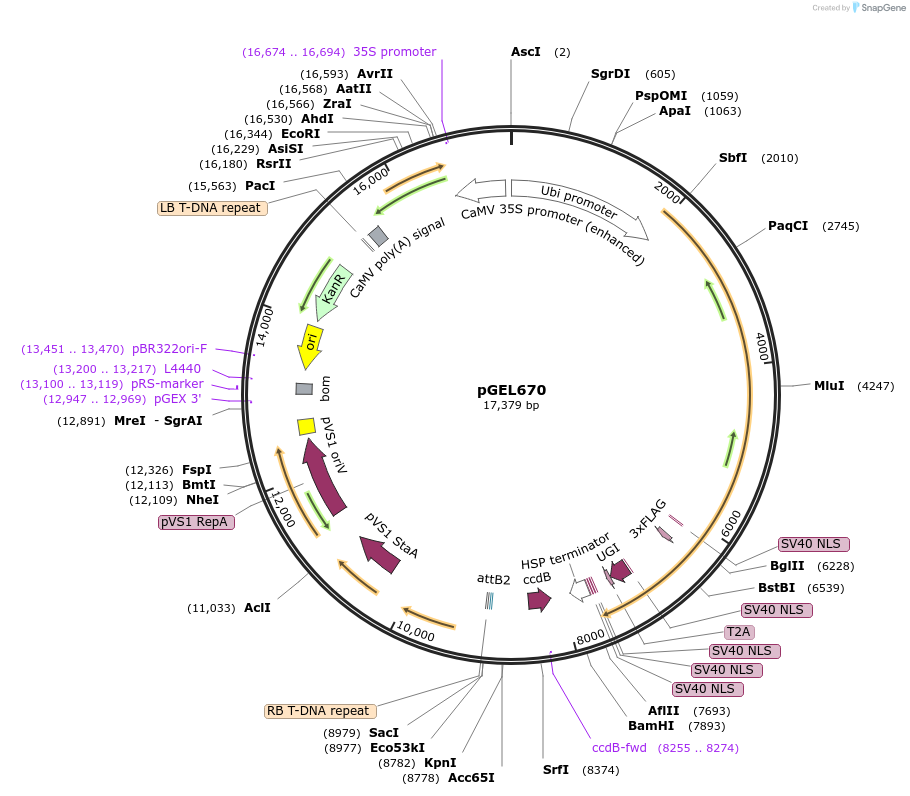
Plasmid#196818PurposeLrCas9 cytosine base editing plasmid in riceDepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionPlantPromoterZmUbiAvailable SinceOct. 12, 2023AvailabilityAcademic Institutions and Nonprofits only
-
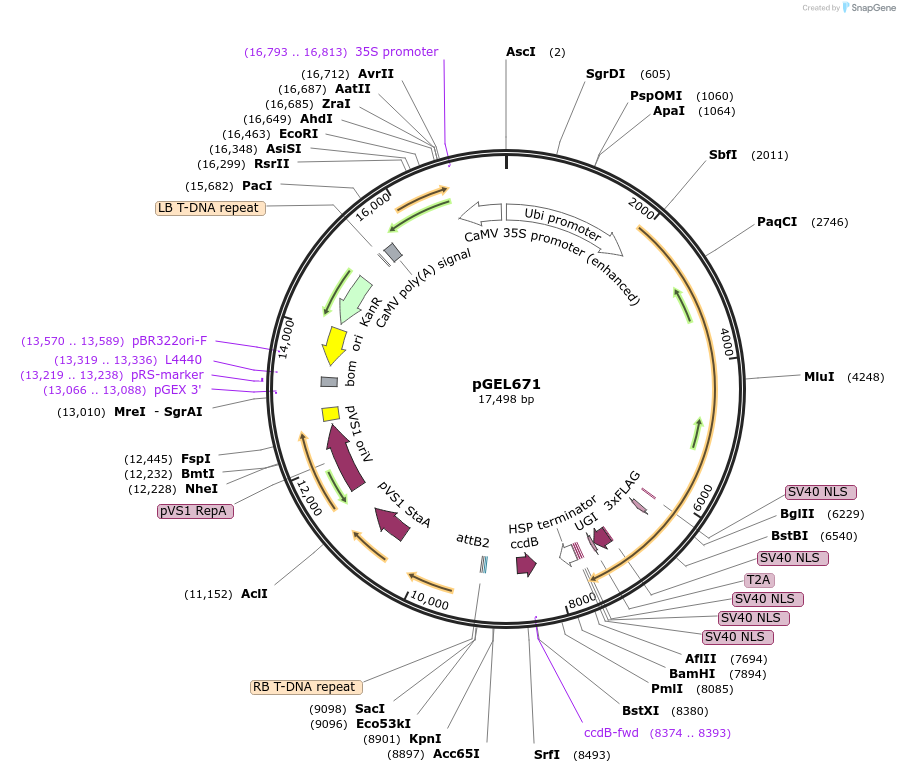
pGEL671
Plasmid#196819PurposeLrCas9 cytosine base editing plasmid in wheatDepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionPlantPromoterZmUbiAvailable SinceOct. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
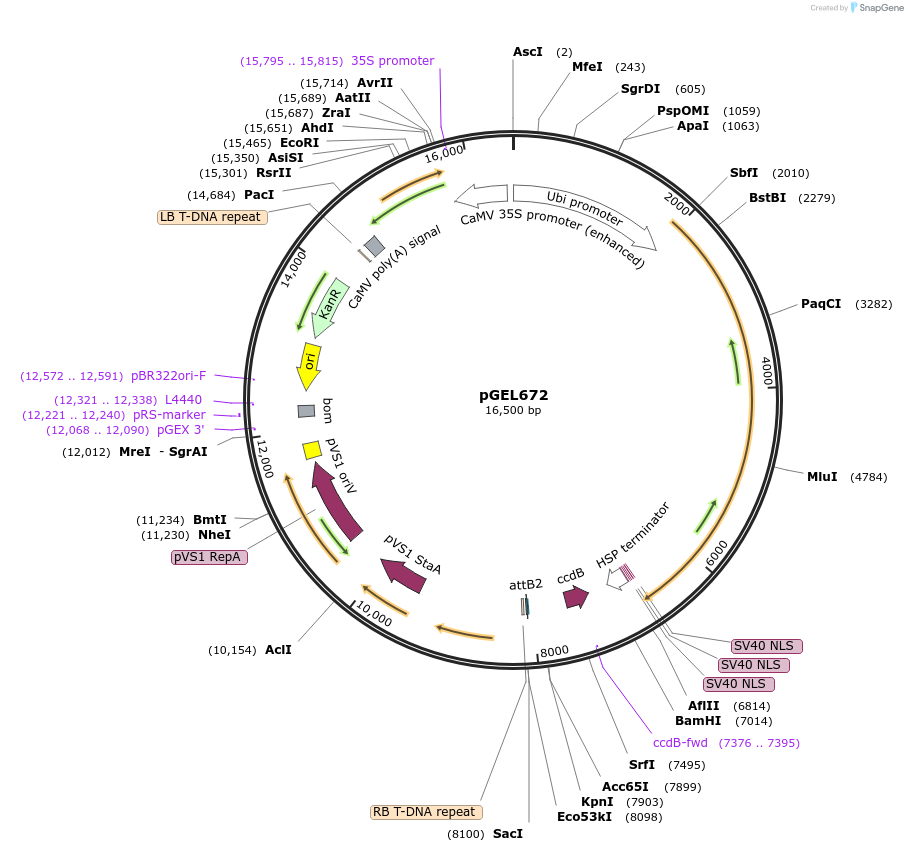
pGEL672
Plasmid#196820PurposeLrCas9 adenine base editing plasmid V1.0 in riceDepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionPlantPromoterZmUbiAvailable SinceOct. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
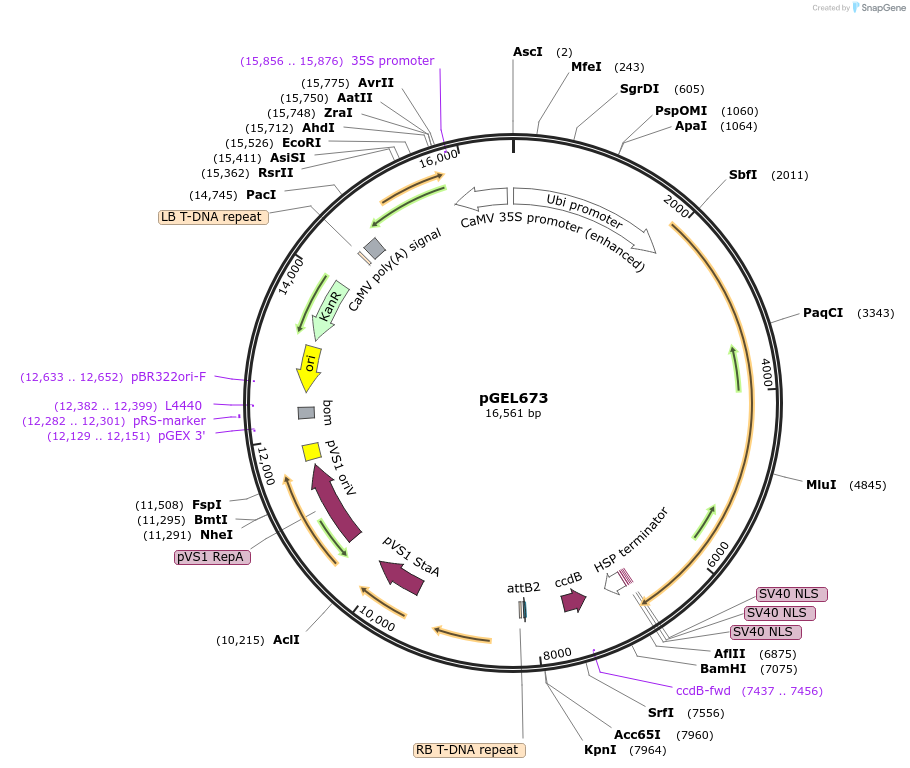
pGEL673
Plasmid#196821PurposeLrCas9 adenine base editing plasmid V2.0 in riceDepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionPlantPromoterZmUbiAvailable SinceOct. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
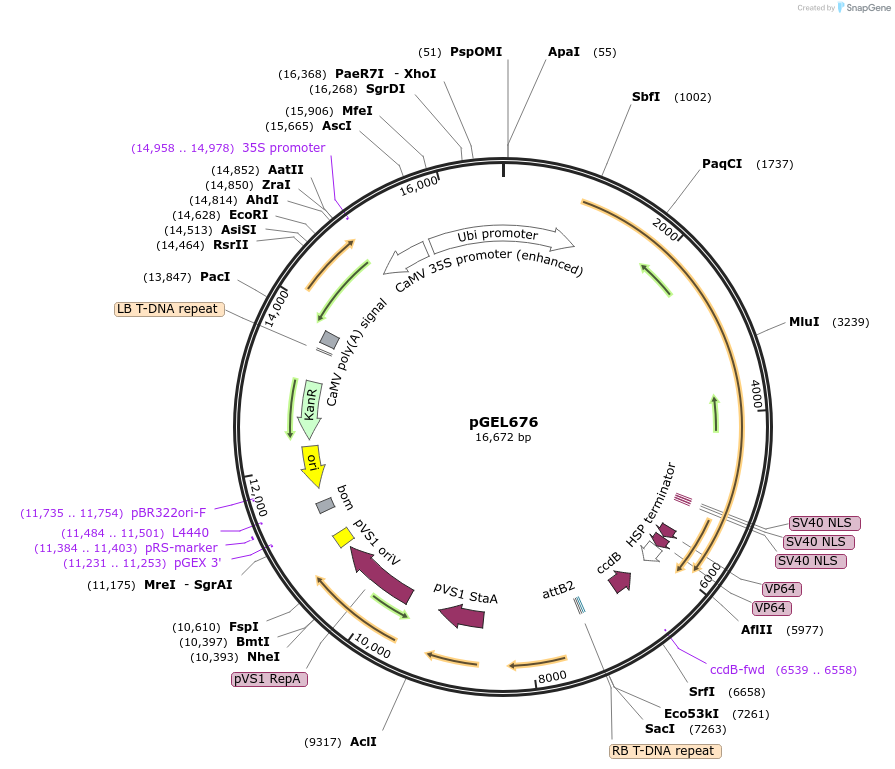
pGEL676
Plasmid#196823PurposedLrCas9 gene expression activation plasmid in riceDepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionPlantPromoterZmUbiAvailable SinceOct. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
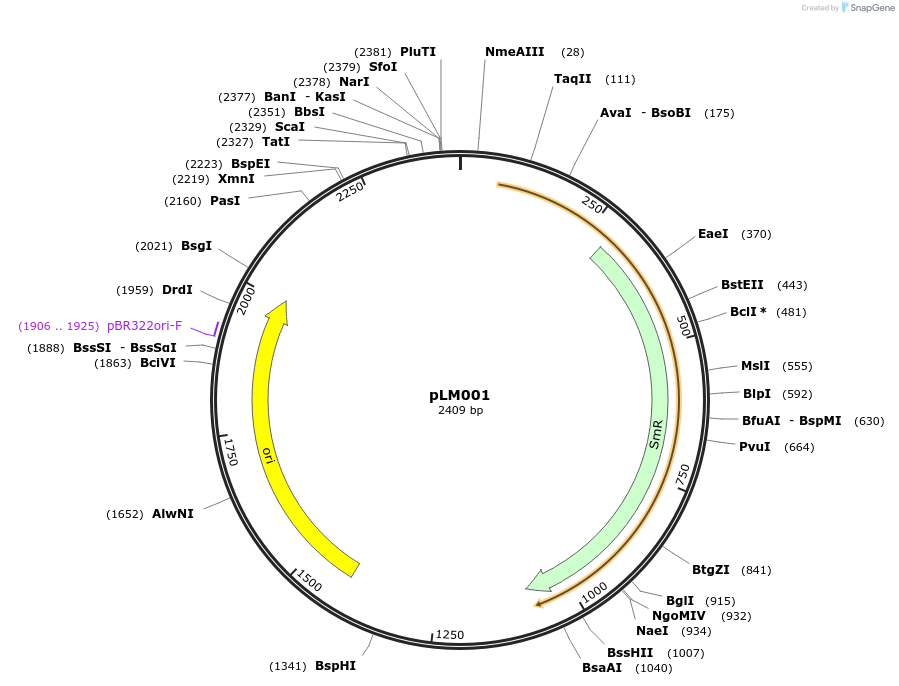
pLM001
Plasmid#201311PurposeAvrSr50 (with stop codon) as Golden Gate compatible MoClo CDS partDepositorInsertAvrSr50
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
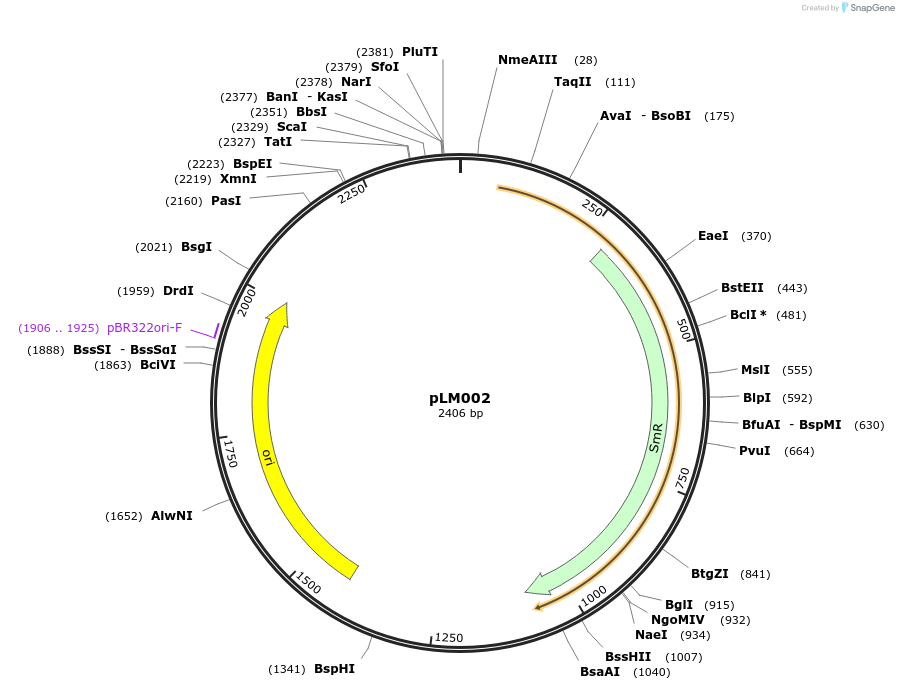
pLM002
Plasmid#201312PurposeAvrSr50 (no stop codon) as Golden Gate compatible MoClo CDS partDepositorInsertAvrSr50
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
