We narrowed to 10,180 results for: yeast
-
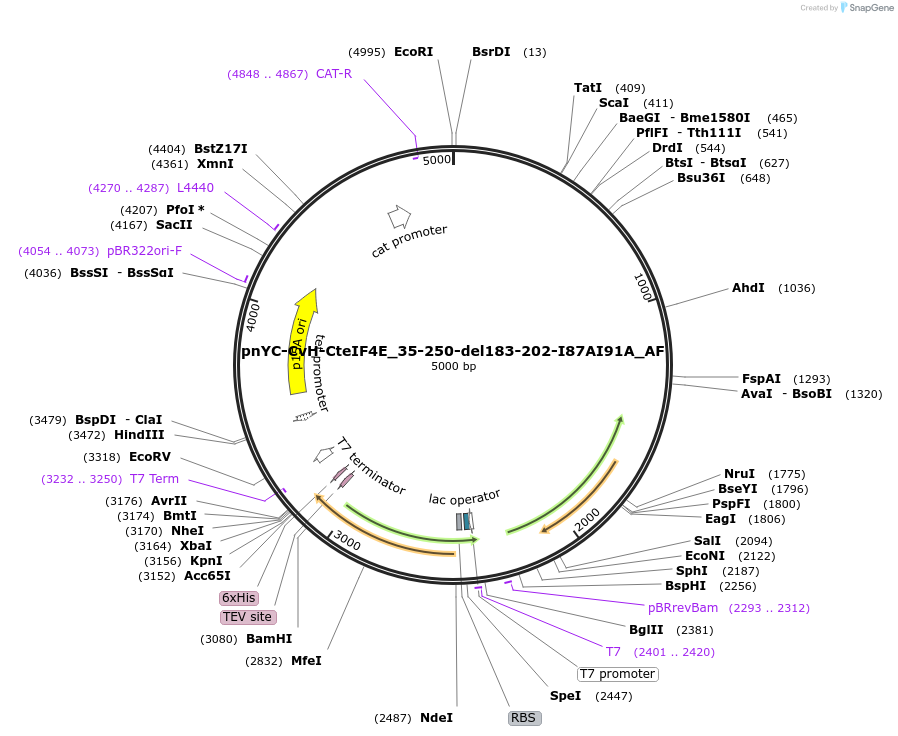
Plasmid#148722PurposeBacterial Expression of CteIF4E_35-250-del183-202-I87AI91ADepositorInsertCteIF4E_35-250-del183-202-I87AI91A
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
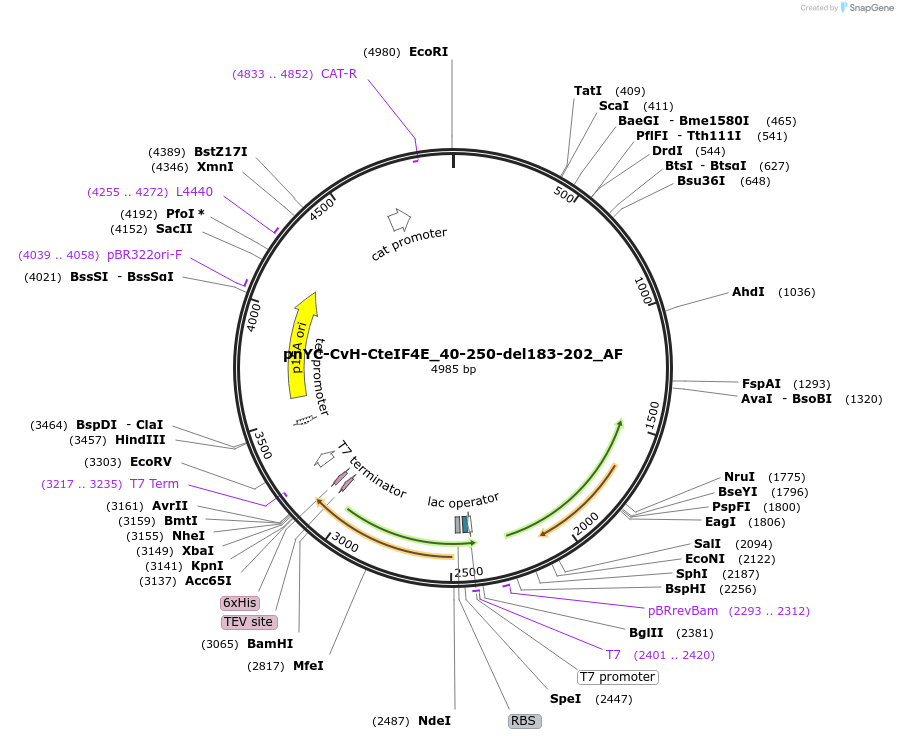
pnYC-CvH-CteIF4E_40-250-del183-202_AF
Plasmid#148723PurposeBacterial Expression of CteIF4E_40-250-del183-202DepositorInsertCteIF4E_40-250-del183-202
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
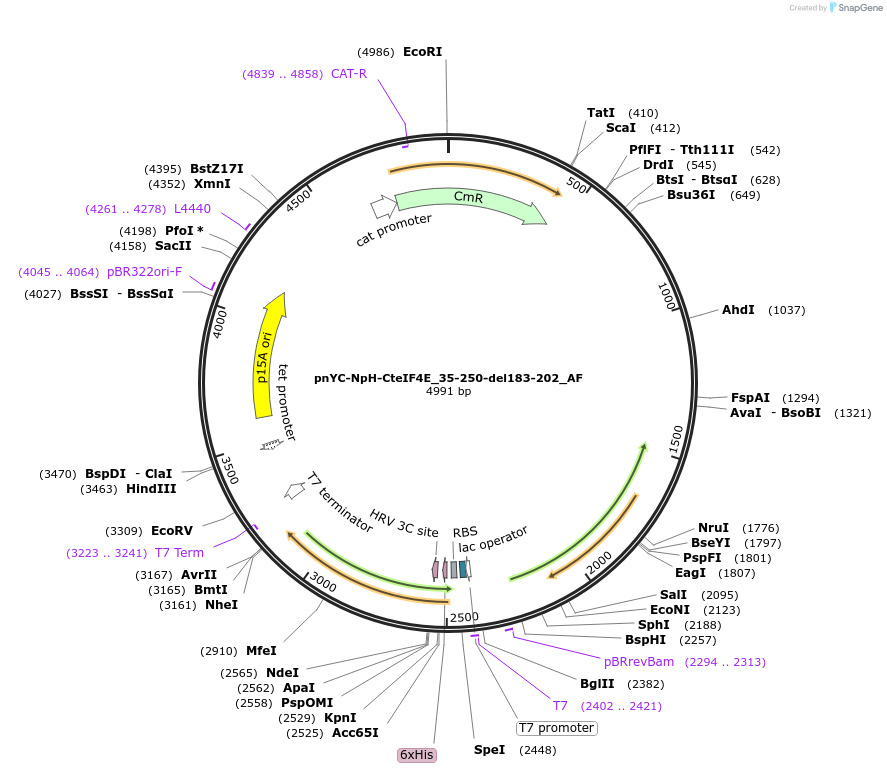
pnYC-NpH-CteIF4E_35-250-del183-202_AF
Plasmid#148725PurposeBacterial Expression of CteIF4E_35-250-del183-202DepositorInsertCteIF4E_35-250-del183-202
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
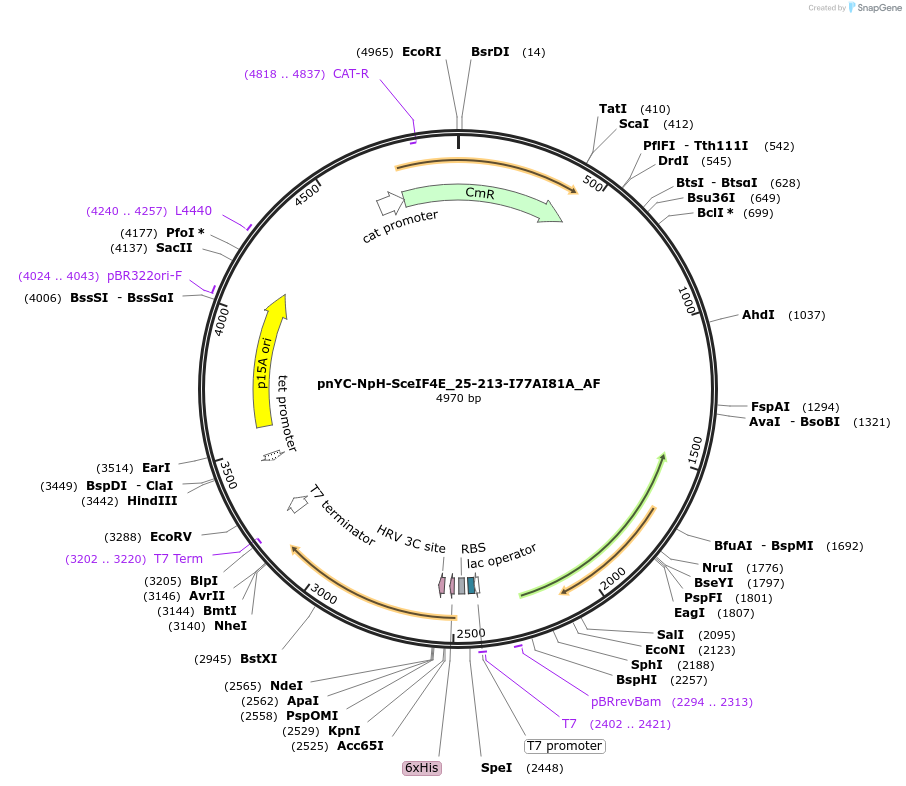
pnYC-NpH-SceIF4E_25-213-I77AI81A_AF
Plasmid#148727PurposeBacterial Expression of SceIF4E_25-213-I77AI81ADepositorInsertSceIF4E_25-213-I77AI81A (CDC33 Budding Yeast)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
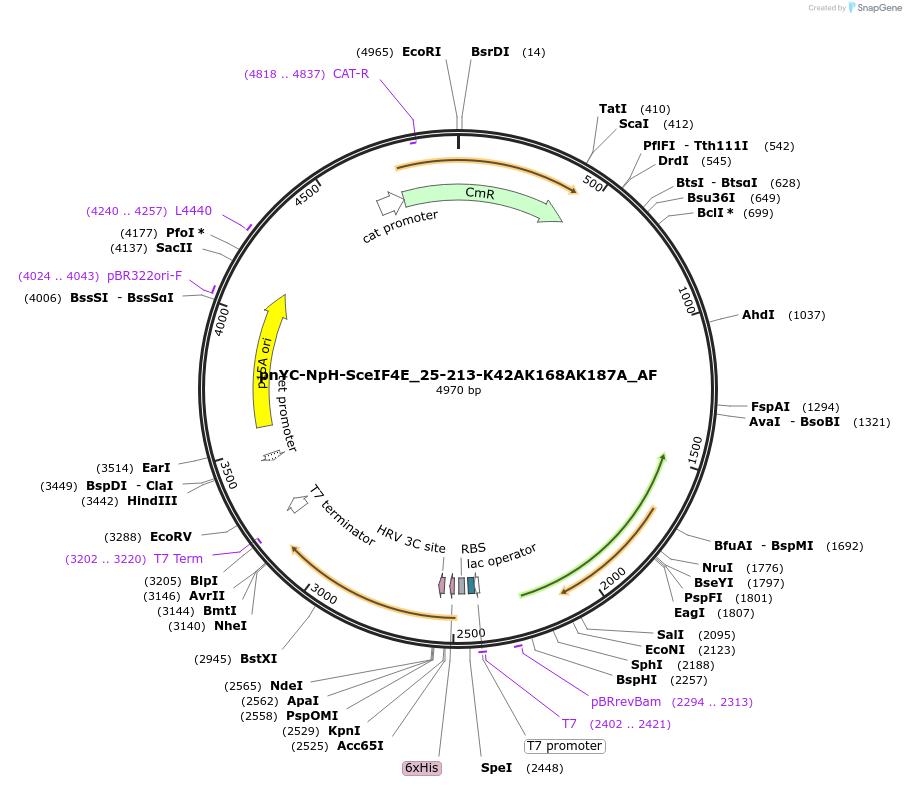
pnYC-NpH-SceIF4E_25-213-K42AK168AK187A_AF
Plasmid#148728PurposeBacterial Expression of SceIF4E_25-213-K42AK168AK187ADepositorInsertSceIF4E_25-213-K42AK168AK187A (CDC33 Budding Yeast)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
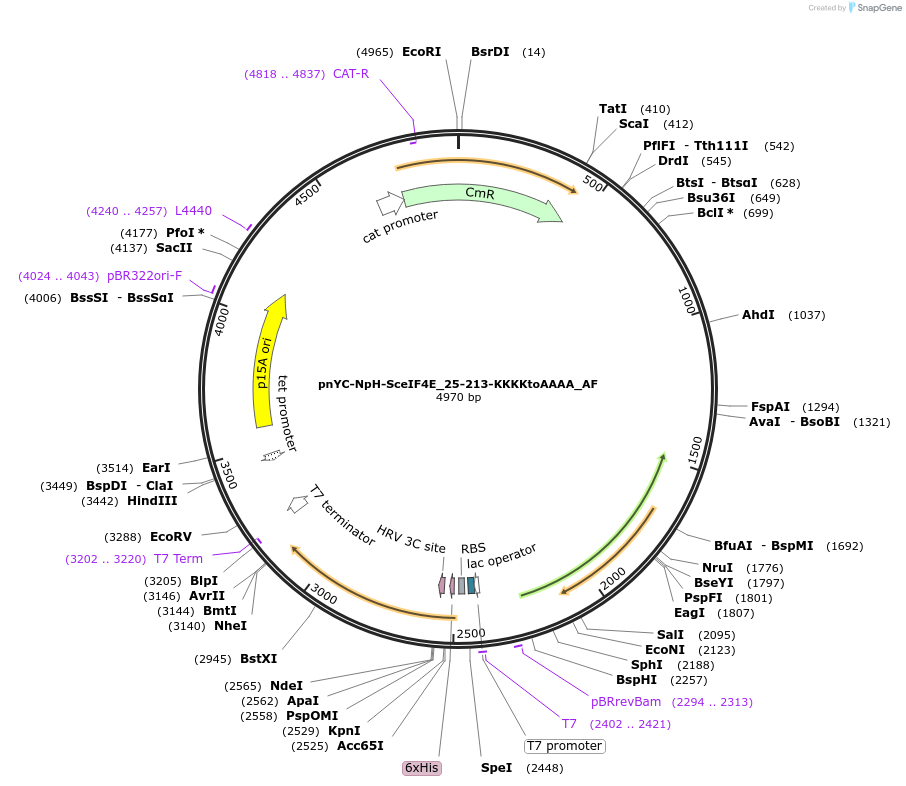
pnYC-NpH-SceIF4E_25-213-KKKKtoAAAA_AF
Plasmid#148729PurposeBacterial Expression of SceIF4E_25-213-KKKKtoAAAADepositorInsertSceIF4E_25-213-KKKKtoAAAA (CDC33 Budding Yeast)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
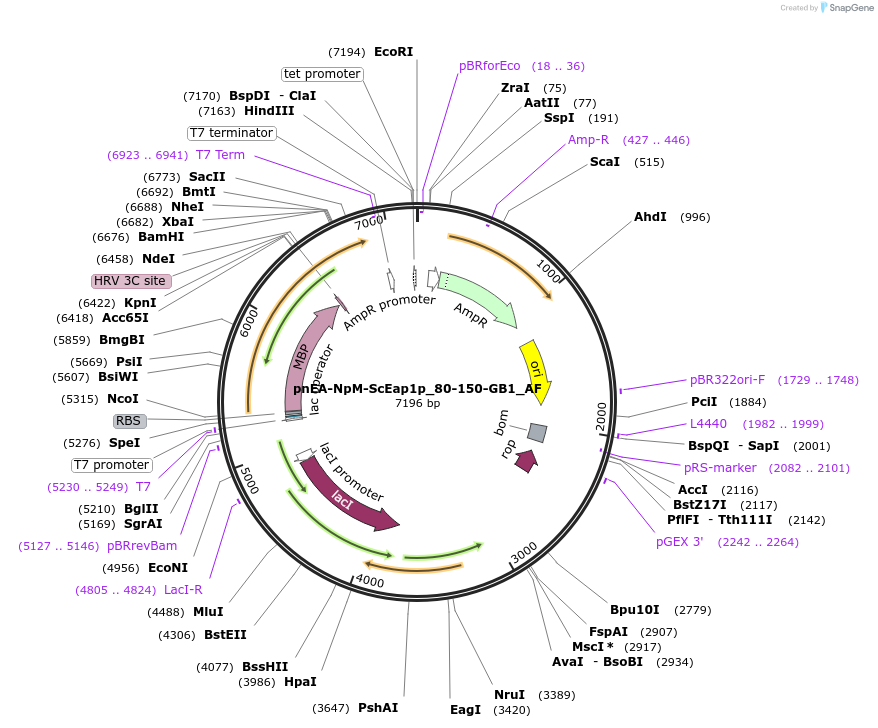
pnEA-NpM-ScEap1p_80-150-GB1_AF
