We narrowed to 14,065 results for: SHI
-
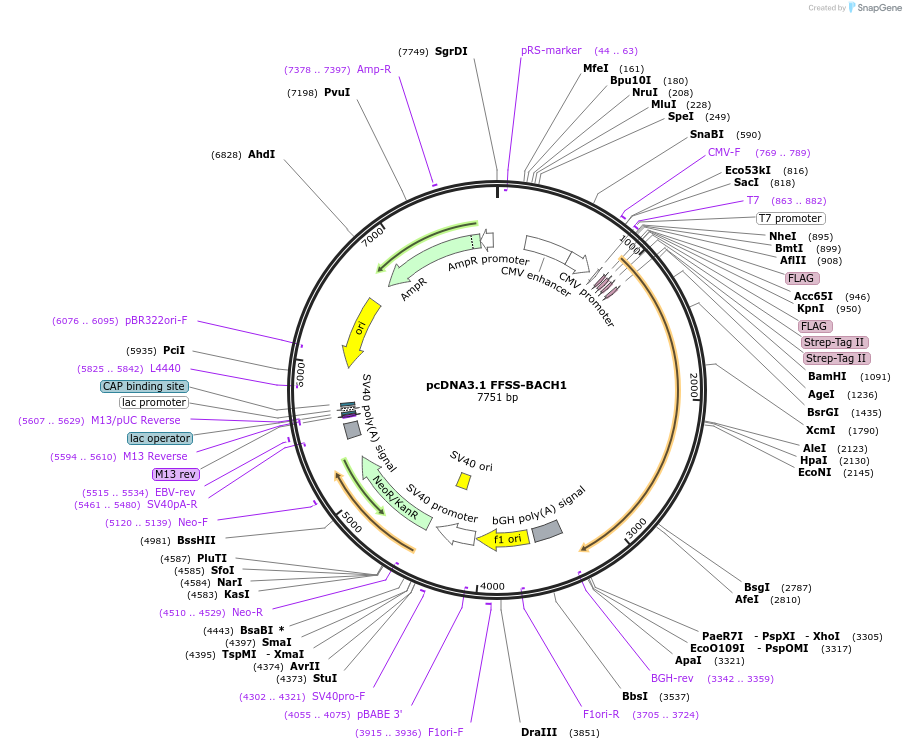
Plasmid#232273PurposeExpression of N-terminal tagged 2xFLAG-2xSTREP-BACH1 (H. sapiens) in human cell linesDepositorAvailable SinceFeb. 20, 2025AvailabilityAcademic Institutions and Nonprofits only
-
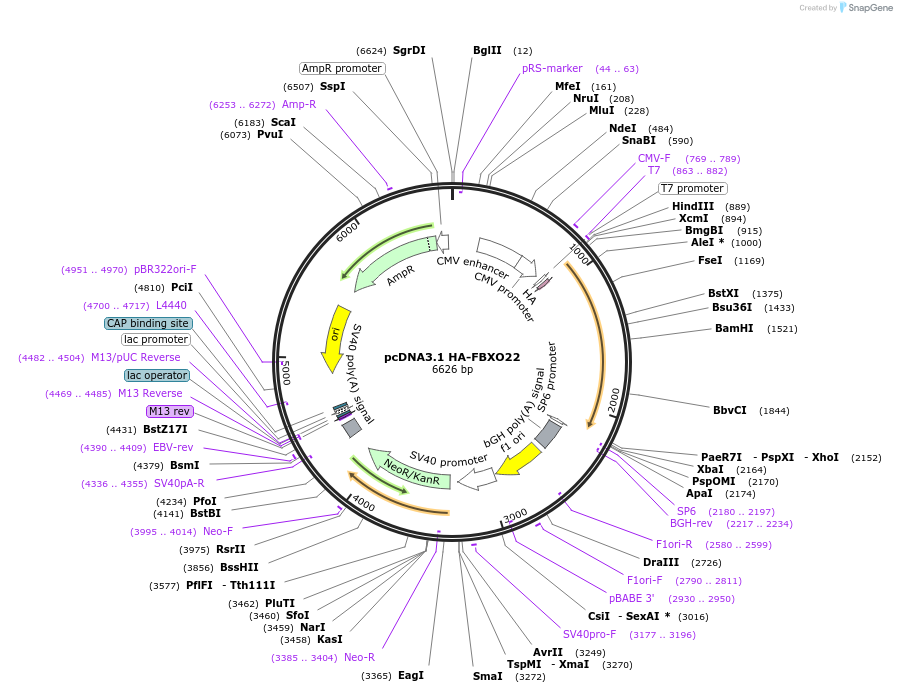
pcDNA3.1 HA-FBXO22
Plasmid#232271PurposeExpression of N-terminal tagged HA-FBXO22 (H. sapiens) in human cell linesDepositorAvailable SinceFeb. 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
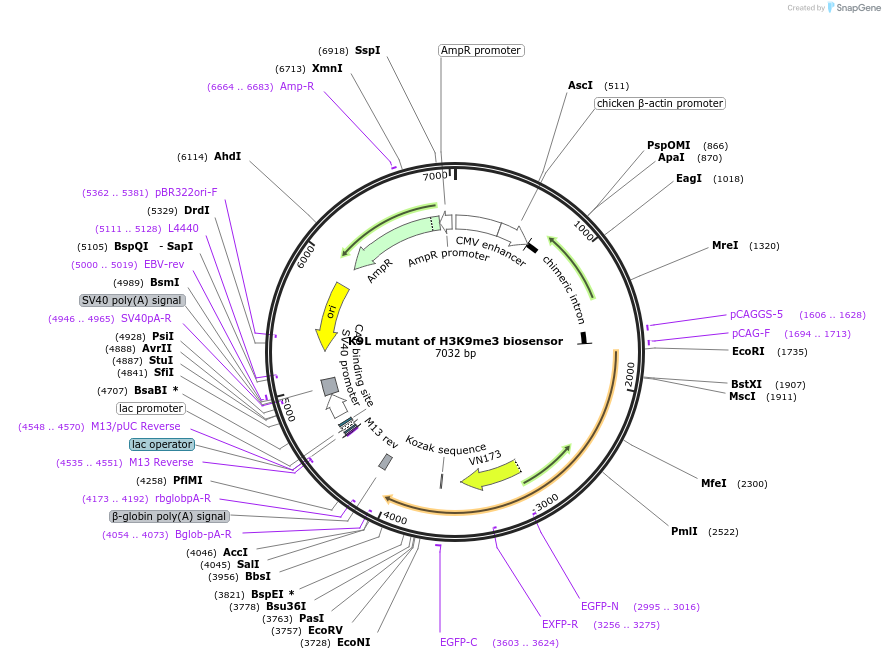
K9L mutant of H3K9me3 biosensor
Plasmid#120807PurposeFRET biosensor. Eliminating the H3 lysine 9 methylation site in H3K9me3 biosensor to abolish the detection capabilityDepositorInsertTruncated YPet-HP1-EV linker-ECFP-mouse histone H3(K9L mutation)