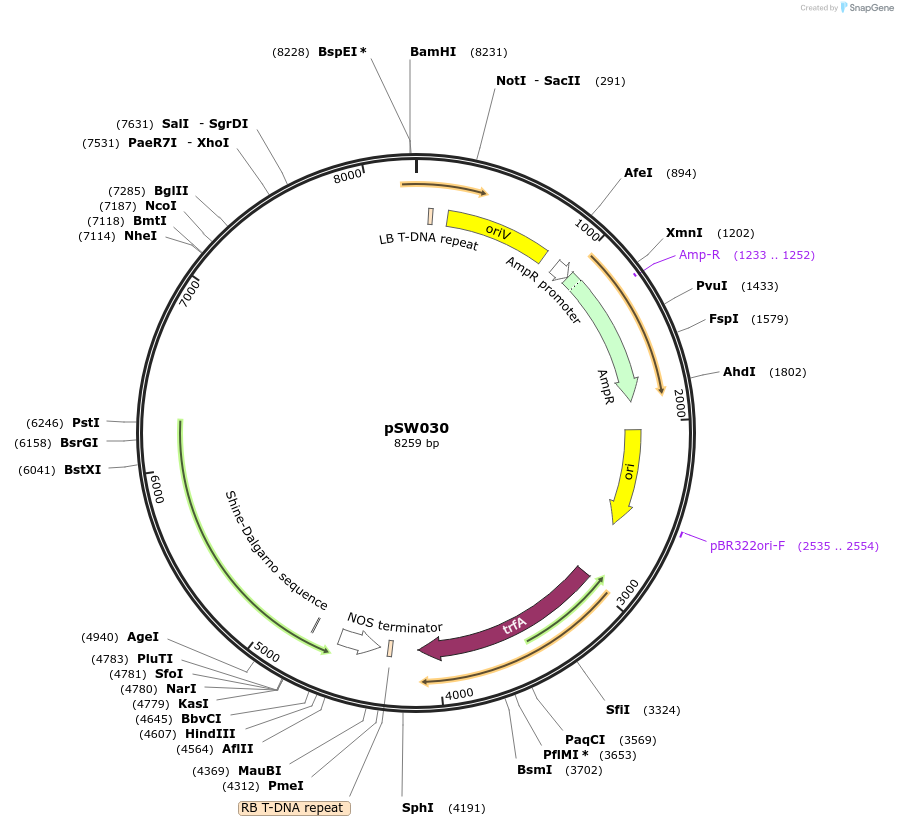
pSW030
Plasmid#201317PurposeUBI:Eluc (green-shifted luciferase with Ubiquitin promoterDepositorInsertEluc
ExpressionPlantMutationremoval of BsaI and BpiI sitesPromoterpICSL12009Available SinceOct. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
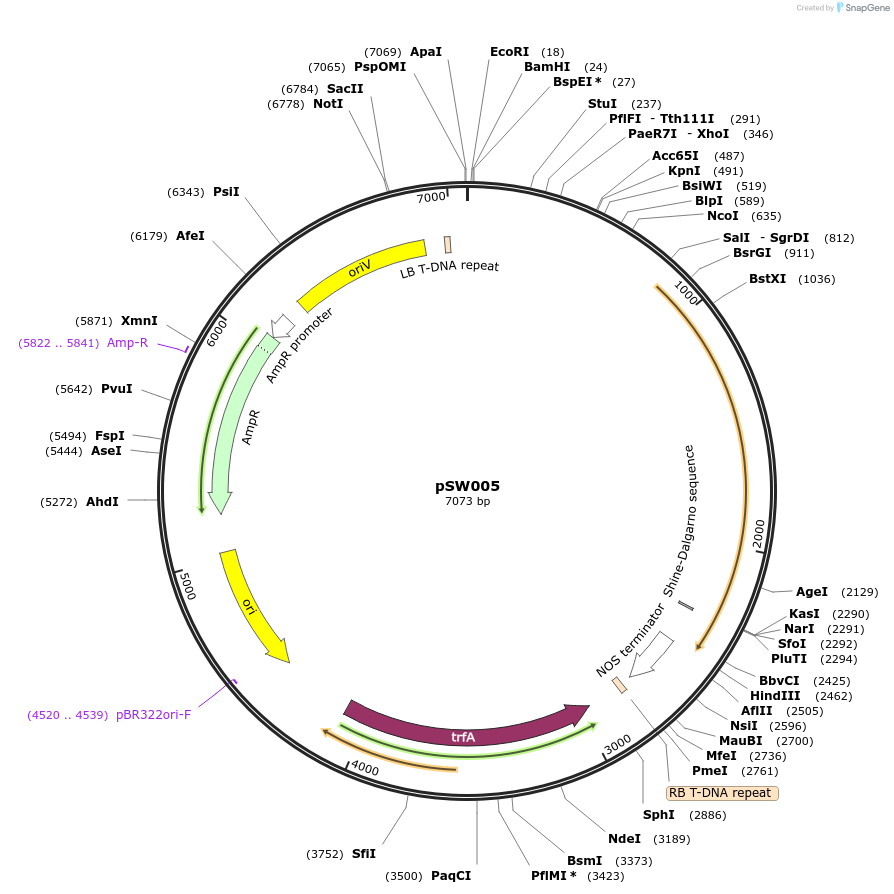
pSW005
Plasmid#201324PurposepDefence5:Eluc (defence-induced promoter with green-shifted luciferase expression cassette)DepositorInsertpDefence5:Eluc
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
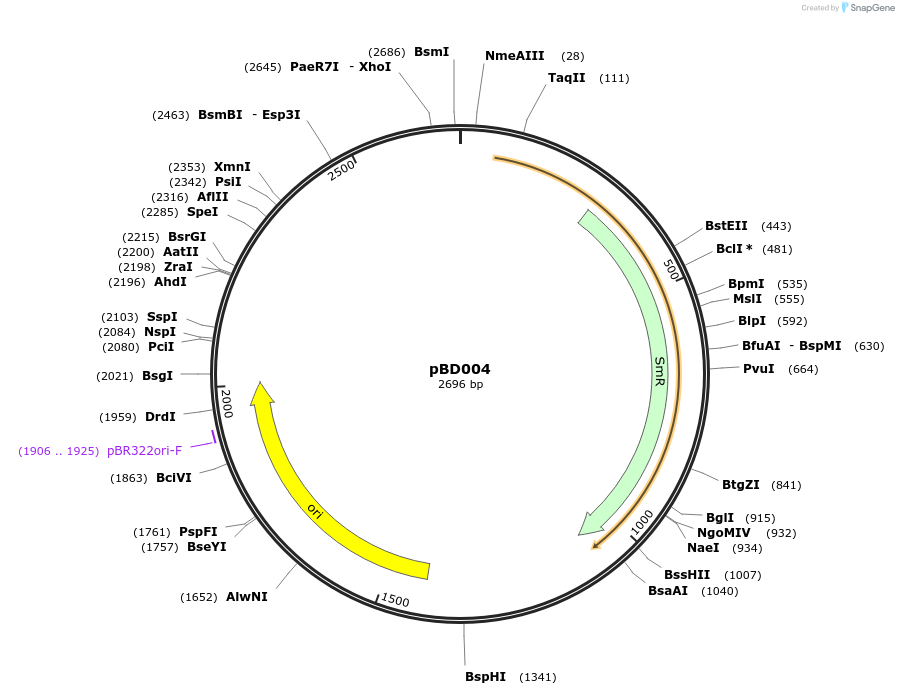
pBD004
Plasmid#201304PurposeD14: Promoter of TraesCS3D02G368800 as Golden Gate compatible MoClo PROM partDepositorInsertD14: Promoter of TraesCS3D02G368800
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
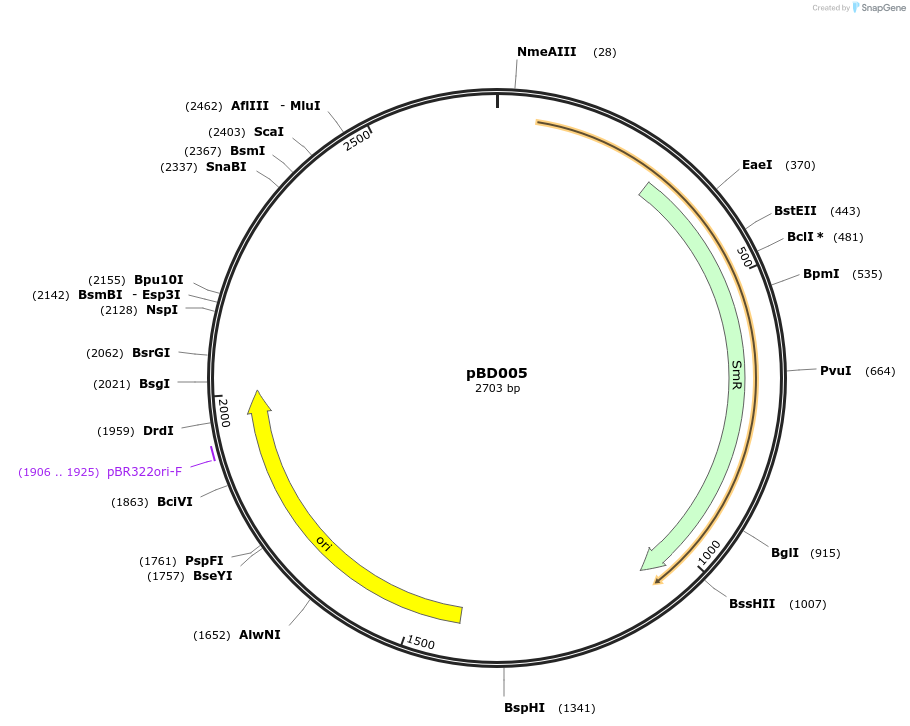
pBD005
Plasmid#201305PurposeD15: Promoter of TraesCS3D02G517500 as Golden Gate compatible MoClo PROM partDepositorInsertD15: Promoter of TraesCS3D02G517500
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
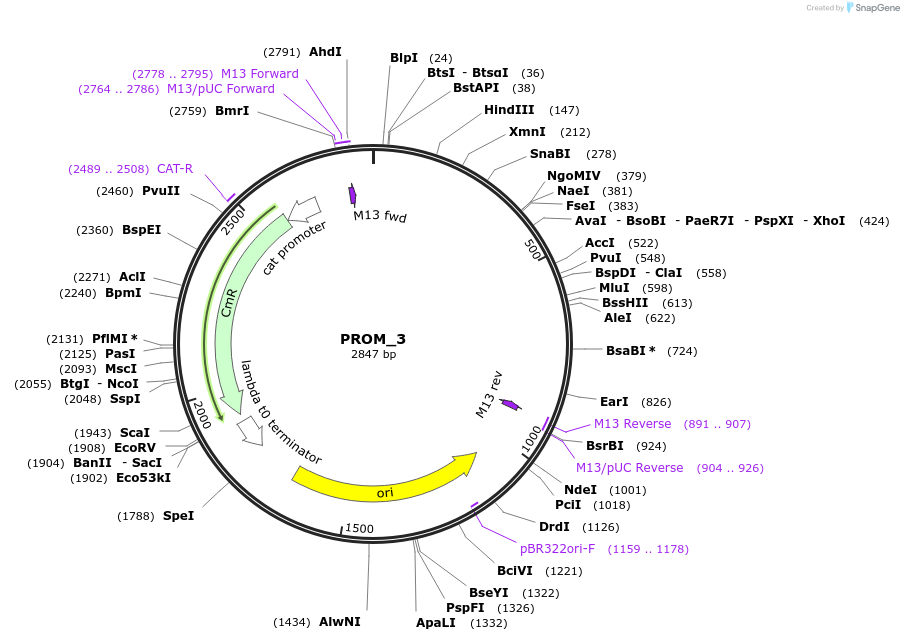
PROM_3
Plasmid#201296PurposeD3: Promoter of TraesCS3A02G510700 as Golden Gate compatible MoClo PROM partDepositorInsertD3: Promoter of TraesCS3A02G510700
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
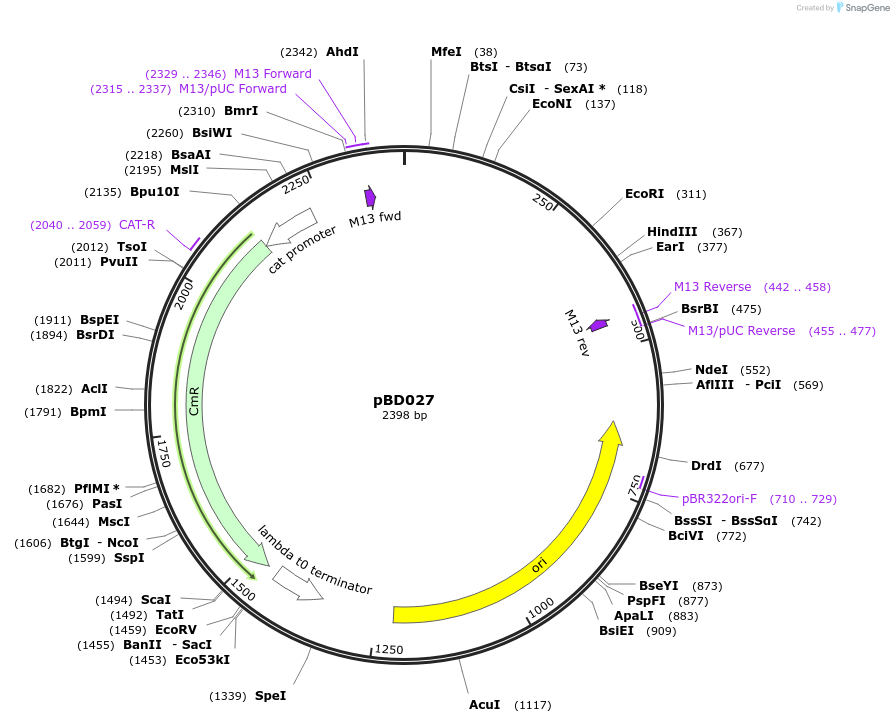
pBD027
Plasmid#201310PurposeavrSr27-3 as Golden Gate compatible MoClo CDS part (no recognition by Sr27)DepositorInsertavrSr27-3
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
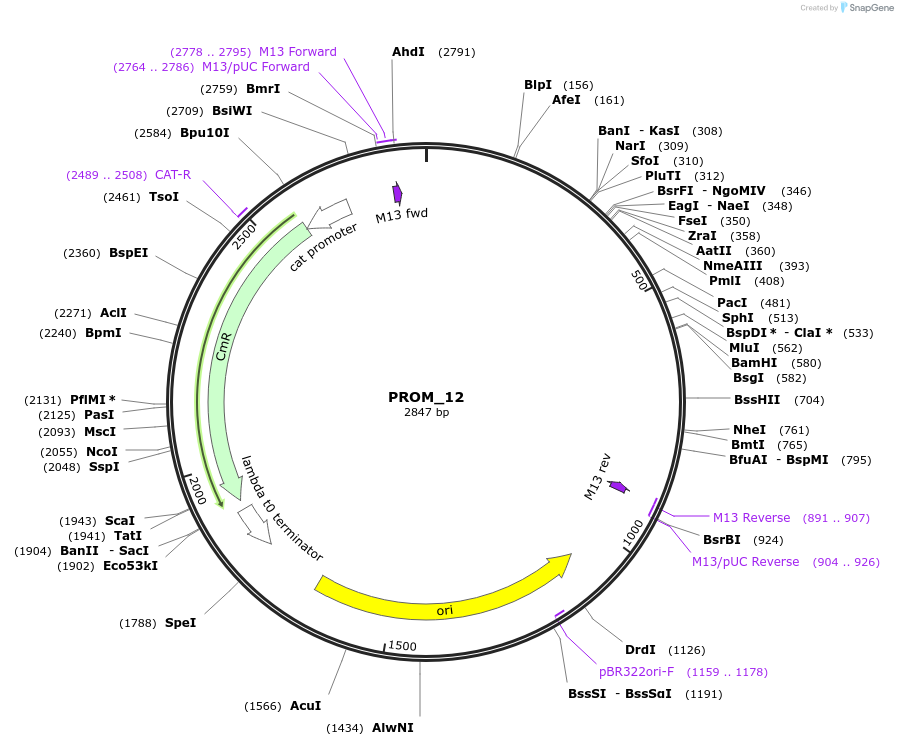
PROM_12
Plasmid#201303PurposeD12: Promoter of TraesCS4D02G131100 as Golden Gate compatible MoClo PROM partDepositorInsertD12: Promoter of TraesCS4D02G131100
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