Plasmid#148708PurposeBacterial Expression of ScEap1p_80-150DepositorInsertScEap1p_80-150 (EAP1 Budding Yeast)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
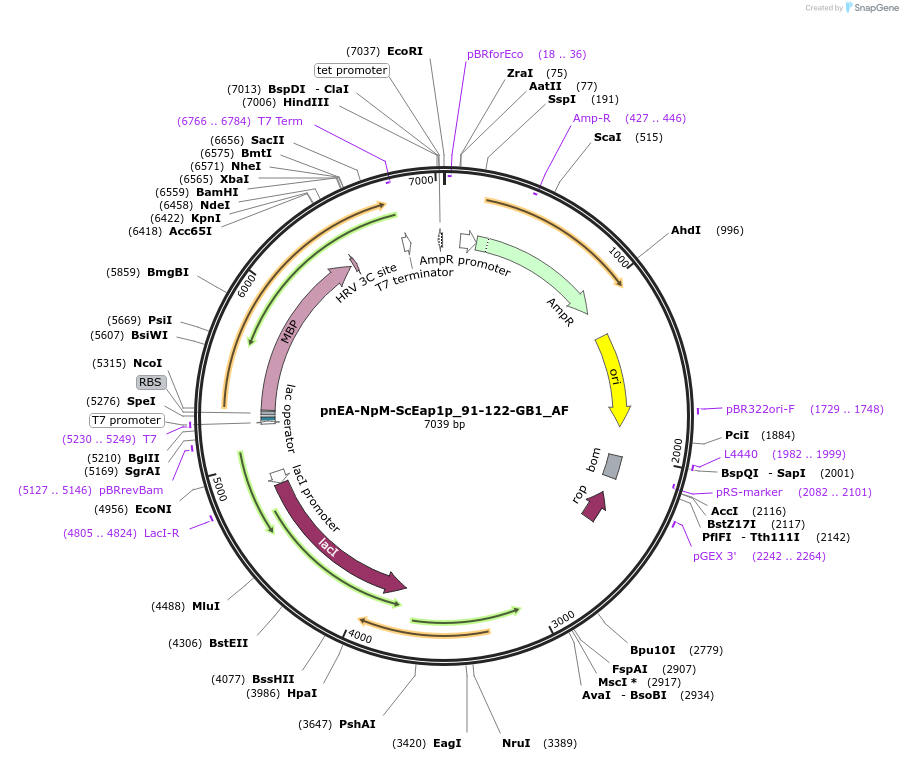
pnEA-NpM-ScEap1p_91-122-GB1_AF
Plasmid#148709PurposeBacterial Expression of ScEap1p_91-122DepositorInsertScEap1p_91-122 (EAP1 Budding Yeast)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
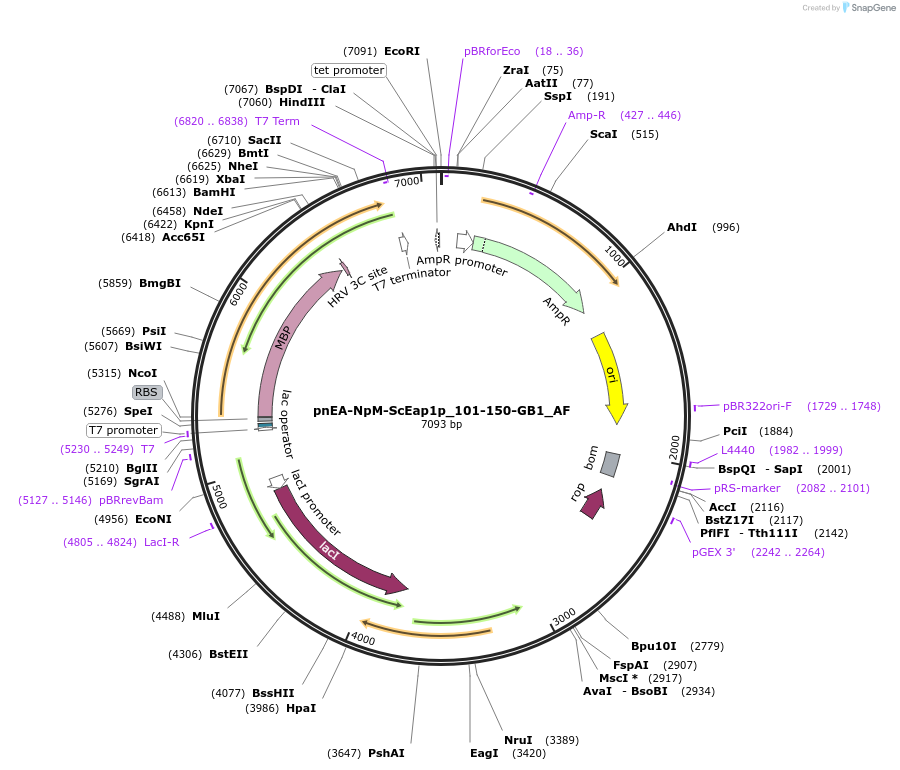
pnEA-NpM-ScEap1p_101-150-GB1_AF
Plasmid#148711PurposeBacterial Expression of ScEap1p_101-150DepositorInsertScEap1p_101-150 (EAP1 Budding Yeast)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
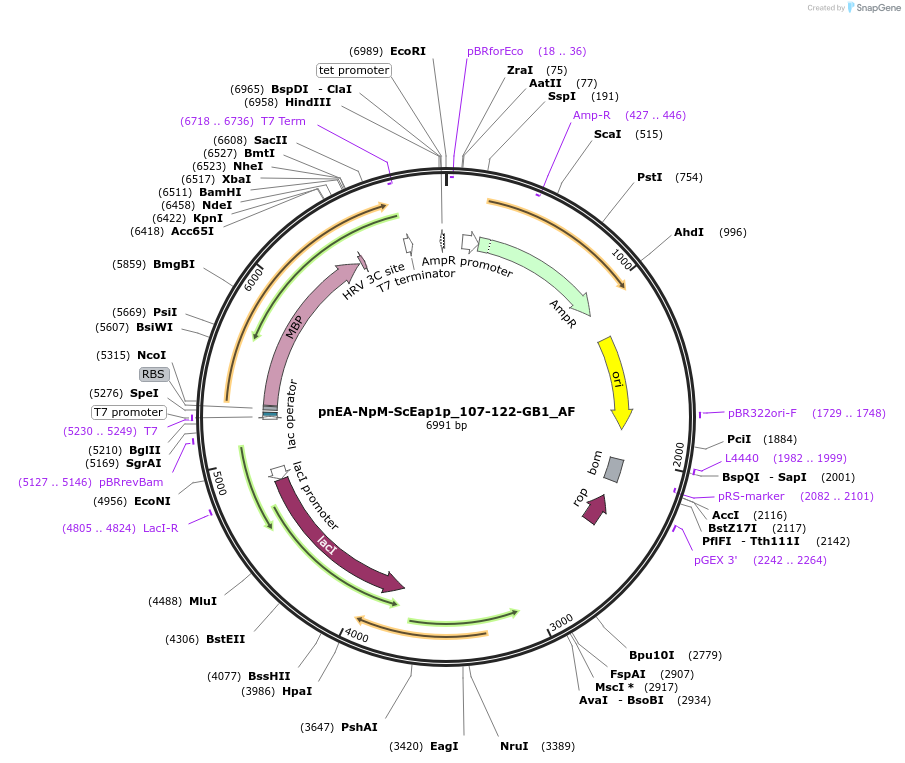
pnEA-NpM-ScEap1p_107-122-GB1_AF
Plasmid#148712PurposeBacterial Expression of ScEap1p_107-122DepositorInsertScEap1p_107-122 (EAP1 Budding Yeast)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
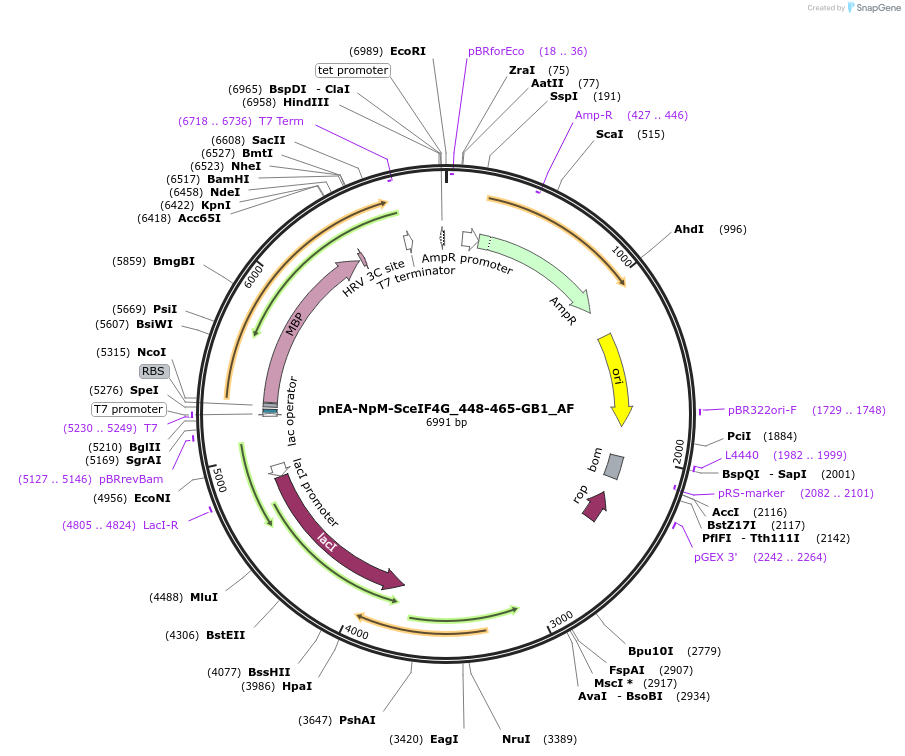
pnEA-NpM-SceIF4G_448-465-GB1_AF
Plasmid#148715PurposeBacterial Expression of SceIF4G_448-465DepositorInsertSceIF4G_448-465 (TIF4631 Budding Yeast)
ExpressionBacterialAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
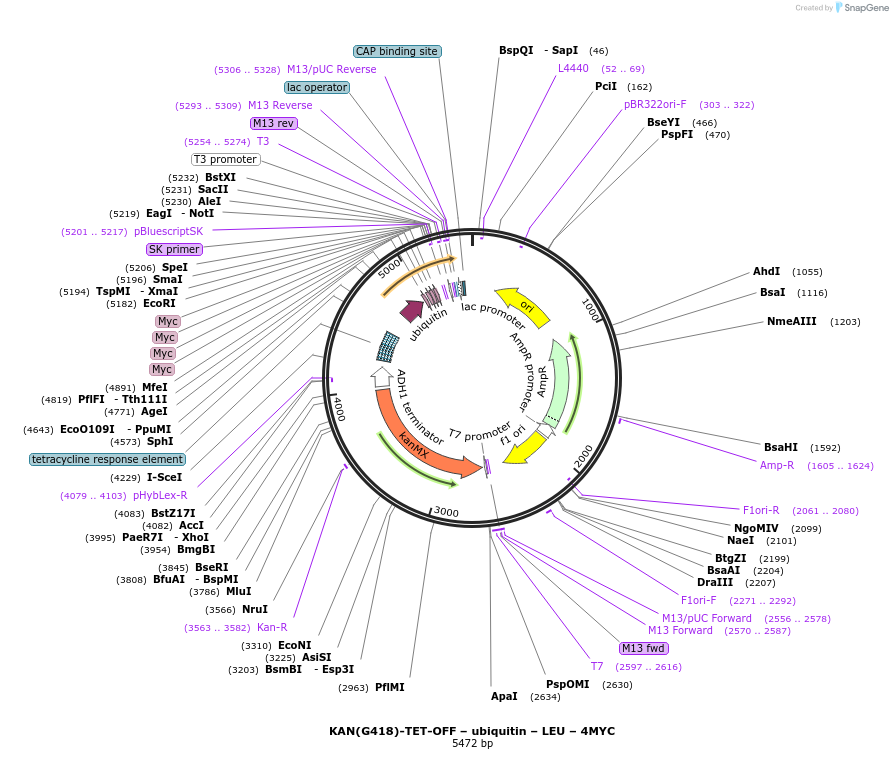
KAN(G418)-TET-OFF – ubiquitin – LEU – 4MYC
Plasmid#193318PurposeDoxycycline-induced transcriptional regulation and Ubiquitin - LEU - 4xMYC N-terminal tagging. Ubiquitin is cleaved off, resulting in an unstable protein with an N-terminal leucine.DepositorInsertUbiquitin - Leucine - 4MYC N-terminal tagging
Tags4MYCExpressionBacterialAvailable SinceFeb. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
KAN(G418)-TET-OFF – ubiquitin – LEU – 4FLAG