ExpressionMammalianMutationlysine 9 is mutated to leucine 9 on histone H3PromoterCMVAvailable SinceFeb. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
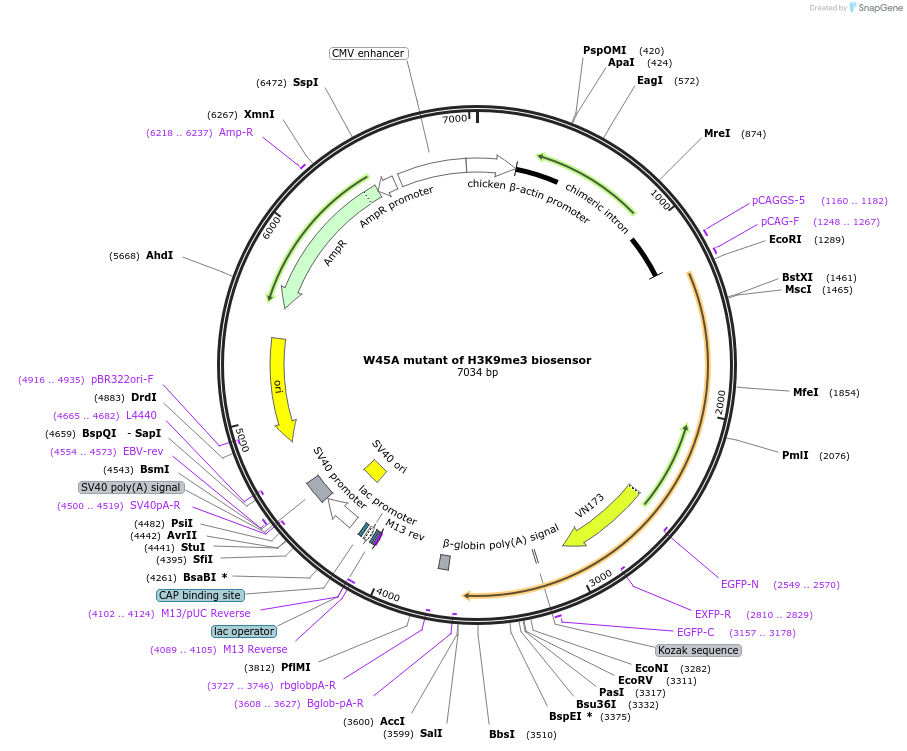
W45A mutant of H3K9me3 biosensor
Plasmid#120808PurposeFRET biosensor. Disrupting HP1 binding capability in H3K9me3 biosensor to abolish the detection capabilityDepositorInsertTruncated YPet-HP1(W45A mutation)-EV linker-ECFP-mouse histone H3
ExpressionMammalianMutationTryptophan 45 is mutated to Alanine 45 on HP1 dom…PromoterCMVAvailable SinceFeb. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
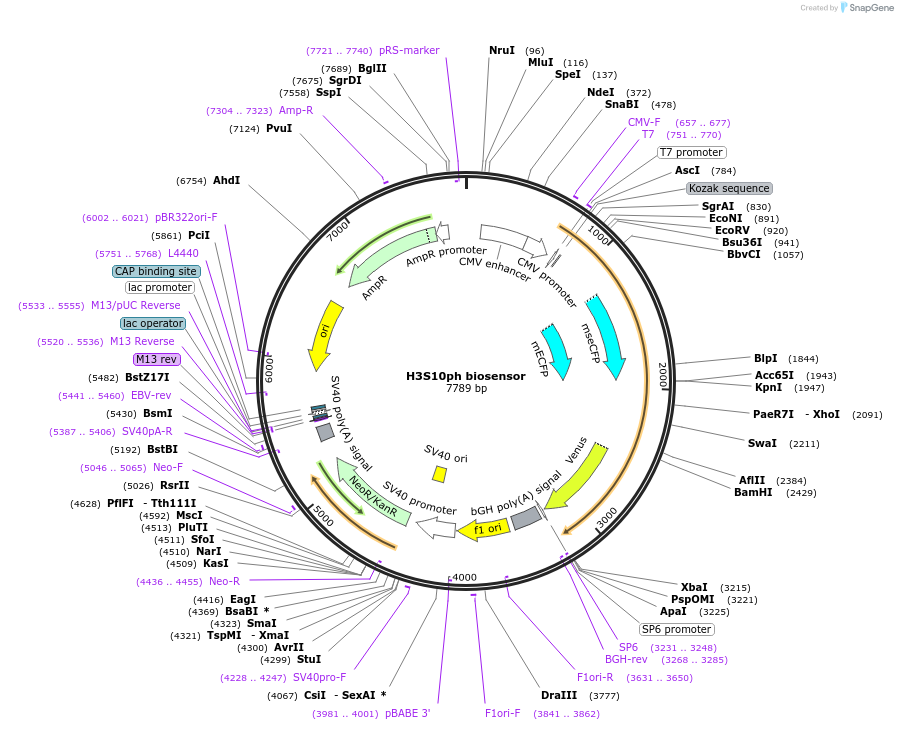
H3S10ph biosensor
Plasmid#120809PurposeFRET biosensor. To monitor histone H3 Serine 10 phosphorylation in mammalian cells by FRETDepositorInsertMouse histone H3-CFP-FHA2-histone H3 peptide (1-14aa)-YFP
