We narrowed to 40,900 results for: Eras
-
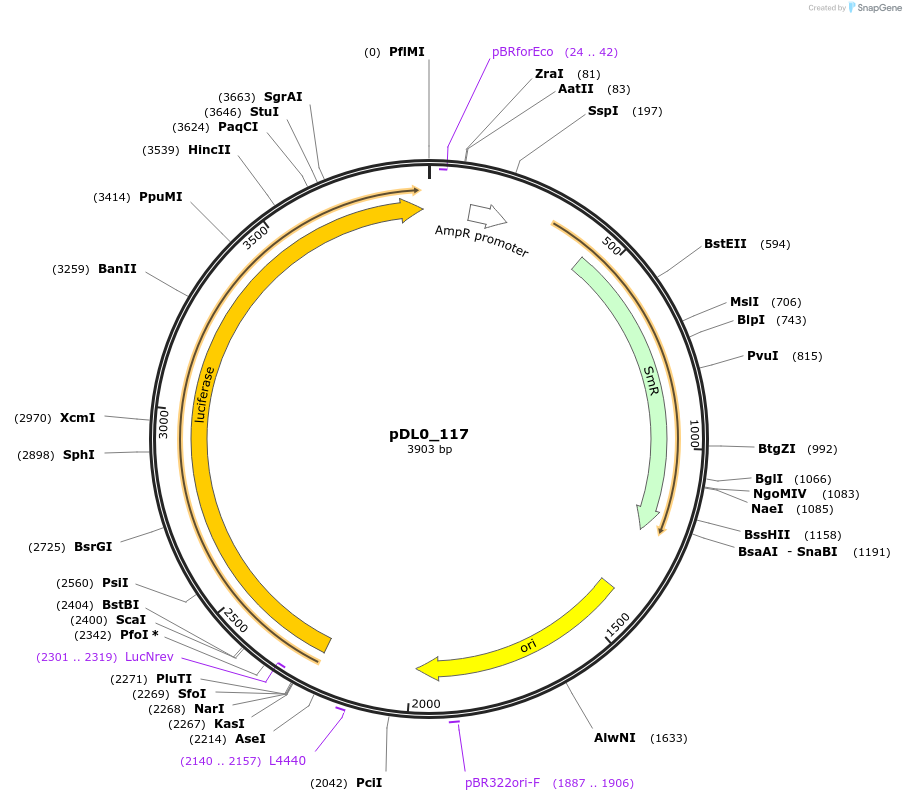
Plasmid#210864PurposeSC_Photinus pryalis Firefly Luciferase coding sequenceDepositorInsertFirefly Luciferase
UseLuciferase and Synthetic BiologyExpressionPlantAvailable SinceJune 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
pDL0_198
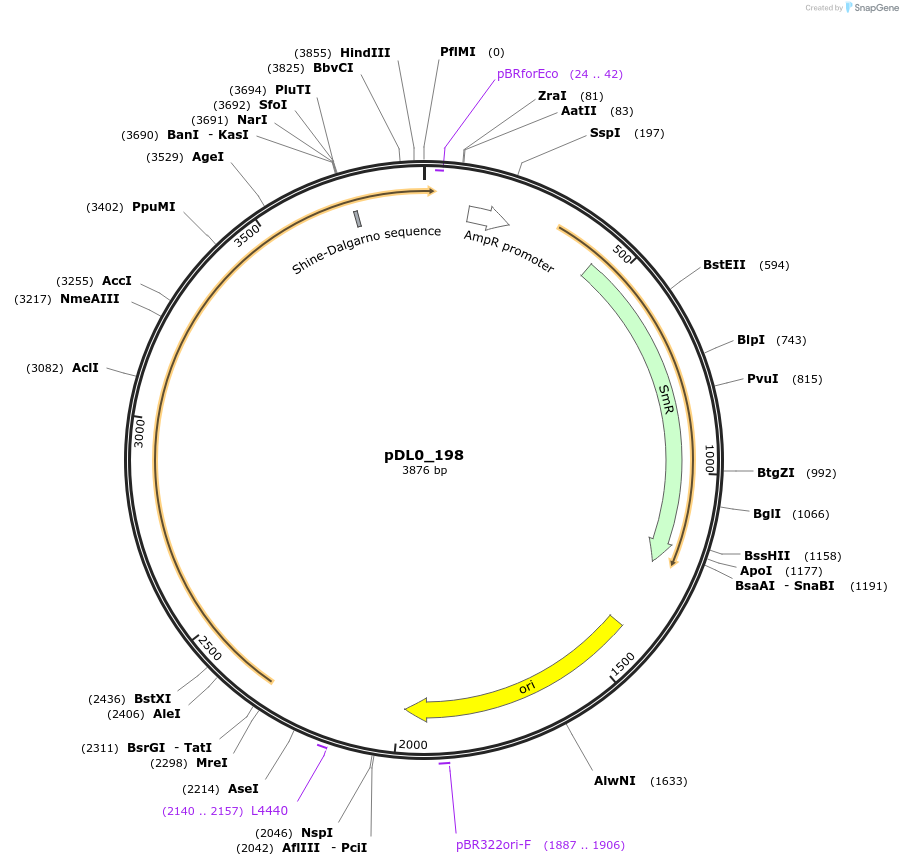
Plasmid#210865PurposeC1_Pyrearinus termitilluminans beetle luciferase ('Eluc' version - human-codon optimized)DepositorInsertPyrearinus termitilluminans beetle luciferase
UseLuciferase and Synthetic BiologyExpressionPlantAvailable SinceJune 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
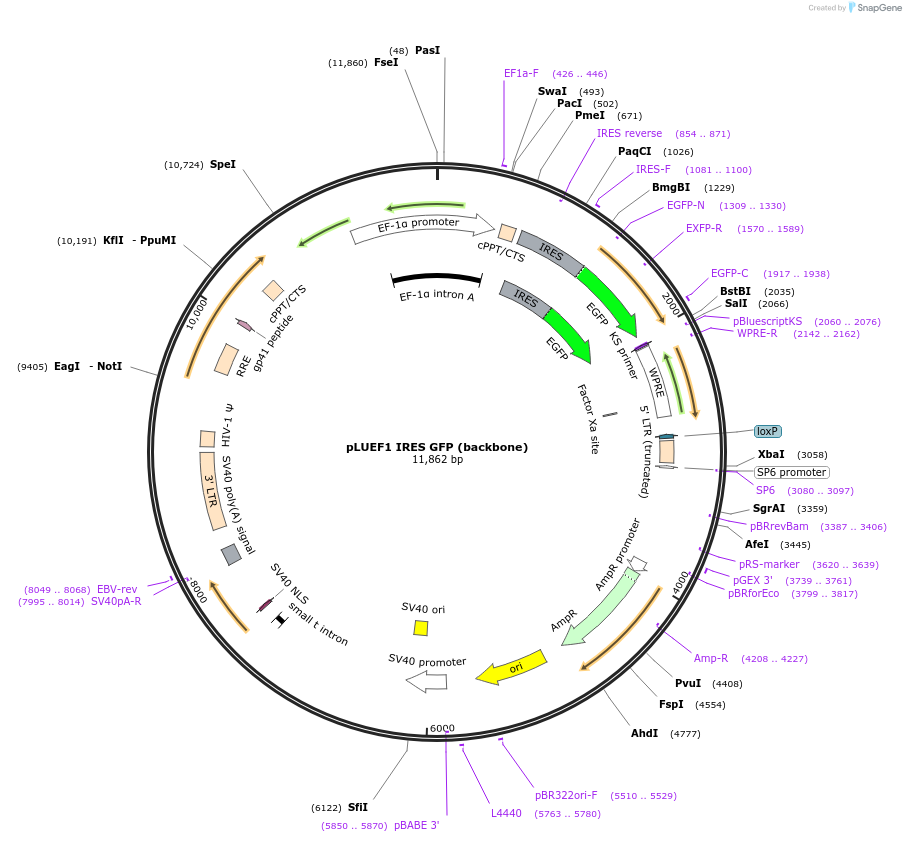
pLUEF1 IRES GFP (backbone)
