We narrowed to 5,133 results for: codon optimized
-
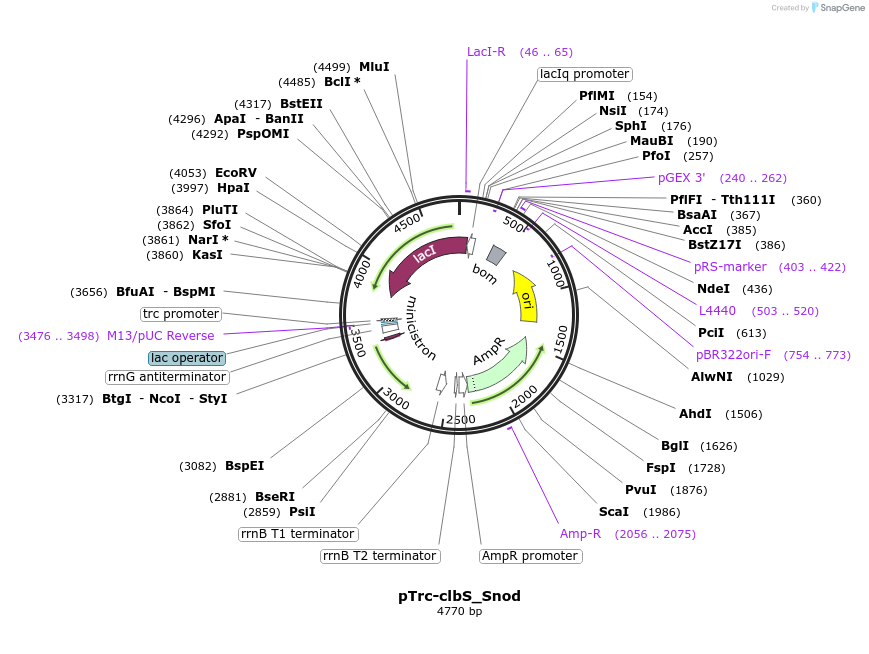
Plasmid#223355PurposeEncoding ClbS from Snodgrassella alvi wkB2DepositorInsertEncoding ClbS from Snodgrassella alvi wkB2 (Codon optimized for E coli)
ExpressionBacterialPromoterpTrcAvailable SinceNov. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
pTrc-clbS_Mixta
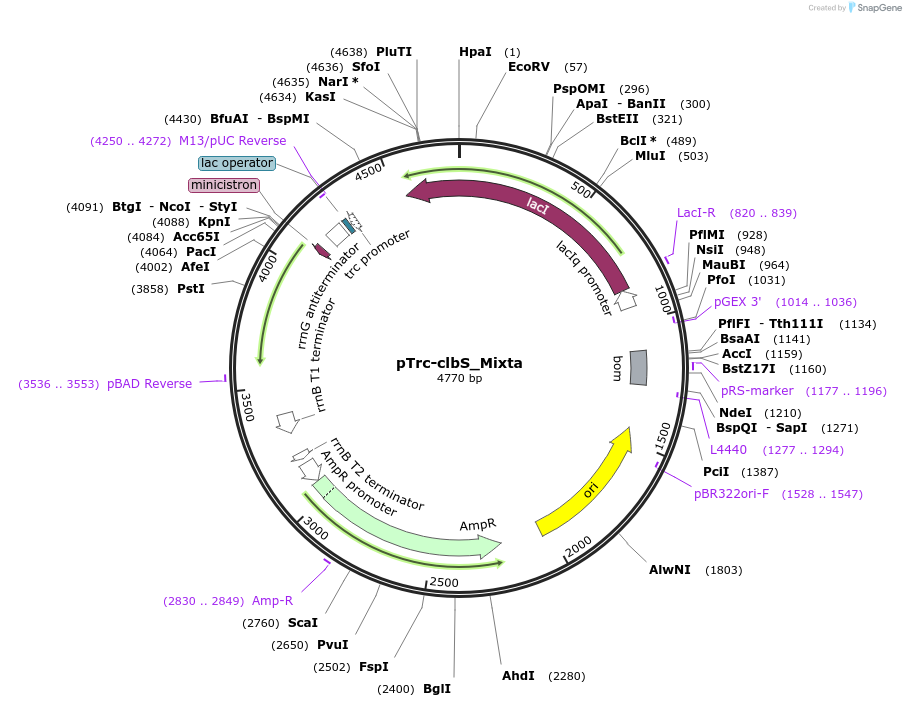
Plasmid#223352PurposeEncoding ClbS from Mixta theicolaDepositorInsertEncoding ClbS from Mixta theicola (Codon optimized for E coli)
ExpressionBacterialPromoterpTrcAvailable SinceNov. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
pTrc-clbS_Dickeya
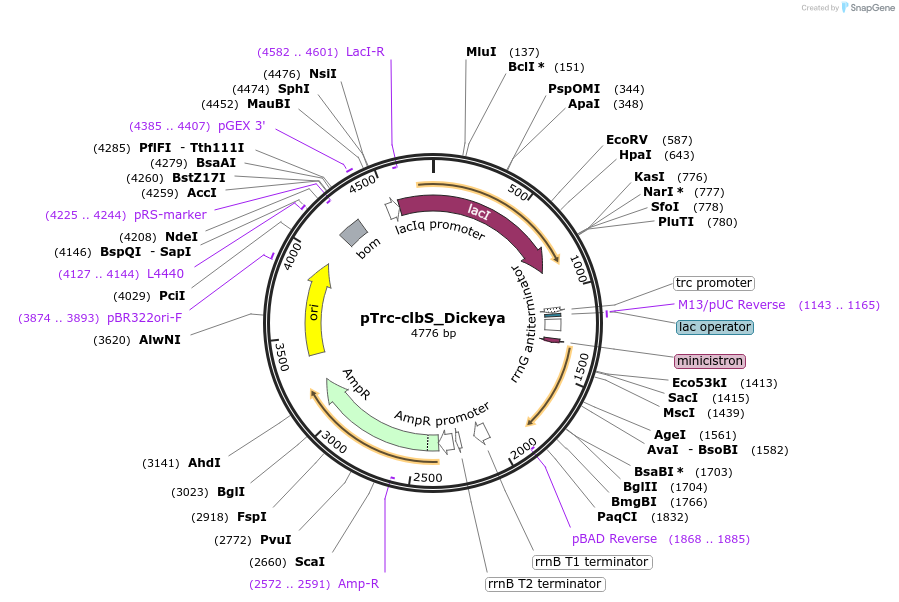
Plasmid#223351PurposeEncoding ClbS from Dickeya dadantiiDepositorInsertEncoding ClbS from Dickeya dadantii (Codon optimized for E coli) (CO076_RS11420 Dickeya dadantii)