ExpressionMammalianMutationThreonine 3, Threonine 6, and Threonine 11 are mu…PromoterCMVAvailable SinceFeb. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
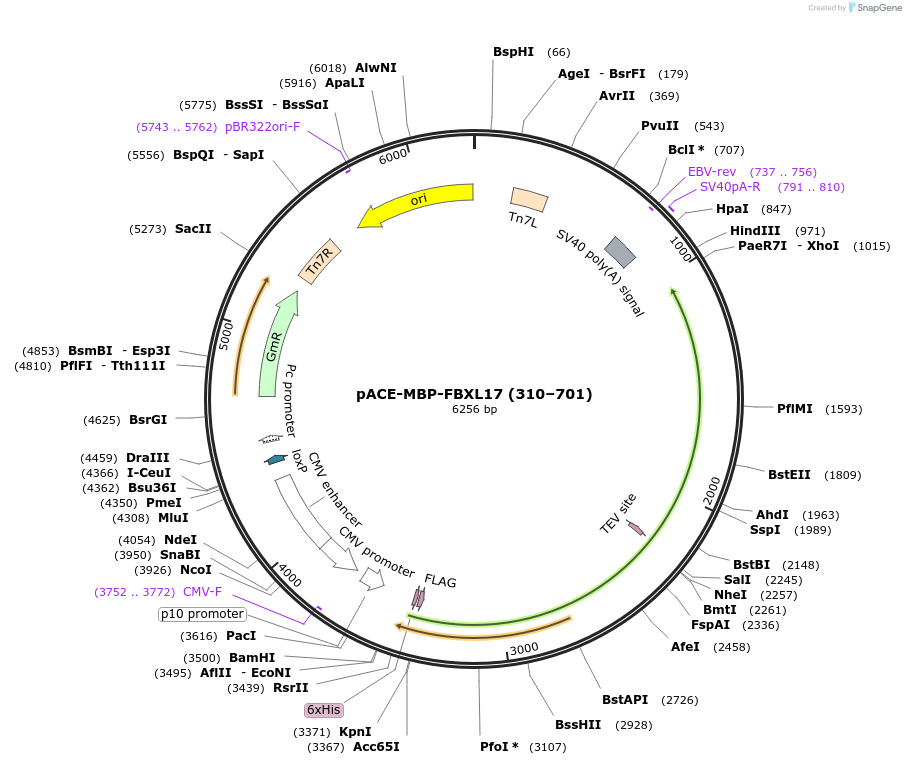
pACE-MBP-FBXL17 (310–701)
Plasmid#228369Purposeexpress human FBXL17 in insect cells, such as Trichoplusia ni Hi5DepositorAvailable SinceMay 15, 2025AvailabilityAcademic Institutions and Nonprofits only -
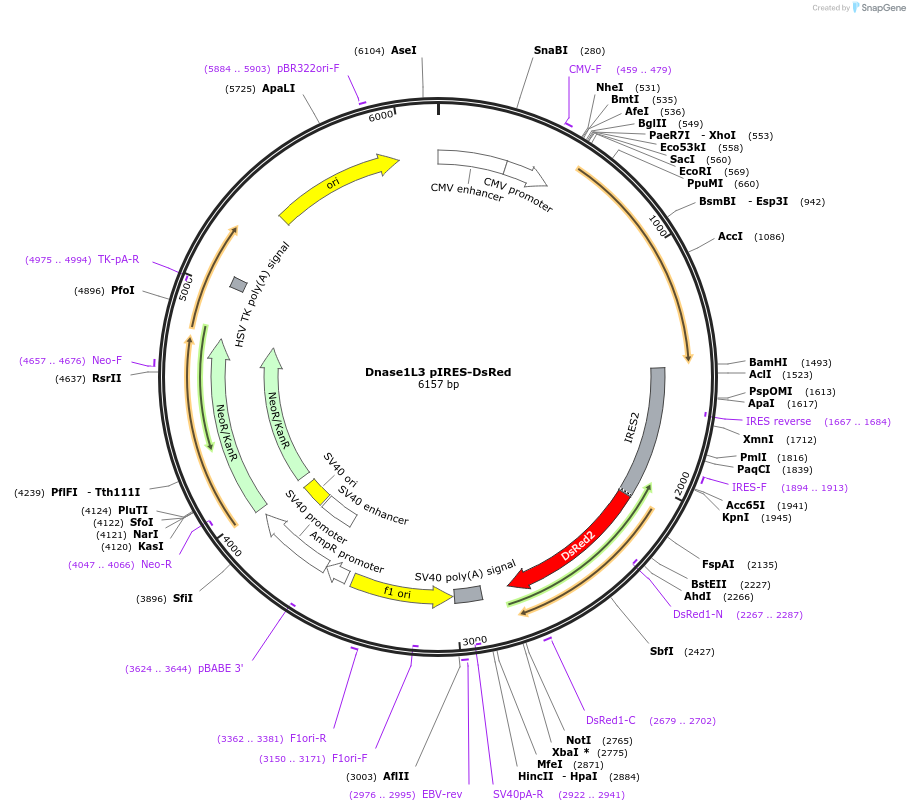
Dnase1L3 pIRES-DsRed
Plasmid#98845PurposeExpresses Human Dnase1L3 in mammalian cells along with DsRedDepositorInsertDnase1L3 (DNASE1L3 Human)
TagsIRES DsRedExpressionMammalianMutationlong isoform, wild typePromoterCMVAvailable SinceAug. 7, 2017AvailabilityAcademic Institutions and Nonprofits only -
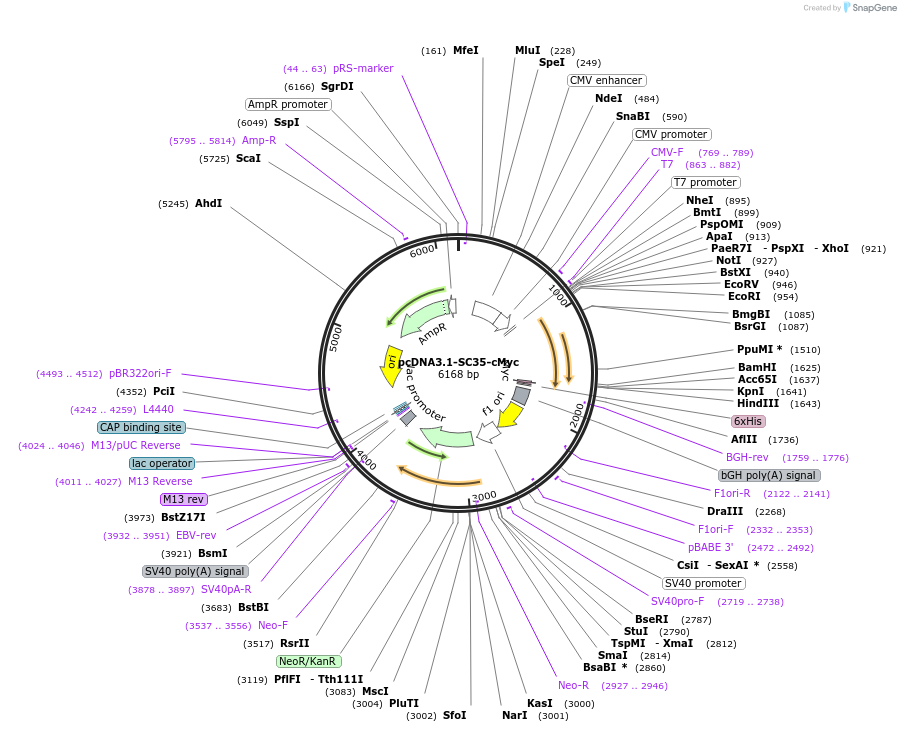
pcDNA3.1-SC35-cMyc
Plasmid#44721DepositorAvailable SinceApril 29, 2013AvailabilityAcademic Institutions and Nonprofits only -