Plasmid#224466PurposeEf1a downstream of a ubiquitous chromatin opening element drives expression of GOI and IRES GFPDepositorTypeEmpty backboneUseLentiviralPromoterEf1aAvailable SinceApril 22, 2025AvailabilityAcademic Institutions and Nonprofits only -
Galt1-HT7-mGold
Plasmid#230027PurposeExpressing fluorescent HaloTag7 fusion protein in mammalian cells with localization in the Golgi apparatus.DepositorAvailable SinceJan. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
-
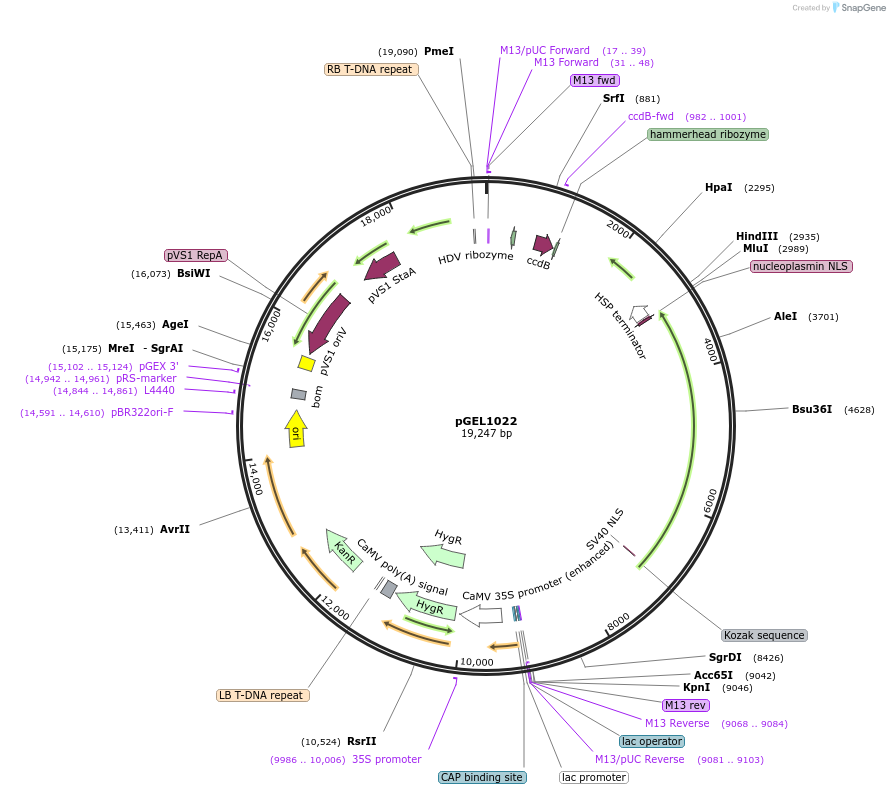
pGEL1022
Plasmid#225593PurposeThe backbone vector for rice genome editing is based on HkCas12aDepositorTypeEmpty backboneUseCRISPRAvailable SinceDec. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
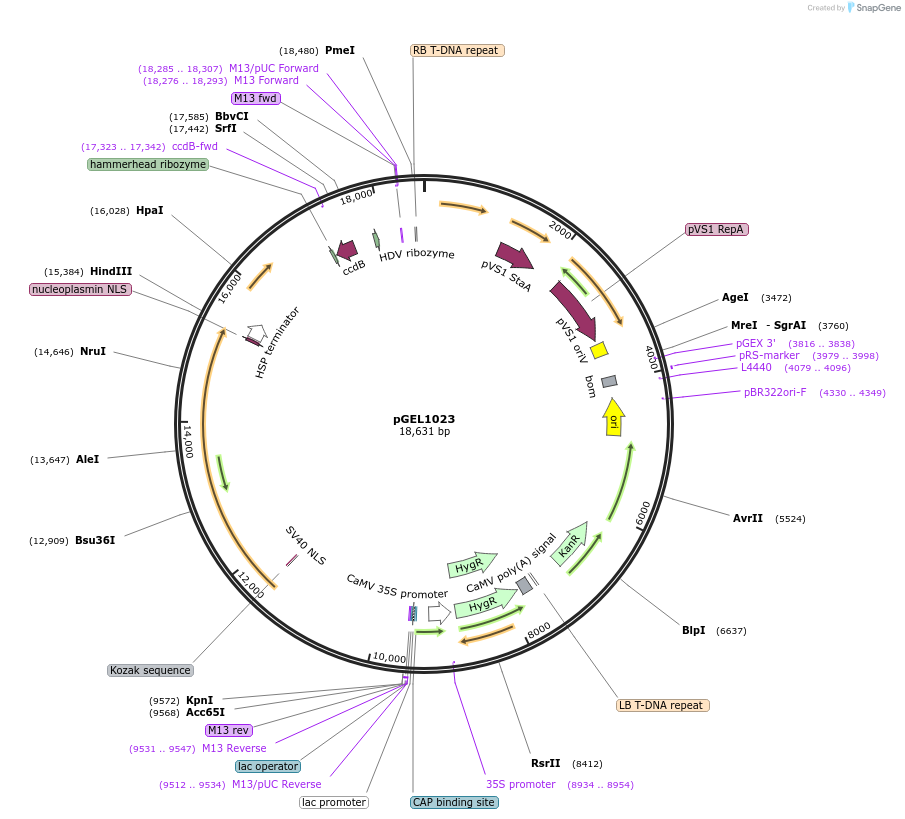
pGEL1023