Plasmid#193317PurposeDoxycycline-induced transcriptional regulation and Ubiquitin - LEU - 4xFLAG N-terminal tagging. Ubiquitin is cleaved off, resulting in an unstable protein with an N-terminal leucine.DepositorInsertUbiquitin - Leucine - 4FLAG N-terminal tagging
Tags4FLAGExpressionBacterialAvailable SinceFeb. 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
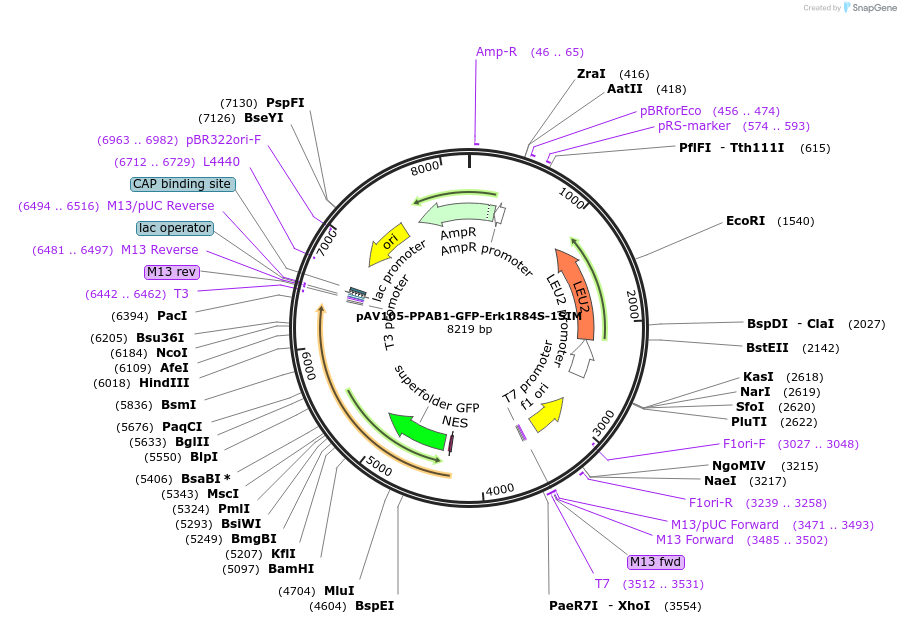
pAV105-PPAB1-GFP-Erk1R84S-1SIM
Plasmid#190253PurposeExpression Erk1-GFP-1SIM in yeast cellsDepositorInsertERK1R84S-SIM
TagsGFPExpressionYeastMutationERK1R84SAvailable SinceNov. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
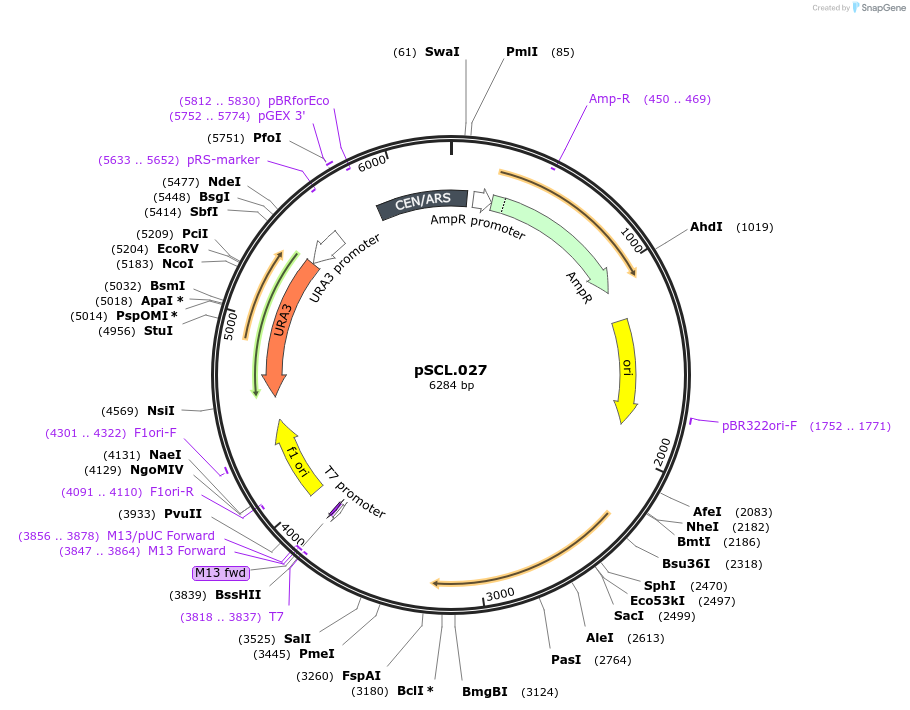
pSCL.027
Plasmid#184964PurposeExpress -Eco1 RT and ncRNA in yeastDepositorInsertEco1: RT and ncRNA(wt), a1/a2 length: 12
ExpressionYeastMutationhuman codon optimized RTPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
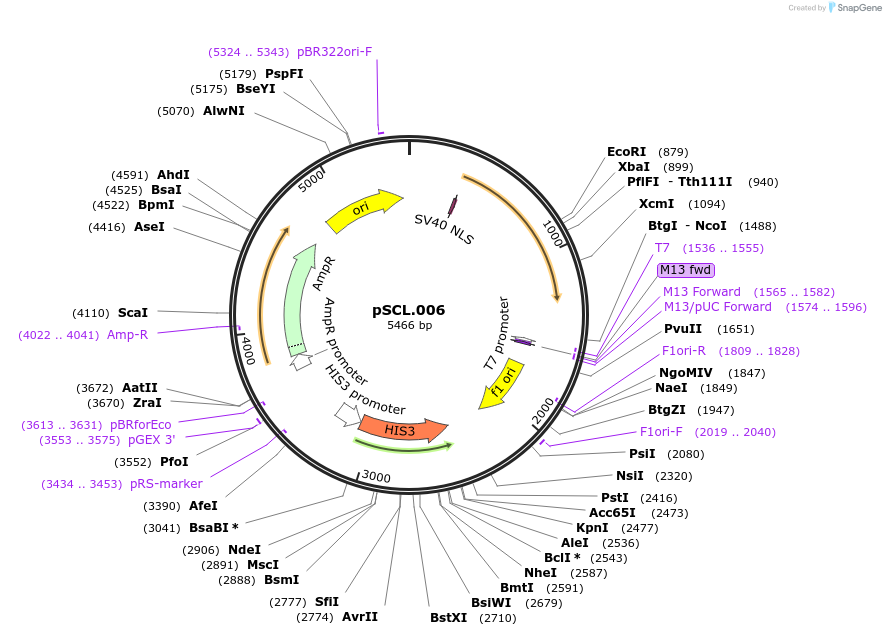
pSCL.006
Plasmid#184970PurposeExpress -Eco1 RT in yeastDepositorInsertIntegrating inducible cassette: Eco1 RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
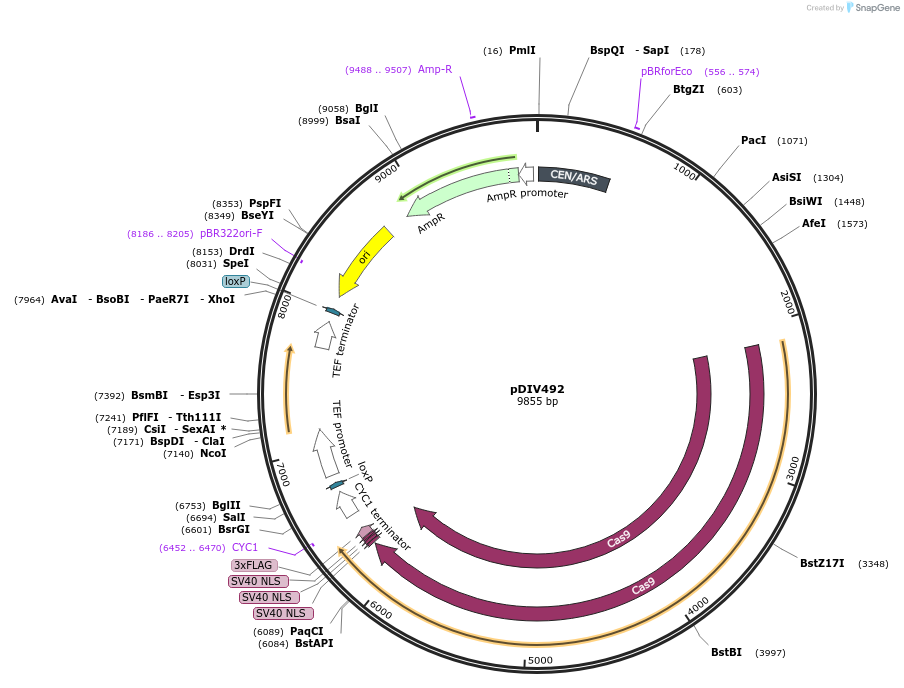
pDIV492
Plasmid#177700PurposePlasmid expressing optimized Cas9 and NAT marker. Compatible with CTG-clade yeast species.DepositorInsertsCas9
Nat
UseCRISPRTagsSV40ExpressionBacterial and YeastPromoterRHR2p (Debaryomyces hansenii ) and TEF1p (Ashbya …Available SinceJuly 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
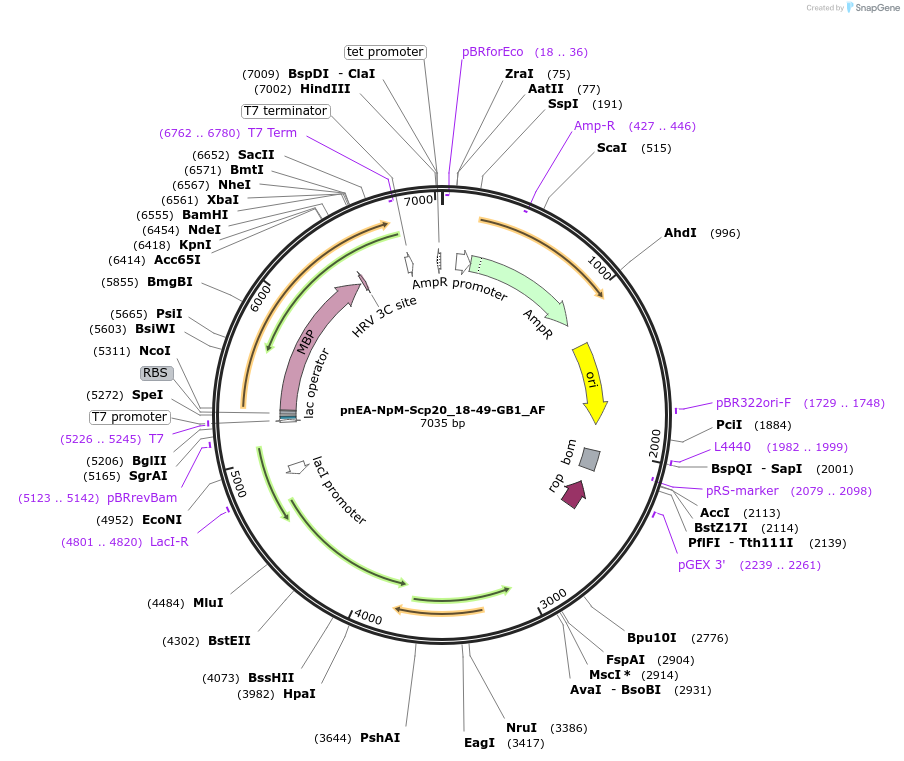
pnEA-NpM-Scp20_18-49-GB1_AF
Plasmid#148720PurposeBacterial Expression of Scp20_18-49DepositorInsertScp20_18-49 (CAF20 Budding Yeast)
ExpressionBacterialAvailable SinceMarch 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