ExpressionBacterialPromoterpTrcAvailable SinceNov. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
pTrc-clbS
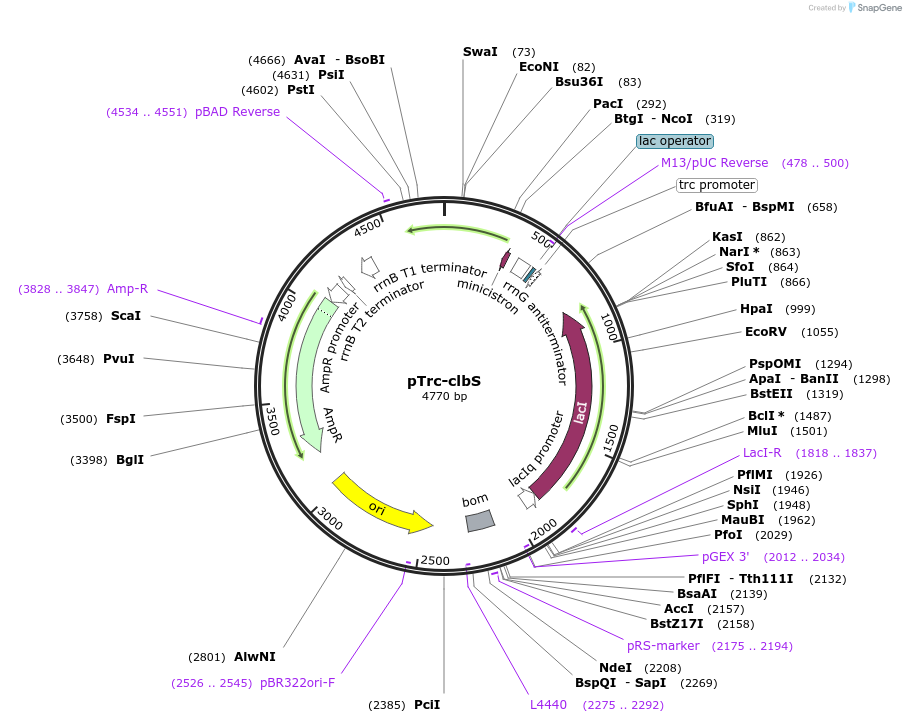
Plasmid#223350PurposeEncoding wild type ClbS from E. coli CFT073DepositorInsertClbS from CFT073 (Codon optimized for E coli)
ExpressionBacterialPromoterpTrcAvailable SinceNov. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
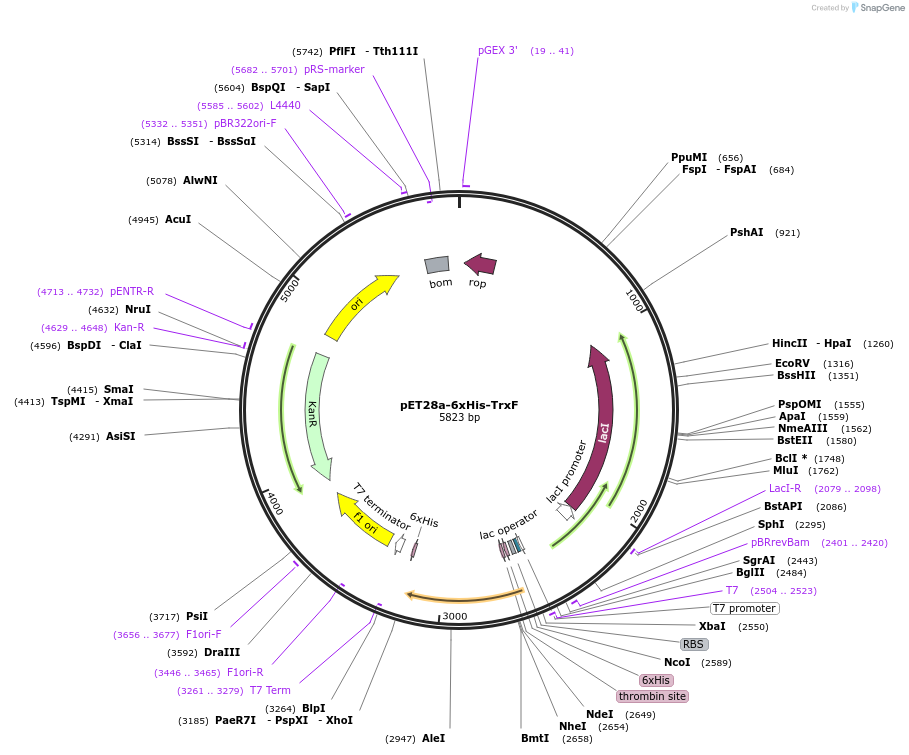
pET28a-6xHis-TrxF
Plasmid#207454PurposeE. coli expression of Mesembryanthemum crystallinum Thioredoxin F with thrombin-cleavable N-terminal 6xHis tagDepositorInsertThioredoxin F
Tags6xHisExpressionBacterialMutationCodon optimized for E coli expressionPromoterT7Available SinceJan. 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
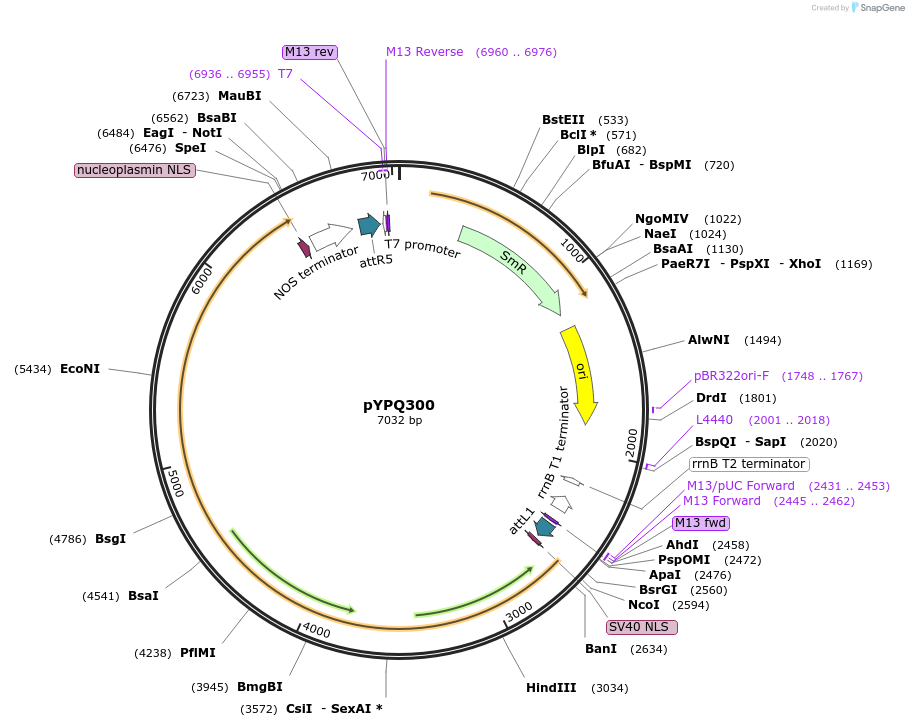