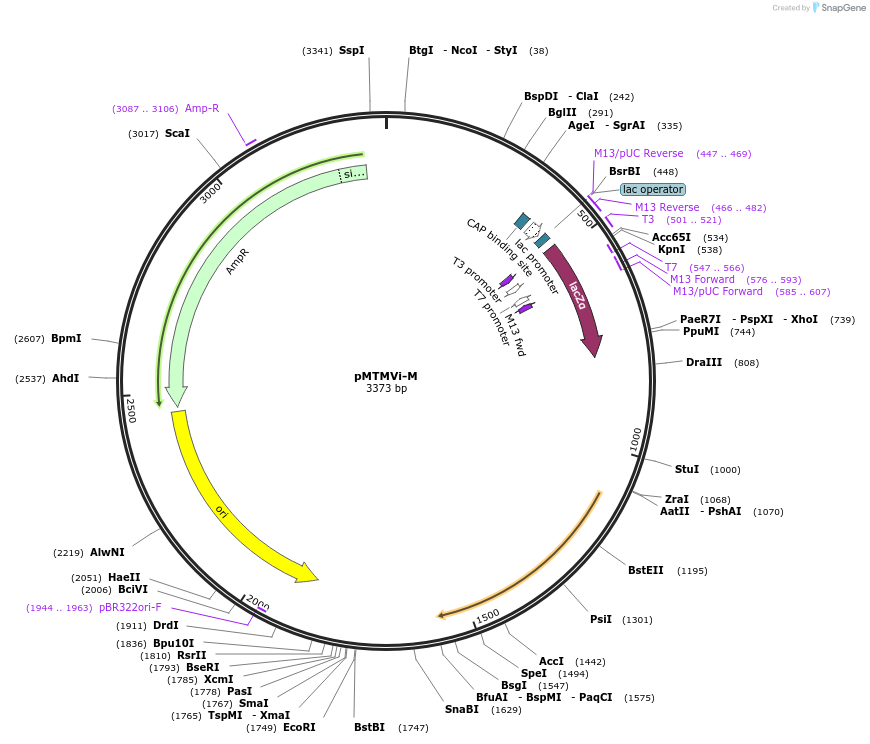
pMTMVi-M
Plasmid#118757PurposeIntermediate plasmid to build tobacco mosaic virus recombinant clones that move systemically throughout the plant.DepositorInsertTobacco mosaic virus cDNA fragment with a cloning polylinker
UseIntermediate to build tobacco mosaic virus recomb…Available SinceDec. 19, 2018AvailabilityAcademic Institutions and Nonprofits only -
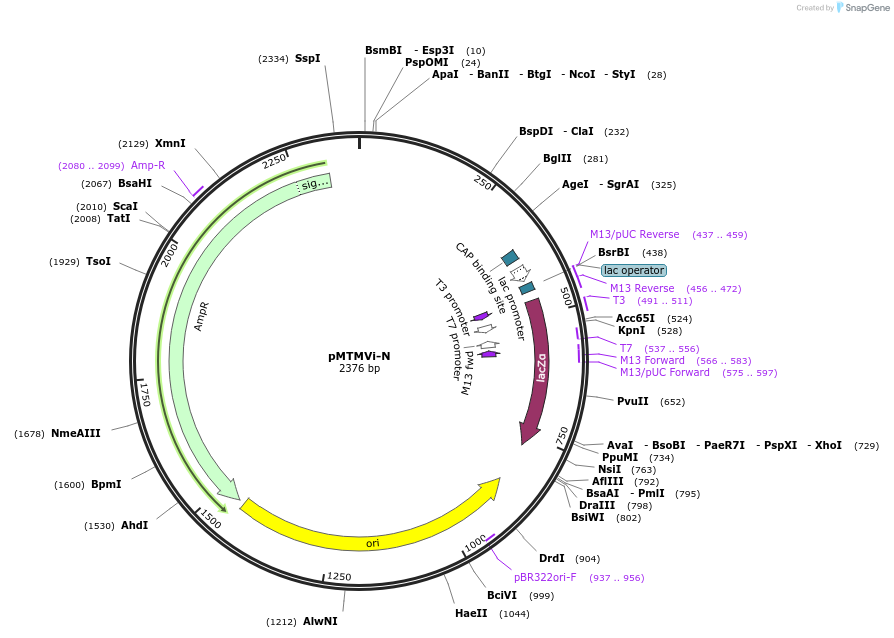
pMTMVi-N
Plasmid#118756PurposeIntermediate plasmid to build tobacco mosaic virus recombinant clones that do not move systemically throughout the plant.DepositorInsertTobacco mosaic virus cDNA fragment with a cloning polylinker
UseIntermediate to build tobacco mosaic virus recomb…Available SinceFeb. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
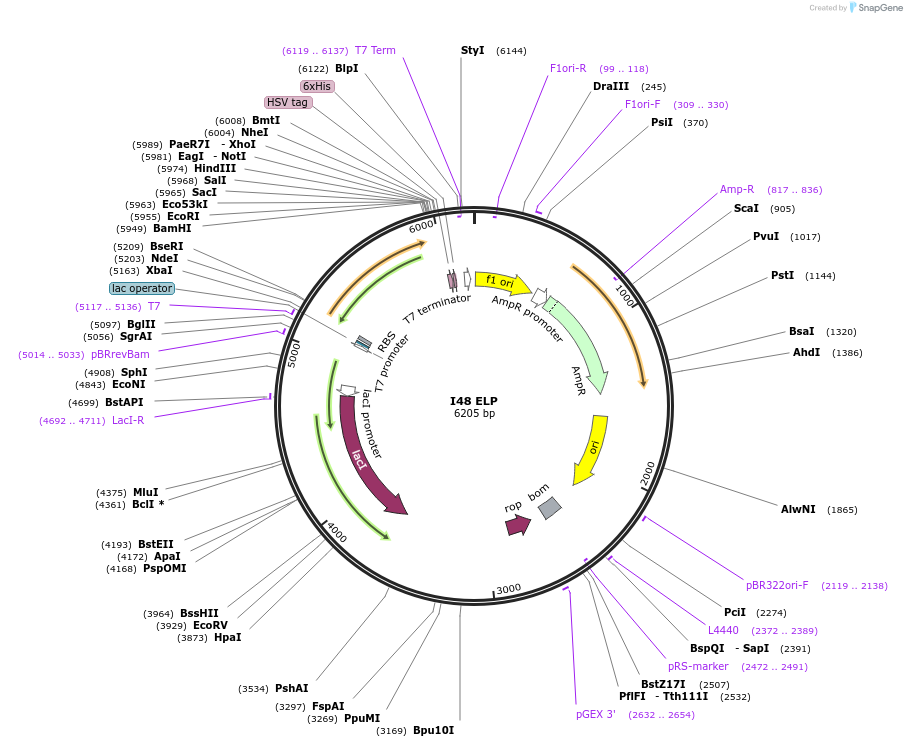
I48 ELP