Plasmid#225594PurposeThe backbone vector for rice genome editing is based on PrCas12aDepositorTypeEmpty backboneUseCRISPRAvailable SinceDec. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
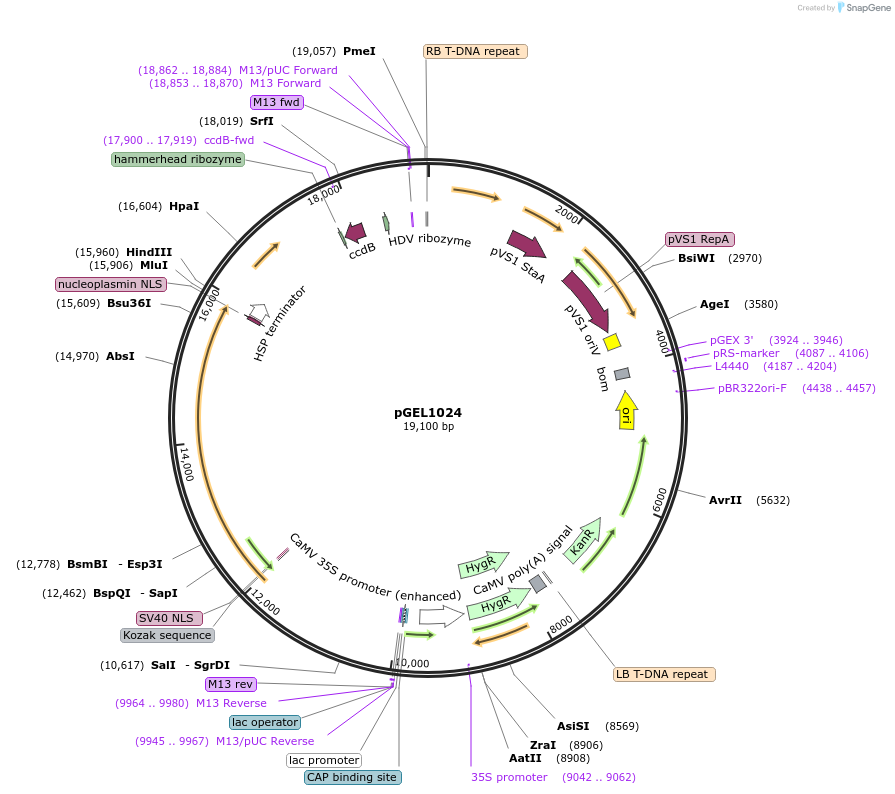
pGEL1024
Plasmid#225595PurposeThe Dual Pol II backbone vector for rice genome editing is based on Mb3Cas12aDepositorTypeEmpty backboneUseCRISPRAvailable SinceDec. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
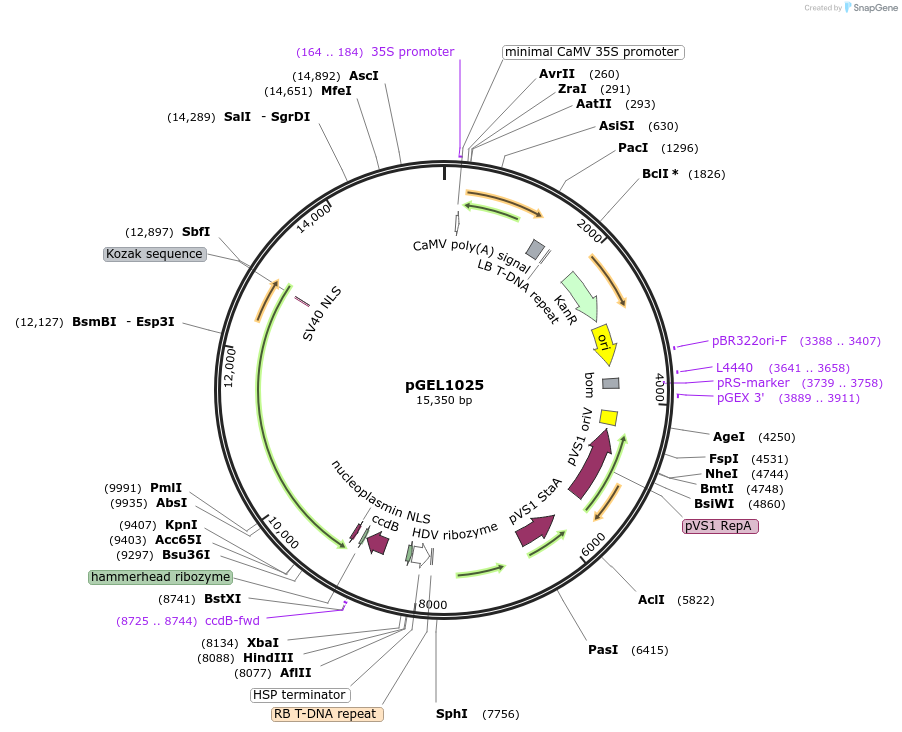
pGEL1025
Plasmid#225596PurposeThe STU backbone vector for rice genome editing is based on Mb3Cas12aDepositorTypeEmpty backboneUseCRISPRAvailable SinceDec. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
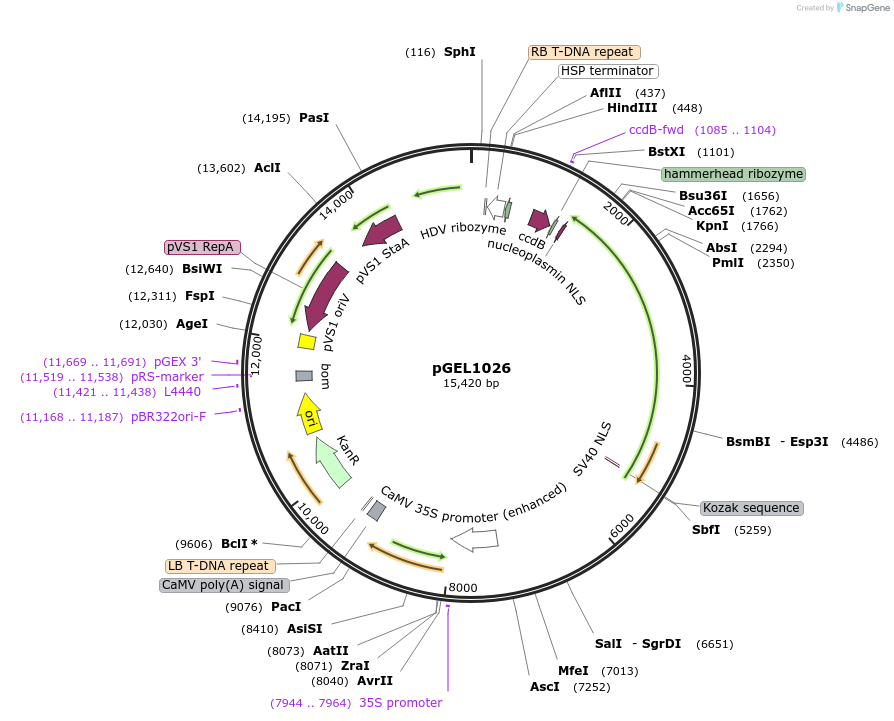
pGEL1026