pYPQ300
Plasmid#205414PurposeGateway entry plasmid (attL1 & attR5) expressing rice codon optimized Ev1Cas12a without promoterDepositorInsertEv1Cas12a
UseCRISPR; Gateway compatible cas12a entry cloneTagsNLS (N terminal on insert) NLS (C terminal on ins…ExpressionPlantAvailable SinceNov. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
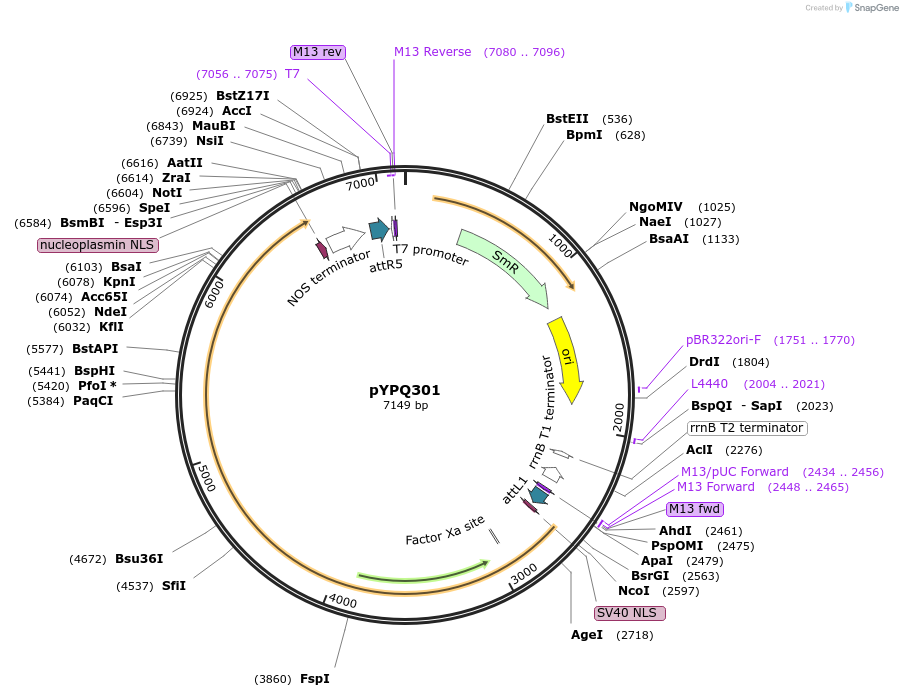
pYPQ301
Plasmid#205415PurposeGateway entry plasmid (attL1 & attR5) expressing rice codon optimized Hs1Cas12a without promoterDepositorInsertHs1Cas12a
UseCRISPR; Gateway compatible cas12a entry cloneExpressionPlantAvailable SinceNov. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
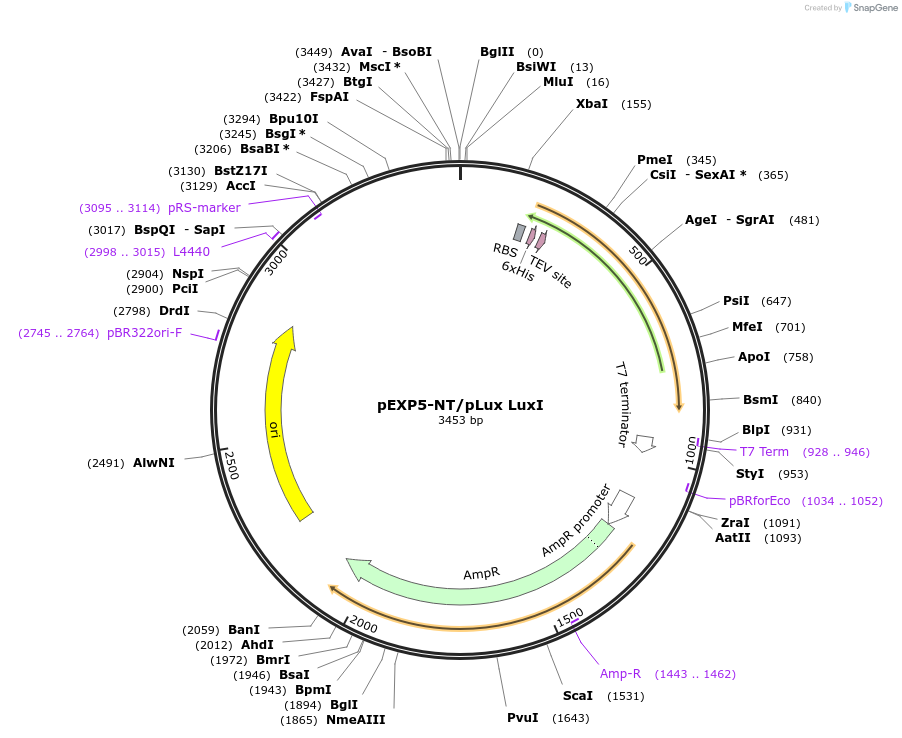
pEXP5-NT/pLux LuxI