Plasmid#68937PurposeElastin-like polypeptide (VPGxG) with 48 repeats of Isoleucine guest residueDepositorInsertI48 ELP
ExpressionBacterialPromoterT7Available SinceOct. 1, 2015AvailabilityAcademic Institutions and Nonprofits only -
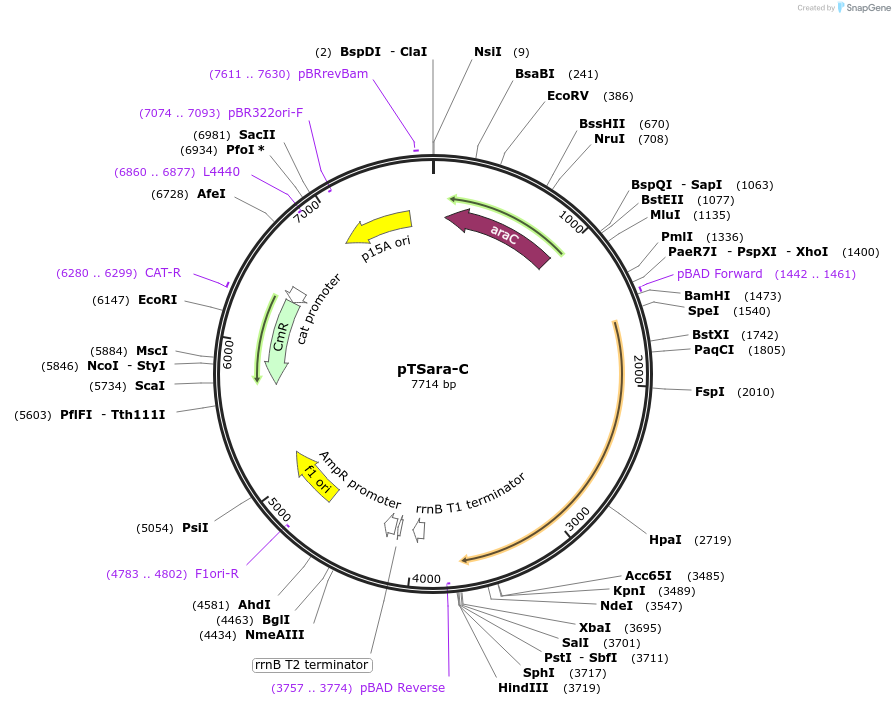
pTSara-C
Plasmid#60722PurposeExpresses the C-terminal fragment of split T7 RNAP split a position 179. Insert is driven by the arabinose inducible promoter PBAD. Contains a constitutive araC ORFDepositorInsertResidues 180-880 of T7 RNAP
UseSynthetic BiologyExpressionBacterialMutationOnly amino acids 180-880 of T7 RNAPPromoterPBADAvailable SinceNov. 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
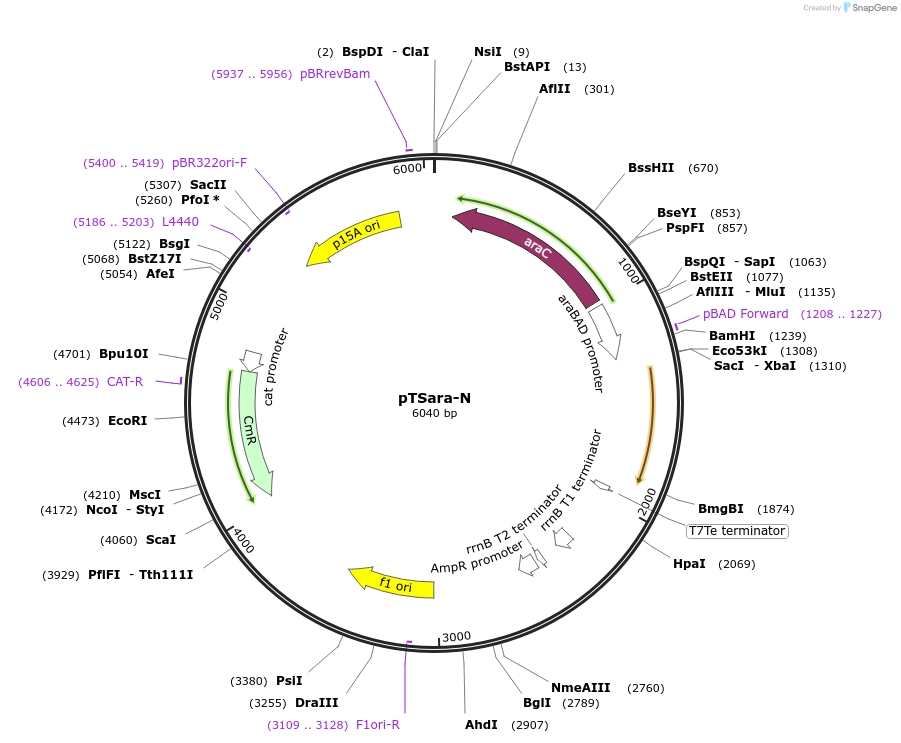
pTSara-N
Plasmid#60721PurposeExpresses the N-terminal fragment of split T7 RNAP split a position 179. Insert is driven by PBAD. Contains a constitutive araC ORFDepositorInsertResidues 1-179 of T7 RNAP