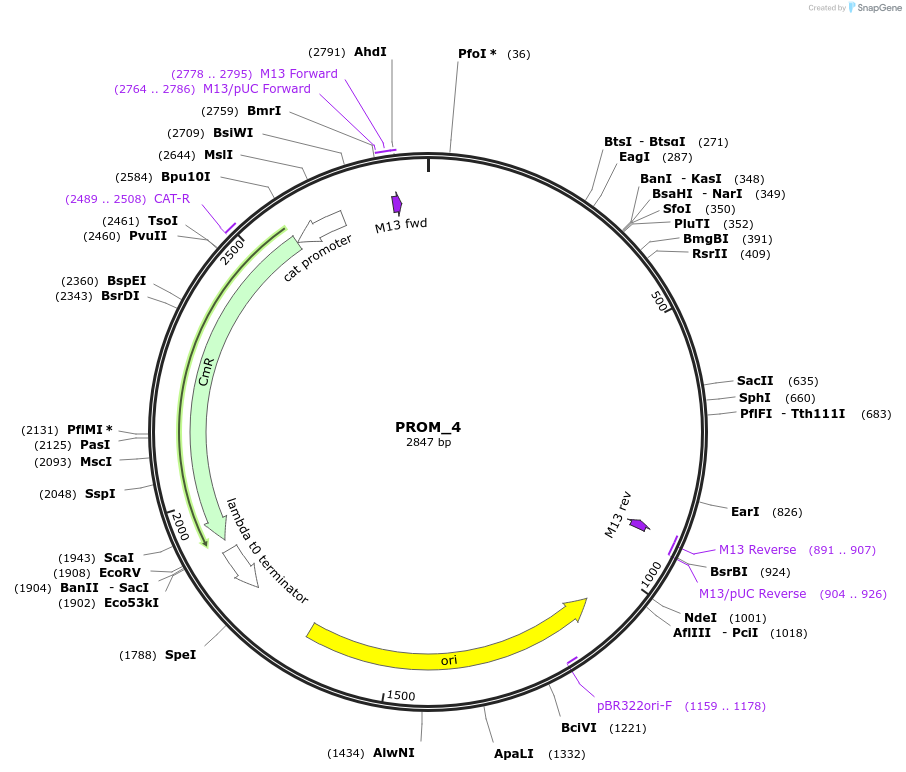
PROM_4
Plasmid#201297PurposeD4: Promoter of TraesCS3B02G552700 as Golden Gate compatible MoClo PROM partDepositorInsertD4: Promoter of TraesCS3B02G552700
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
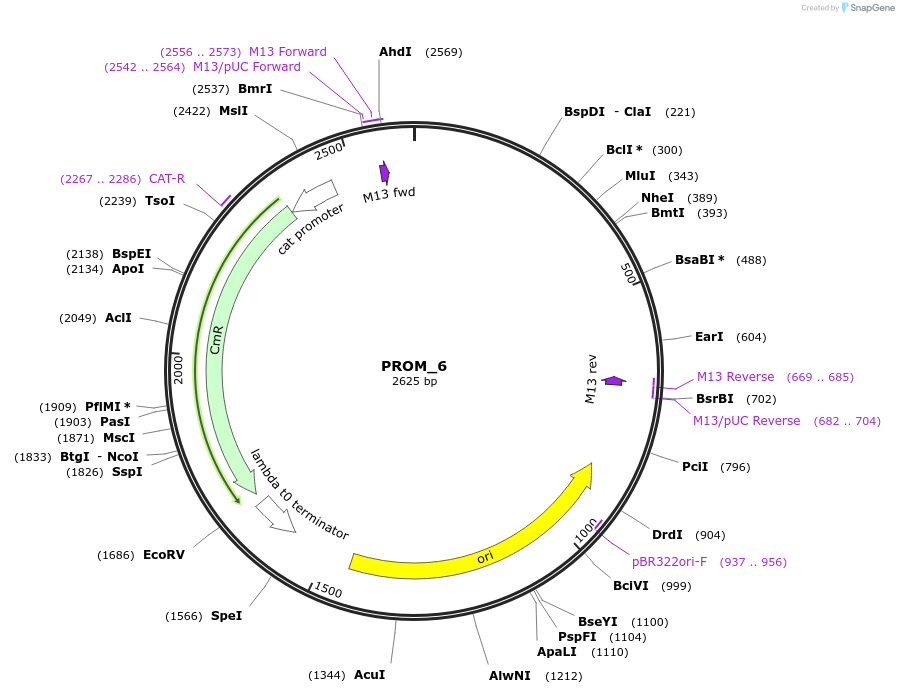
PROM_6
Plasmid#201299PurposeD6: Promoter of TraesCS3B02G578800 as Golden Gate compatible MoClo PROM partDepositorInsertD6: Promoter of TraesCS3B02G578800
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
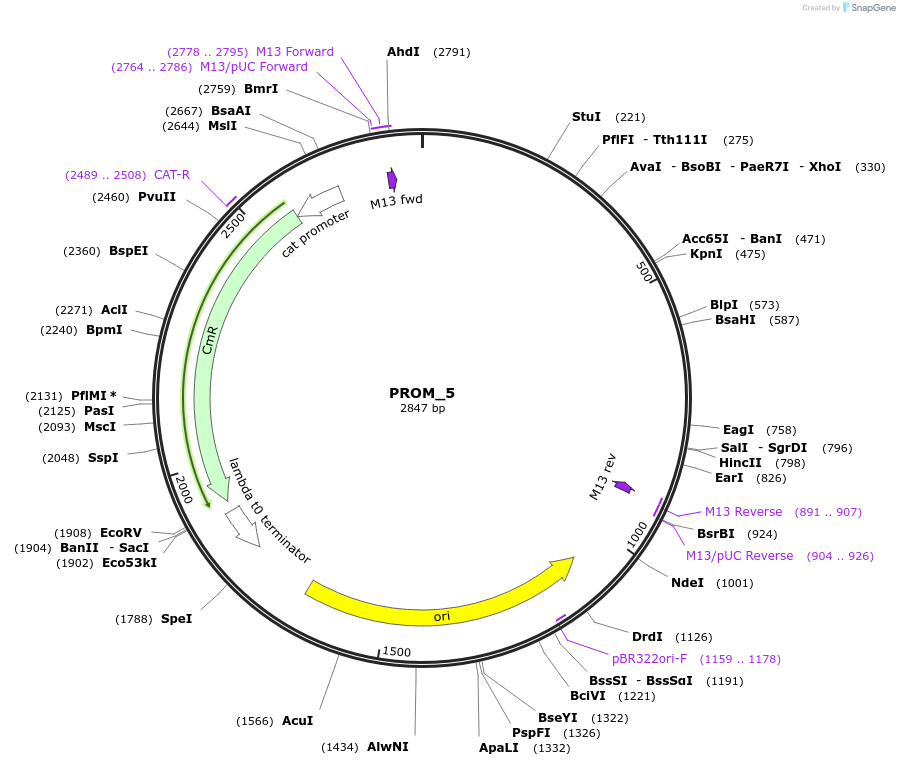
PROM_5
Plasmid#201298PurposeD5: Promoter of TraesCS3B02G553900 as Golden Gate compatible MoClo PROM partDepositorInsertD5: Promoter of TraesCS3B02G553900
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
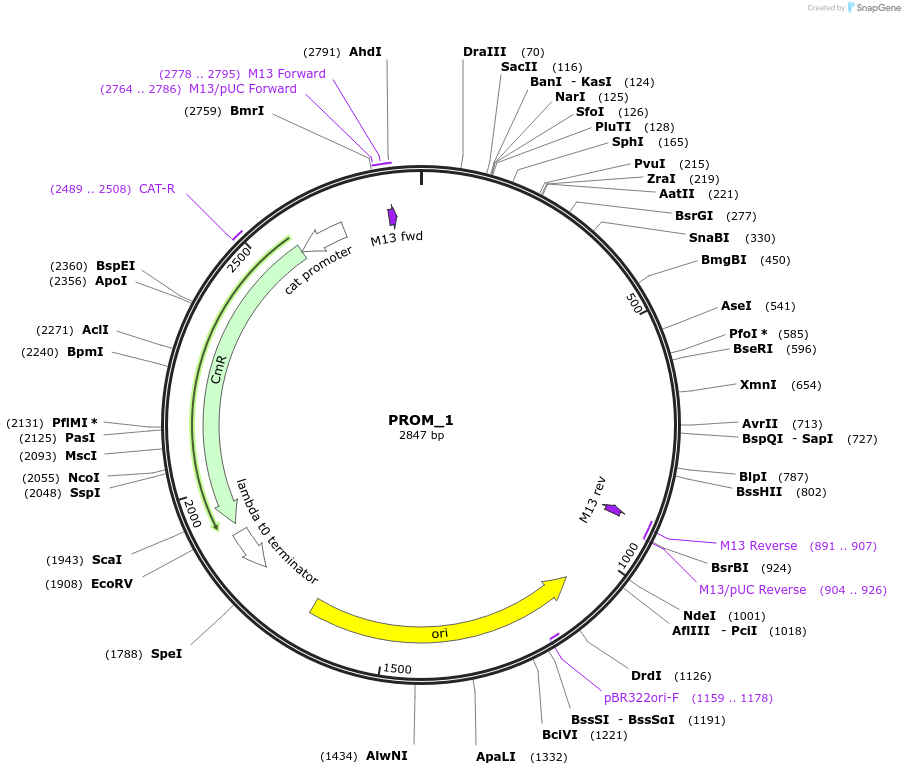
PROM_1
Plasmid#201294PurposeD1: Promoter of TraesCS1A02G332800 as Golden Gate compatible MoClo PROM partDepositorInsertD1: Promoter of TraesCS1A02G332800
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
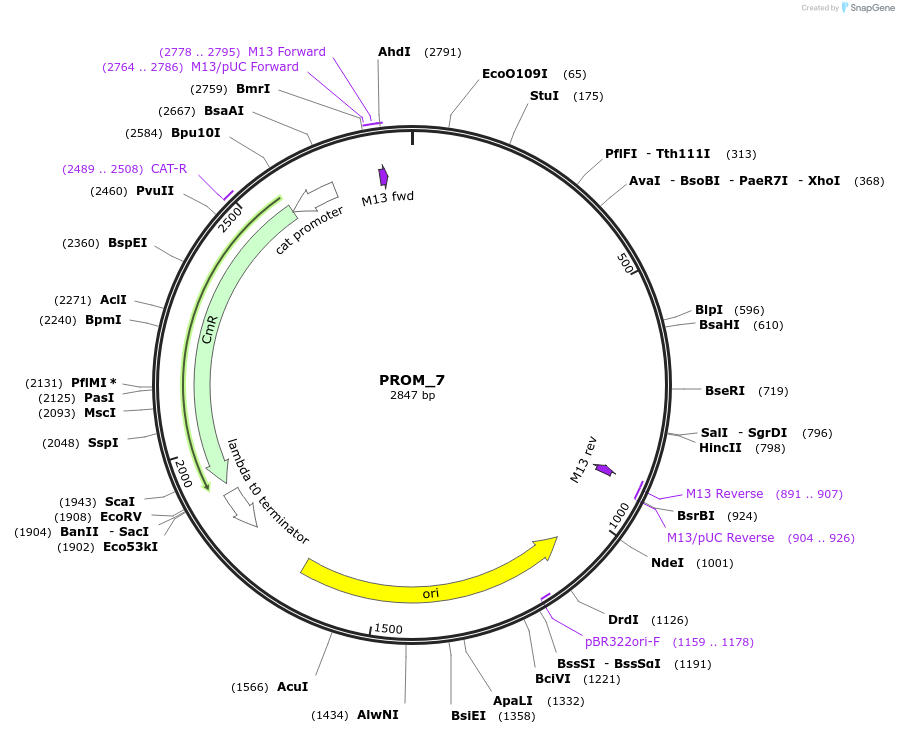
PROM_7
Plasmid#201300PurposeD7: Promoter of TraesCS3D02G316100 as Golden Gate compatible MoClo PROM partDepositorInsertD7: Promoter of TraesCS3D02G316100
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
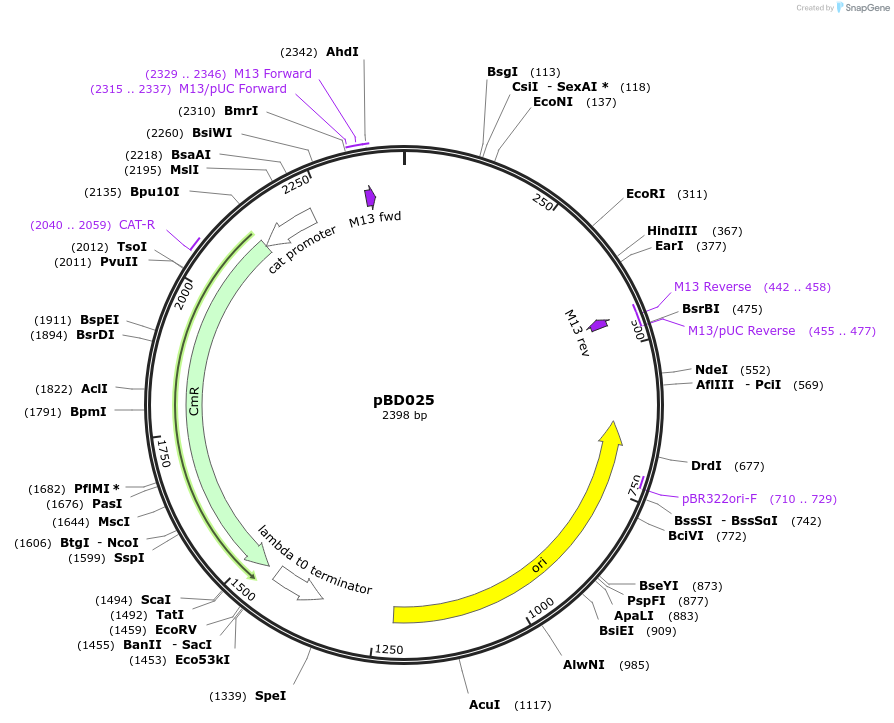
pBD025
Plasmid#201308PurposeAvrSr27-1 as Golden Gate compatible MoClo CDS partDepositorInsertAvrSr27-1
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
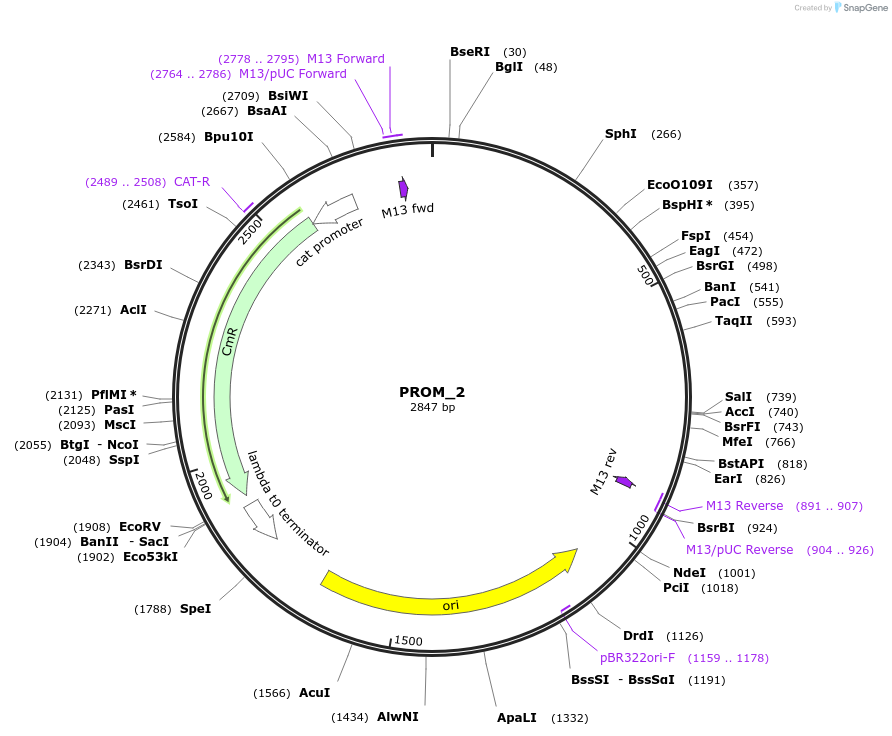
PROM_2
Plasmid#201295PurposeD2: Promoter of TraesCS2B02G533000 as Golden Gate compatible MoClo PROM partDepositorInsertD2: Promoter of TraesCS2B02G533000
ExpressionPlantMutationremoval of BsaI and BpiI sitesAvailable SinceOct. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