Plasmid#225597PurposeThe STU backbone vector for rice genome editing is based on Mb3Cas12a-R (D172R)DepositorTypeEmpty backboneUseCRISPRAvailable SinceDec. 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
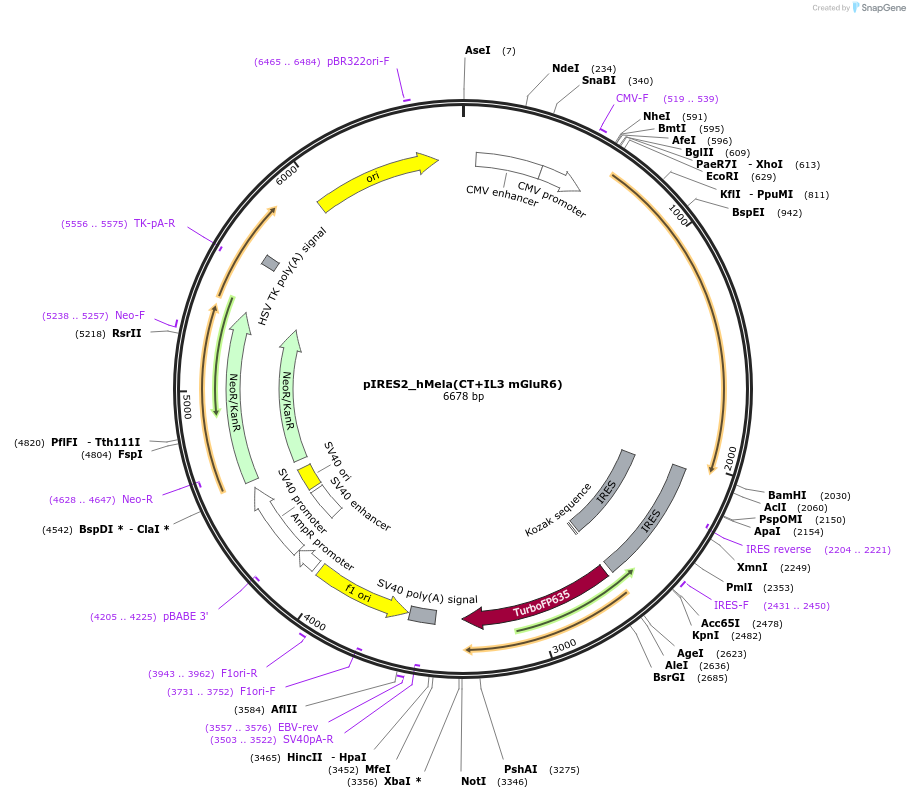
pIRES2_hMela(CT+IL3 mGluR6)
Plasmid#191343PurposeOptogenetic toolDepositorInserthMela(CT+IL3 mGluR6)
TagsIRES_TurboExpressionMammalianAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
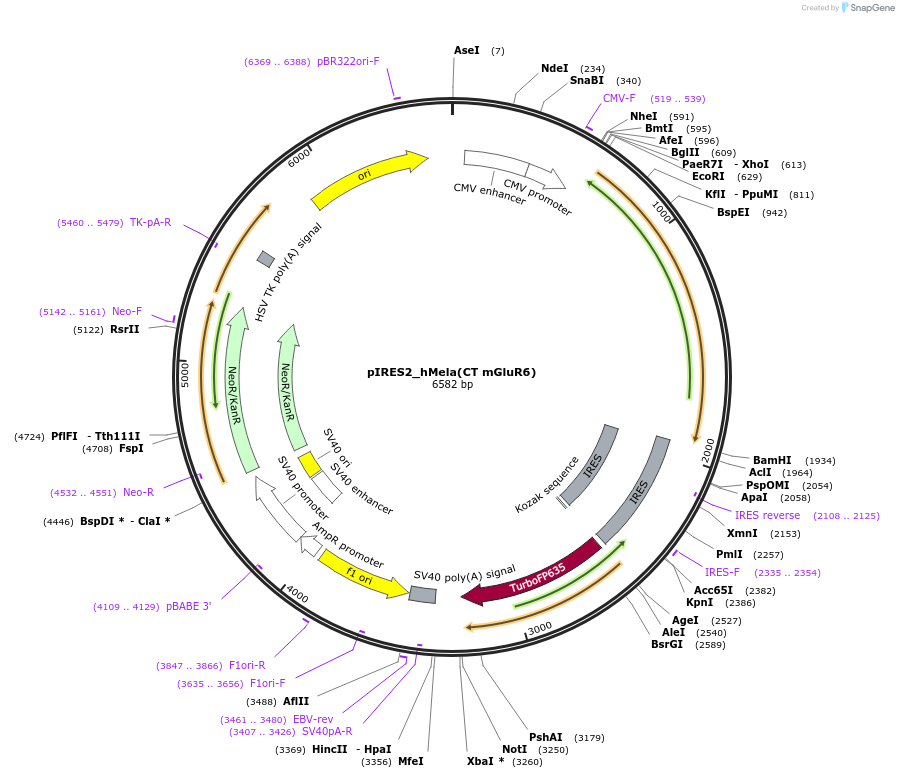
pIRES2_hMela(CT mGluR6)
Plasmid#191344PurposeOptogenetic toolDepositorInserthMela(CTmGluR6)
TagsIRES_TurboExpressionMammalianPromoterCMVAvailable SinceDec. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
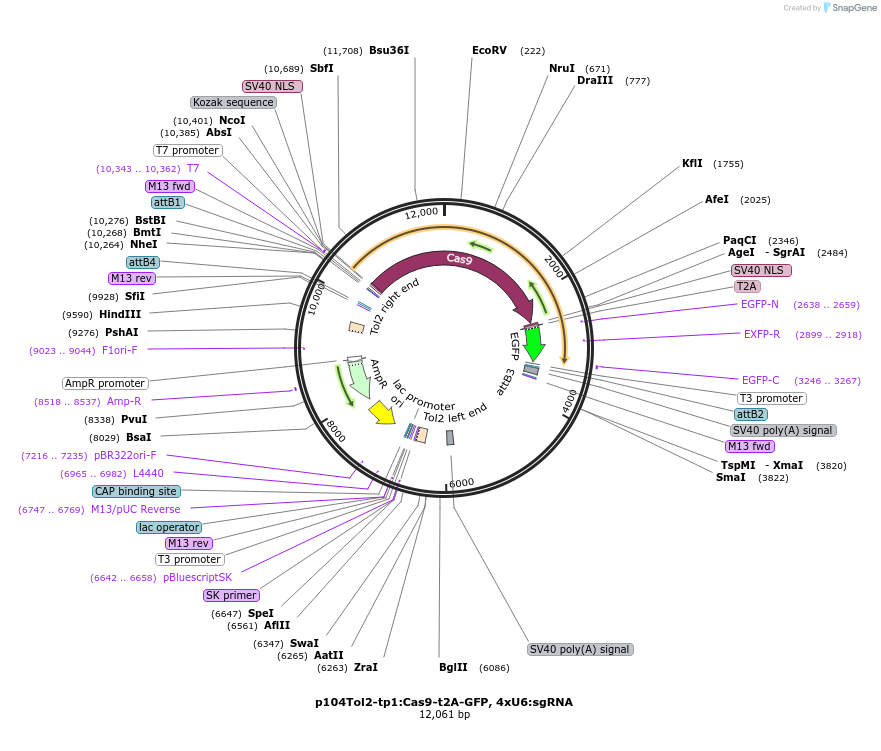
p104Tol2-tp1:Cas9-t2A-GFP, 4xU6:sgRNA