UseSynthetic BiologyExpressionBacterialMutationOnly amino acids 1-179 of T7 RNAPPromoterPBADAvailable SinceNov. 4, 2014AvailabilityAcademic Institutions and Nonprofits only -
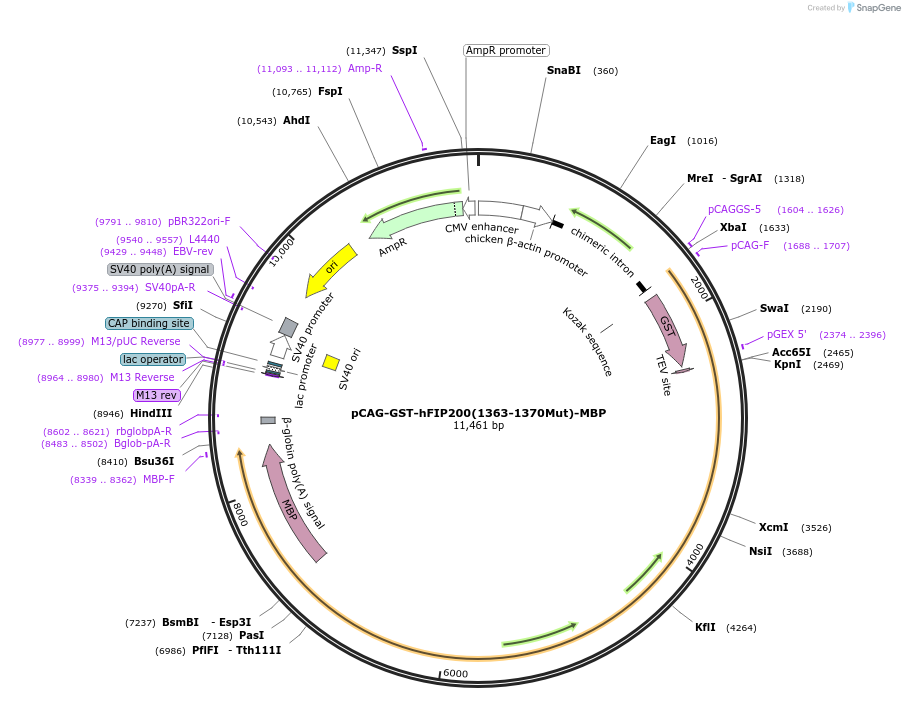
pCAG-GST-hFIP200(1363-1370Mut)-MBP
Plasmid#190038PurposeExpression of FIP200 mutantDepositorInsertFIP200 (RB1CC1 Human)
TagsGST and MBPExpressionMammalianMutation1363 RDKDLIES 1370 to GSSGGSSGPromoterCMVAvailable SinceNov. 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
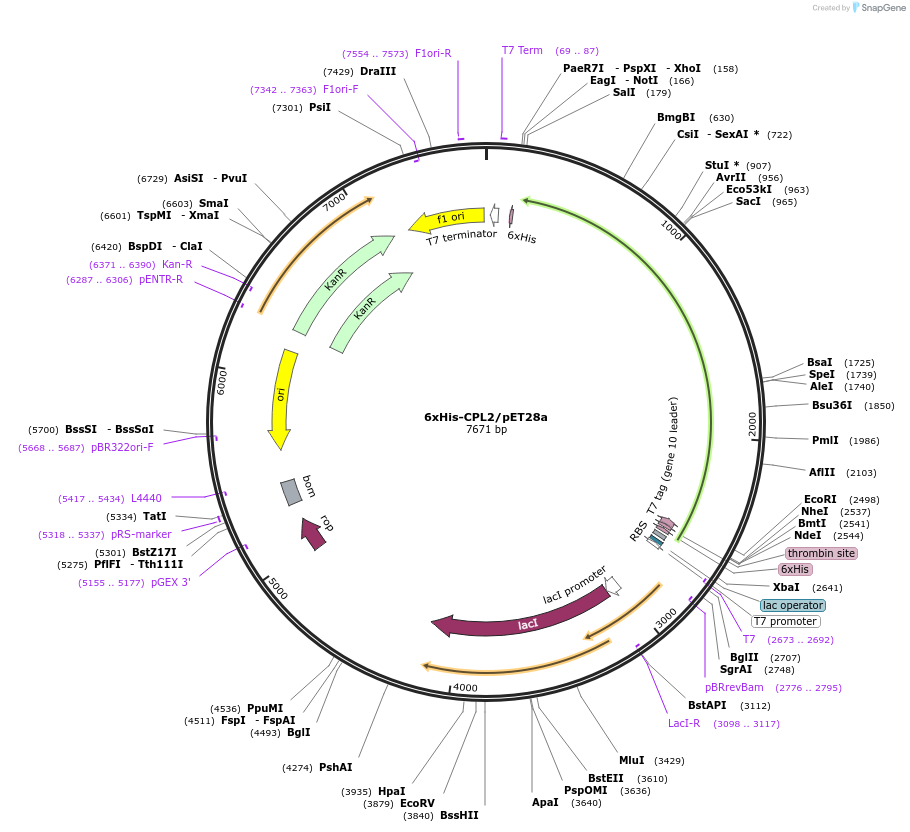
6xHis-CPL2/pET28a
Plasmid#172095PurposecDNA of CER11/CPL2 cloned into pET28a for protein expressionDepositorAvailable SinceOct. 20, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
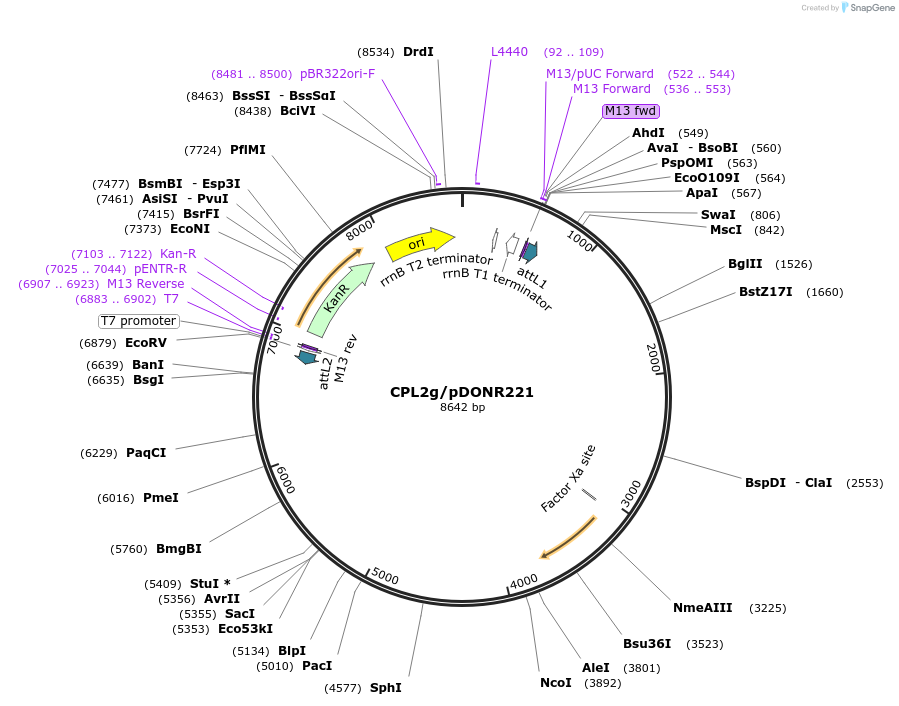
CPL2g/pDONR221