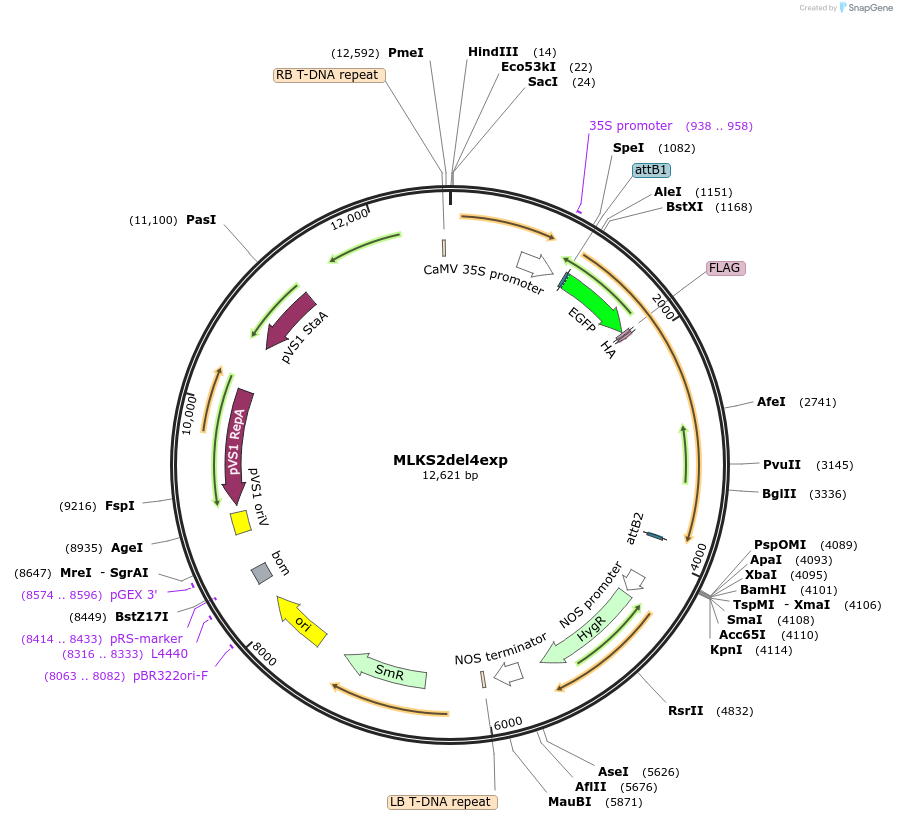
MLKS2del4exp
Plasmid#130861PurposeExpression of plant nuclear envelope LINC protein fused to fluorescent protein and affinity tags for cell biologyDepositorInsertMLKS2
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of terminal four amino acidsAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
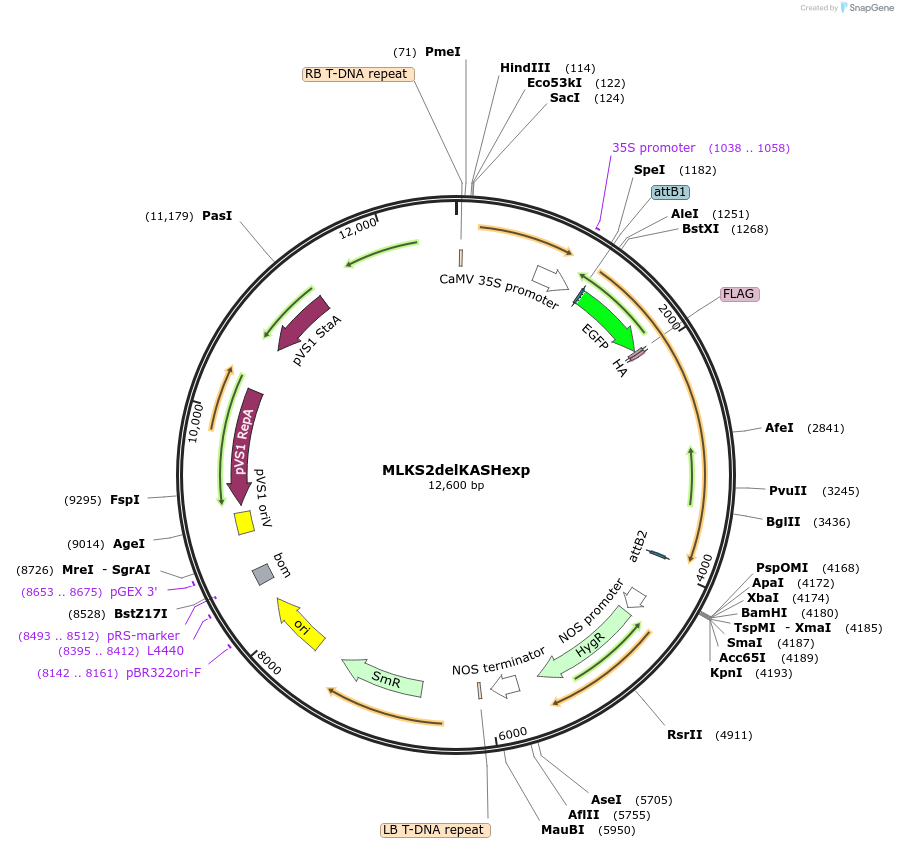
MLKS2delKASHexp
Plasmid#130862PurposeExpression of plant nuclear envelope LINC protein fused to fluorescent protein and affinity tags for cell biologyDepositorInsertMLKS2
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of KASH domainAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
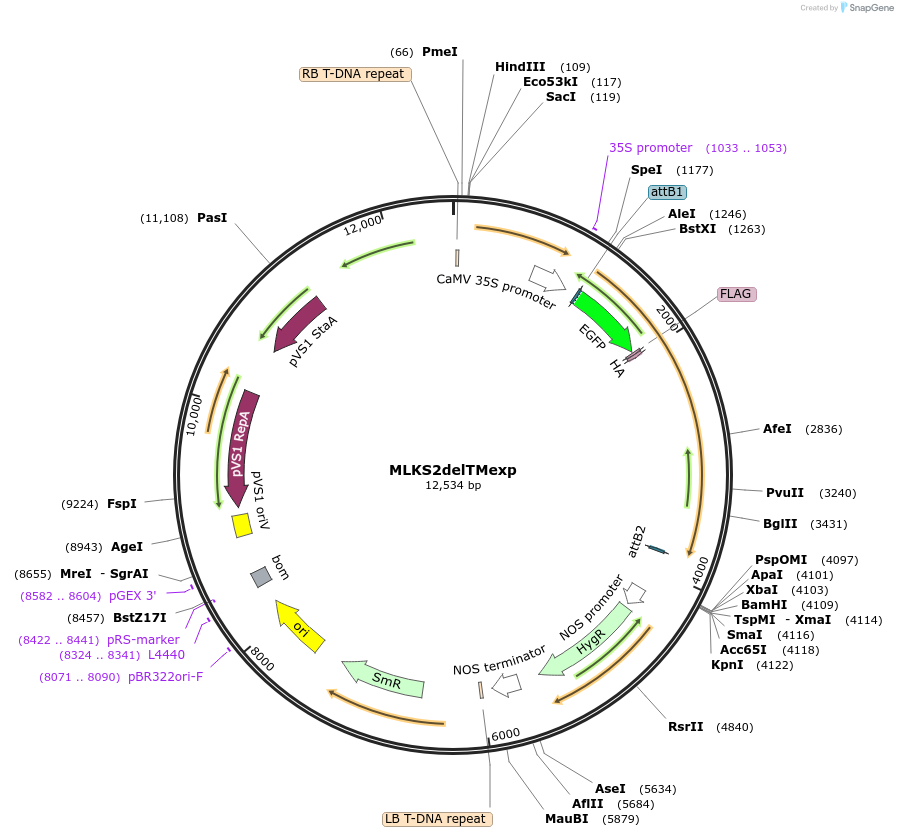
MLKS2delTMexp
Plasmid#130863PurposeExpression of plant nuclear envelope LINC protein fused to fluorescent protein and affinity tags for cell biologyDepositorInsertMLKS2
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of transmembrane domainAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
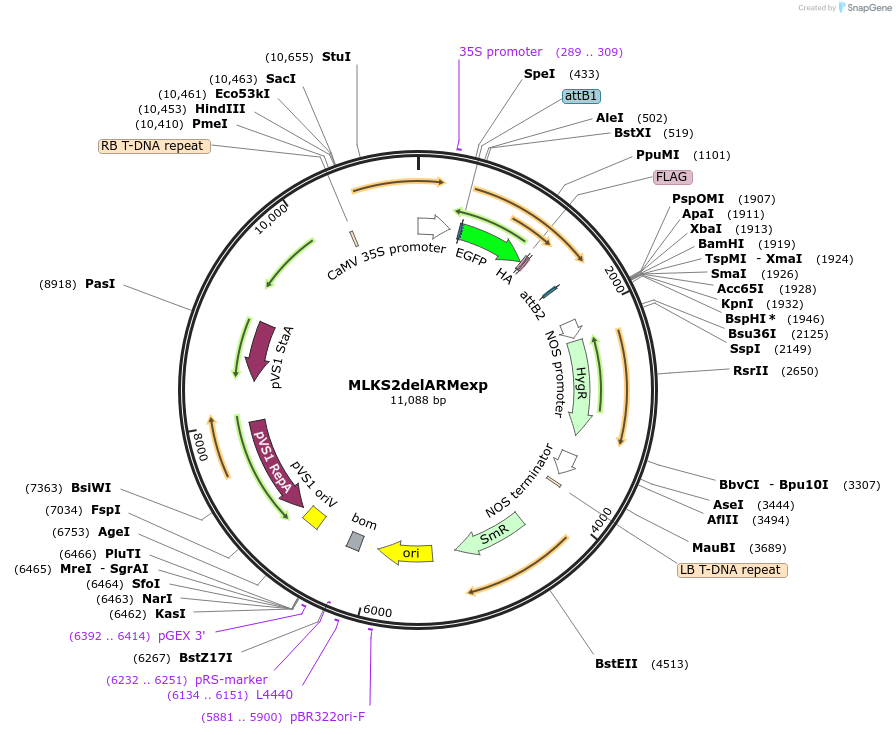
MLKS2delARMexp
Plasmid#130864PurposeExpression of plant nuclear envelope LINC protein fused to fluorescent protein and affinity tags for cell biologyDepositorInsertMLKS2
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of armadillo domainAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
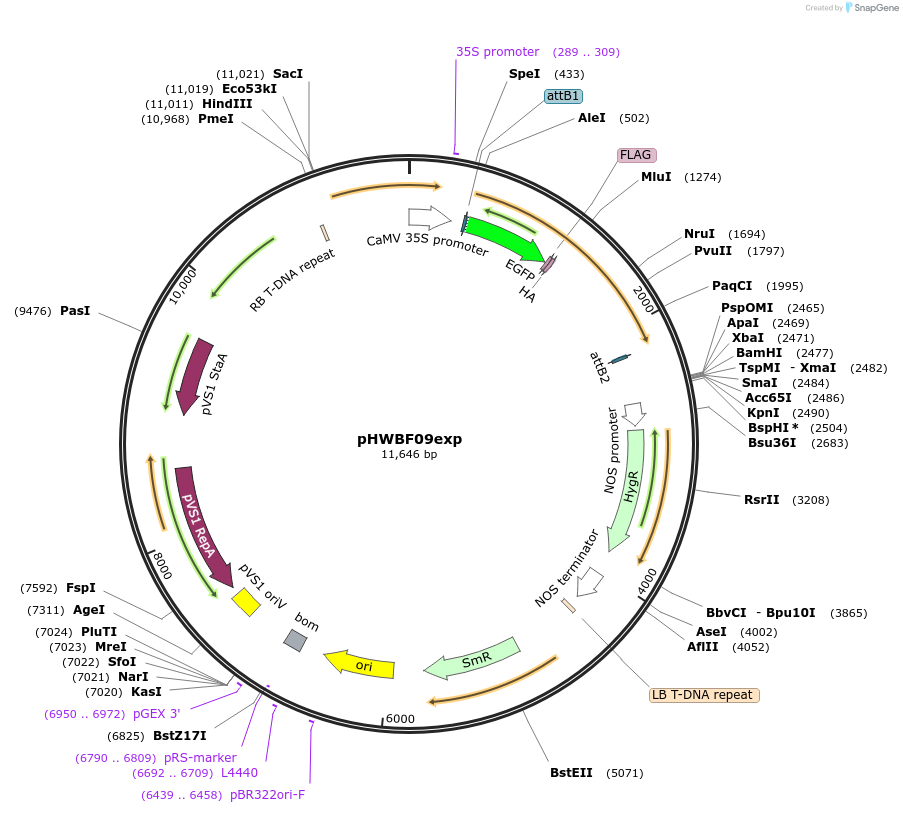
pHWBF09exp
Plasmid#130833PurposeExpression of plant LINC fluorescent fusion protein for cell biologyDepositorInsertMLKP1
TagseGFP-FLAG-HAExpressionPlantPromoterCaMV 35SAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
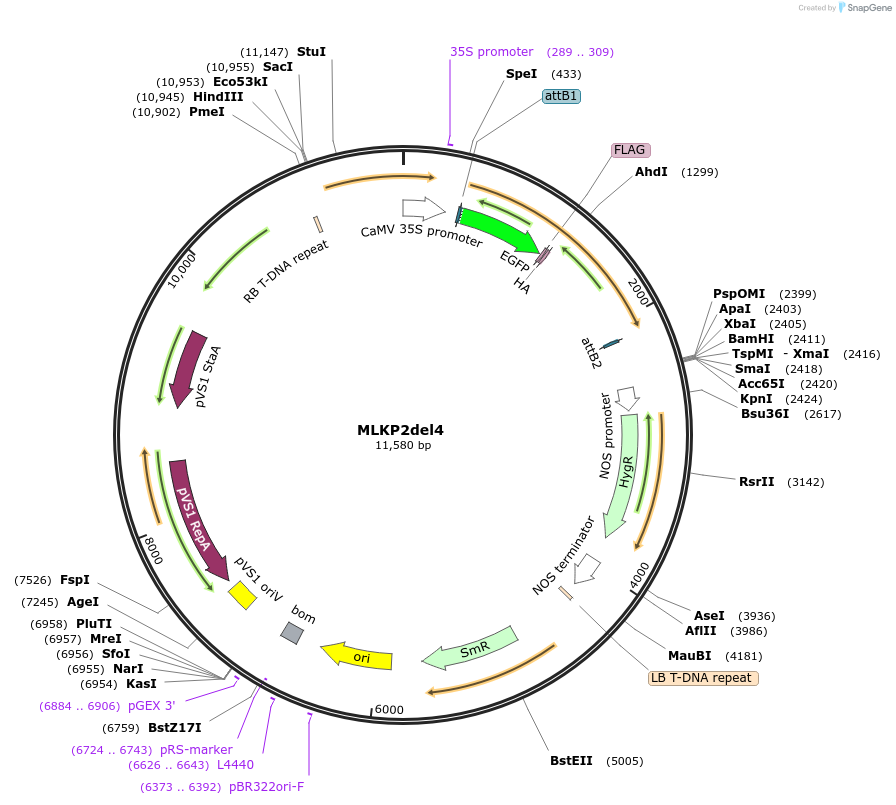
MLKP2del4
Plasmid#130837PurposeExpression of plant LINC fluorescent fusion protein for cell biologyDepositorInsertMLKP2