Plasmid#193627PurposeInducible expression of LuxI by the transcription factor complex LuxR-3OC6 homoserine lactone (HSL). LuxI is codon optimized for E. coli.DepositorInsertLuxI
Tags6xHisExpressionBacterialPromoterLux promoterAvailable SinceJuly 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
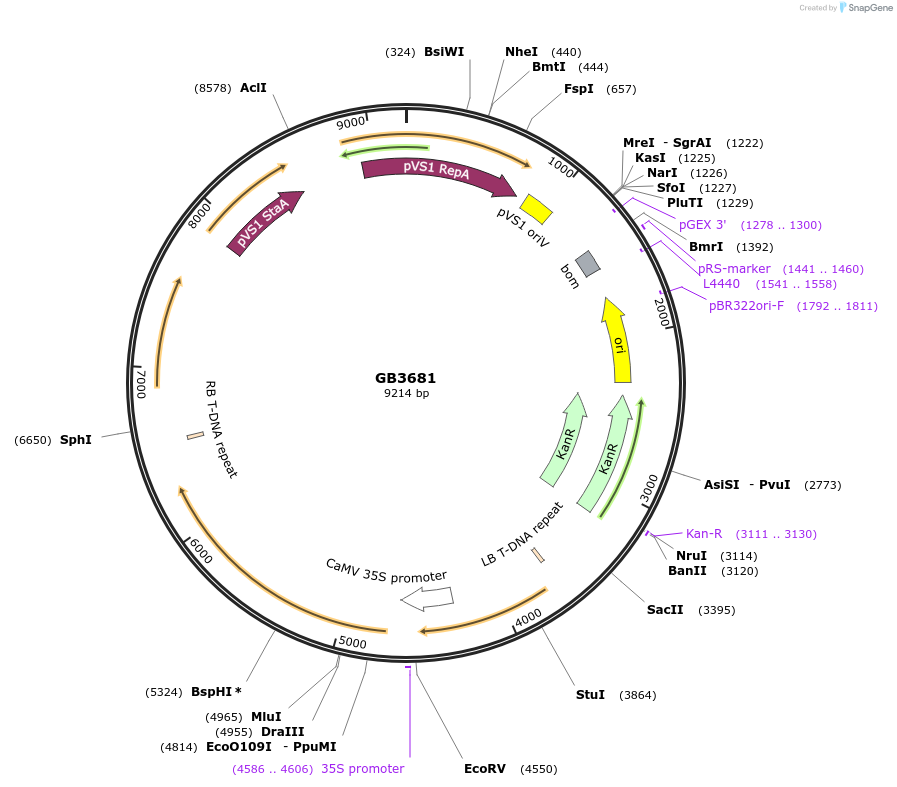
GB3681
Plasmid#187806PurposeTranscriptional unit for expression of alcohol O-acetyltransferase from Saccharomyces cerevisiae S288C, codon optimized for Nicotiana.DepositorInsertScATF1
ExpressionPlantAvailable SinceJune 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
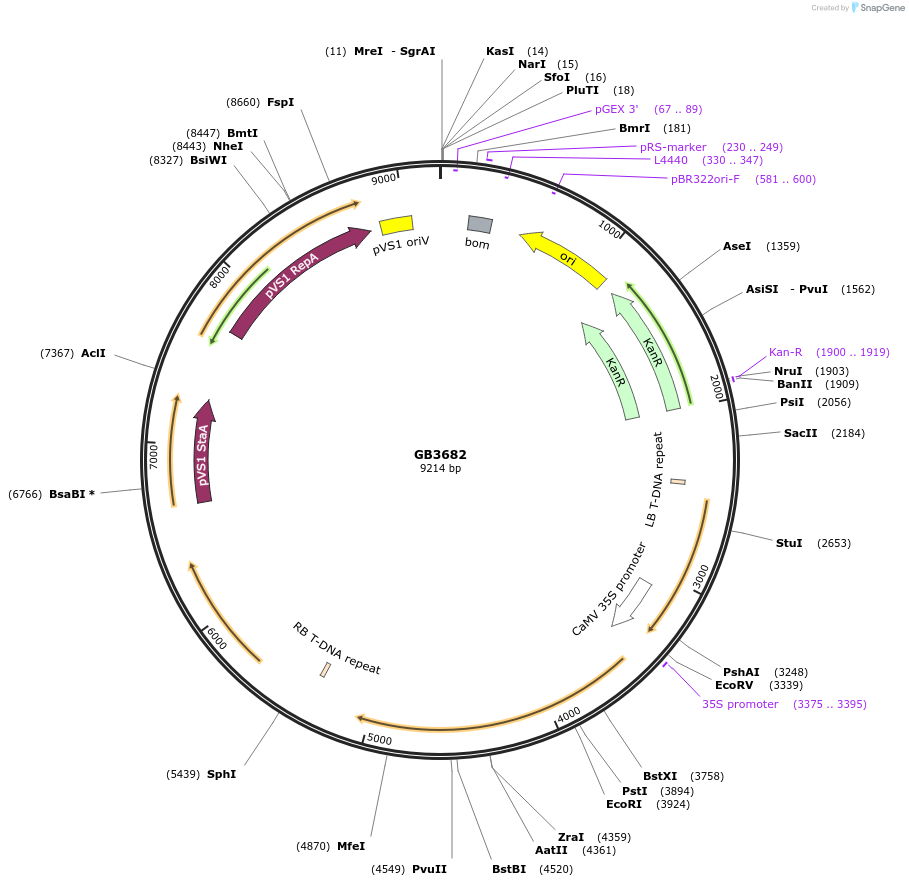
GB3682
Plasmid#187807PurposeTranscriptional unit for expression of alcohol O-acetyltransferase from Saccharomyces pastorianus strain CBS 1483 chromosome SeVIII-SeXV, codon optimized for Nicotiana.DepositorInsertSpATF1-2