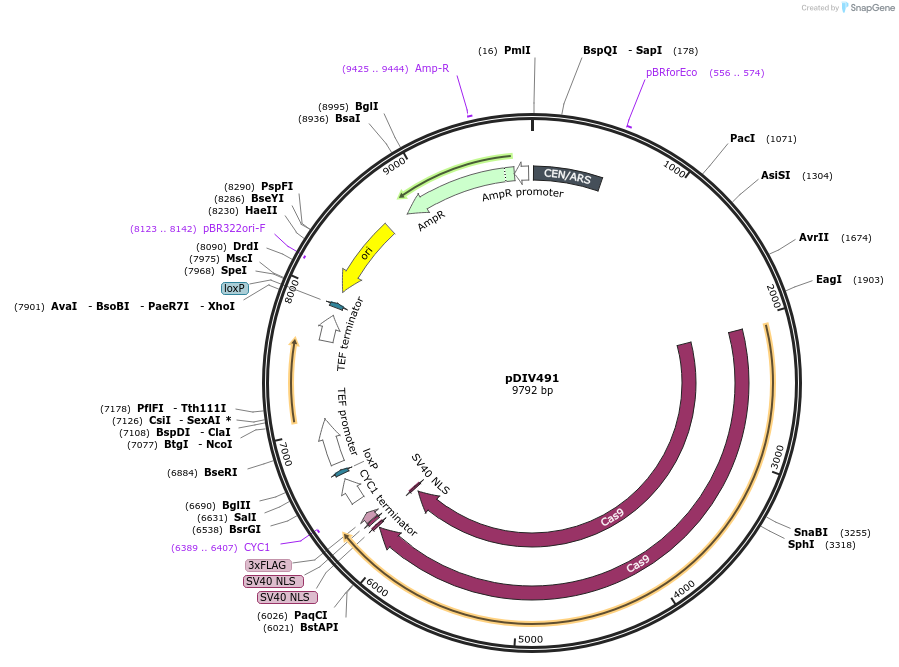
pDIV491
Plasmid#177699PurposePlasmid expressing optimized Cas9 and NAT marker. Compatible with CTG-clade yeast species.DepositorInsertsCas9
Nat
UseCRISPRTagsSV40ExpressionBacterial and YeastPromoterRNR2p (Debaryomyces hansenii ) and TEF1p (Ashbya …Available SinceMarch 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
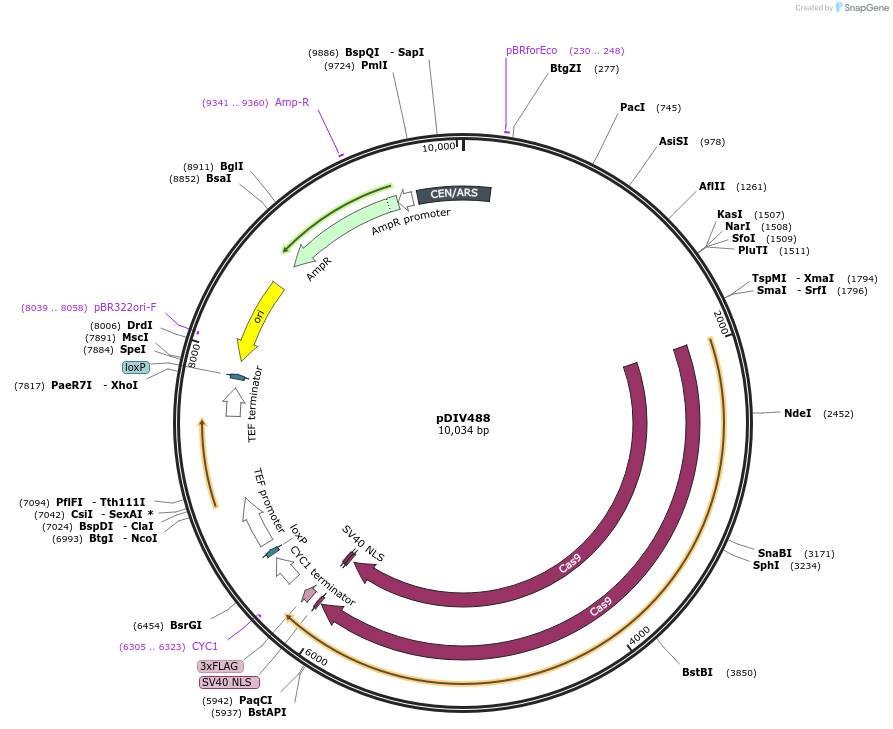
pDIV488
Plasmid#177696PurposePlasmid expressing optimized Cas9 and NAT marker. Compatible with CTG-clade yeast speciesDepositorInsertsCas9
Nat
UseCRISPRTagsSV40ExpressionBacterial and YeastPromoterTDH3p (Candida lusitaniae) and TEF1p (Ashbya goss…Available SinceMarch 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
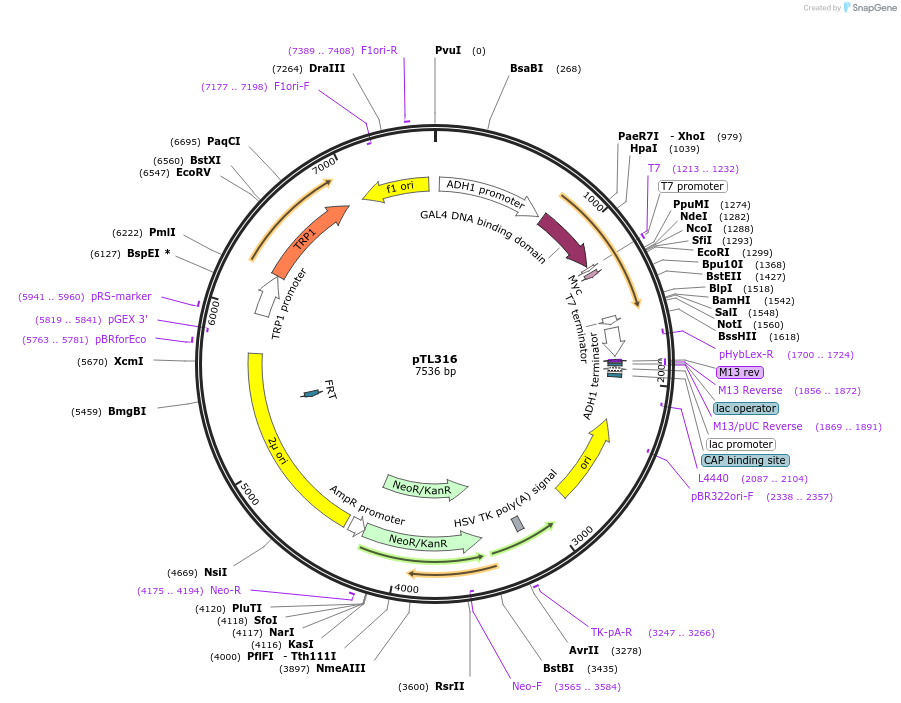
pTL316
Plasmid#168377PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os08g40130
ExpressionYeastAvailable SinceDec. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
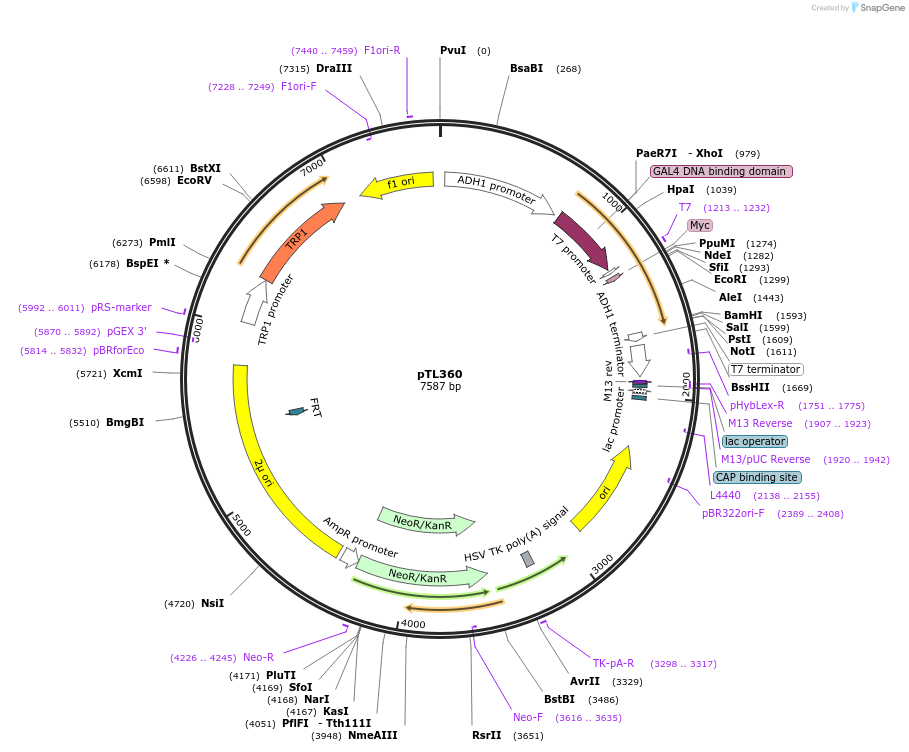
pTL360
Plasmid#168421PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os01g70240
ExpressionYeastAvailable SinceDec. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
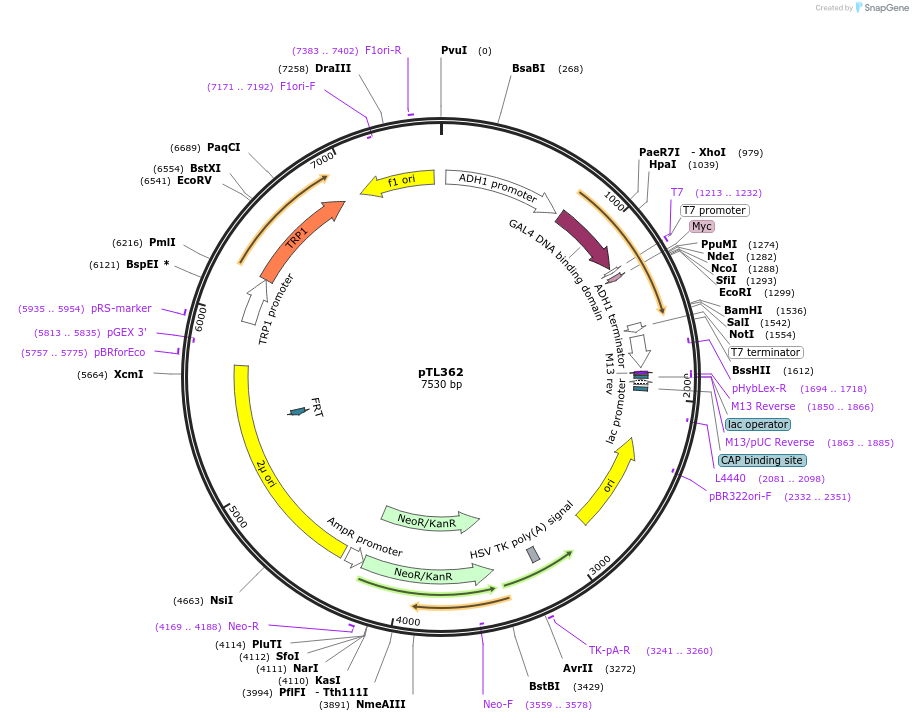
pTL362
Plasmid#168423PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os04g32860
ExpressionYeastAvailable SinceDec. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
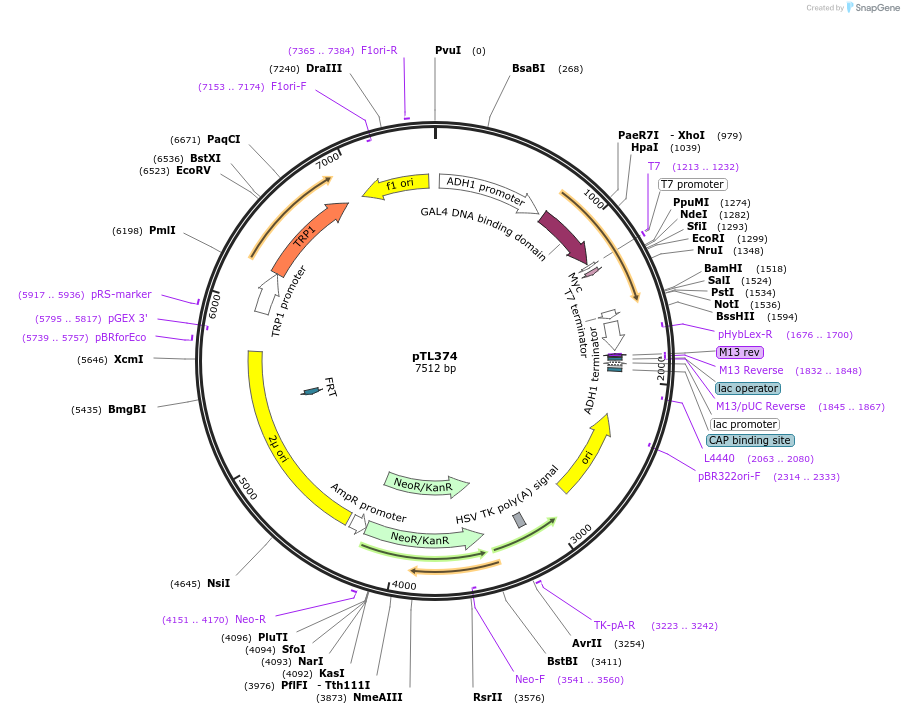
pTL374
Plasmid#168435PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os02g48170
ExpressionYeastAvailable SinceDec. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
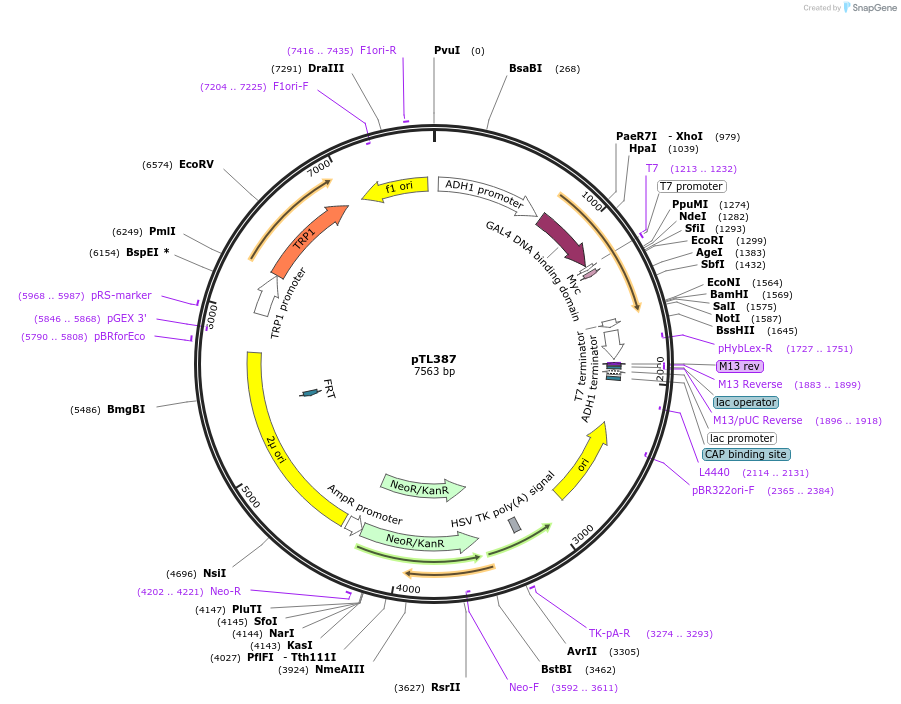