Plasmid#227773PurposeExpression of tp1 (Notch) dependent Cas9-GFP and U6-driven 4 sgRNAs in zebrafishDepositorInsertsCas9-t2A-GFP
4xU6:sgRNA
UseCRISPRPromoterU6 and tp1Available SinceDec. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
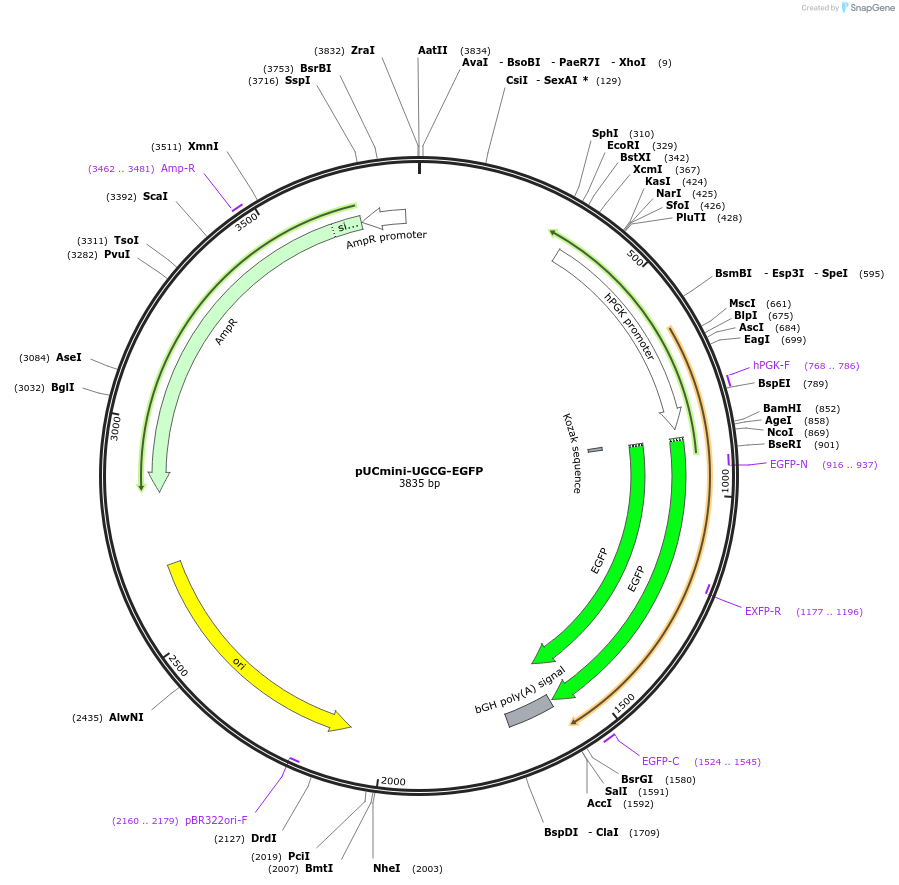
pUCmini-UGCG-EGFP
Plasmid#225878PurposePlasmid donor template for CRISPR/Cas9 mediated UGCG knockout by EGFP insertionDepositorInsertEGFP
UseCRISPRExpressionMammalianAvailable SinceOct. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
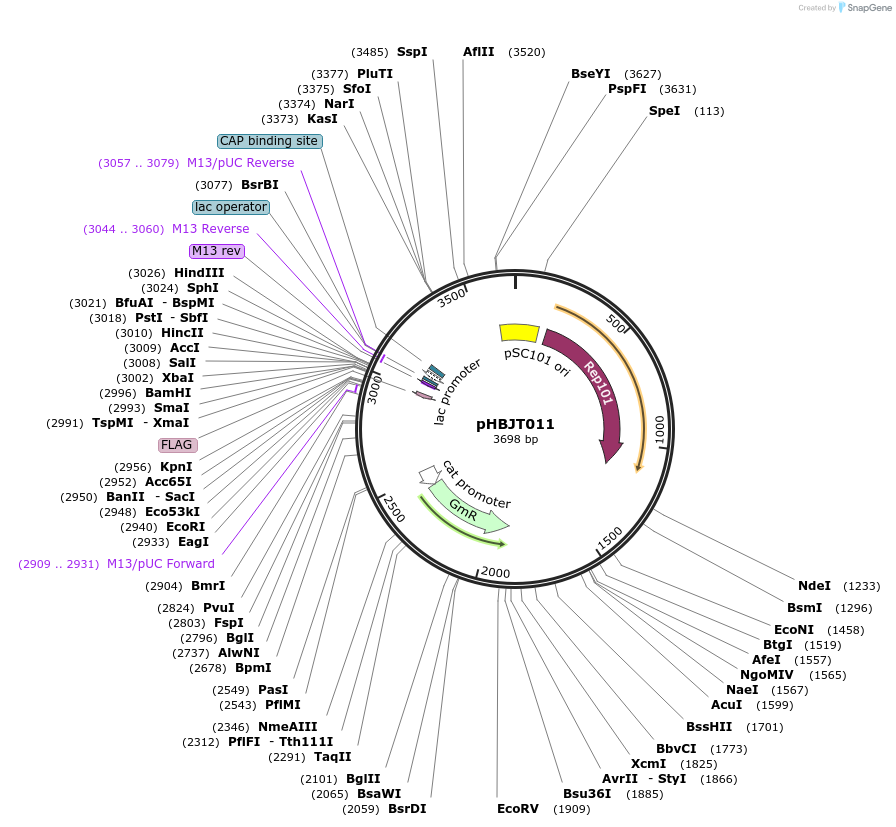
pHBJT011
Plasmid#225162PurposeLow copy cloning vector for integration of C-terminal FLAG epitope tag (GmR). SmaI restriction site immediately upstream of the FLAG epitope. FLAG epitope in same orientation as lac promoter.DepositorTypeEmpty backboneUseSynthetic BiologyTagsFLAG epitope tag (35 bp)ExpressionBacterialAvailable SinceSept. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
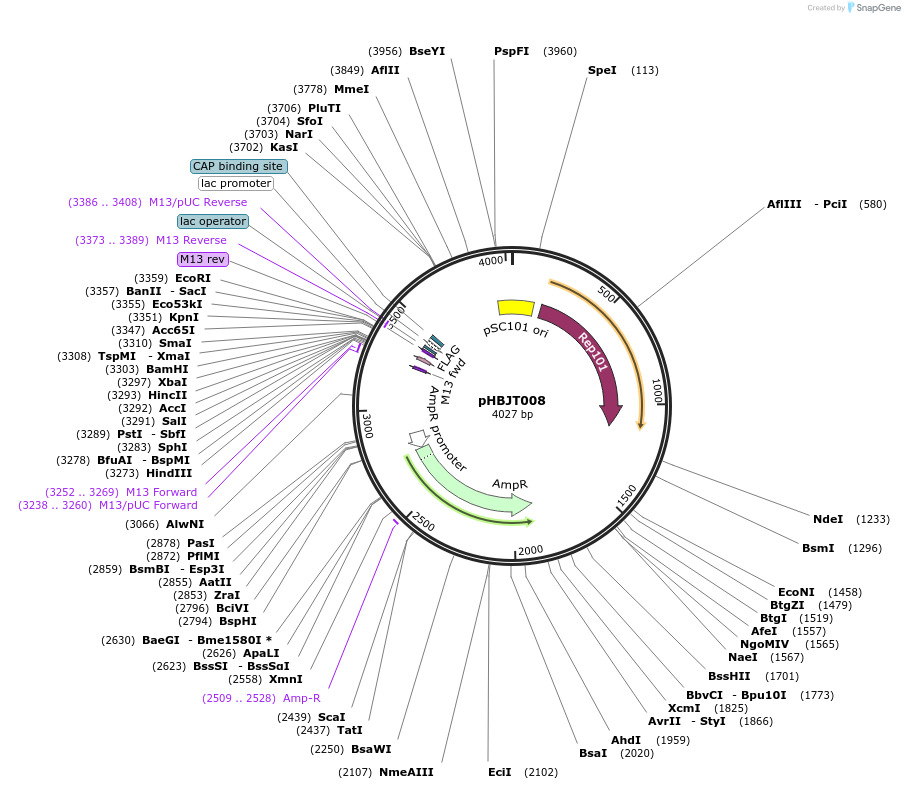
pHBJT008
Plasmid#225159PurposeLow copy cloning vector for integration of C-terminal FLAG epitope tag (AmpR). SmaI restriction site immediately upstream of the FLAG epitope. FLAG epitope in opposite orientation of lac promoter.DepositorTypeEmpty backboneUseSynthetic BiologyTagsFLAG epitope tag (35 bp)ExpressionBacterialAvailable SinceSept. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
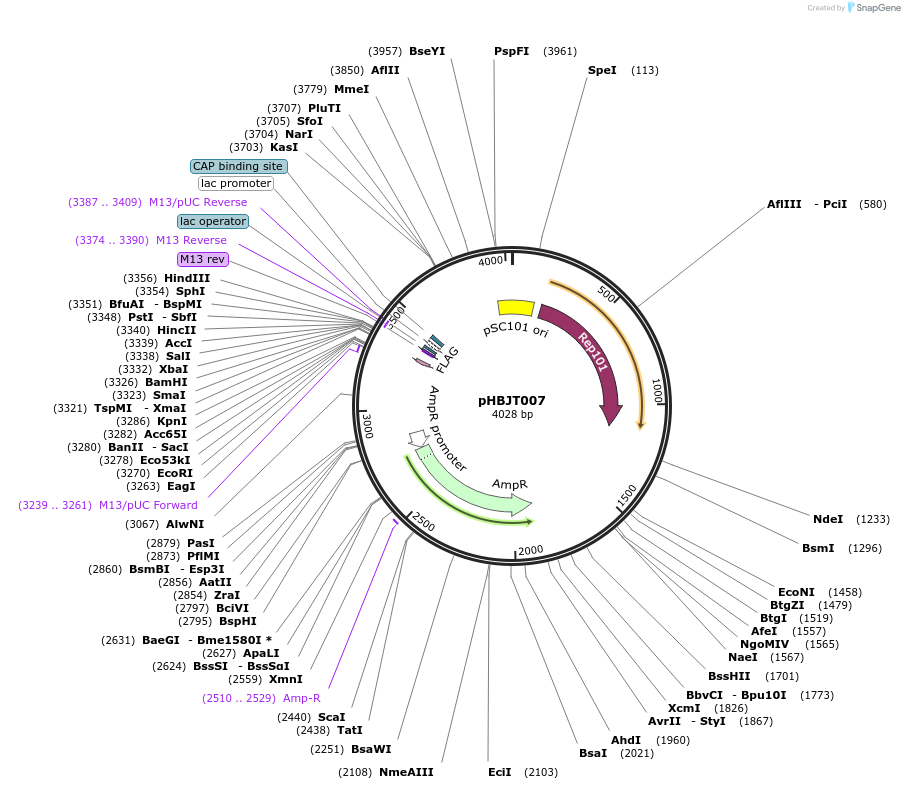
pHBJT007
Plasmid#225158PurposeLow copy cloning vector for integration of C-terminal FLAG epitope tag (AmpR). SmaI restriction site immediately upstream of the