ExpressionPlantAvailable SinceJune 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
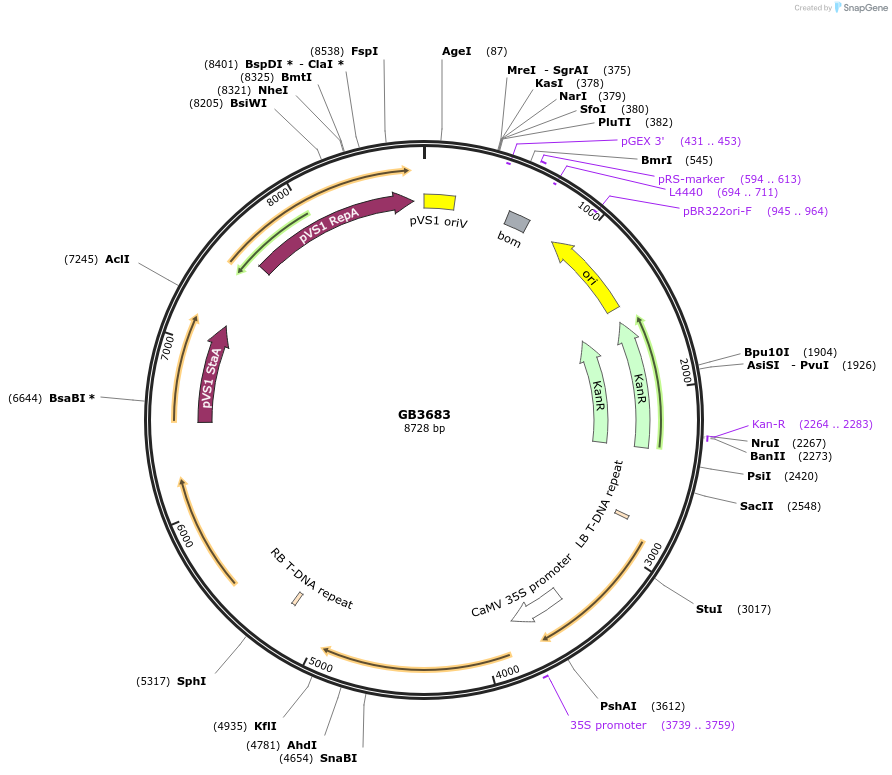
GB3683
Plasmid#187808PurposeTranscriptional unit for expression of 1,2-diacyl-sn-glycerol:acetyl-CoA acetyltransferase from Euonymus fortunei, codon optimized for Nicotiana.DepositorInsertEfDAct
ExpressionPlantAvailable SinceJune 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
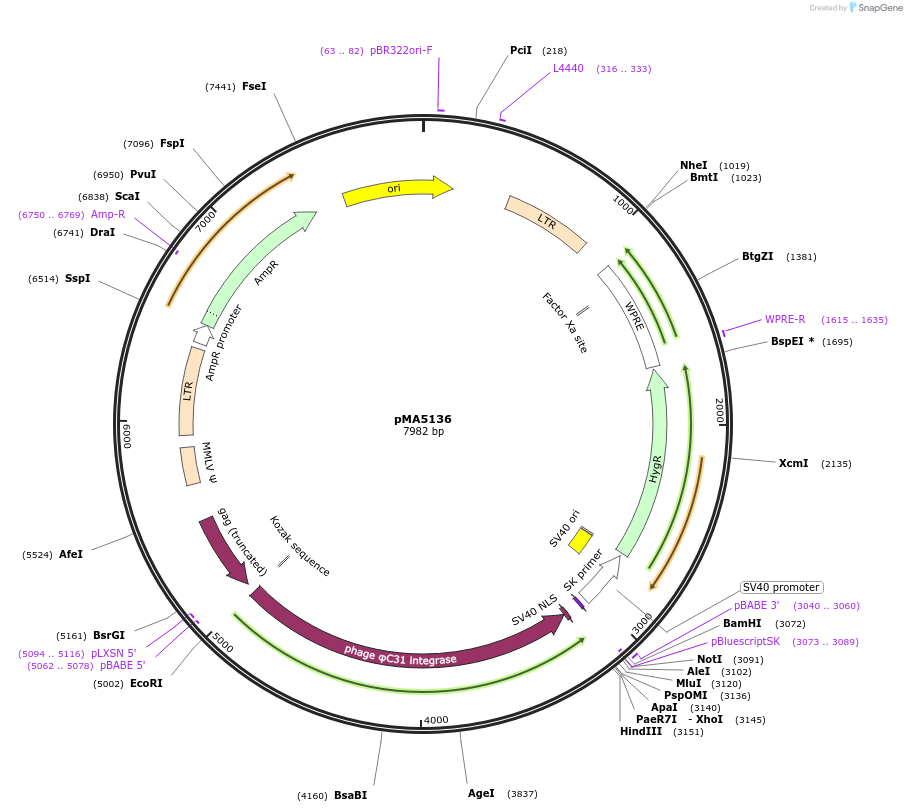
pMA5136
Plasmid#184853PurposeA retroviral vector encoding hygromycin resistance and PhiC31 recombinaseDepositorInsertPhiC31o recombinase
UseRetroviral; Phic31 attp-attb recombination/excisi…ExpressionMammalianMutationCodon-optimized for E. coli expressionPromoterLTRAvailable SinceAug. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
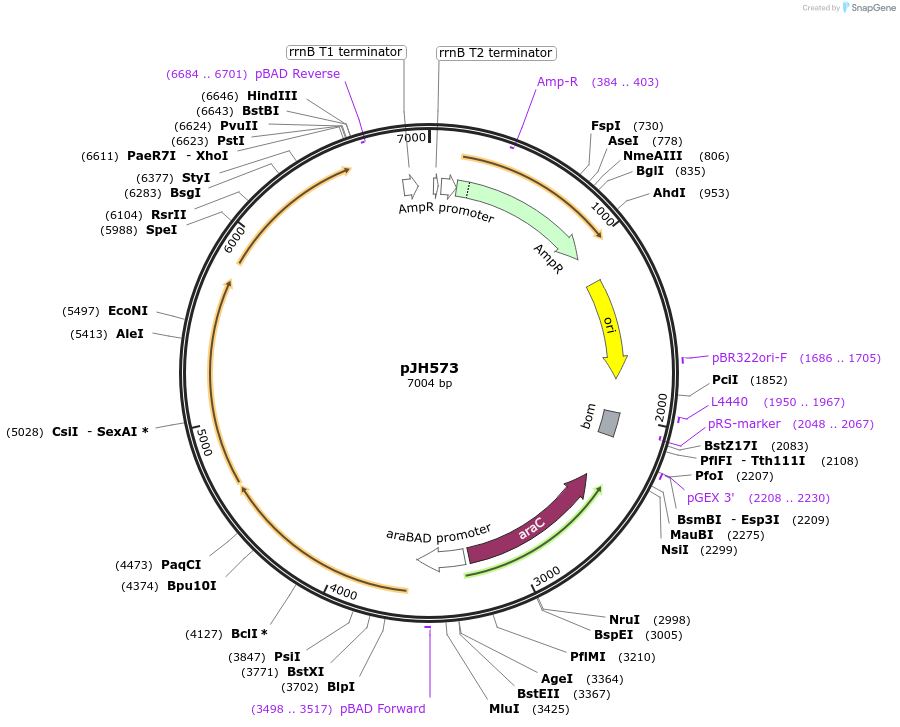
pJH573