pTL387
Plasmid#168448PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os04g39370
ExpressionYeastAvailable SinceDec. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
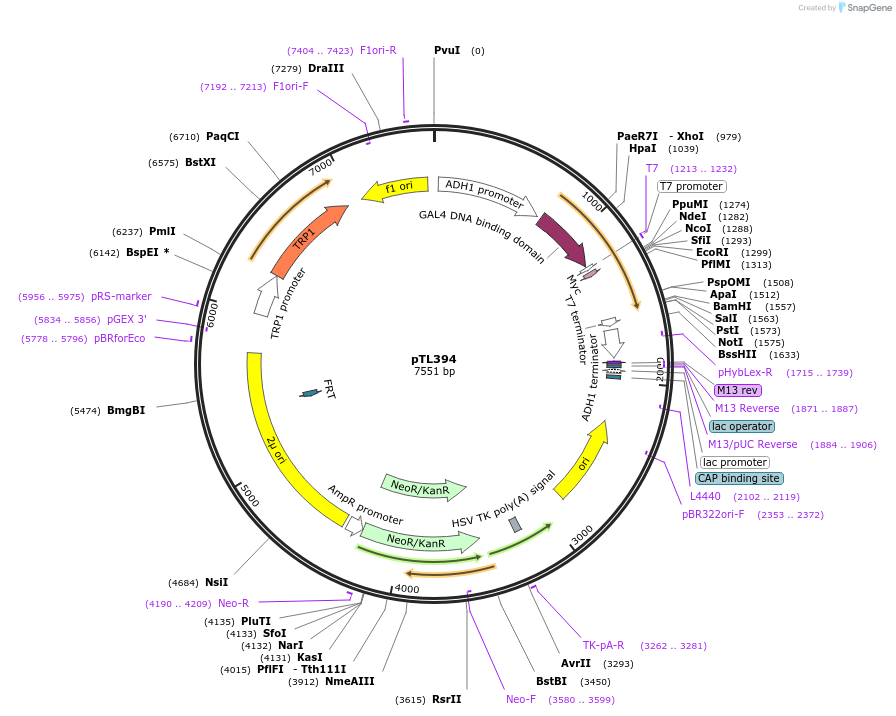
pTL394
Plasmid#168455PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os03g02070
ExpressionYeastAvailable SinceDec. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
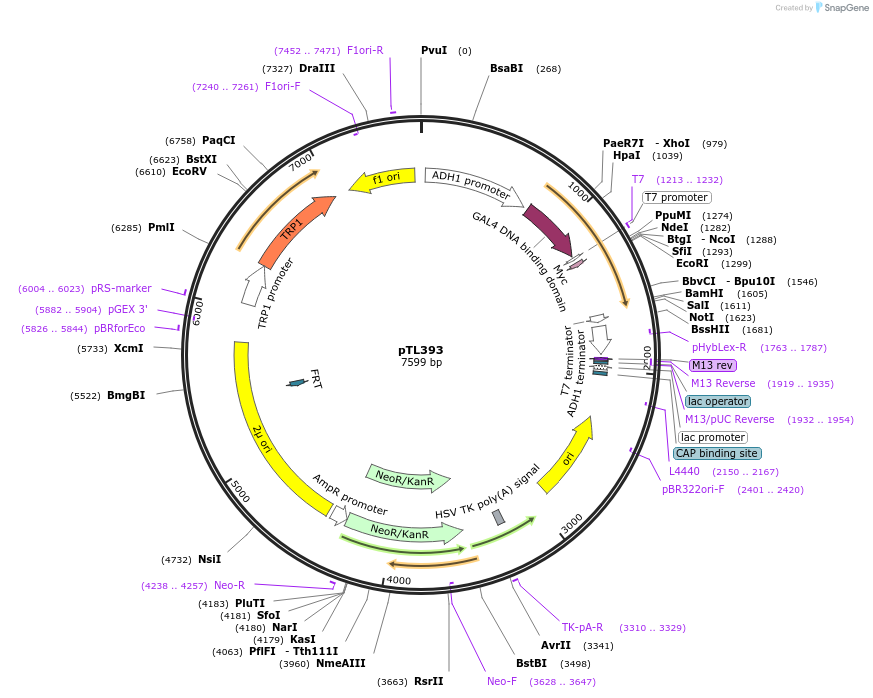
pTL393
Plasmid#168454PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertOsaHIP05 (Pi21)
ExpressionYeastAvailable SinceDec. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
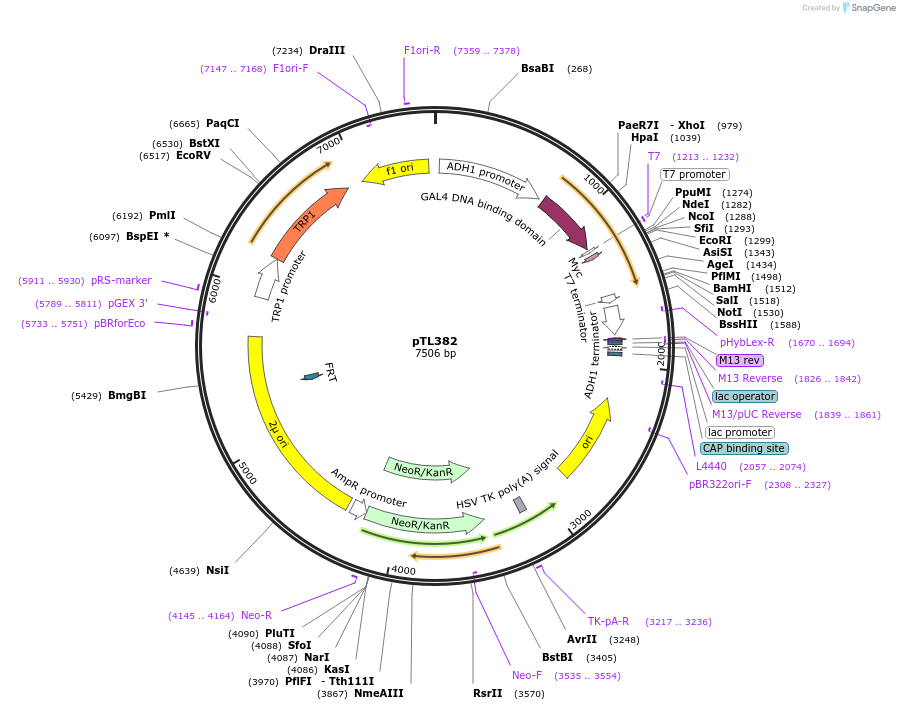
pTL382
Plasmid#168443PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os01g61070
ExpressionYeastAvailable SinceDec. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
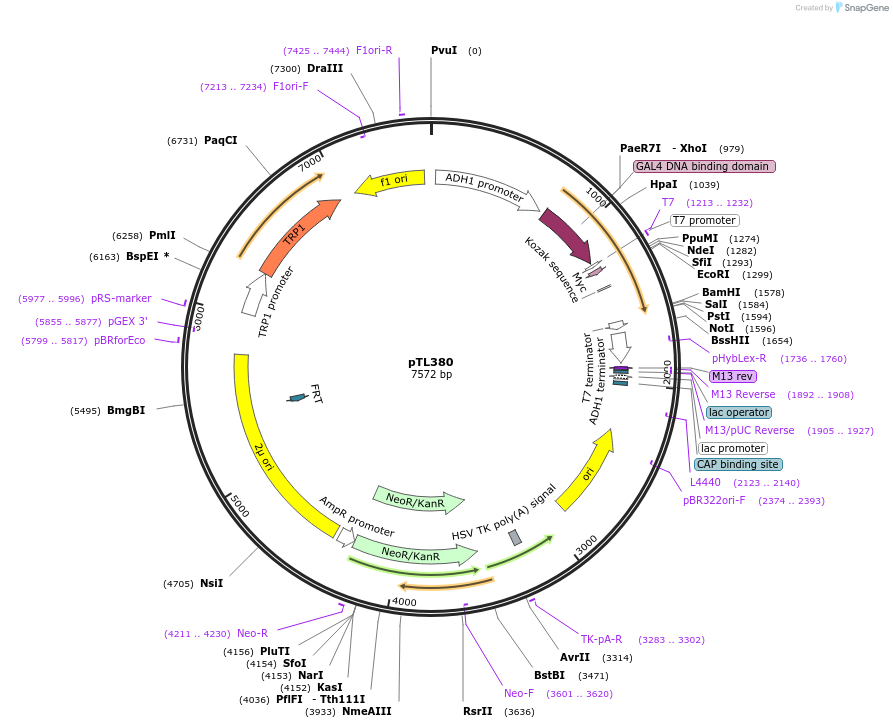
pTL380
Plasmid#168441PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os02g37180
ExpressionYeastAvailable SinceDec. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
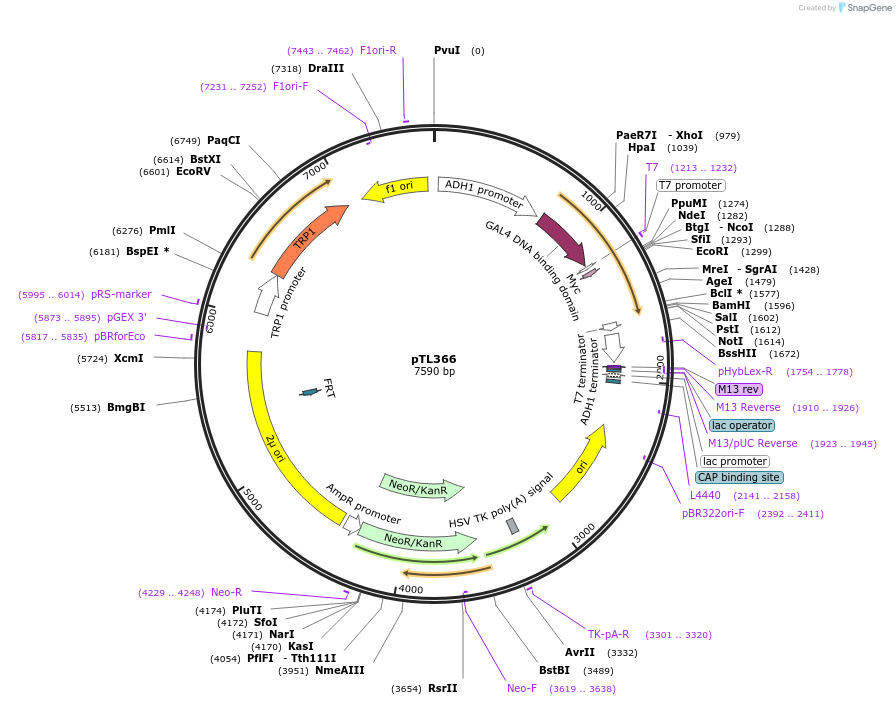
pTL366
Plasmid#168427PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os04g42350
ExpressionYeastAvailable SinceDec. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
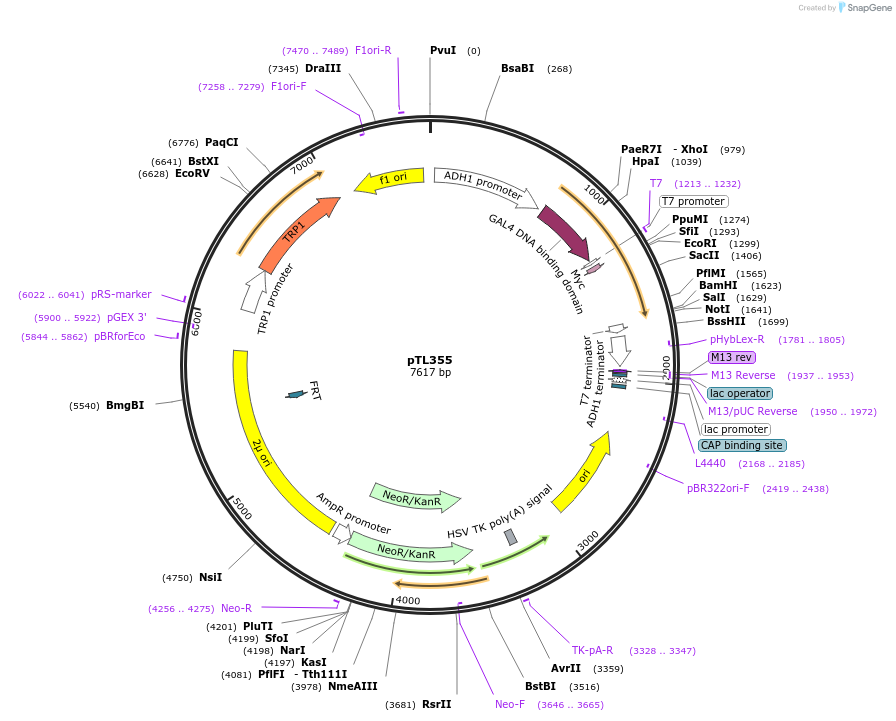
pTL355
Plasmid#168416PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os06g30480
ExpressionYeastAvailable SinceDec. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
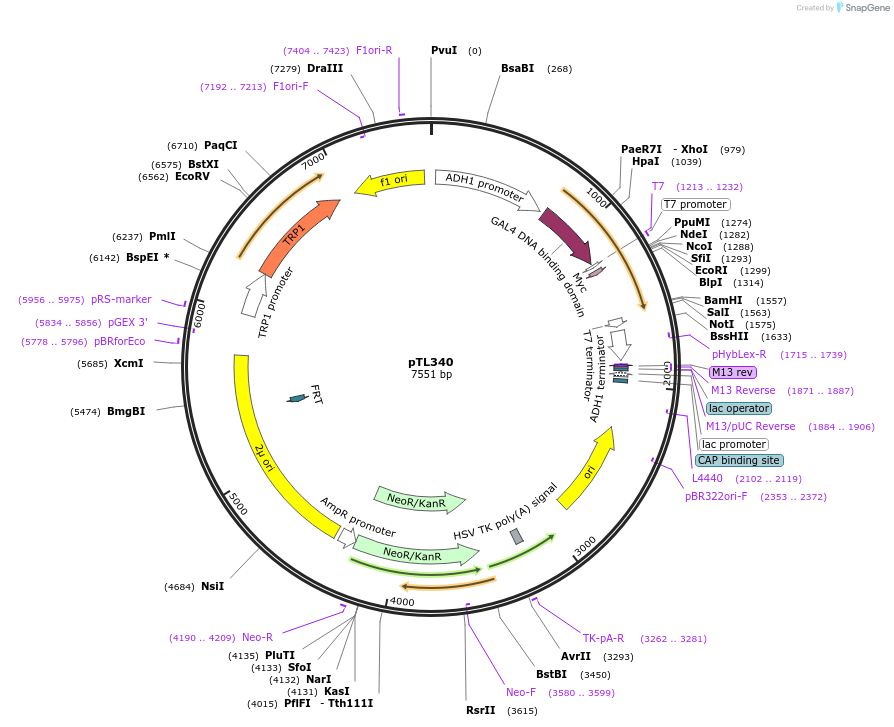
pTL340
Plasmid#168401PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os01g09660