FLAG epitope. FLAG epitope in same orientation as lac promoter.DepositorTypeEmpty backboneUseSynthetic BiologyTagsFLAG epitope tag (35 bp)ExpressionBacterialAvailable SinceSept. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
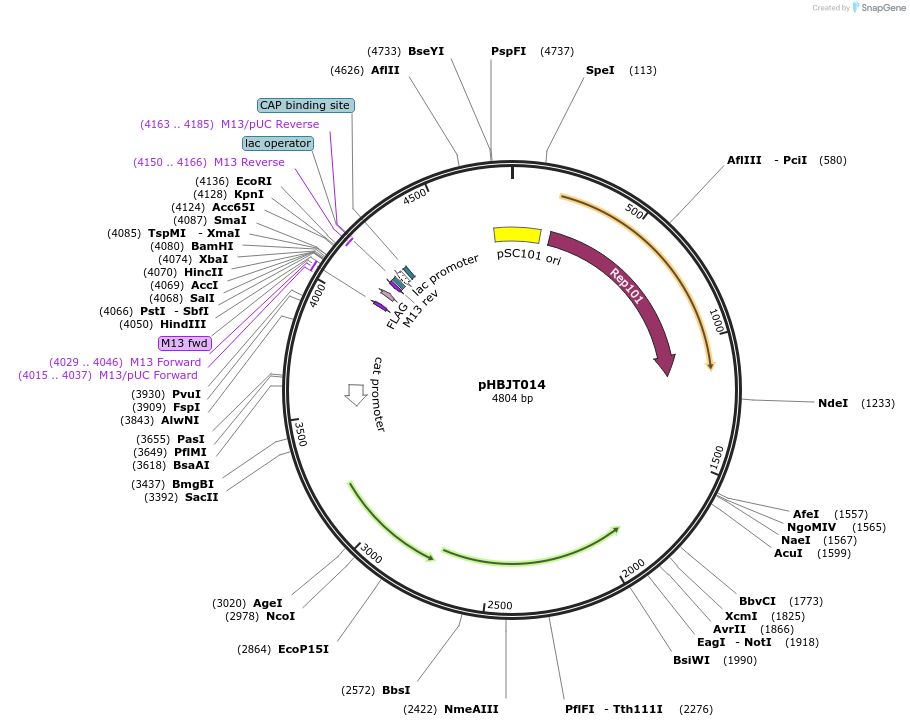
pHBJT014
Plasmid#225165PurposeLow copy cloning vector for integration of C-terminal FLAG epitope tag (SmR). SmaI restriction site immediately upstream of the FLAG epitope. FLAG epitope in opposite orientation of lac promoter.DepositorTypeEmpty backboneUseSynthetic BiologyTagsFLAG epitope tag (35 bp)ExpressionBacterialAvailable SinceSept. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
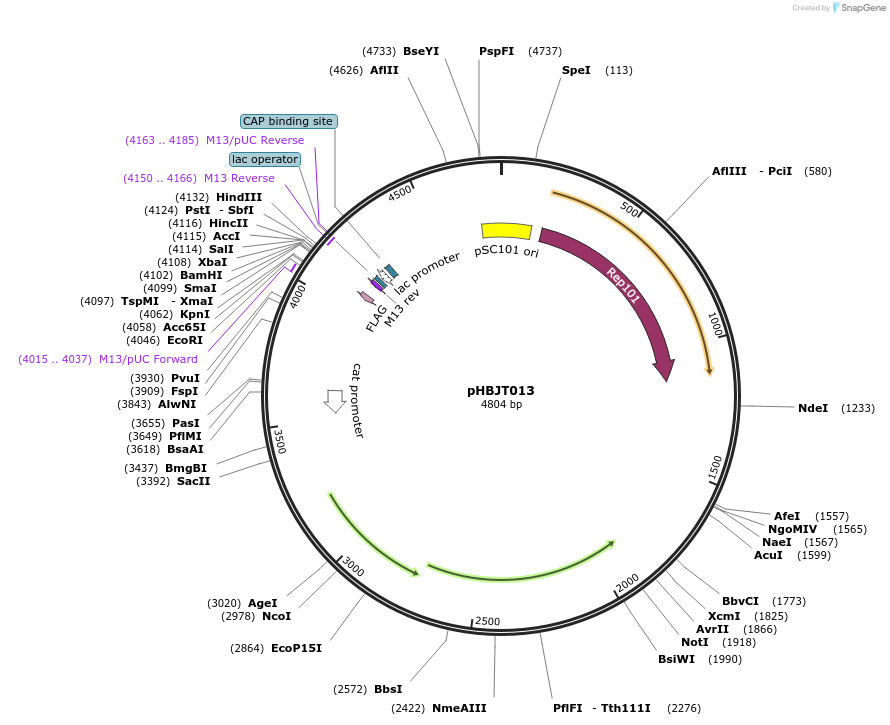
pHBJT013