Plasmid#172157PurposepylBCD variant in pBAD backboneDepositorInsertPara (Untagged pylBCD); AraC
ExpressionBacterialMutationCodon-optimized Methanosarcina acetivorans pylBCDAvailable SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
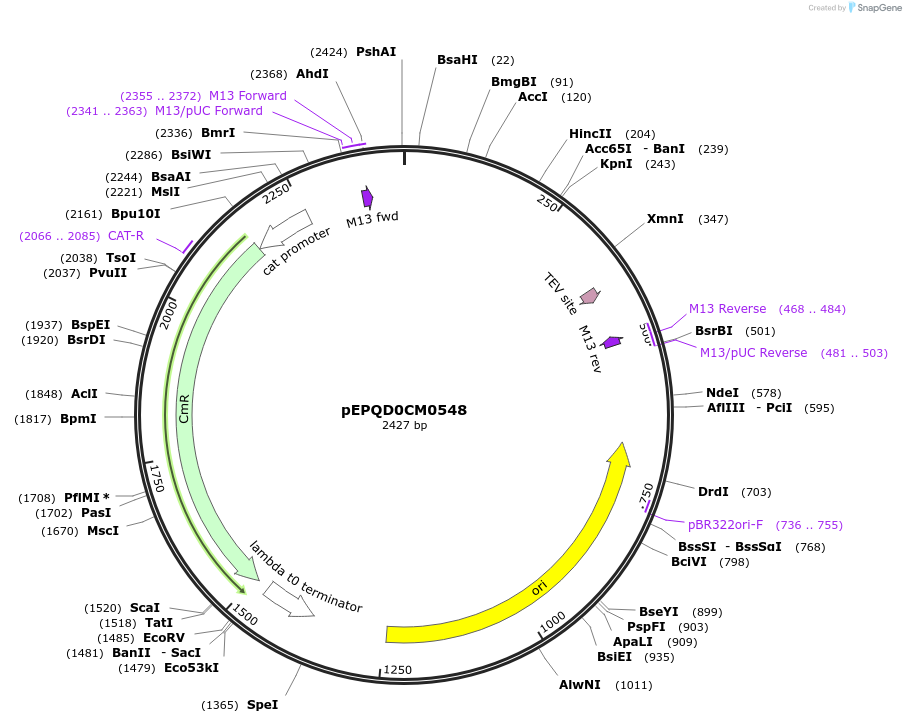
pEPQD0CM0548
Plasmid#162299PurposeLevel 0 N-terminal tag part for MoClo assembly, CCAT-AATGDepositorInsertN-terminal tag, HiBiT-TrxA-[TEV cleavage site]
UseSynthetic BiologyMutationcodon optimized for E. coliAvailable SinceJan. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
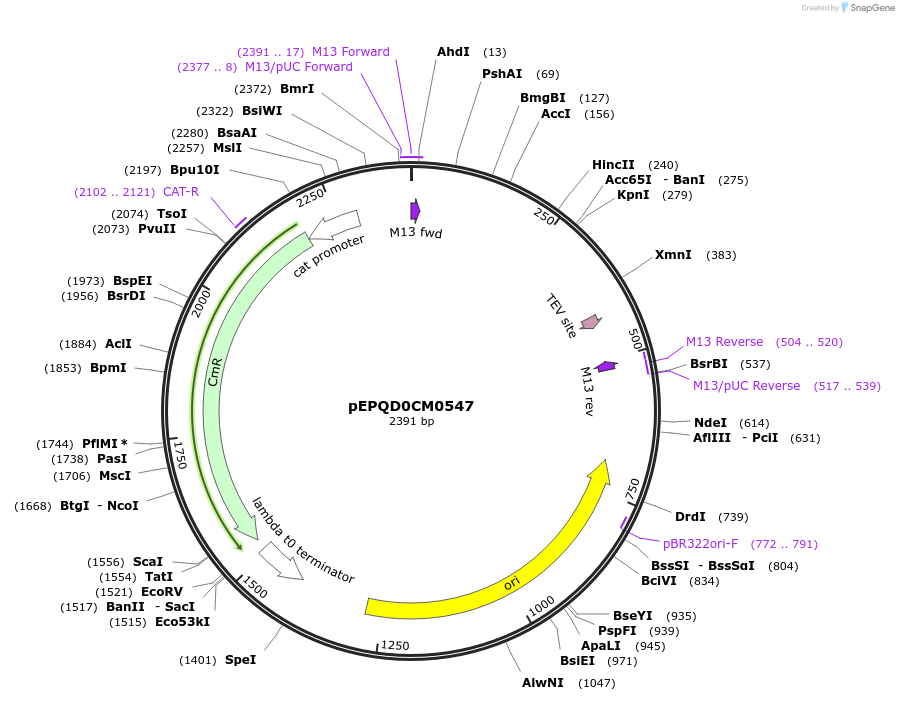
pEPQD0CM0547
Plasmid#162298PurposeLevel 0 N-terminal tag part for MoClo assembly, CCAT-AATGDepositorInsertN-terminal tag, TrxA-[TEV cleavage site]
UseSynthetic BiologyMutationcodon optimized for E. coliAvailable SinceDec. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