ExpressionYeastAvailable SinceDec. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
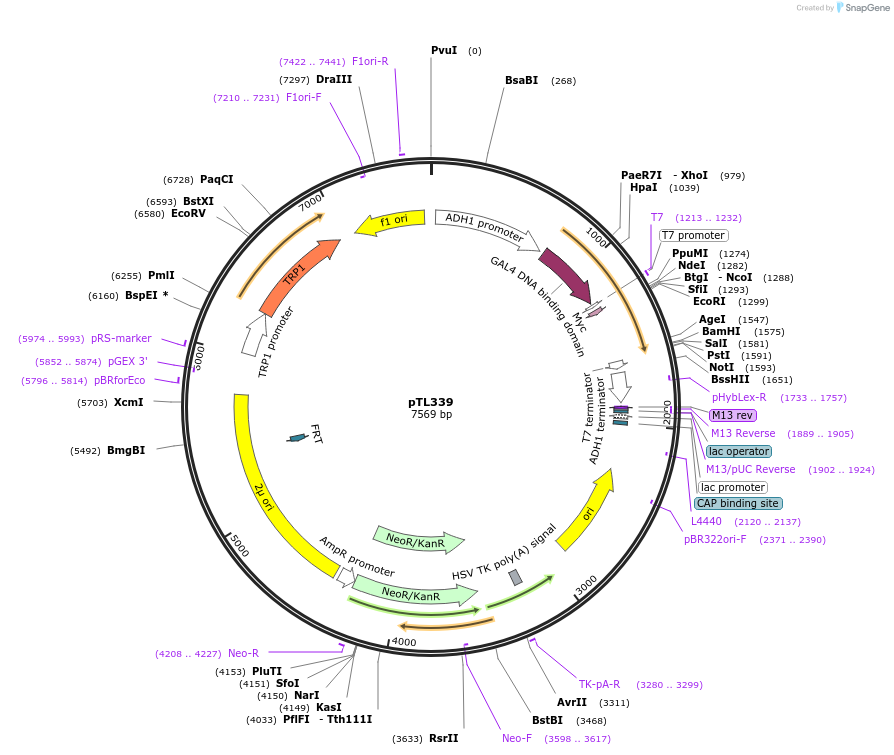
pTL339
Plasmid#168400PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os04g32830
ExpressionYeastAvailable SinceDec. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
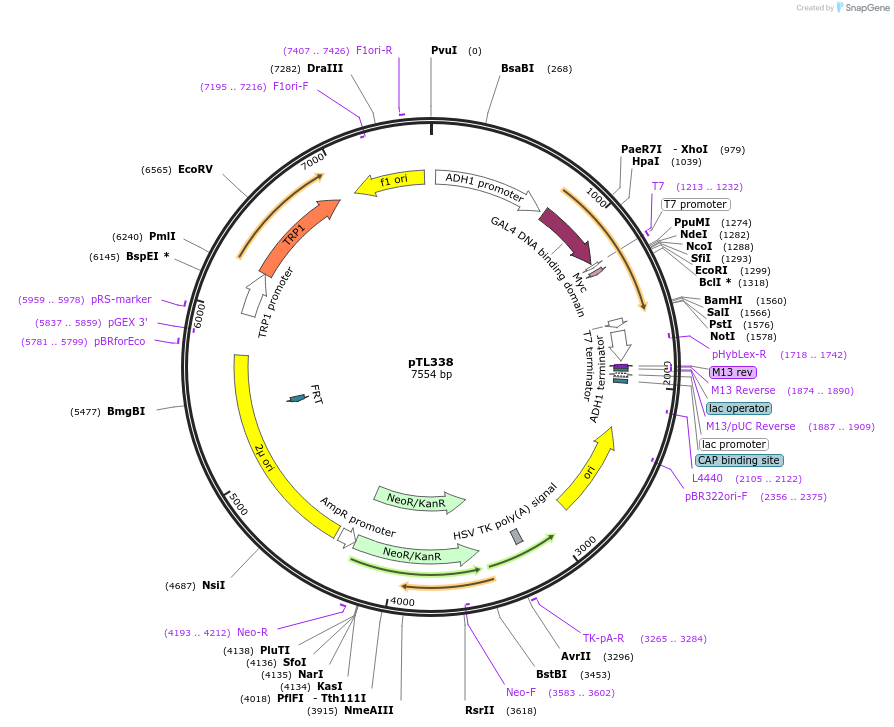
pTL338
Plasmid#168399PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os03g25610
ExpressionYeastAvailable SinceDec. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
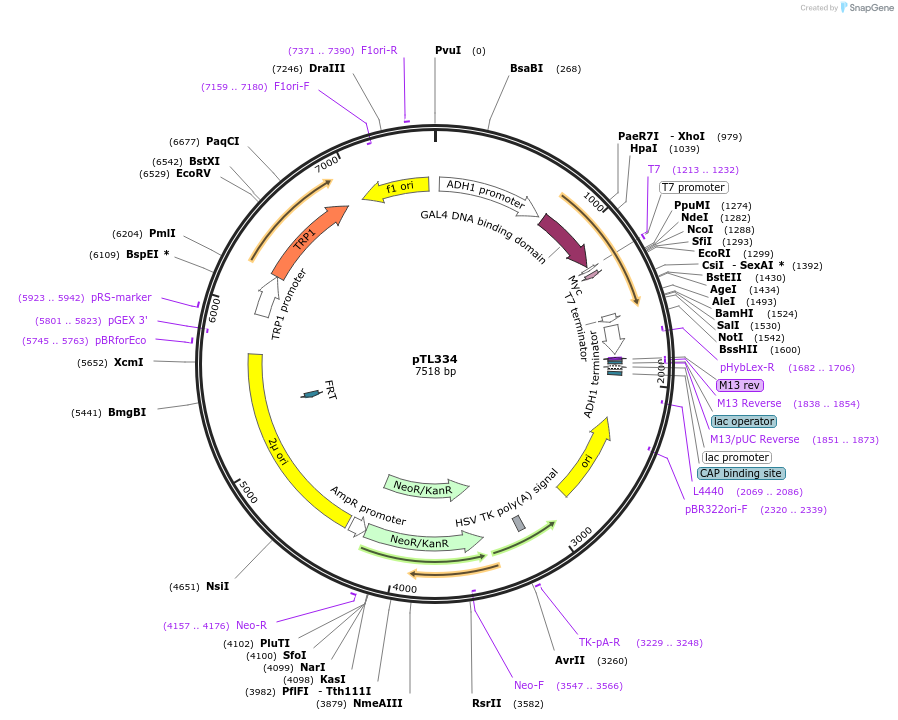
pTL334
Plasmid#168395PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os05g45820
ExpressionYeastAvailable SinceDec. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
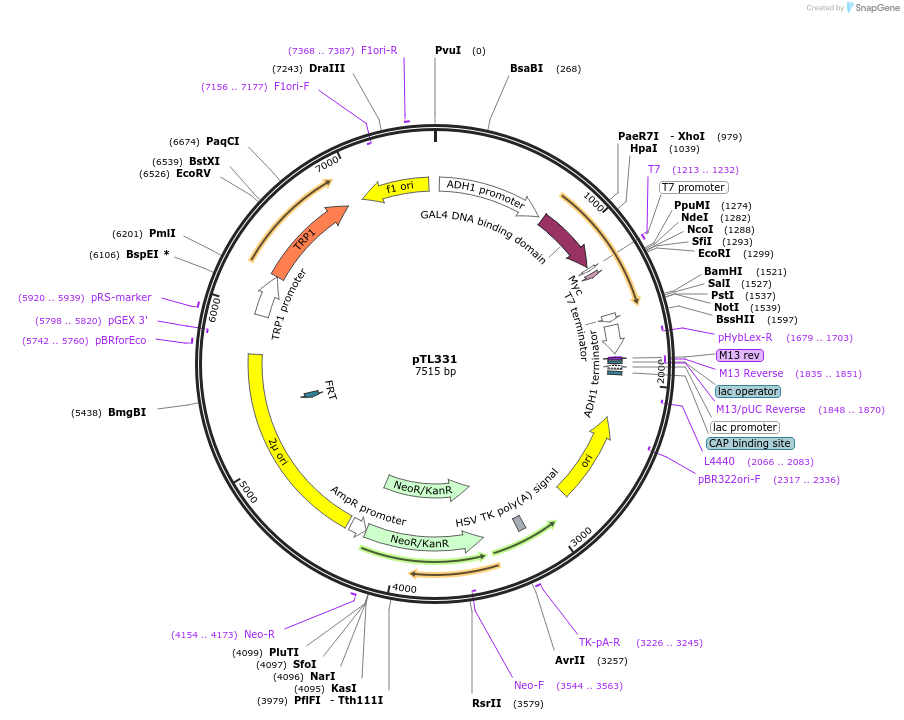
pTL331
Plasmid#168392PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os01g52160
ExpressionYeastAvailable SinceDec. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
pTL323
Plasmid#168384PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os09g37730
ExpressionYeastAvailable SinceDec. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
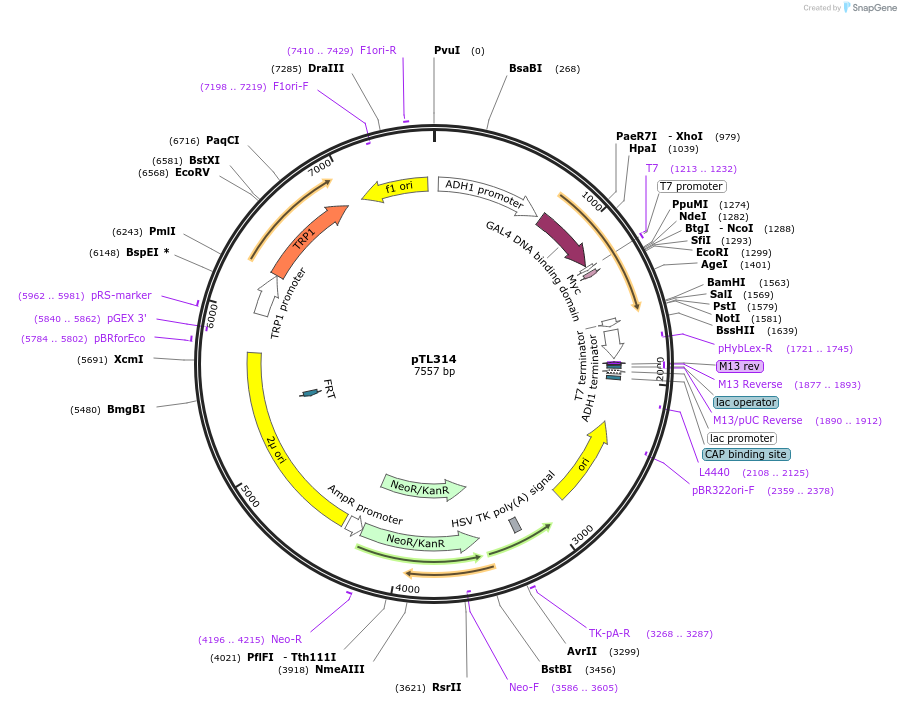
pTL314
Plasmid#168375PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os09g15860
ExpressionYeastAvailable SinceDec. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
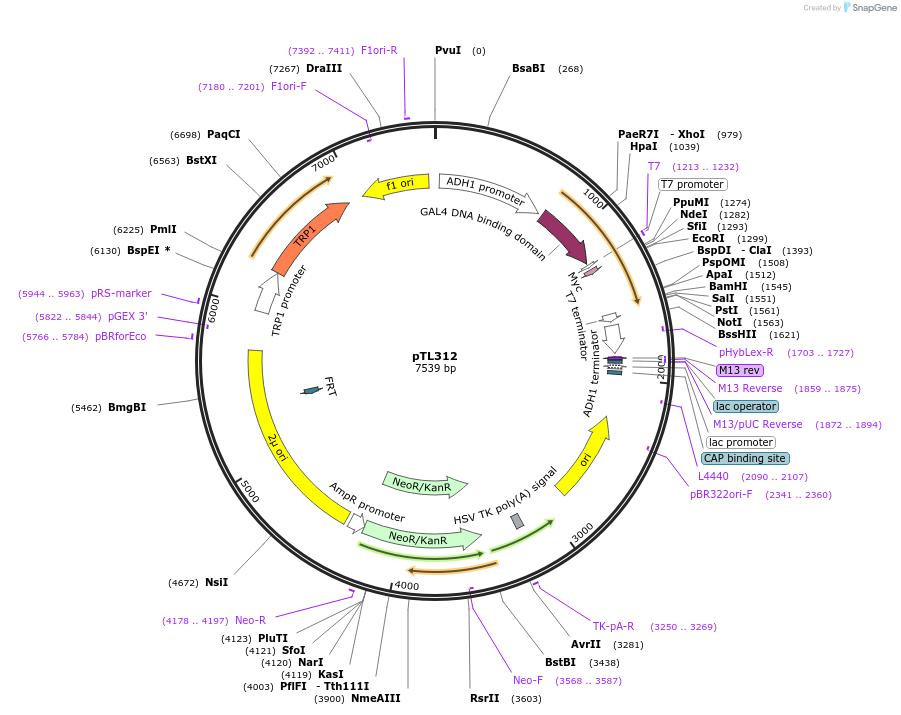
pTL312
Plasmid#168373PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os04g45130
ExpressionYeastAvailable SinceDec. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
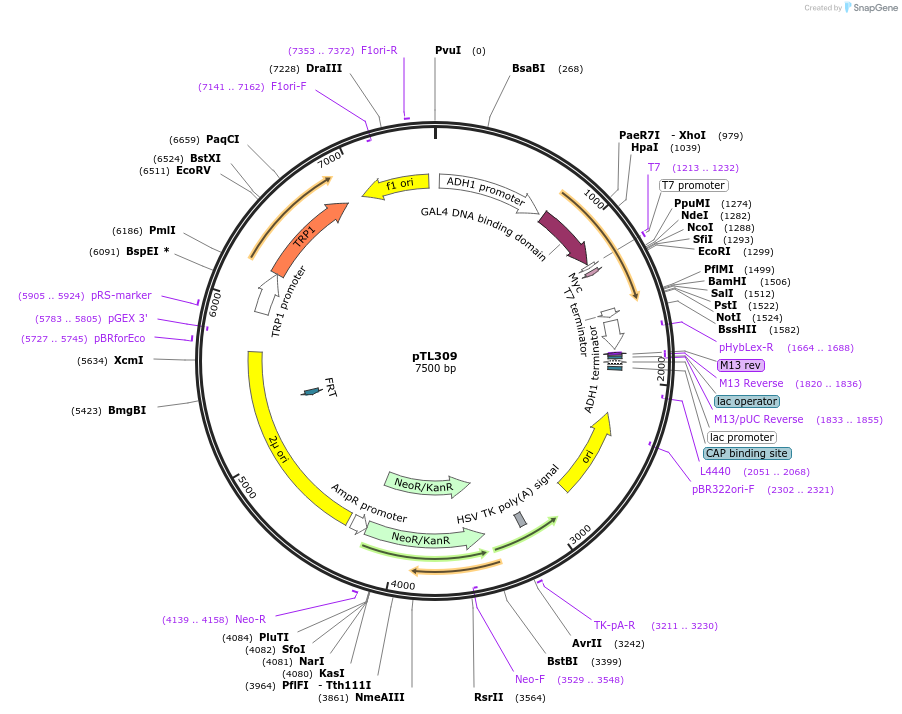
pTL309
Plasmid#168370PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os01g41200