Plasmid#172093PurposeGenomic fragment containing the upstream region and coding region of CER11/CPL2 cloned into pDONR221DepositorAvailable SinceOct. 20, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
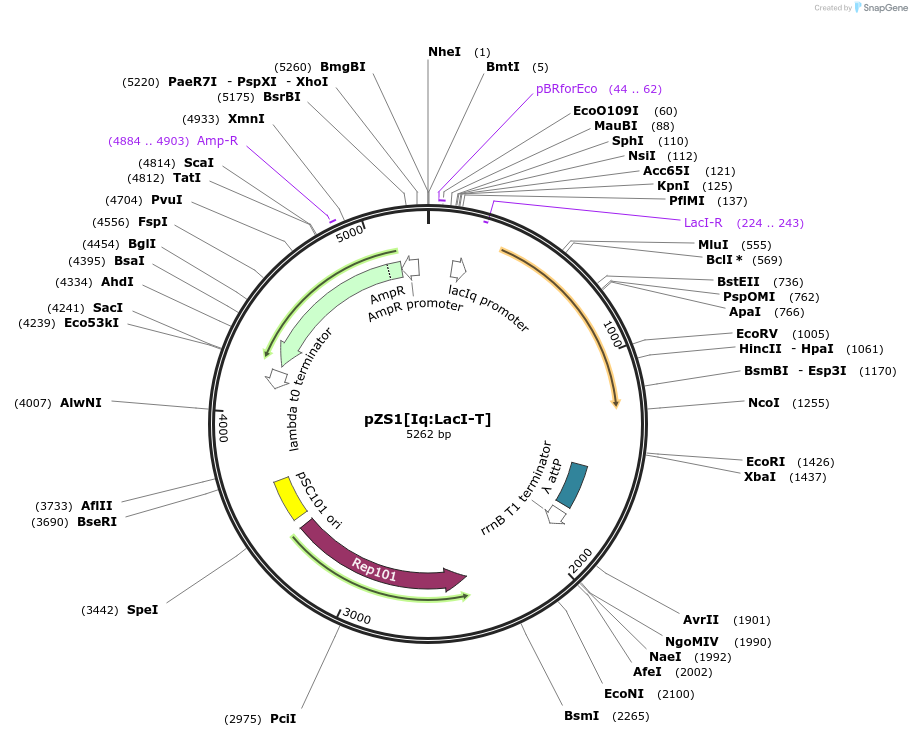
pZS1[Iq:LacI-T]
Plasmid#60756PurposeContains PIq driving expression LacI-T, the Lactose inducible chimera with the TAN DBD.DepositorInsertLacI-T
UseSynthetic BiologyExpressionBacterialMutationLacI without the tetramerization domain and the T…Available SinceJune 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
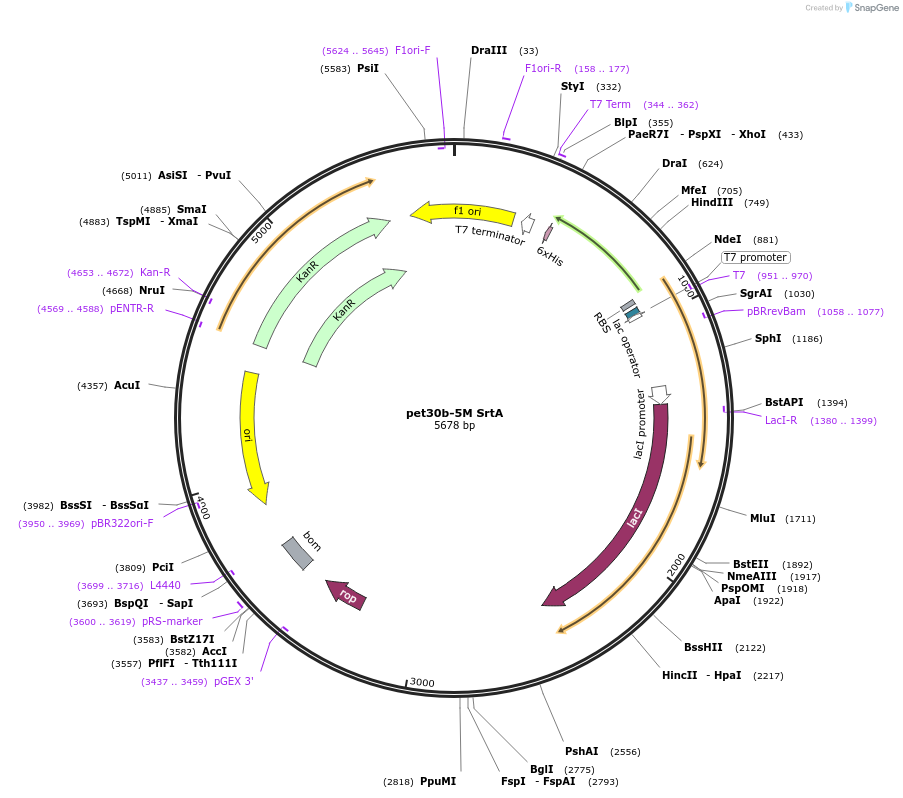
pet30b-5M SrtA
Plasmid#51140Purposeexpression plasmid for C-terminal his tagged S. aureus SrtA with enhanced catalytic activityDepositorInsertS.aureus SrtA 5M
Tags6x HisExpressionBacterialMutationP94R, D160N, D165A, K190E, K196TPromoterT7Available SinceFeb. 24, 2014AvailabilityAcademic Institutions and Nonprofits only -