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of terminal four amino acid residuesPromoterCaMV 35SAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
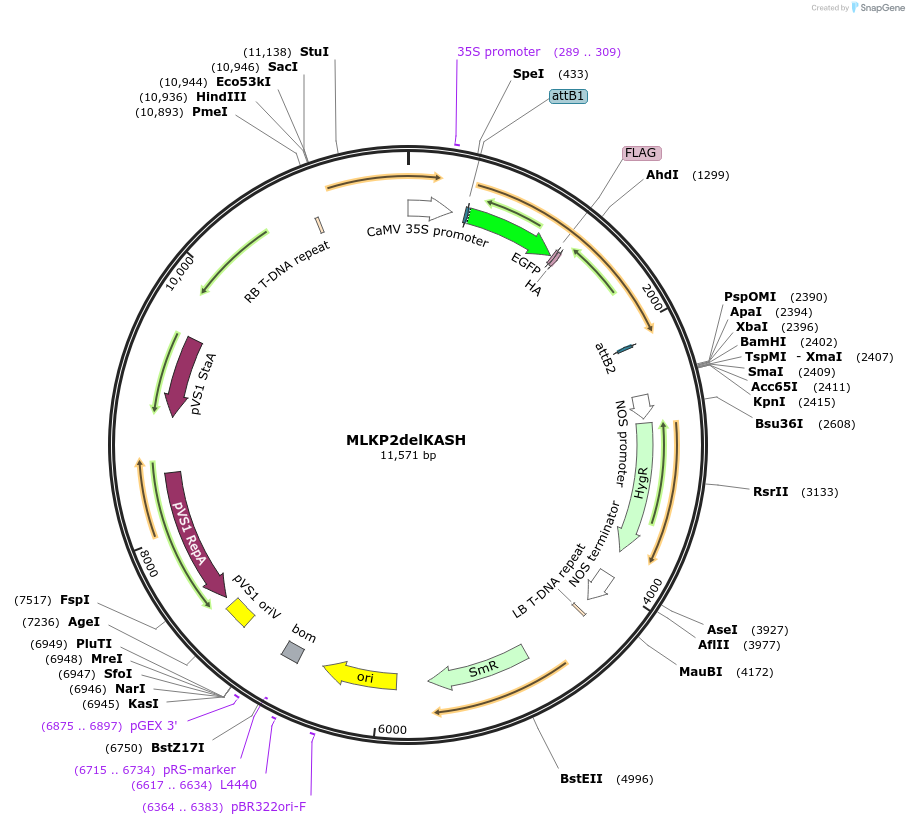
MLKP2delKASH
Plasmid#130838PurposeExpression of plant LINC fluorescent fusion protein for cell biologyDepositorInsertMLKP2
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of KASH domain, addition of three alani…PromoterCaMV 35SAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
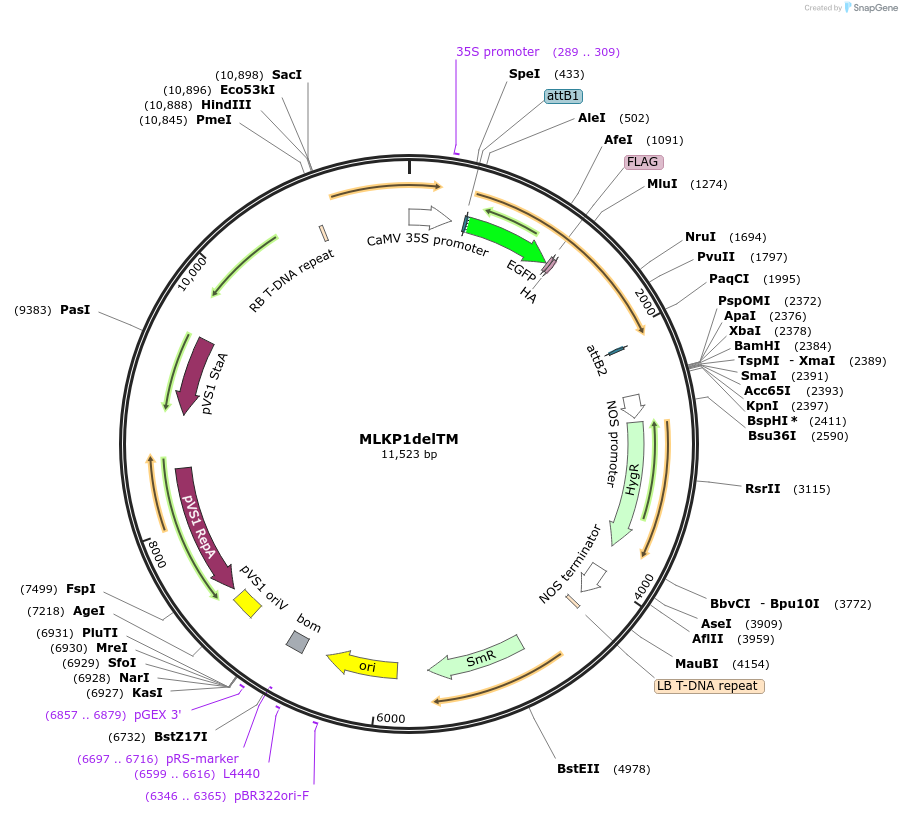
MLKP1delTM
Plasmid#130841PurposeExpression of plant LINC fluorescent fusion protein for cell biologyDepositorInsertMLKP1
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of transmembrane domainPromoterCaMV 35SAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
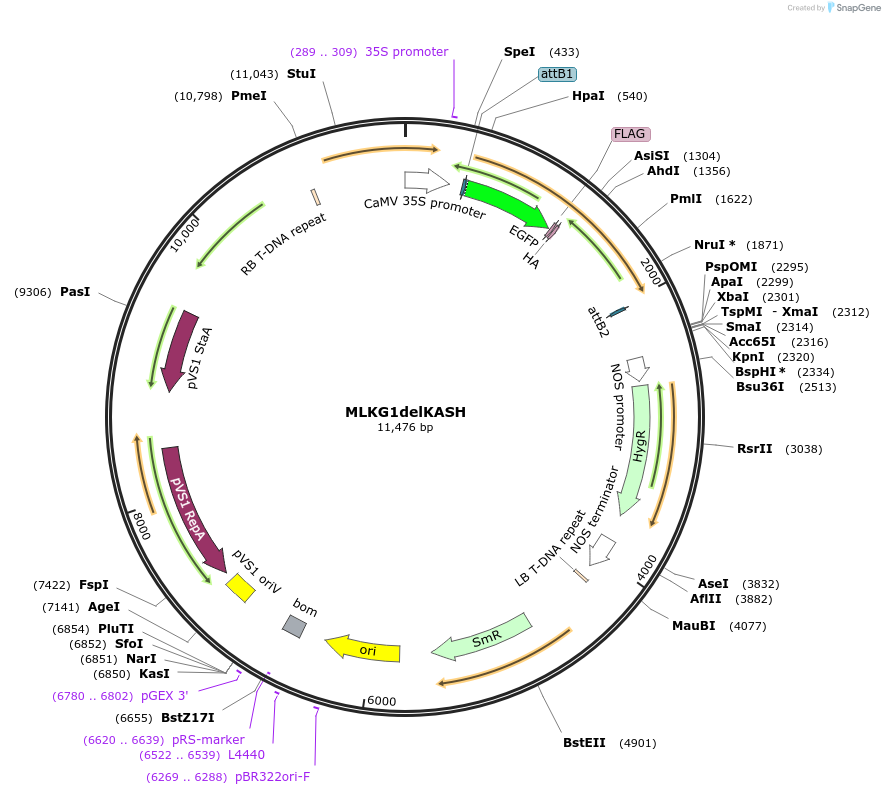
MLKG1delKASH
Plasmid#130842PurposeExpression of plant LINC fluorescent fusion protein for cell biologyDepositorInsertMLKG1
TagseGFP-FLAG-HAExpressionPlantMutationDeletion of KASH domainPromoterCaMV 35SAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
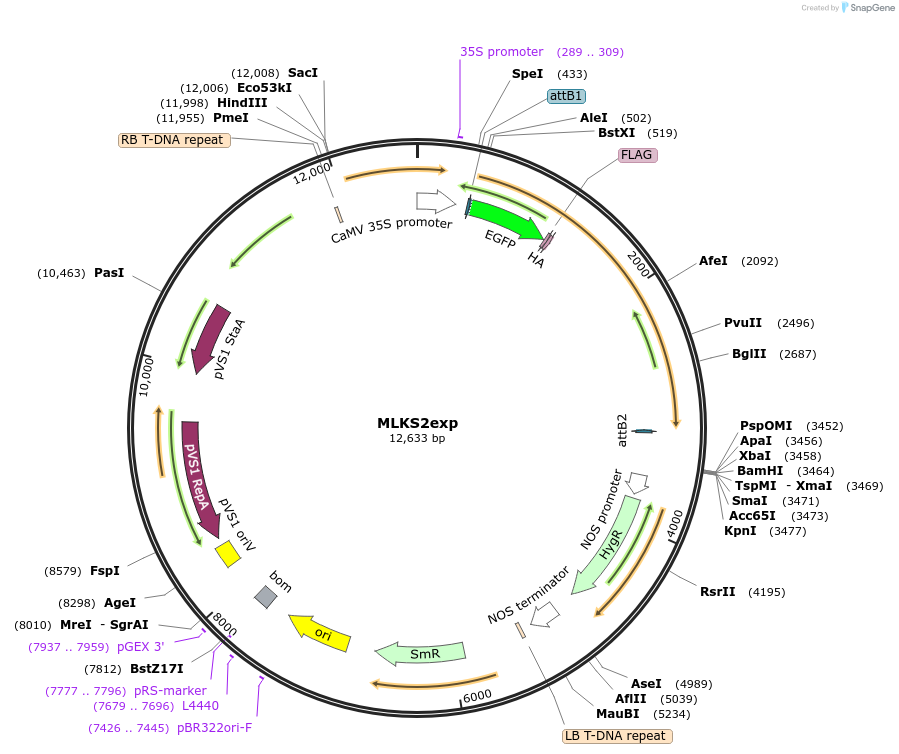
MLKS2exp
Plasmid#130856PurposeExpression of plant nuclear envelope LINC protein fused to fluorescent protein and affinity tags for cell biologyDepositorInsertMLKS2
TagseGFP-FLAG-HAExpressionPlantAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
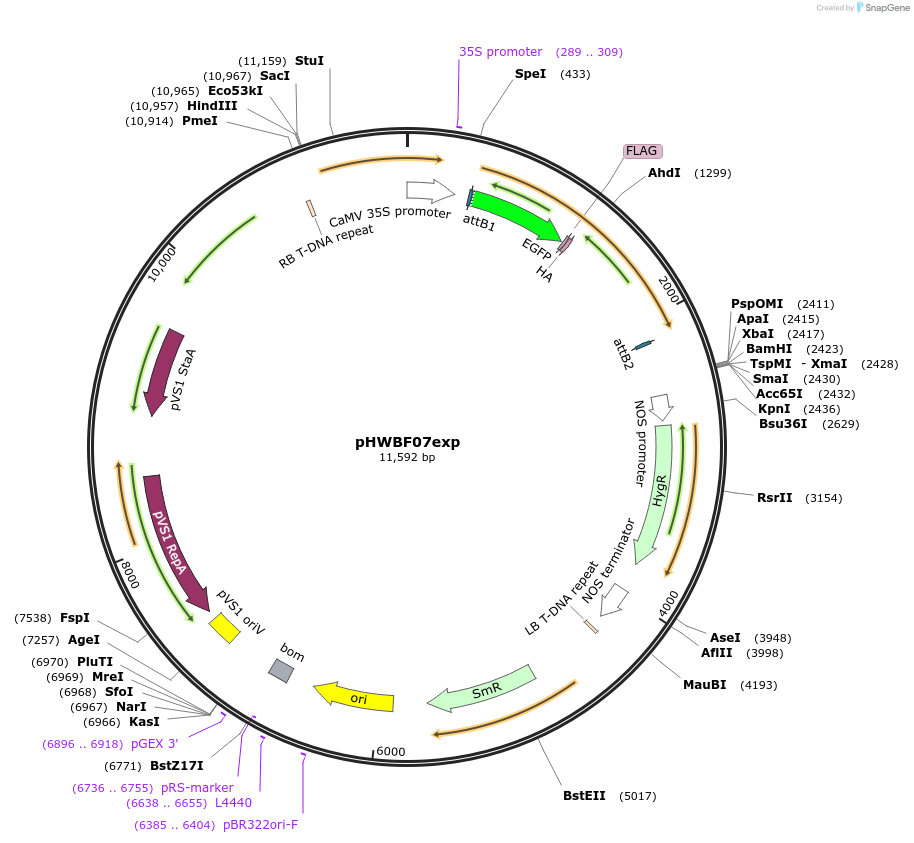
pHWBF07exp
Plasmid#130832PurposeExpression of plant LINC fluorescent fusion protein for cell biologyDepositorInsertMLKP2
TagseGFP-FLAG-HAExpressionPlantPromoterCaMV 35SAvailable SinceSept. 28, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
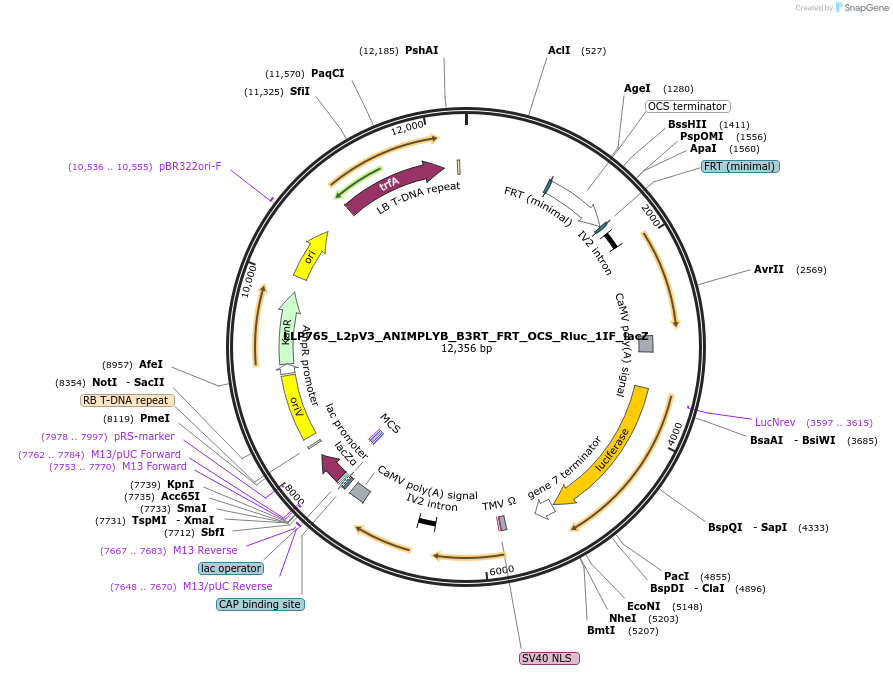