ExpressionYeastAvailable SinceDec. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
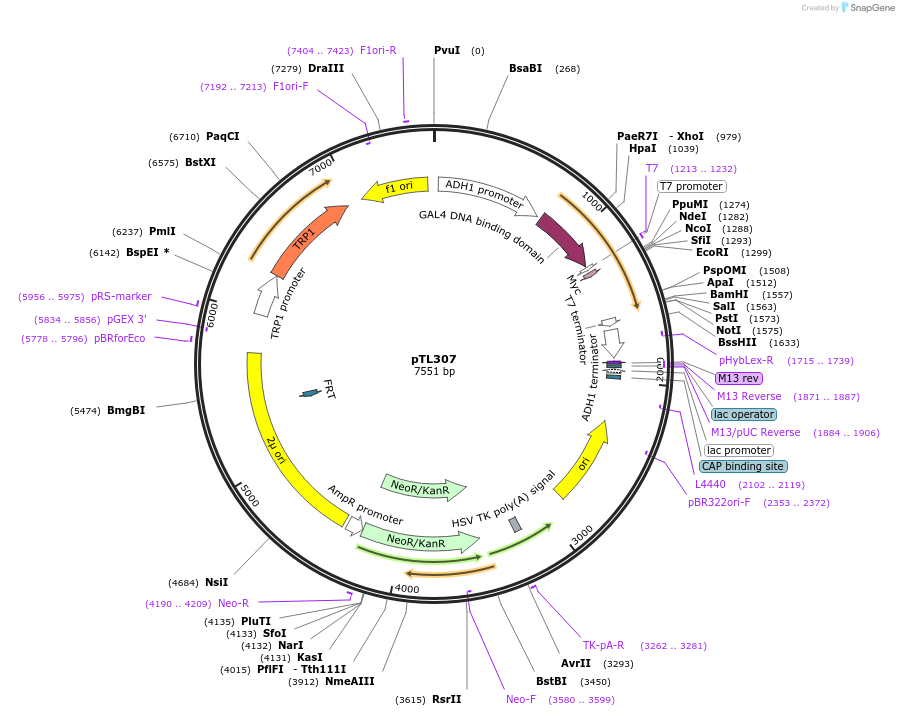
pTL307
Plasmid#168368PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertLOC_ Os10g36200
ExpressionYeastAvailable SinceDec. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
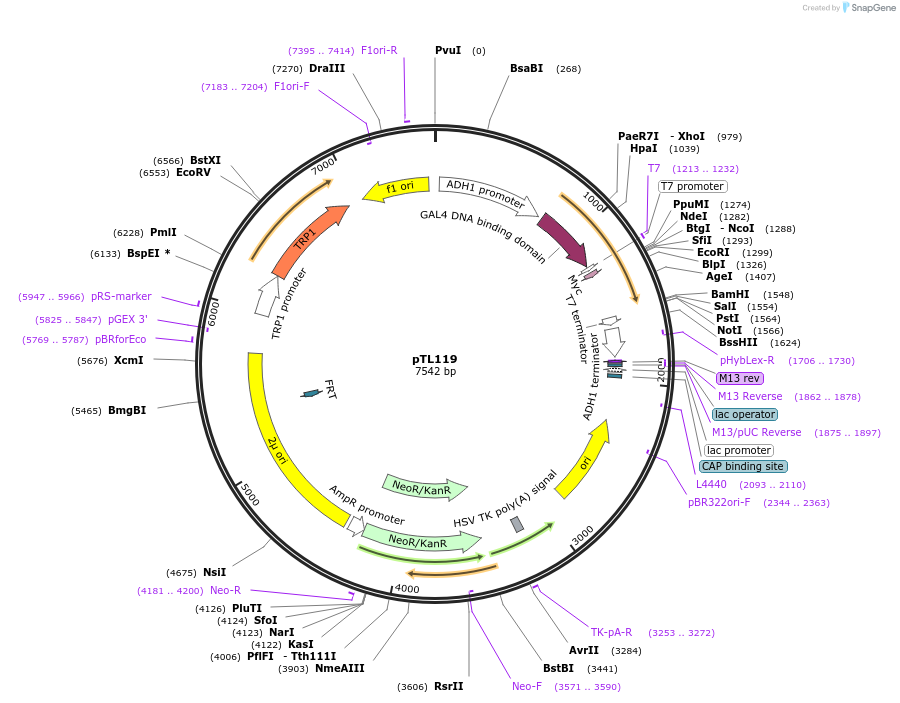
pTL119
Plasmid#168353PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertTraes_2BL_FFF4186AF.1
ExpressionYeastAvailable SinceDec. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
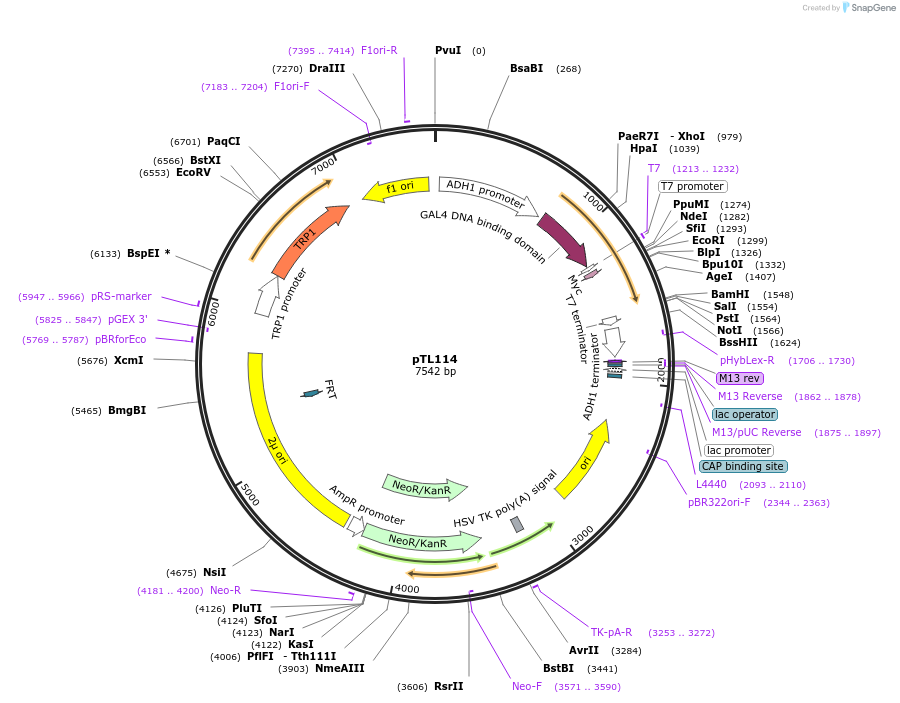
pTL114
Plasmid#168348PurposepGBKT7 yeast two-hybrid plasmidDepositorInsertTraes_2AL_0C4FF568B.1
ExpressionYeastAvailable SinceDec. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
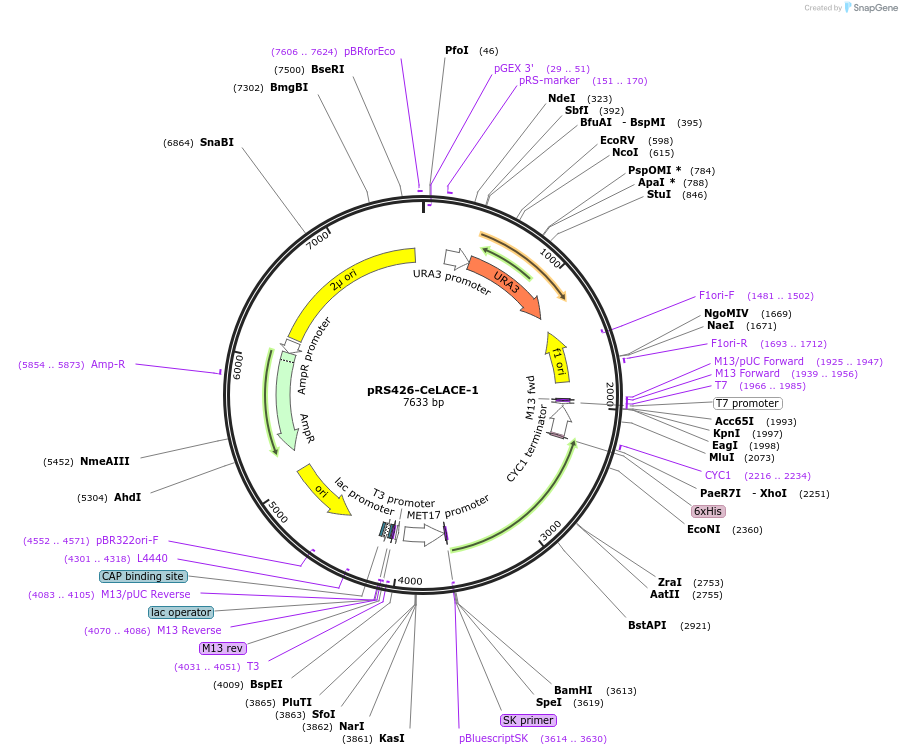
pRS426-CeLACE-1
Plasmid#174523PurposeExpress C.elegans LACE-1 in yeast cellsDepositorAvailable SinceSept. 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
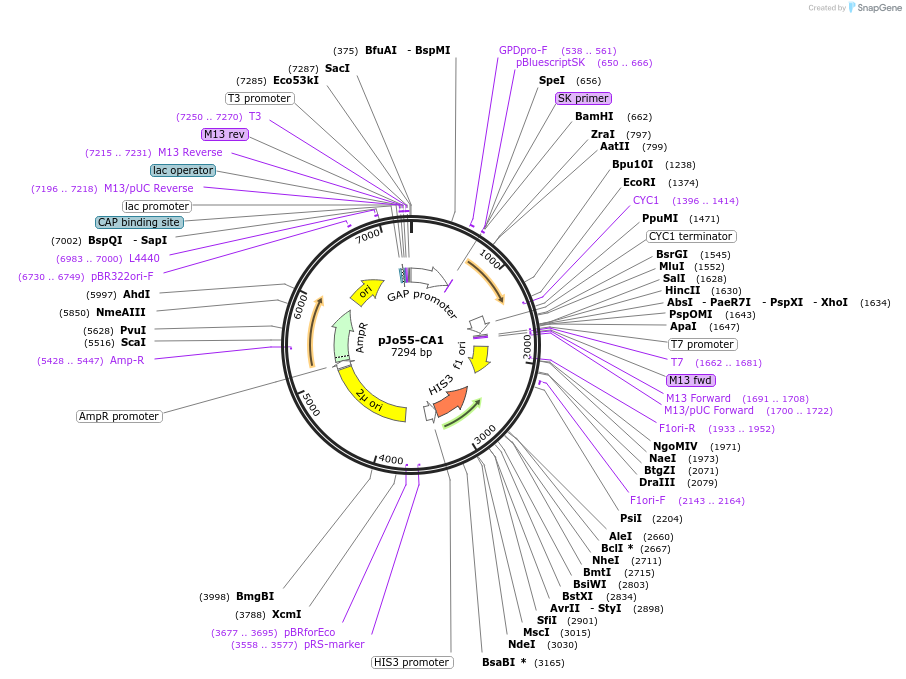
pJo55-CA1
Plasmid#159624PurposePlasmid pJo55, which contains positive control (C. annuum eIF4E-1) for complementation test in Jo55 yeasts.DepositorInsertC. annuum eIF4E-1
ExpressionBacterial and YeastAvailable SinceSept. 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
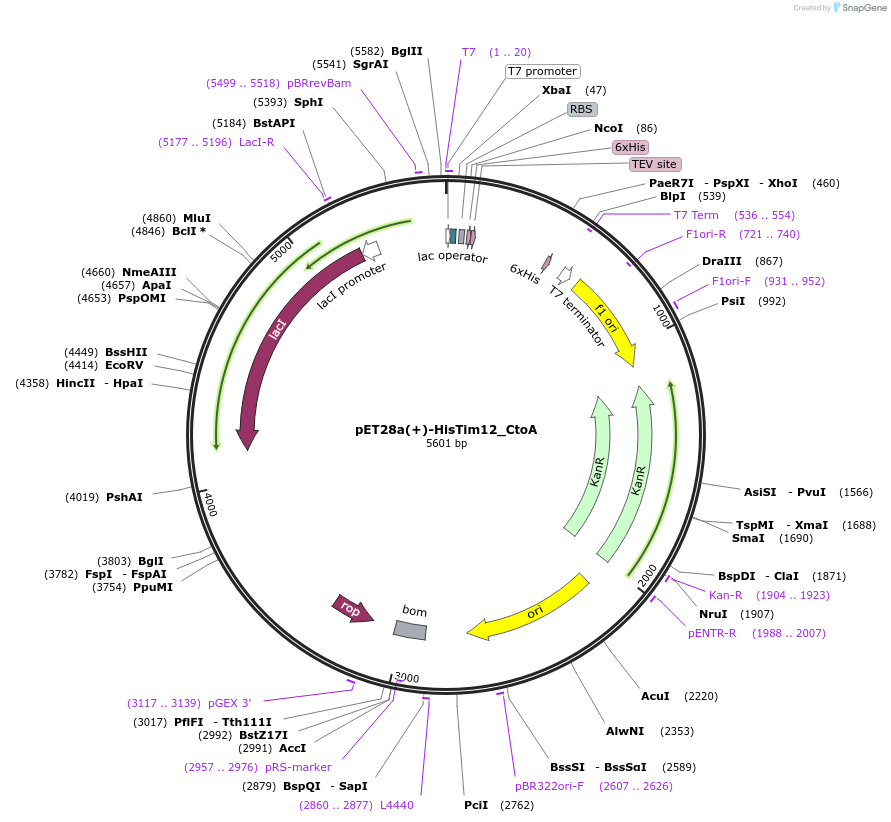
pET28a(+)-HisTim12_CtoA
Plasmid#170281PurposeExpresses yeast Tim12 (the latter with cleavable N-ter His-tag), codon-optimized for E. coli productionDepositorInsertyeast Tim12 (TIM12 Budding Yeast)
TagsTEV-cleavable N-terminal His tag (on backbone)ExpressionBacterialMutationmutated two non-conserved Cys to AlaAvailable SinceJune 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