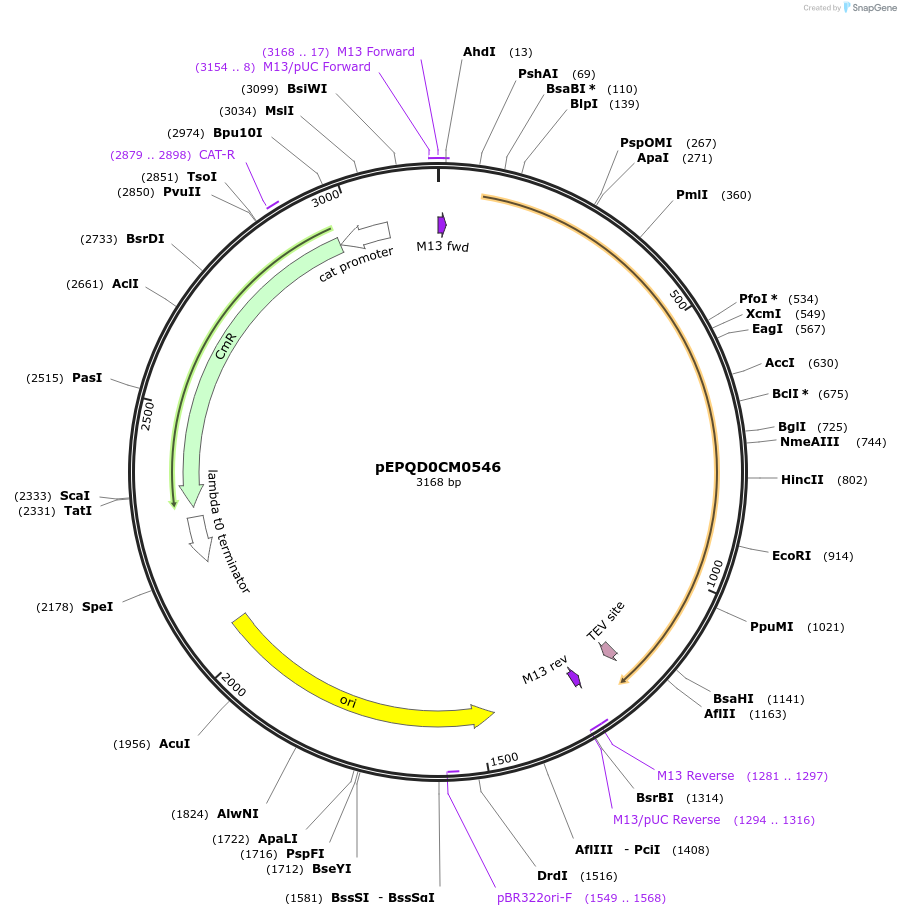
pEPQD0CM0546
Plasmid#162297PurposeLevel 0 N-terminal tag part for MoClo assembly, CCAT-AATGDepositorInsertN-terminal tag, MBP-[TEV cleavage site]
UseSynthetic BiologyMutationcodon optimized for E. coliAvailable SinceDec. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
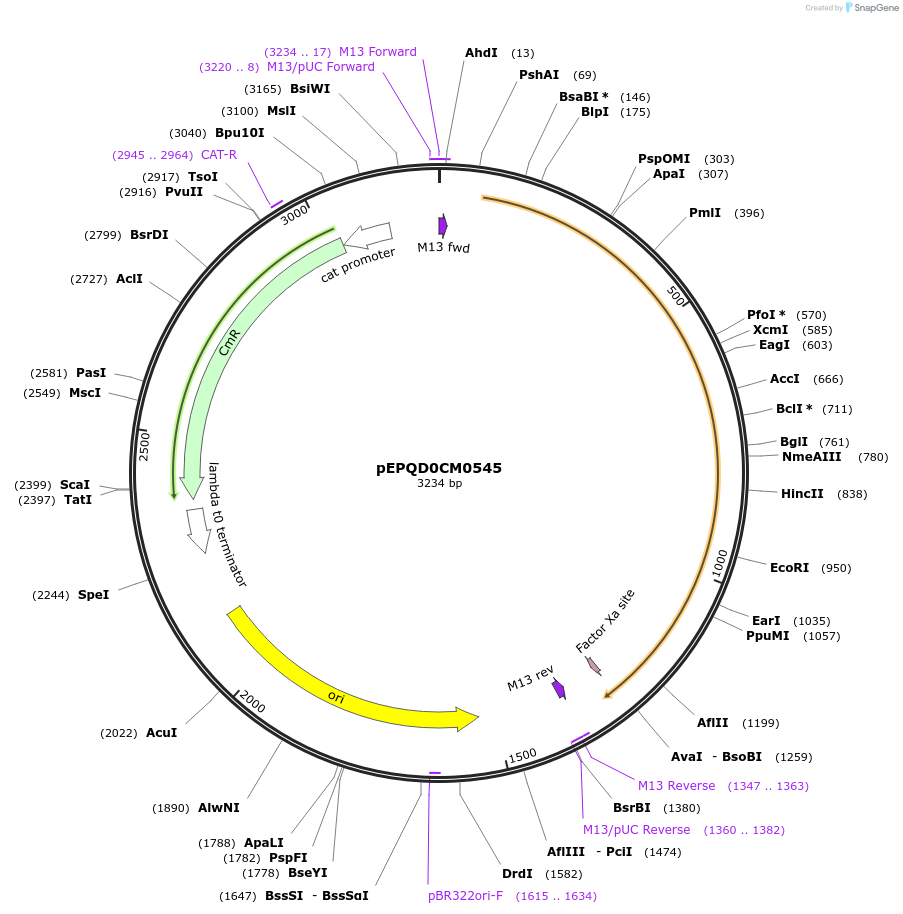
pEPQD0CM0545
Plasmid#162296PurposeLevel 0 N-terminal tag part for MoClo assembly, CCAT-AATGDepositorInsertN-terminal tag, HiBiT-MBP-[Factor Xa cleavage site]
UseSynthetic BiologyMutationcodon optimized for E. coliAvailable SinceDec. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
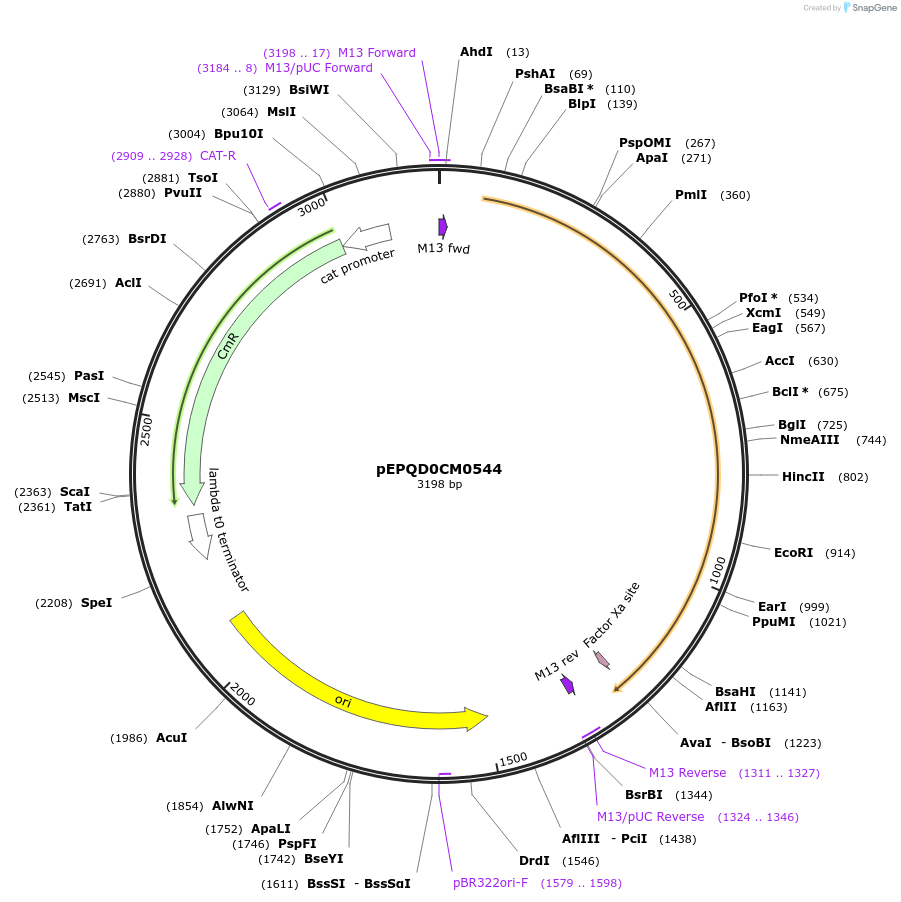
pEPQD0CM0544
Plasmid#162295PurposeLevel 0 N-terminal tag part for MoClo assembly, CCAT-AATGDepositorInsertN-terminal tag, MBP-[Factor Xa cleavage site]