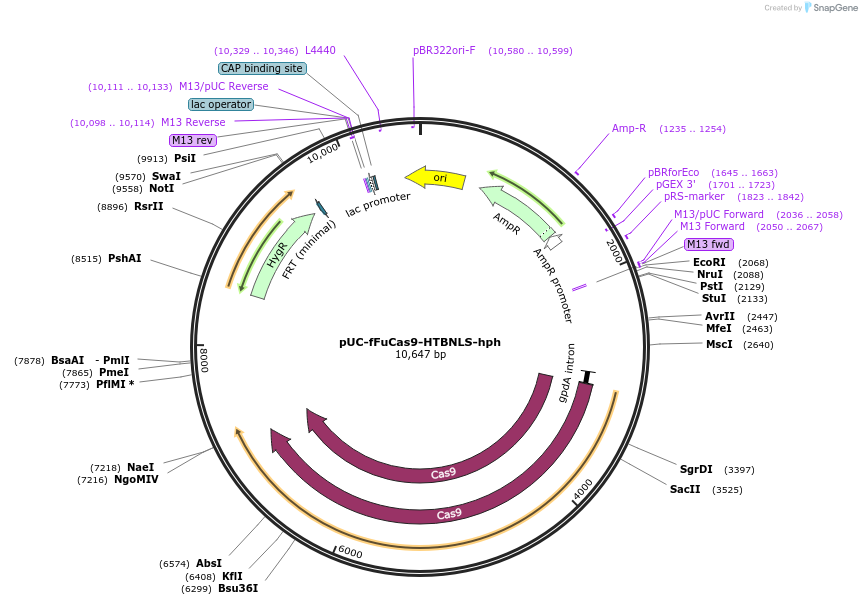
pUC-fFuCas9-HTBNLS-hph
Plasmid#121092PurposeMultigene editing in the Fusarium fujikuroi genome using the CRISPR-Cas9 systemDepositorInsertCas9 and hygromycin resist gene
UseCRISPRExpressionBacterialPromotergpdA;trpCAvailable SinceJuly 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
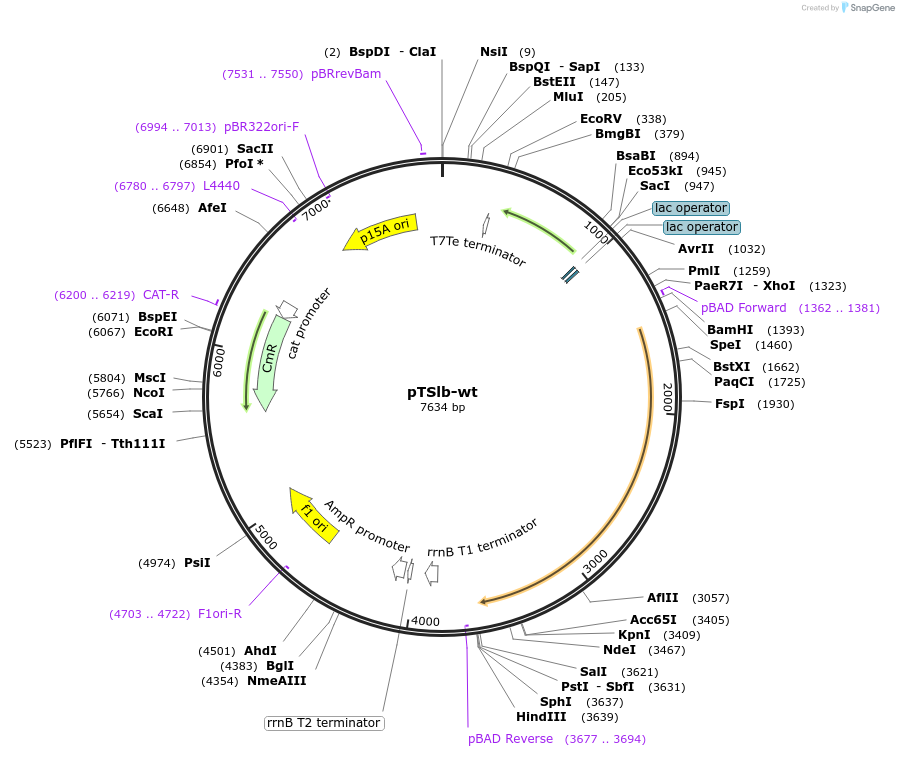
pTSlb-wt
Plasmid#60728PurposeExpresses both fragments of T7 RNAP split at position 179. The N-terminal fragment is driven by PLac while the C-terminal fragment is driven by PBAD.DepositorInsertsResidues 1-179 of split T7 RNAP
Residues 180-880 of split T7 RNAP
UseSynthetic BiologyExpressionBacterialMutationResidues 1-180 of split T7 RNAP and Residues 180-…Available SinceNov. 4, 2014AvailabilityAcademic Institutions and Nonprofits only