LLP765_L2pV3_ANIMPLYB_B3RT_FRT_OCS_Rluc_1IF_lacZ
Plasmid#192409PurposeTo act as a 1-input state of the A NIMPLY B gate.DepositorInsertB3RT-Act2-B3RT::FRT-OCS-FRT::Rluc
UseSynthetic BiologyTagsPESTExpressionPlantAvailable SinceSept. 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
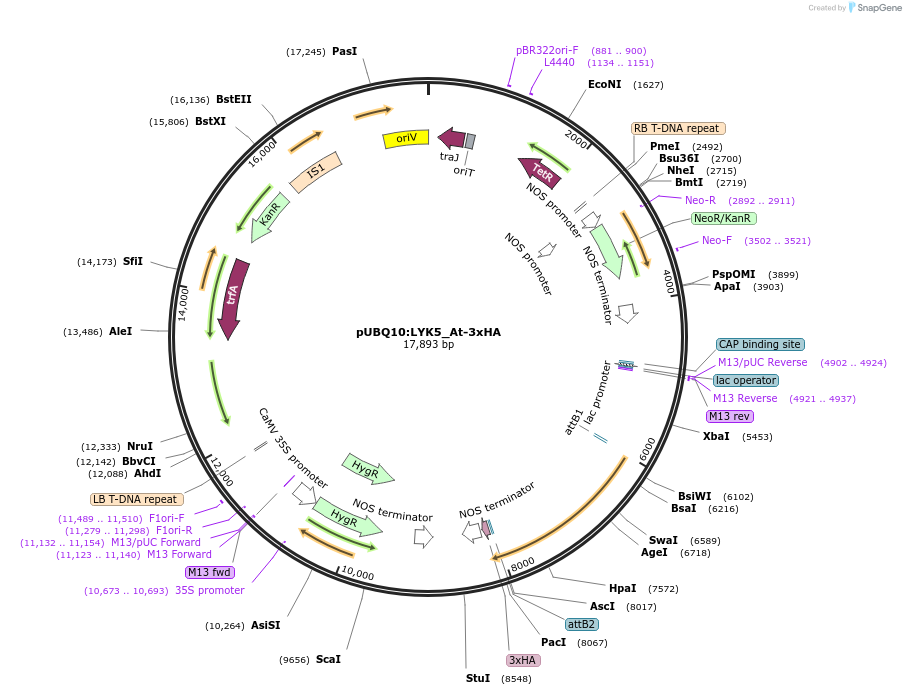
pUBQ10:LYK5_At-3xHA
Plasmid#202192PurposeExpress Arabidopsis thaliana LYK5 gene under UBQ10 promoterDepositorInsertLYSM-CONTAINING RECEPTOR-LIKE KINASE 5 (LYK5 Mustard Weed)
TagsHAExpressionPlantPromoterAtUBQ10Available SinceSept. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
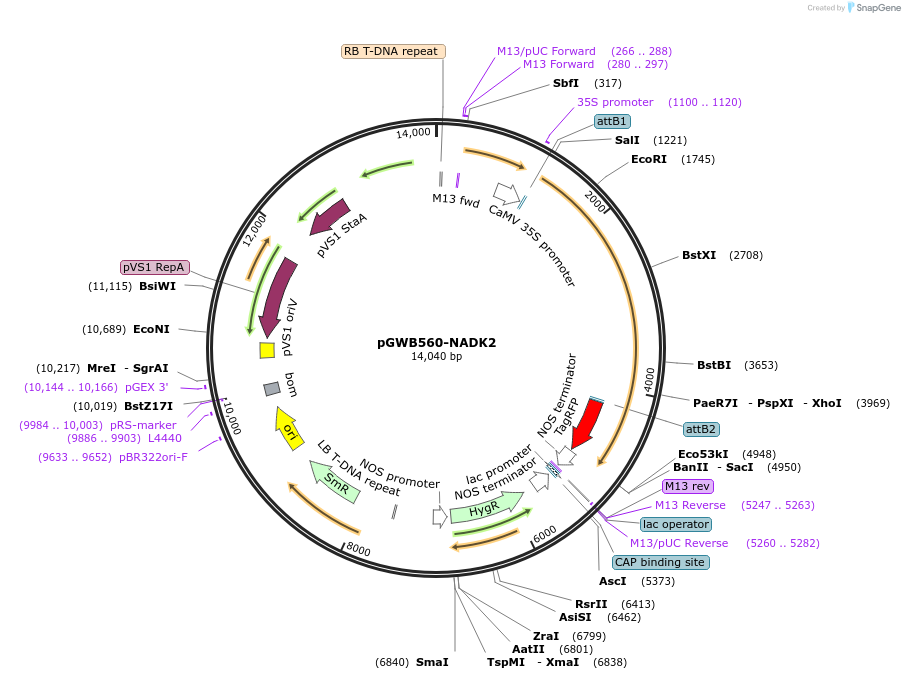
pGWB560-NADK2
Plasmid#203760PurposeFor Agrobacterium transformation. tagRFP fused Arabidopsis thaliana NAD KINASE2 (NADK2) under the control of the cauliflower mosaic virus (CaMV) 35S promoter.DepositorInsertNAD KINASE2
ExpressionBacterial and PlantAvailable SinceAug. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
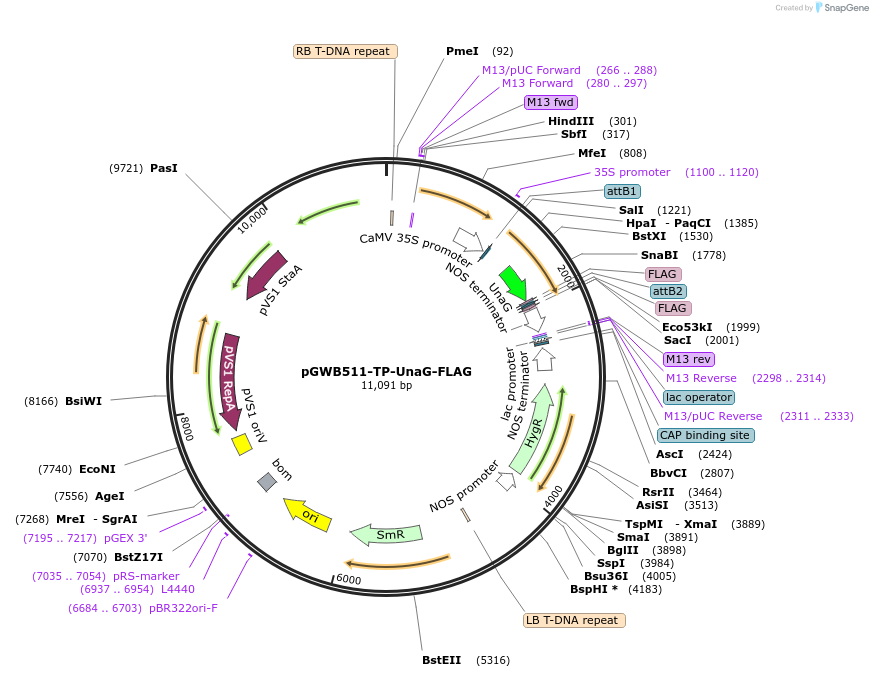
pGWB511-TP-UnaG-FLAG
Plasmid#203479PurposeFor Agrobacterium transformation. A plastid transit peptide fused UnaG-FLAG under the control of the cauliflower mosaic virus (CaMV) 35S promoter.DepositorInsertPlastid transit peptide fused UnaG-FLAG
ExpressionBacterial and PlantAvailable SinceAug. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
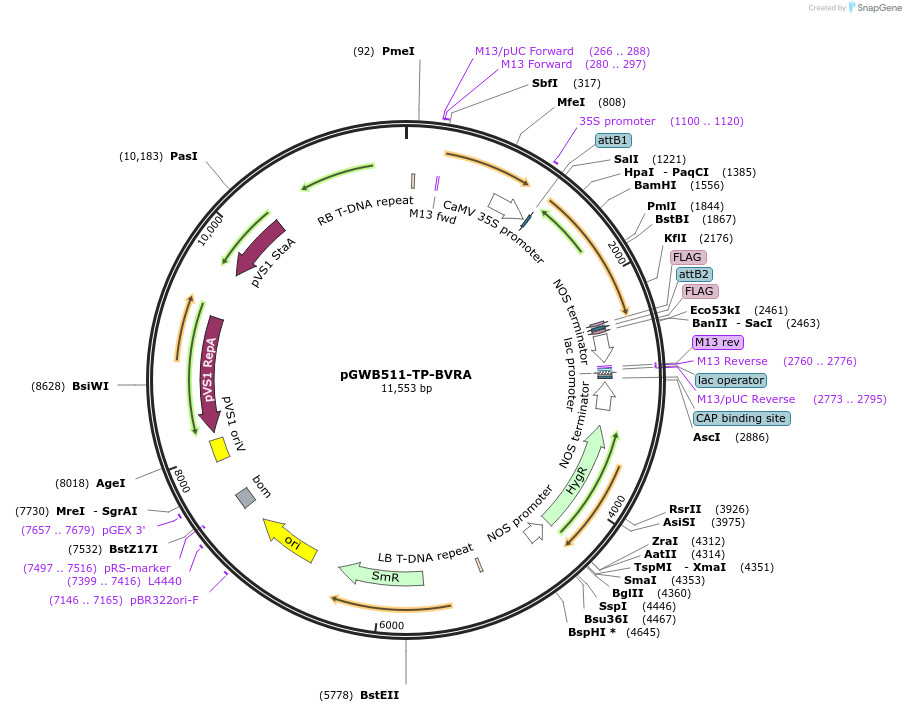
pGWB511-TP-BVRA
Plasmid#203480PurposeFor Agrobacterium transformation. Rattus rattus biliverdin reductase A (BVRA) flanked with an N-terminal plastid transit peptide and a C-terminal FLAG tag under the control of the 35S promoter.DepositorInsertRattus rattus biliverdin reductase A flanked with an N-terminal plastid transit peptide and a C-terminal FLAG tag.