UseSynthetic BiologyMutationcodon optimized for E. coliAvailable SinceDec. 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
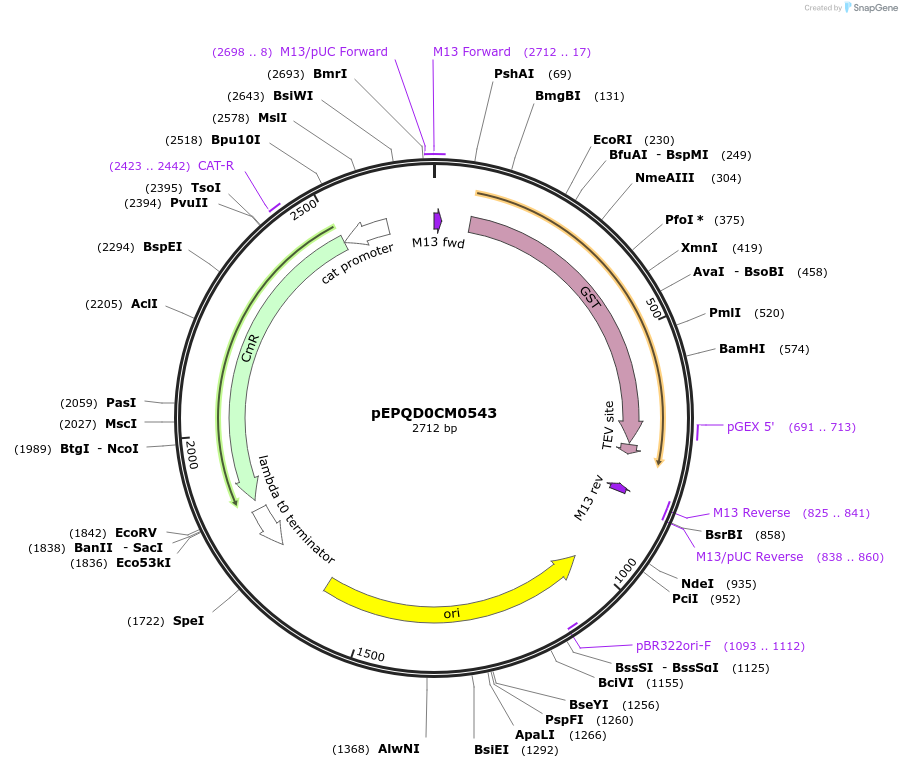
pEPQD0CM0543
Plasmid#162294PurposeLevel 0 N-terminal tag part for MoClo assembly, CCAT-AATGDepositorInsertN-terminal tag, GST-[TEV cleavage site]
UseSynthetic BiologyMutationcodon optimized for E. coliAvailable SinceDec. 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
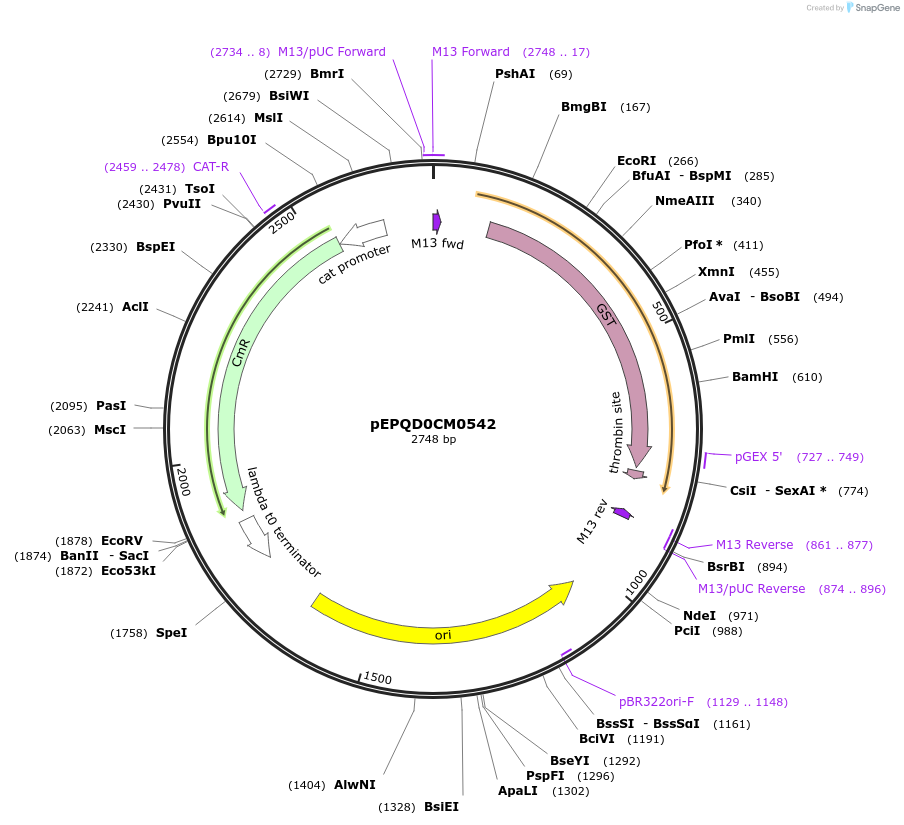
pEPQD0CM0542
Plasmid#162293PurposeLevel 0 N-terminal tag part for MoClo assembly, CCAT-AATGDepositorInsertN-terminal tag, HiBiT-GST-[thrombin cleavage site]
UseSynthetic BiologyMutationcodon optimized for E. coliAvailable SinceDec. 2, 2020AvailabilityAcademic Institutions and Nonprofits only