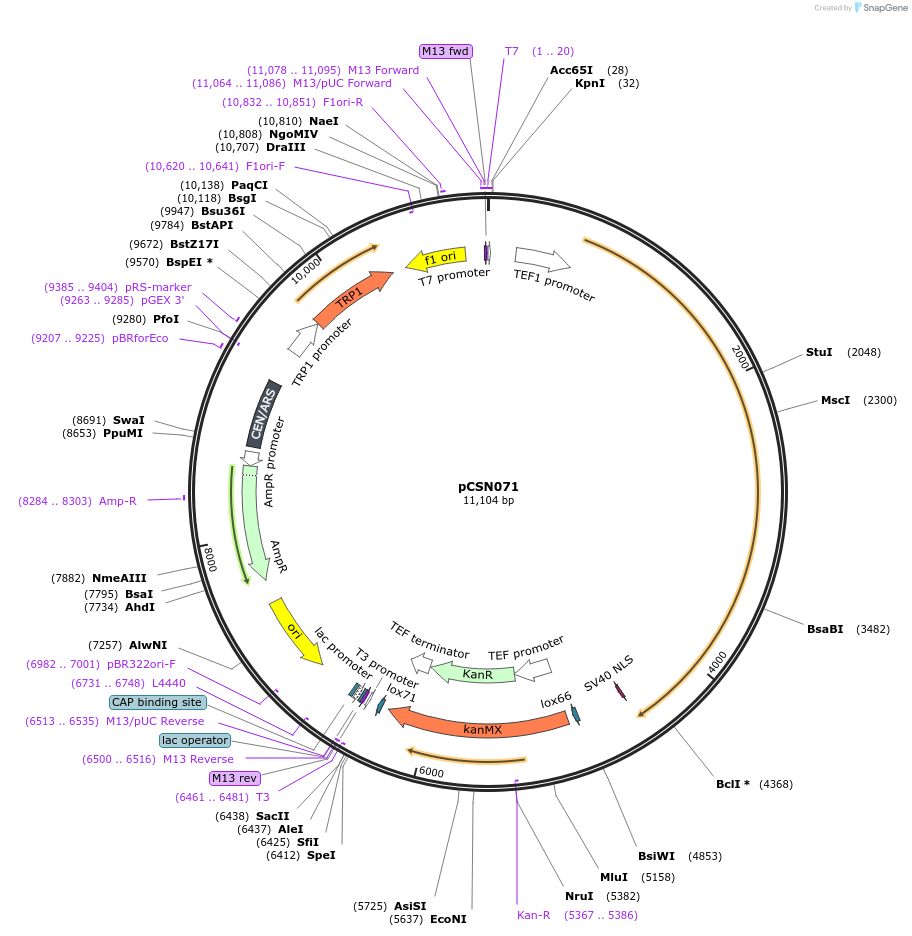
pCSN071
Plasmid#166729PurposeSingle copy yeast plasmid expressing DNase-dead dLbCas12a E925A fused to Mxi1 and NLS, codon optimized for Saccharomyces cerevisiaeDepositorInsertdCas12a-Mxi1-NLS
UseCRISPRTagsMxi1 and SV40 NLSExpressionYeastMutationE925APromoterS. cerevisiae TEF1Available SinceJune 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
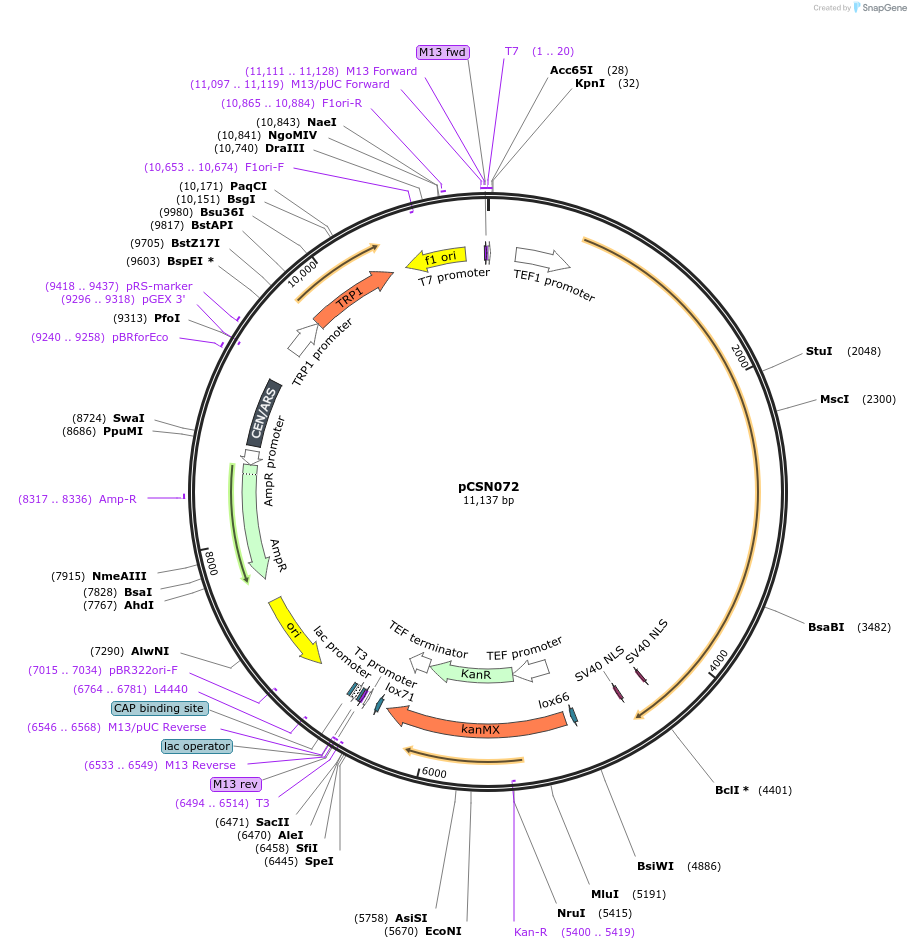
pCSN072
Plasmid#166730PurposeSingle copy yeast plasmid expressing DNase-dead dLbCas12a E925A fused to NLS, Mxi1 and NLS, codon optimized for Saccharomyces cerevisiaeDepositorInsertdCas12a-NLS-Mxi1-NLS
UseCRISPRTagsMxi1 and SV40 NLSExpressionYeastMutationE925APromoterS. cerevisiae TEF1Available SinceJune 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
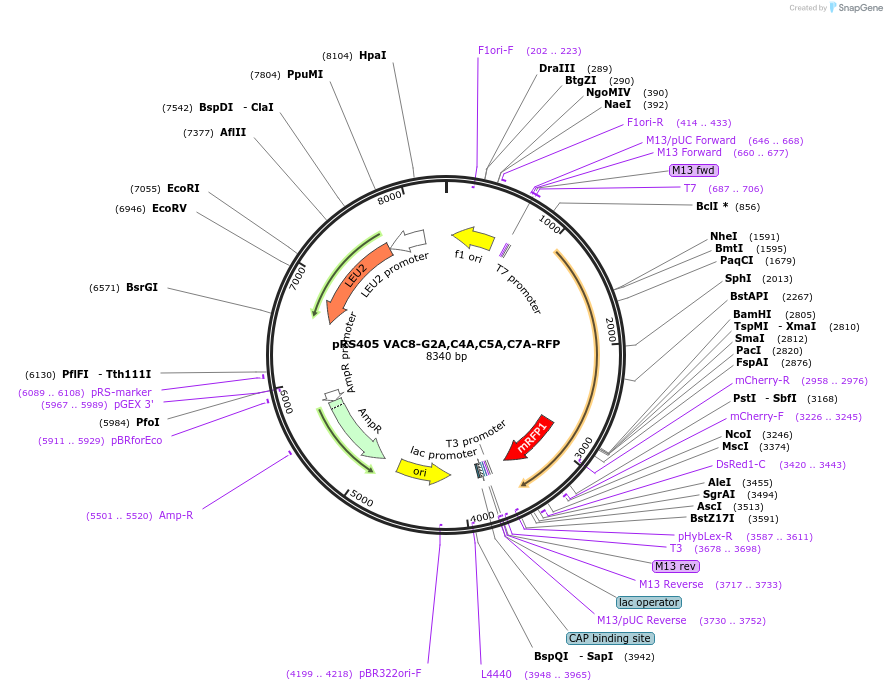
pRS405 VAC8-G2A,C4A,C5A,C7A-RFP
Plasmid#166842PurposeExpresses a myristoylation and palmitoylation mutant Vac8-RFP in yeasts, mutations are Gly2Ala, Cys4Ala, Cys5Ala, Cys7AlaDepositorInsertVAC8 (VAC8 Budding Yeast)
TagsRFPExpressionYeastMutationG2A, C4A, C5A, C7APromoterVAC8 promoterAvailable SinceApril 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
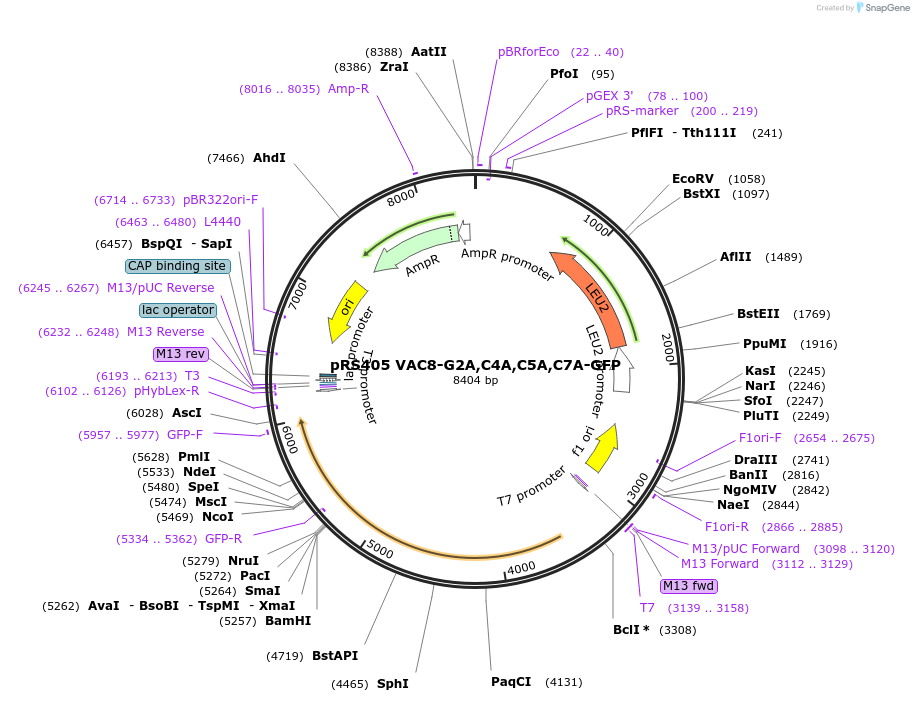
pRS405 VAC8-G2A,C4A,C5A,C7A-GFP
Plasmid#166839PurposeExpresses a myristoylation and palmitoylation mutant Vac8-GFP in yeasts, mutations are Gly2Ala, Cys4Ala, Cys5Ala, Cys7AlaDepositorInsertVAC8 (VAC8 Budding Yeast)
TagsGFPExpressionYeastMutationG2A, C4A, C5A, C7APromoterVAC8 promoterAvailable SinceApril 2, 2021AvailabilityAcademic Institutions and Nonprofits only