ExpressionBacterial and PlantAvailable SinceAug. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
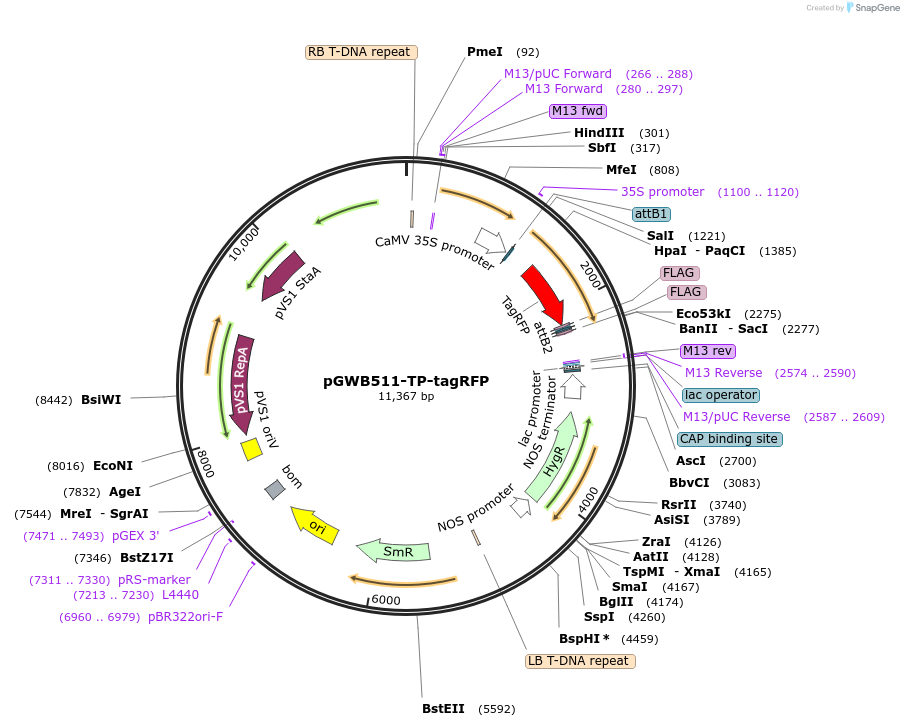
pGWB511-TP-tagRFP
Plasmid#203482PurposeFor Agrobacterium transformation. tagRFP flanked with an N-terminal plastid transit peptide and a C-terminal FLAG tag under the control of the cauliflower mosaic virus (CaMV) 35S promoter.DepositorInsertTagRFP flanked with an N-terminal plastid transit peptide and a C-terminal FLAG tag.
ExpressionBacterial and PlantAvailable SinceAug. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
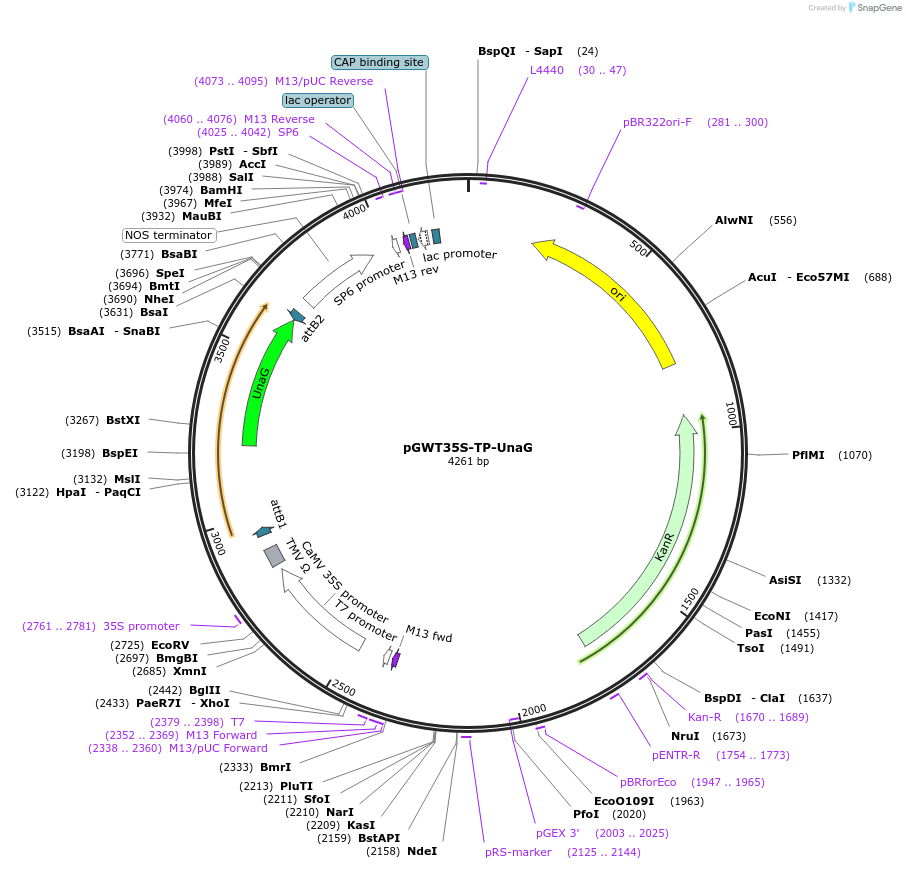
pGWT35S-TP-UnaG
Plasmid#203484PurposeFor particle bombardment. A plastid transit peptide fused UnaG under the control of the cauliflower mosaic virus (CaMV) 35S promoter.DepositorInsertPlastid transit peptide fused UnaG
ExpressionBacterial and PlantAvailable SinceAug. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
pDEST17-HO1(51-231)
Plasmid#203486PurposeFor recombinant protein production in Escherichia coli. His-tagged Arabidopsis thaliana HEME OXYGENASE 1 (HO1) without the plastid transit peptide under the control of the T7 promoter.DepositorInsertArabidopsis thaliana HEME OXYGENASE1 (51 aa – 231 aa)
ExpressionBacterial and PlantAvailable SinceAug. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
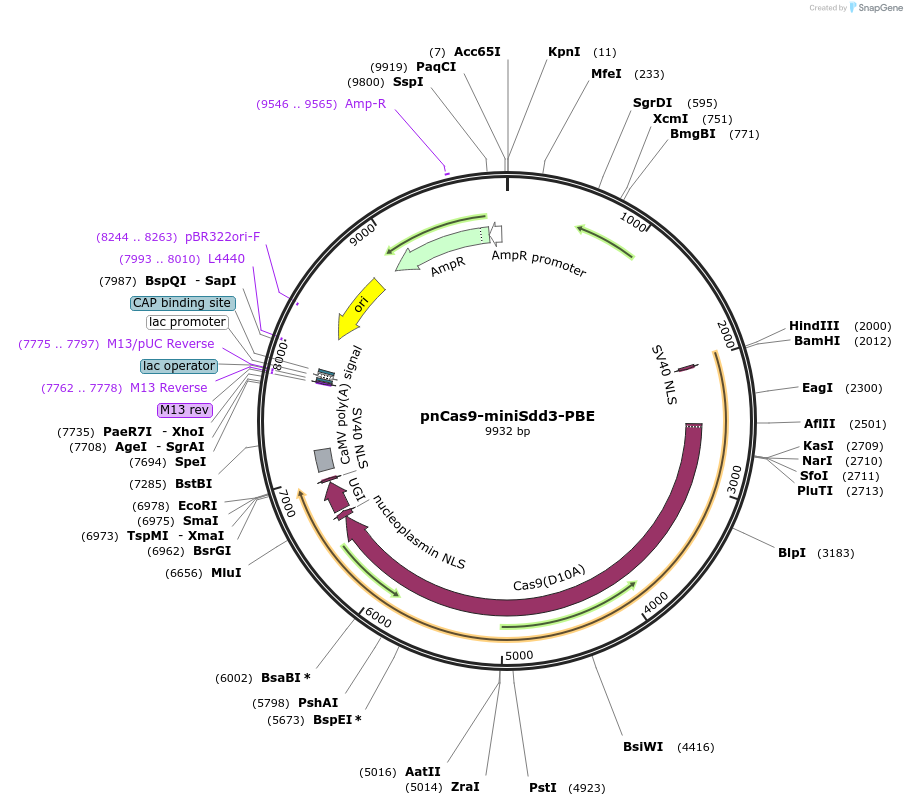
pnCas9-miniSdd3-PBE
Plasmid#204857PurposeFor cytosine base editing using miniSdd3 in monocotyledon plants (transient transformation)DepositorInsertminiSdd3-nSpCas9(D10A)-UGI
UseCRISPRExpressionPlantMutationD10A in SpCas9PromoterZmUbi-1Available SinceAug. 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
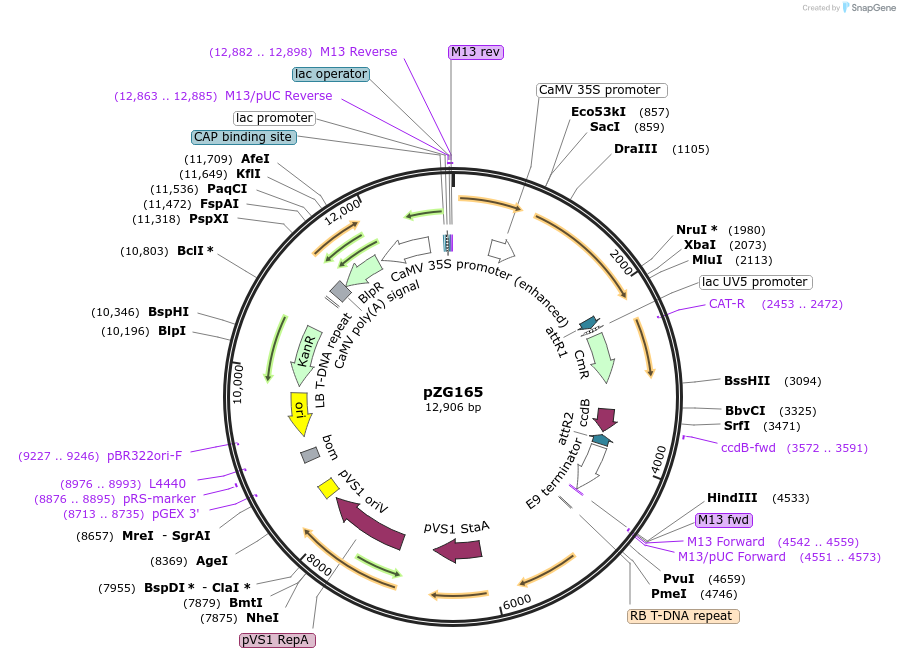
pZG165
Plasmid#202584PurposeRNA editing in plants. Plasmid contains 35S-dADARcd-LIC1-ccdB-LIC2-35S-barDepositorInsertHyperADARcd
ExpressionPlantMutationE488QAvailable SinceAug. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
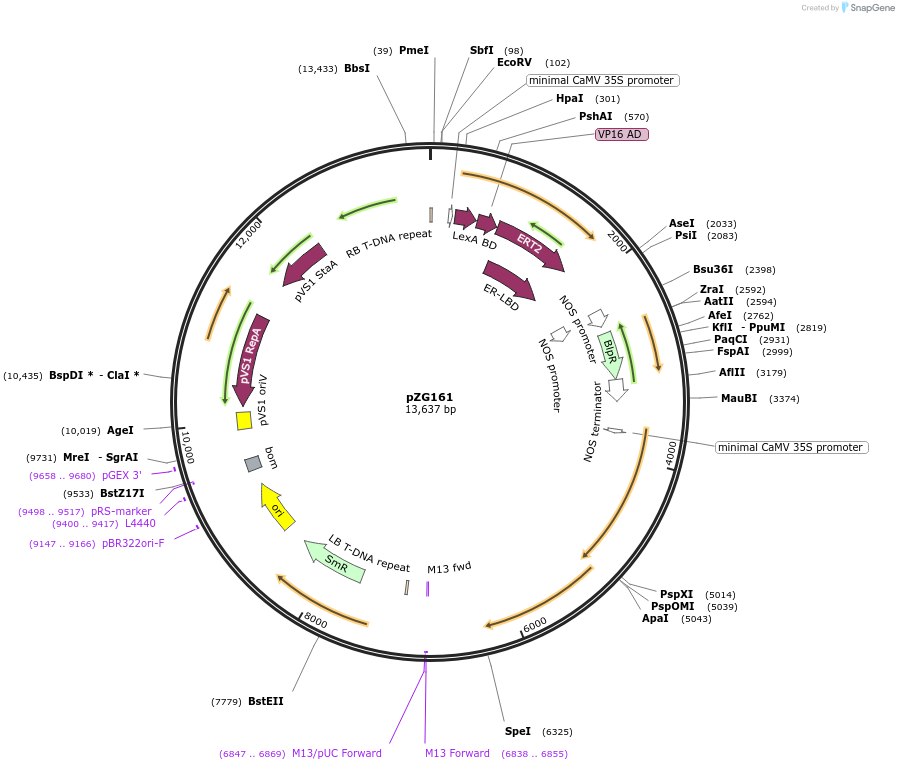
pZG161