Plasmid#225164PurposeLow copy cloning vector for integration of C-terminal FLAG epitope tag (SmR). SmaI restriction site immediately upstream of the FLAG epitope. FLAG epitope in same orientation as lac promoter.DepositorTypeEmpty backboneUseSynthetic BiologyTagsFLAG epitope tag (35 bp)ExpressionBacterialAvailable SinceSept. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
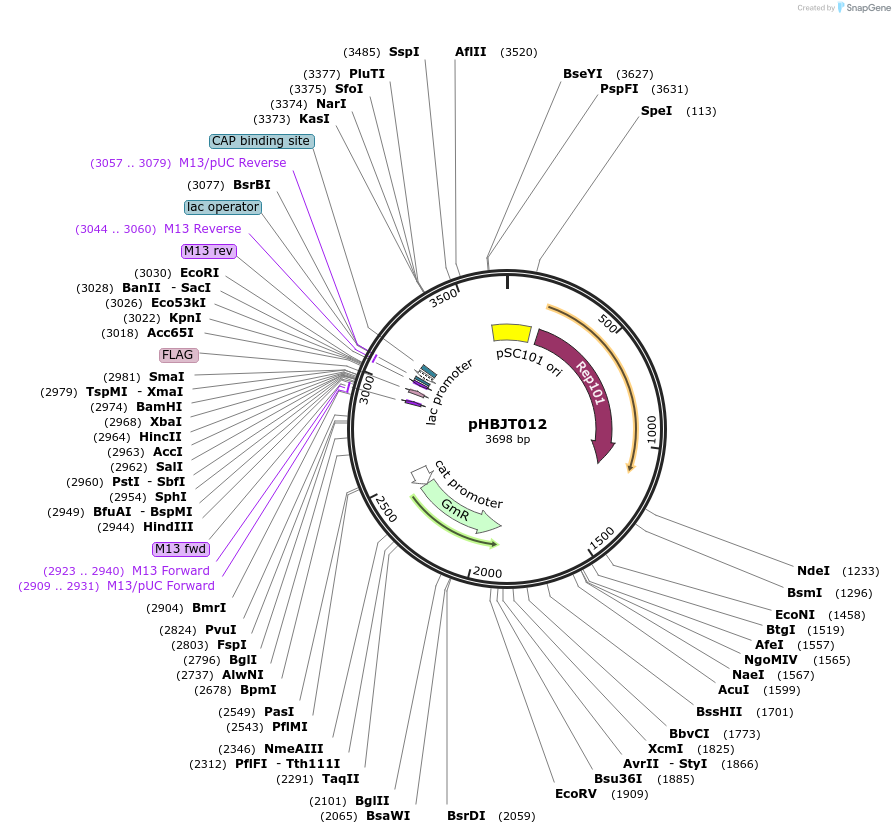
pHBJT012
Plasmid#225163PurposeLow copy cloning vector for integration of C-terminal FLAG epitope tag (GmR). SmaI restriction site immediately upstream of the FLAG epitope. FLAG epitope in opposite orientation of lac promoter.DepositorTypeEmpty backboneUseSynthetic BiologyTagsFLAG epitope tag (35 bp)ExpressionBacterialAvailable SinceSept. 10, 2024AvailabilityAcademic Institutions and Nonprofits only