Plasmid#202580PurposeRNA editing in plants. Plasmid contains extradiol-dADARcd-LIC cassette-dADARcd-35S-barDepositorInsertHyperADARcd
ExpressionPlantMutationE488QAvailable SinceAug. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
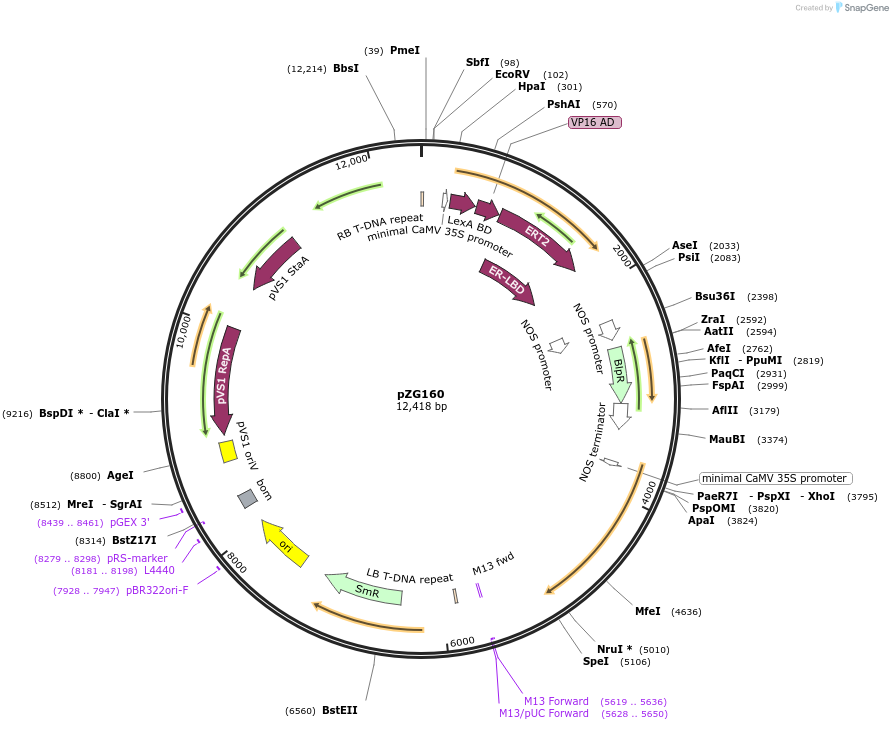
pZG160
Plasmid#202579PurposeRNA editing in plants. Plasmid contains extradiol-LIC cassette-dADARcd-35S-barDepositorInsertHyperADARcd
ExpressionPlantMutationE488QAvailable SinceAug. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
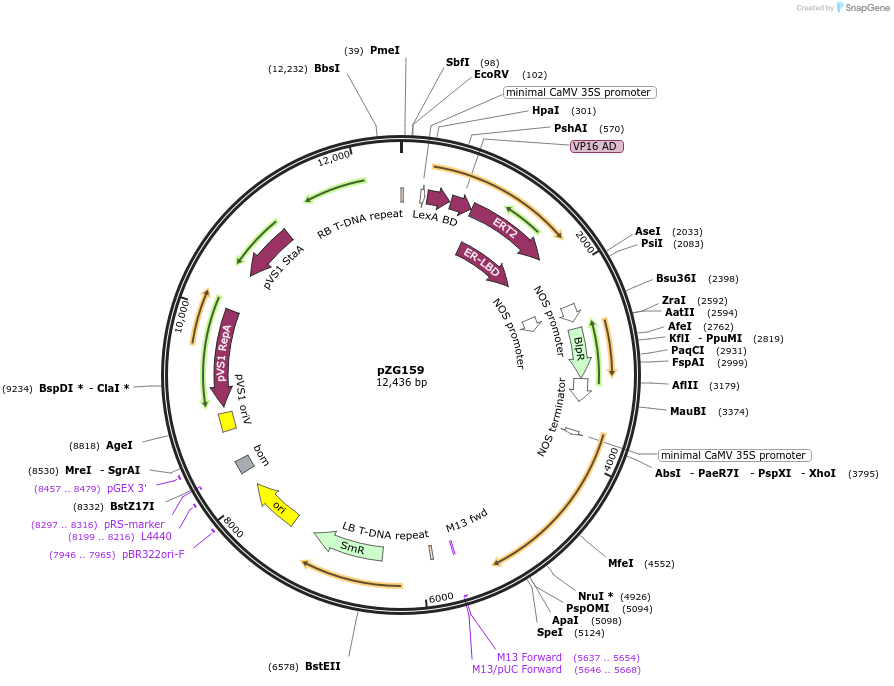
pZG159
Plasmid#202578PurposeRNA editing in plants. Plasmid contains extradiol-dADARcd-LIC cassette-35S-barDepositorInsertHyperADARcd
ExpressionPlantMutationE488QAvailable SinceAug. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
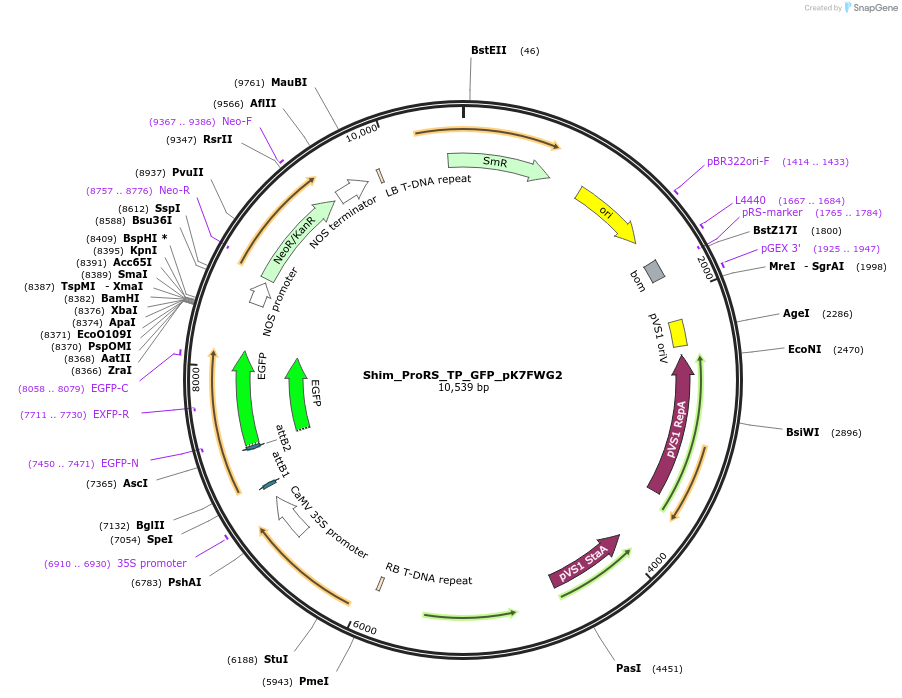
Shim_ProRS_TP_GFP_pK7FWG2
Plasmid#202651PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from putative cytosolic ProRS
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
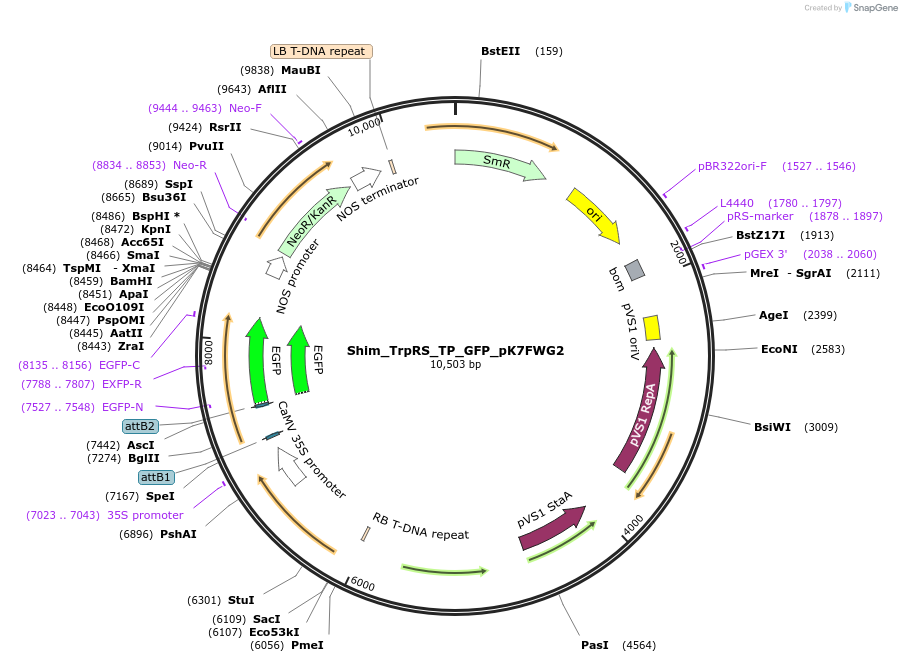
Shim_ThrRS_TP_GFP_pK7FWG2
Plasmid#202652PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from putative cytosolic ThrRS
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
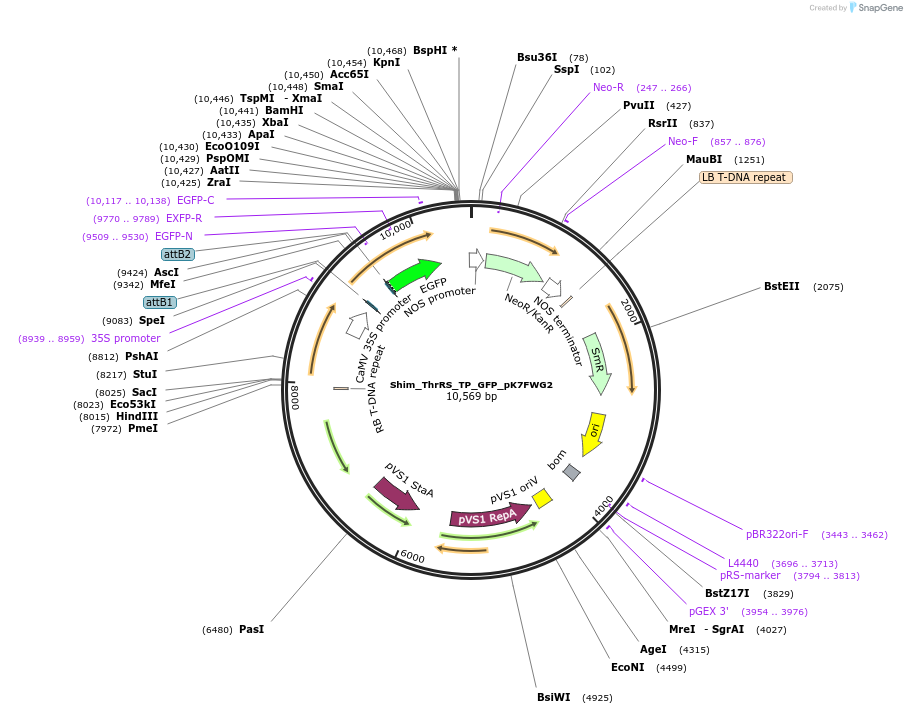
Shim_TrpRS_TP_GFP_pK7FWG2
Plasmid#202653PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from putative cytosolic TrpRS
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
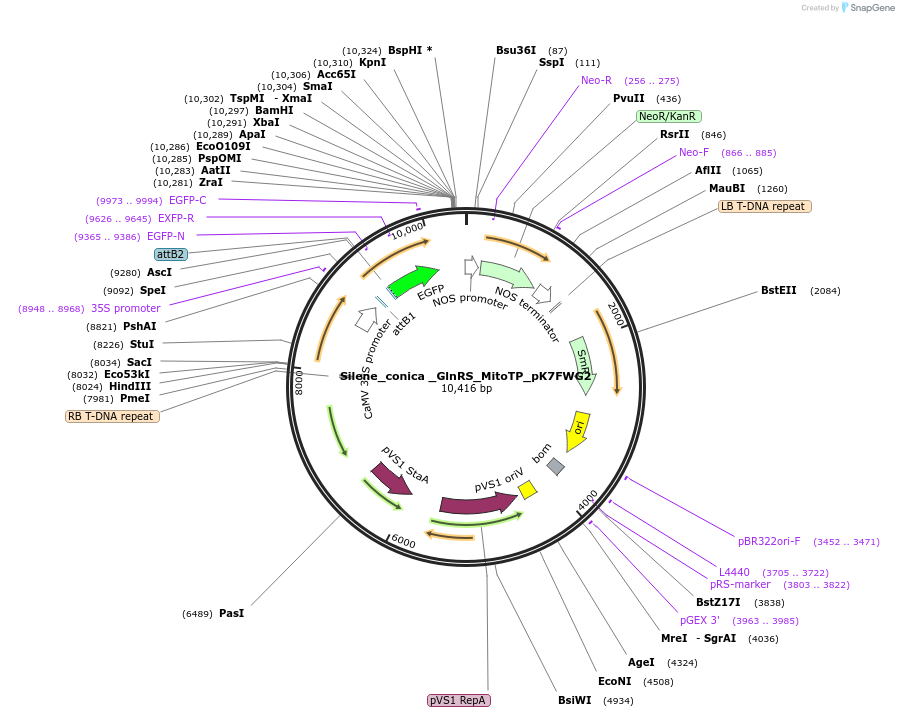
Silene_conica _GlnRS_MitoTP_pK7FWG2
Plasmid#202654PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from Silene conica cytosolic GlnRS predicted to be re-targeted to mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits -
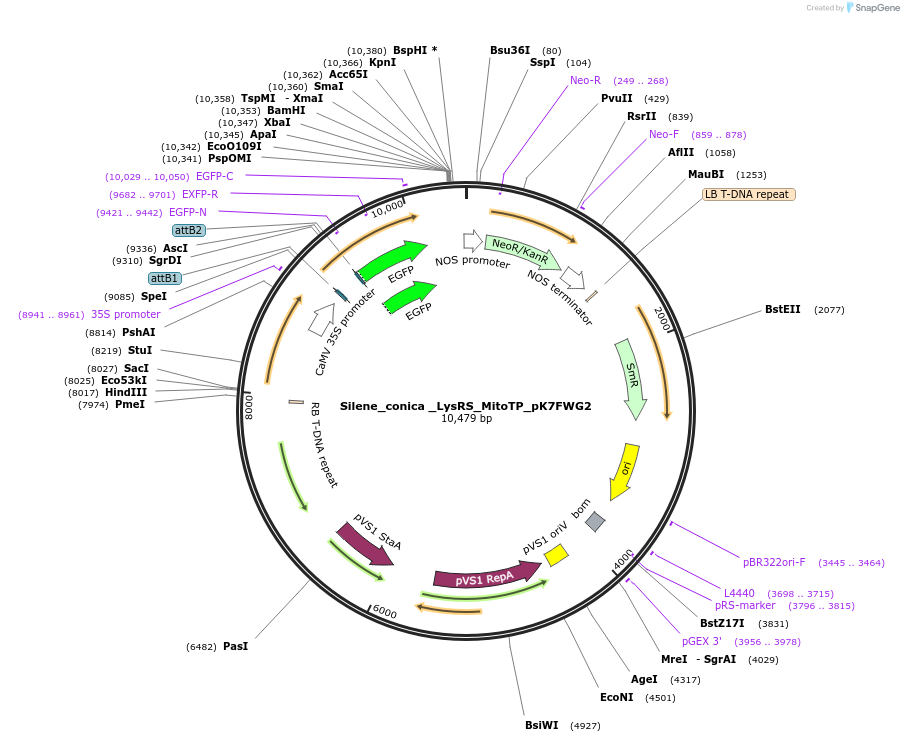
Silene_conica _LysRS_MitoTP_pK7FWG2
Plasmid#202655PurposeDetermine where transit peptide targets GFP in N. benthamianaDepositorInsertN-terminal transit peptide from Silene conica cytosolic LysRS predicted to be re-targeted to mitochondria
TagsGreen Florescent ProtienExpressionPlantAvailable SinceAug. 10, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits