We narrowed to 8,624 results for: FIE
-
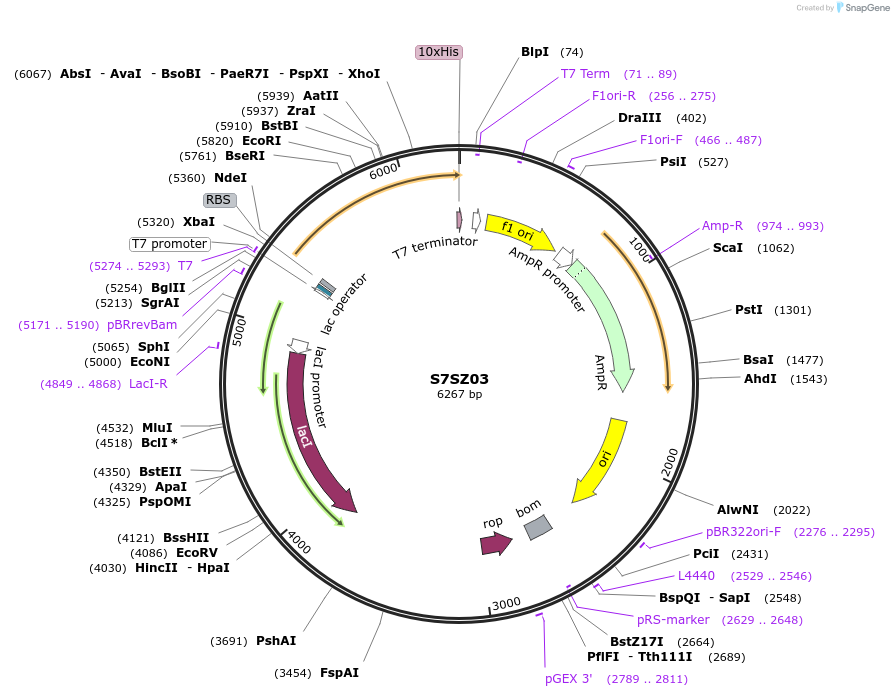
Plasmid#163199PurposeInducible expression of UniProt identifier S7SZ03DepositorInsertS7SZ03
Tags10xHis and T7ExpressionBacterialPromoterT7Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
A0A085FLP7
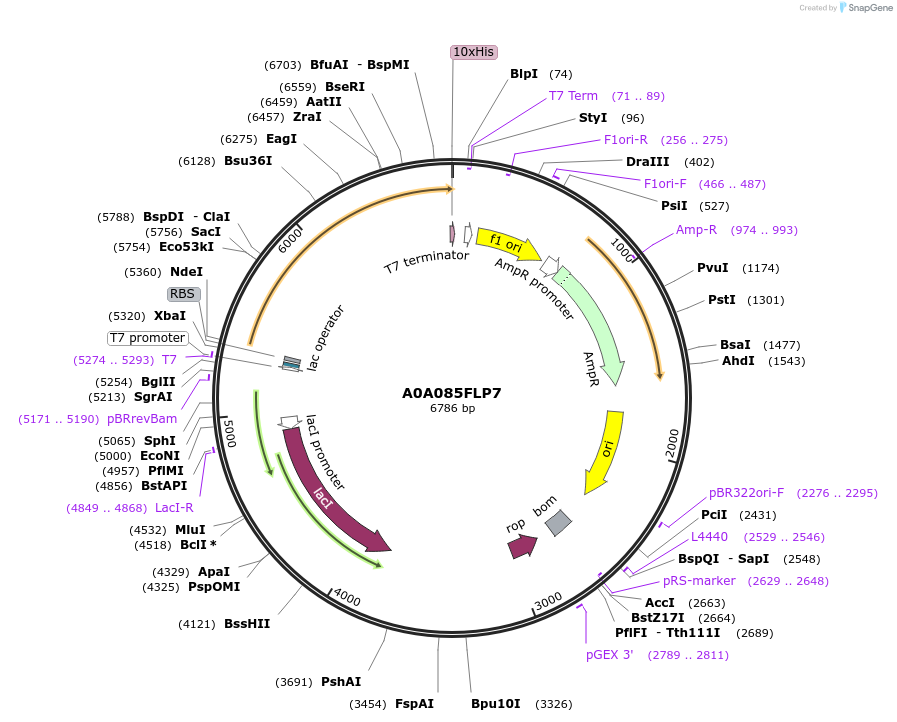
Plasmid#163205PurposeInducible expression of UniProt identifier A0A085FLP7DepositorInsertA0A085FLP7
Tags10xHis and T7ExpressionBacterialPromoterT7Available SinceFeb. 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
M5FIH9
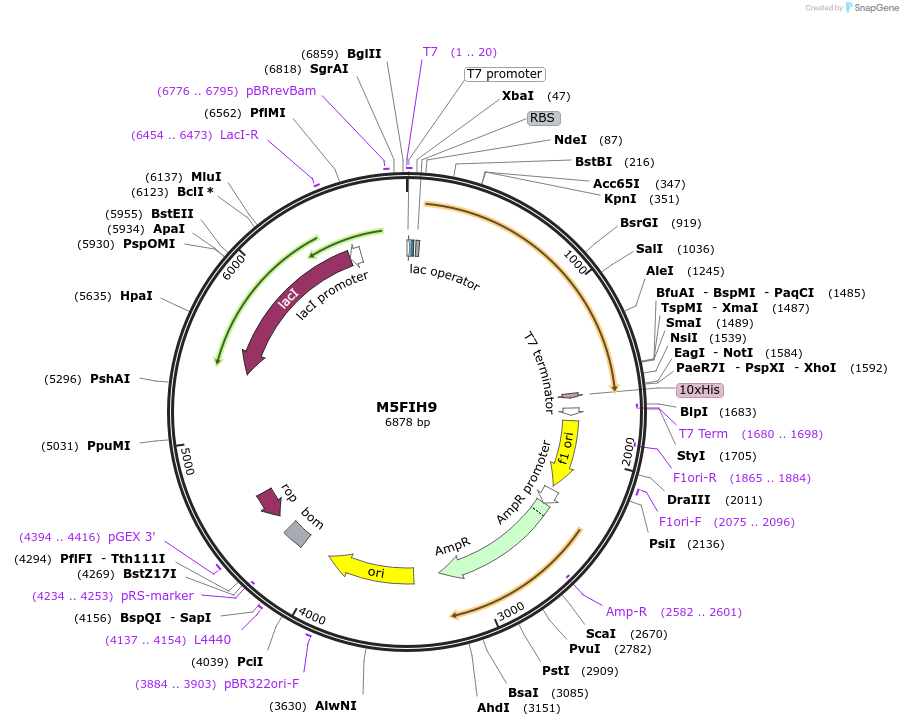
Plasmid#163299PurposeInducible expression of UniProt identifier M5FIH9DepositorInsertM5FIH9
Tags10xHis and T7ExpressionBacterialPromoterT7Available SinceFeb. 14, 2022AvailabilityAcademic Institutions and Nonprofits only -
Q0I8G2
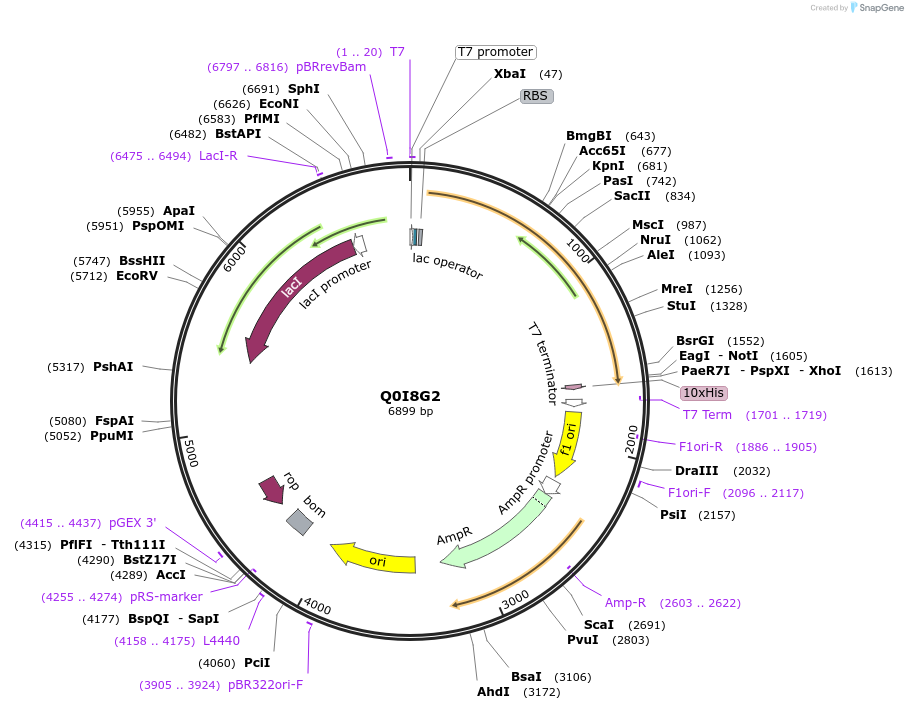
Plasmid#163283PurposeInducible expression of UniProt identifier Q0I8G2DepositorInsertQ0I8G2
Tags10xHis and T7ExpressionBacterialPromoterT7Available SinceFeb. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
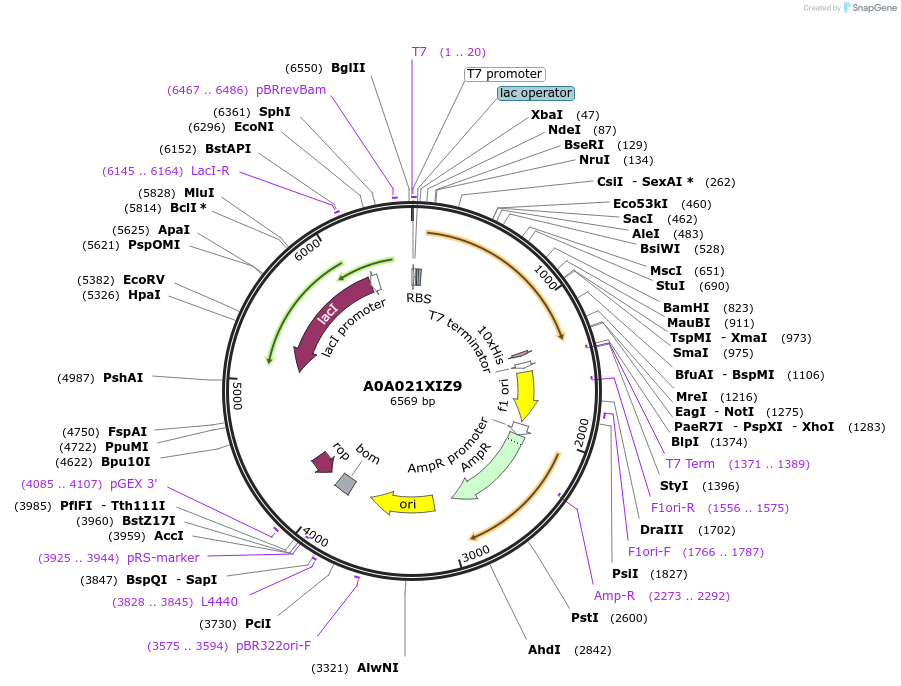
A0A021XIZ9
Plasmid#163296PurposeInducible expression of UniProt identifier A0A021XIZ9DepositorInsertA0A021XIZ9
Tags10xHis and T7ExpressionBacterialPromoterT7Available SinceFeb. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
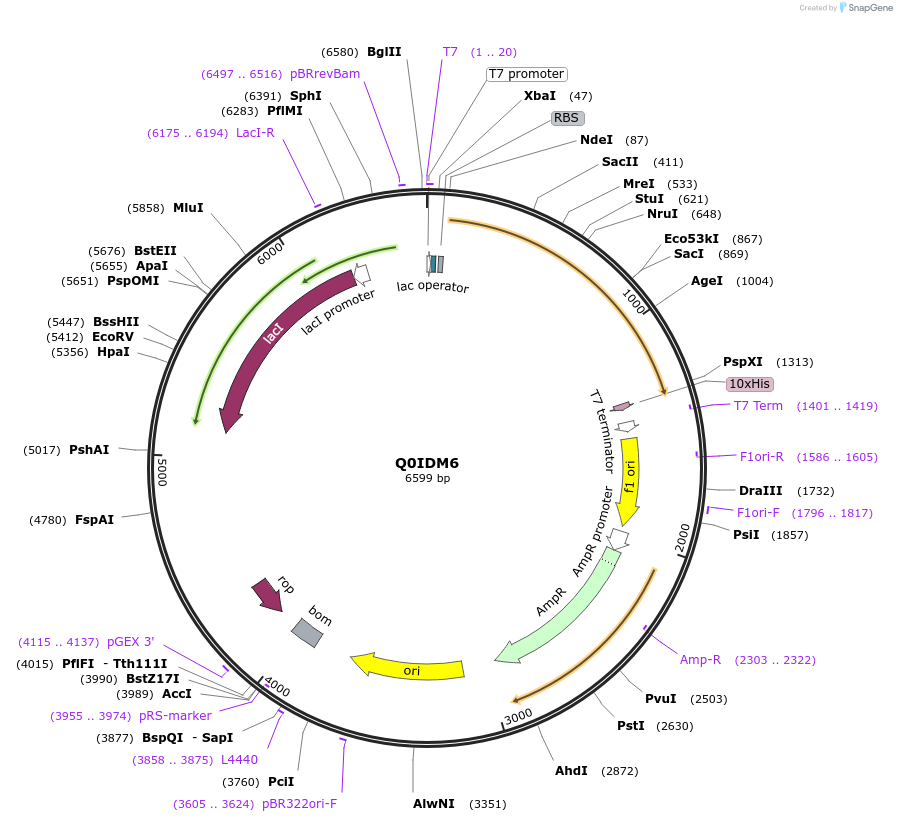
Q0IDM6
Plasmid#163284PurposeInducible expression of UniProt identifier Q0IDM6DepositorInsertQ0IDM6
Tags10xHis and T7ExpressionBacterialPromoterT7Available SinceFeb. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
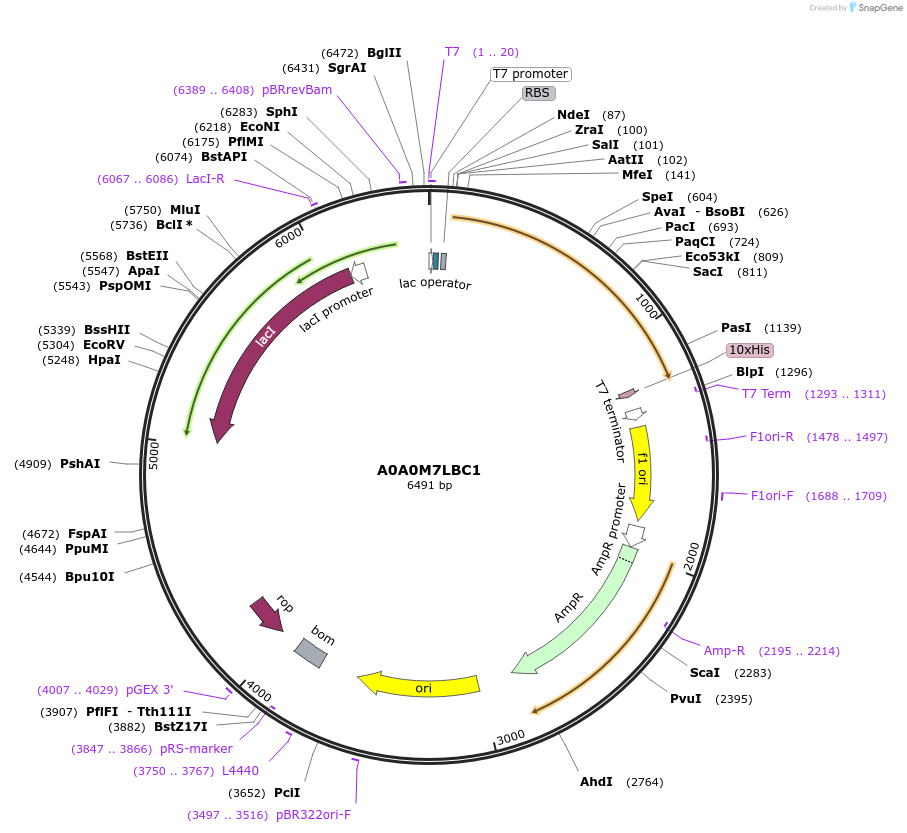
A0A0M7LBC1
Plasmid#163181PurposeInducible expression of UniProt identifier A0A0M7LBC1DepositorInsertA0A0M7LBC1
Tags10xHis and T7ExpressionBacterialPromoterT7Available SinceFeb. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
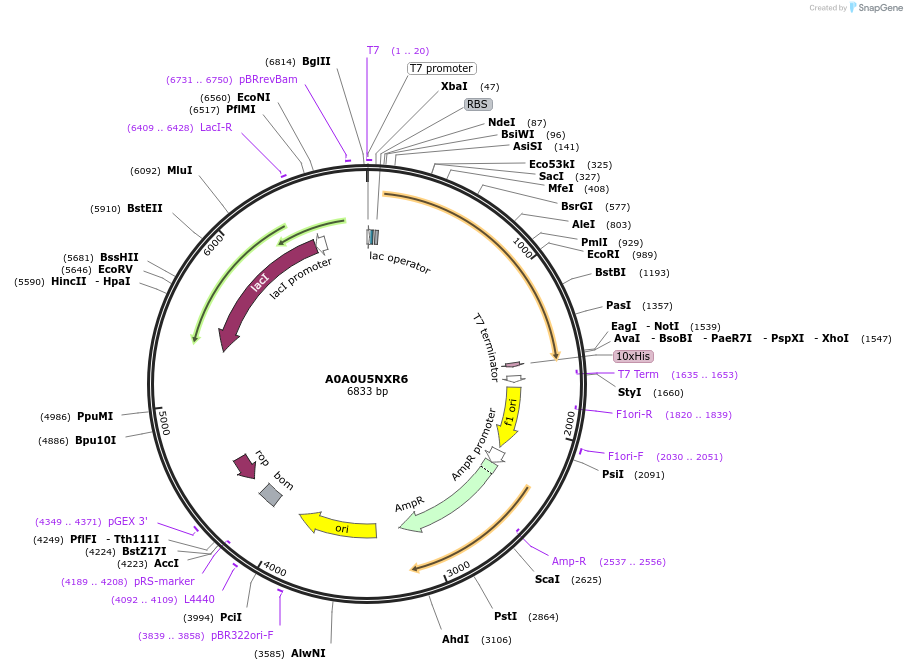
A0A0U5NXR6
Plasmid#163265PurposeInducible expression of UniProt identifier A0A0U5NXR6DepositorInsertA0A0U5NXR6
Tags10xHis and T7ExpressionBacterialPromoterT7Available SinceFeb. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
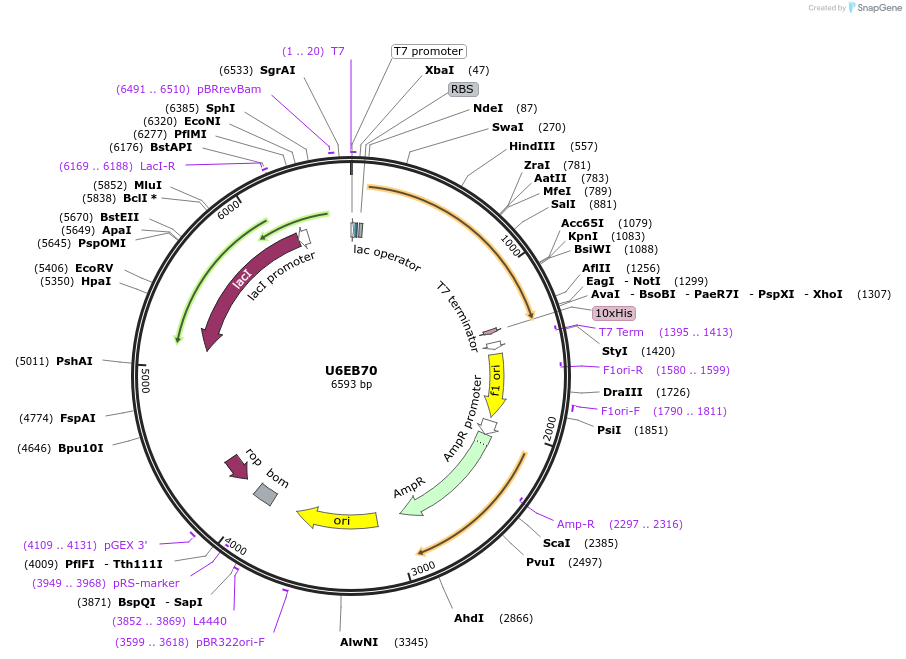
U6EB70
Plasmid#163293PurposeInducible expression of UniProt identifier U6EB70DepositorInsertU6EB70
Tags10xHis and T7ExpressionBacterialPromoterT7Available SinceFeb. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
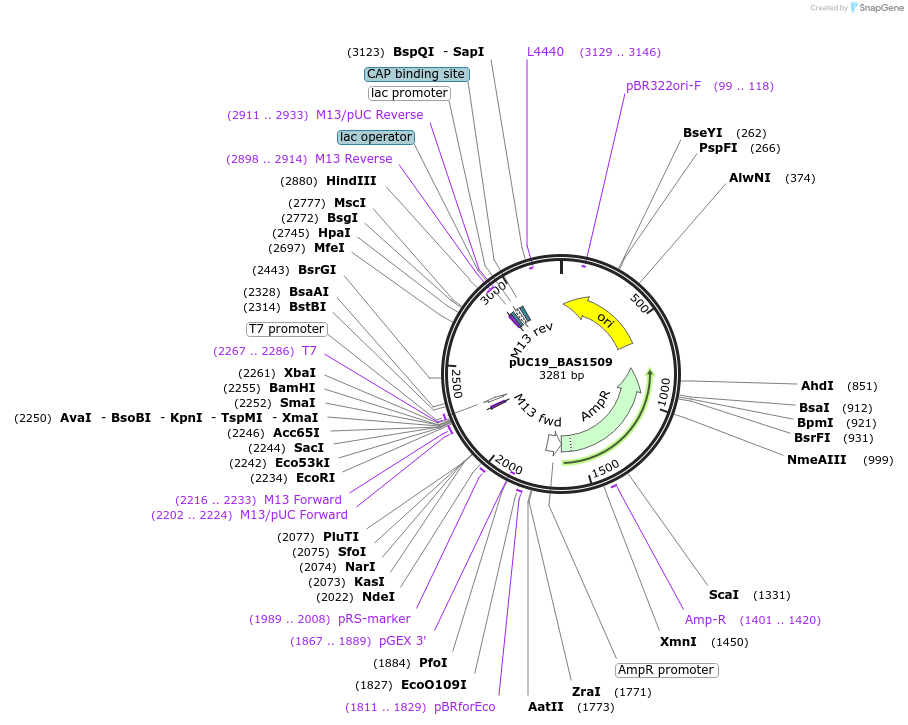
pUC19_BAS1509
Plasmid#82354PurposeT7 in vitro transcription template for Bas mRNADepositorInsertBAS1509 hypothetical protein
UseUnspecifiedPromoterT7Available SinceFeb. 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
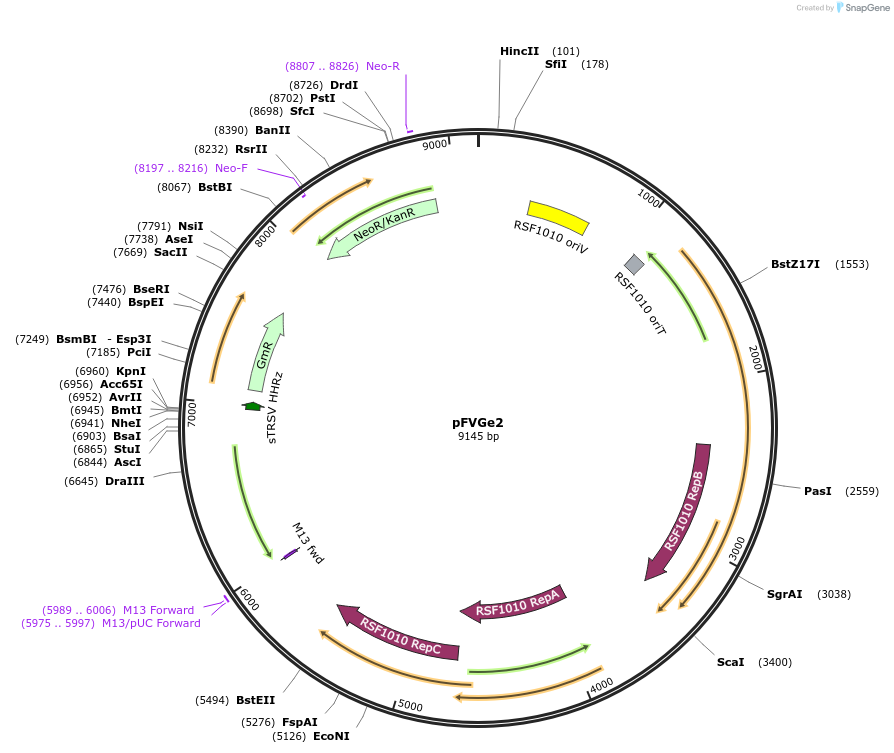
pFVGe2
Plasmid#149500PurposeBroad-Host range expression vector, RSF1010 origin, VanR(AM) repressor, PVanCC promoter, GmR, Kanamycin Resistance Marker.DepositorInsertVanR(AM)-GmR
UseSynthetic BiologyExpressionBacterialPromoterPVanCCAvailable SinceMarch 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
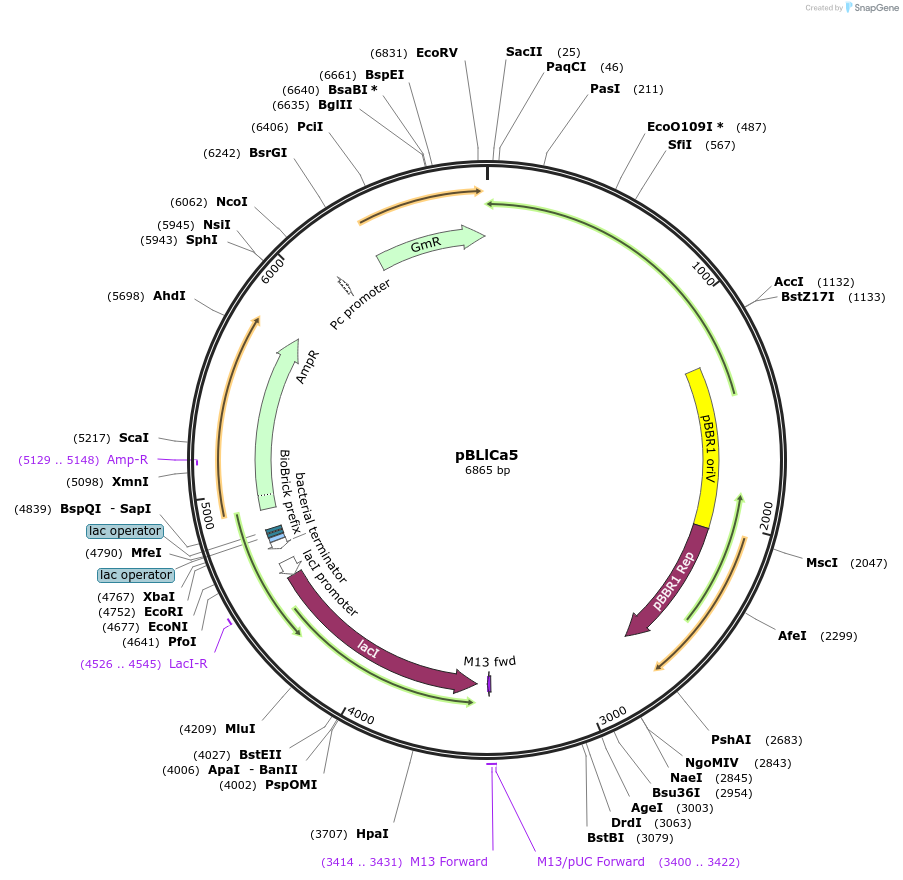
pBLlCa5
Plasmid#149501PurposeBroad-Host range expression vector, pBBR origin, LacI repressor, PLlacO-1 promoter, AmpR, Gentamicin Resistance Marker.DepositorInsertLacI-AmpR
UseSynthetic BiologyExpressionBacterialPromoterPLlacO-1Available SinceMarch 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
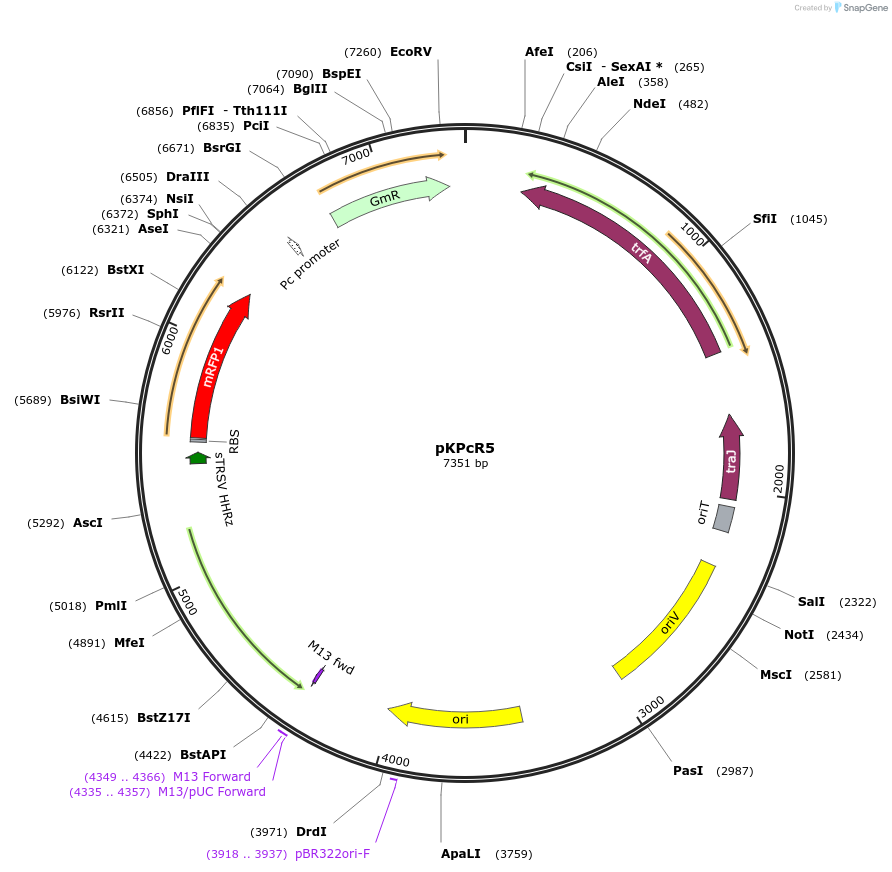
pKPcR5
Plasmid#149467PurposeBroad-Host range expression vector, RK2 and pUC origin, PcaU(AM) repressor, P3B5B promoter, mRFP, Gentamicin Resistance Marker.DepositorInsertPcaU(AM)-mRFP
UseSynthetic BiologyExpressionBacterialPromoterP3B5BAvailable SinceMarch 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
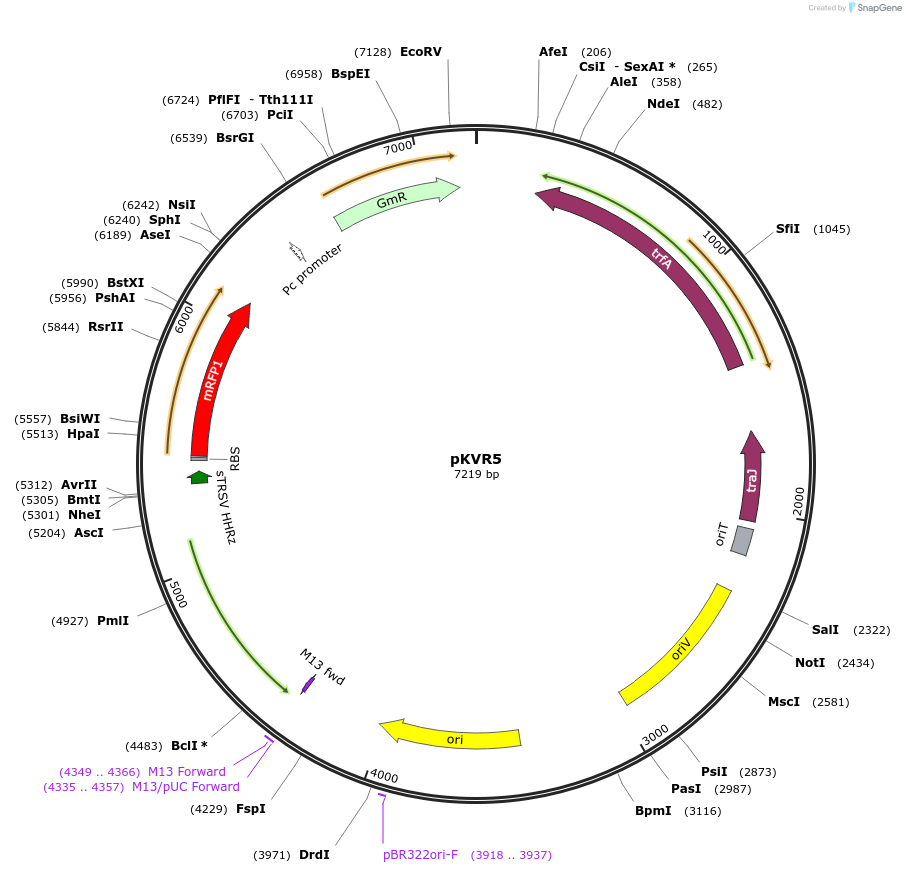
pKVR5
Plasmid#149469PurposeBroad-Host range expression vector, RK2 and pUC origin, VanR(AM) repressor, PVanCC promoter, mRFP, Gentamicin Resistance Marker.DepositorInsertVanR(AM)-mRFP
UseSynthetic BiologyExpressionBacterialPromoterPVanCCAvailable SinceMarch 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
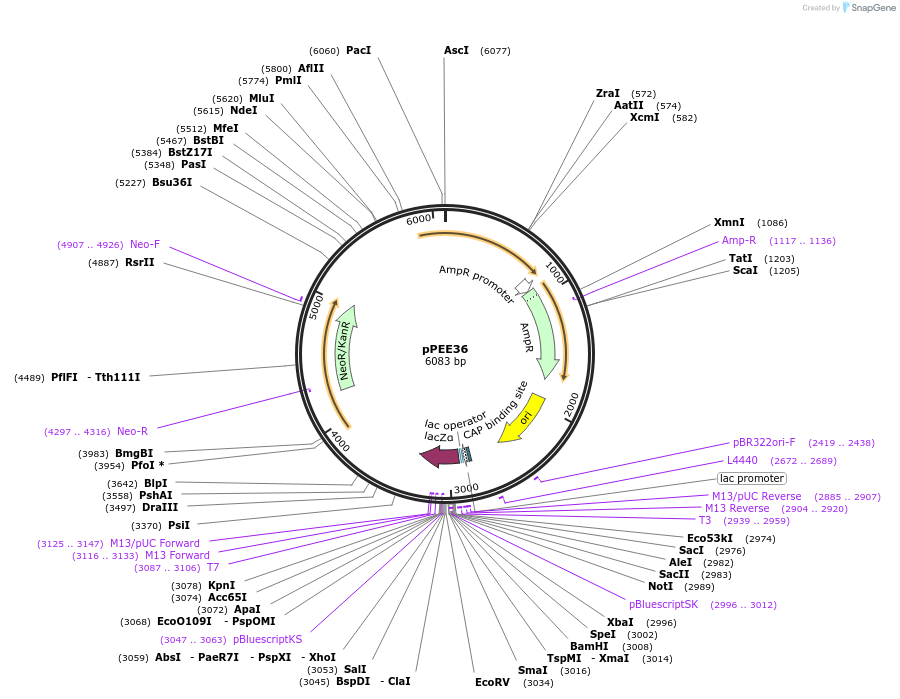
pPEE36
Plasmid#128376PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceApril 28, 2020AvailabilityAcademic Institutions and Nonprofits only -
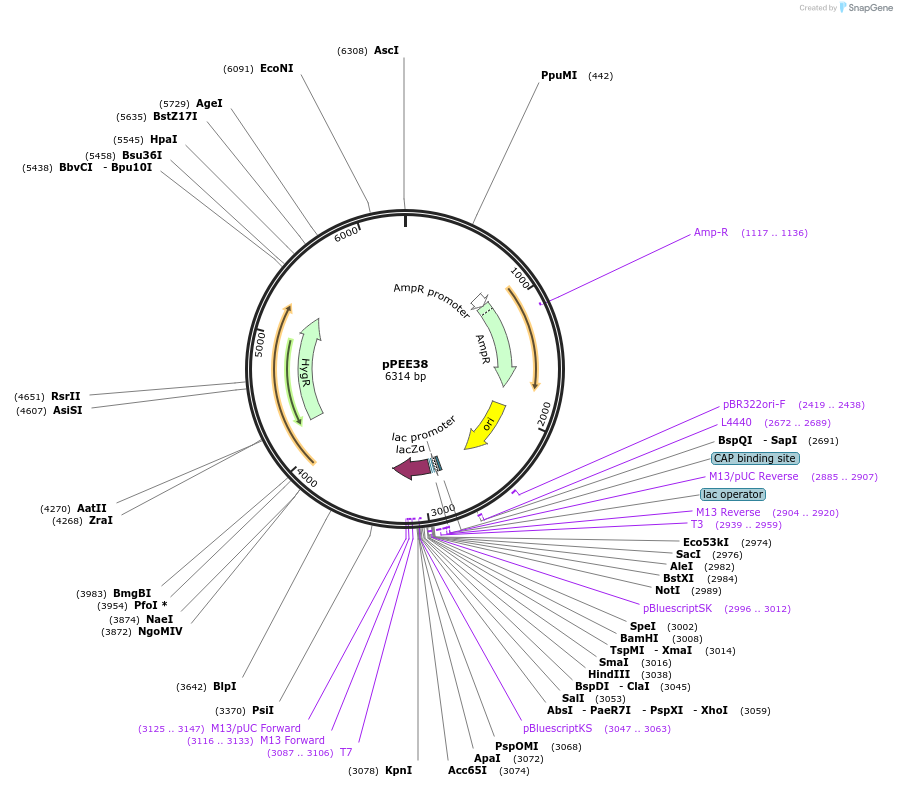
pPEE38
Plasmid#128378PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
pPEE32
Plasmid#128372PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
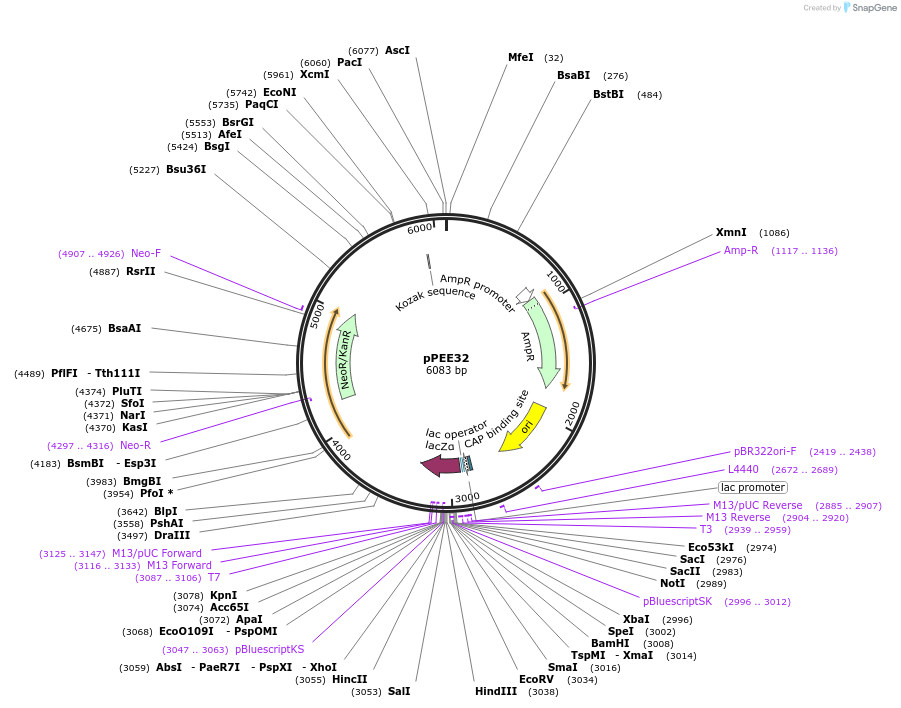
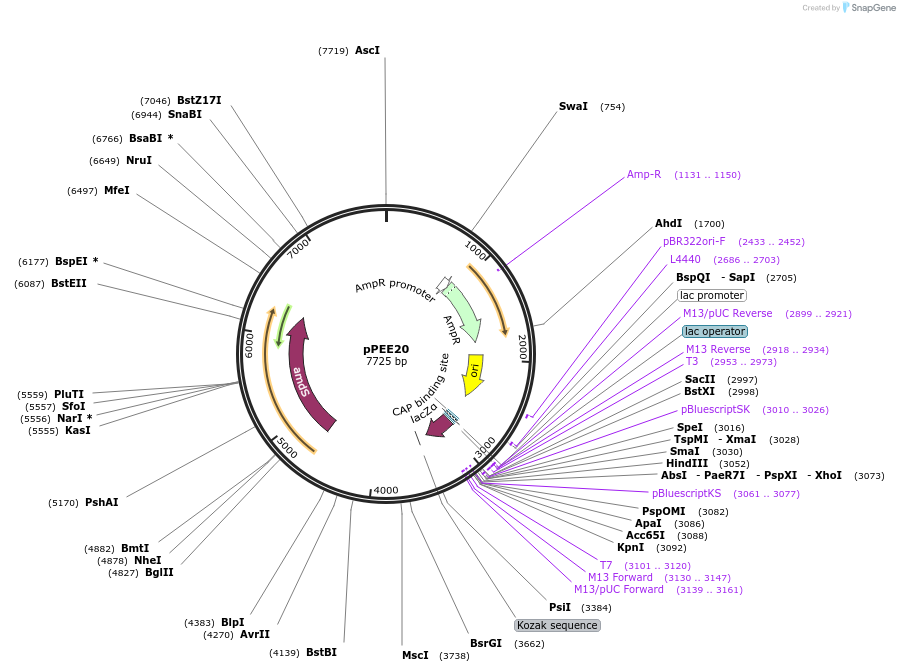
pPEE20
Plasmid#128363PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceOct. 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
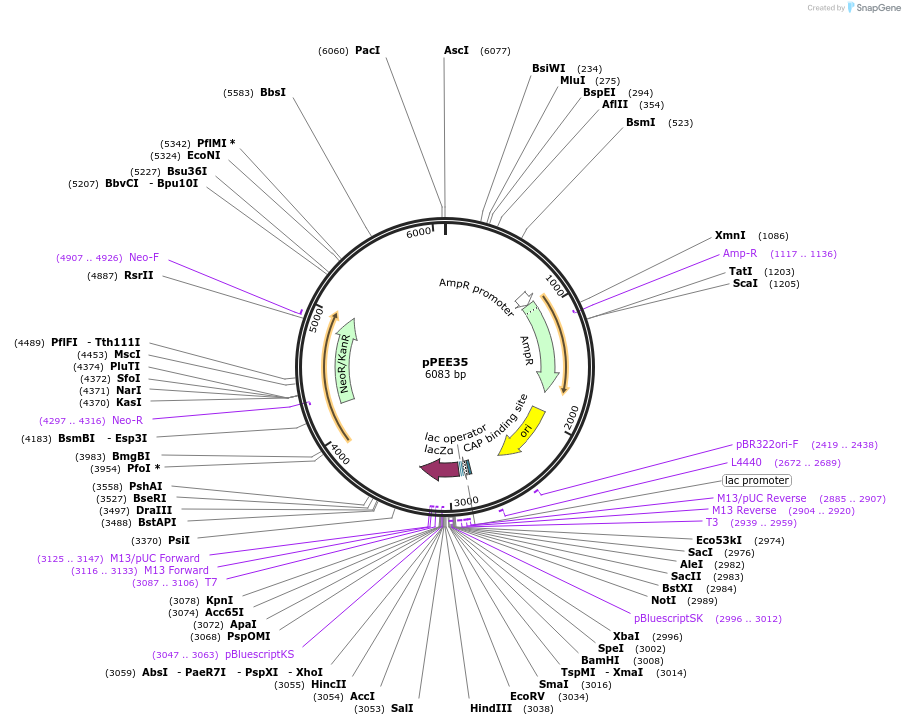
pPEE35
Plasmid#128375PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
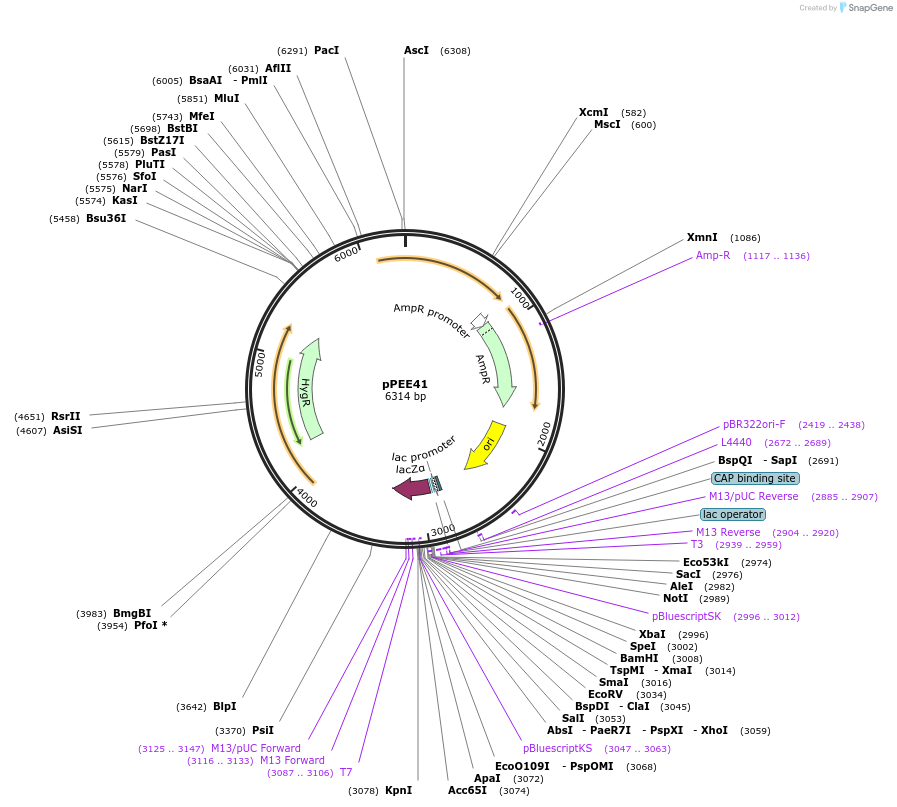
pPEE41
Plasmid#128381PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
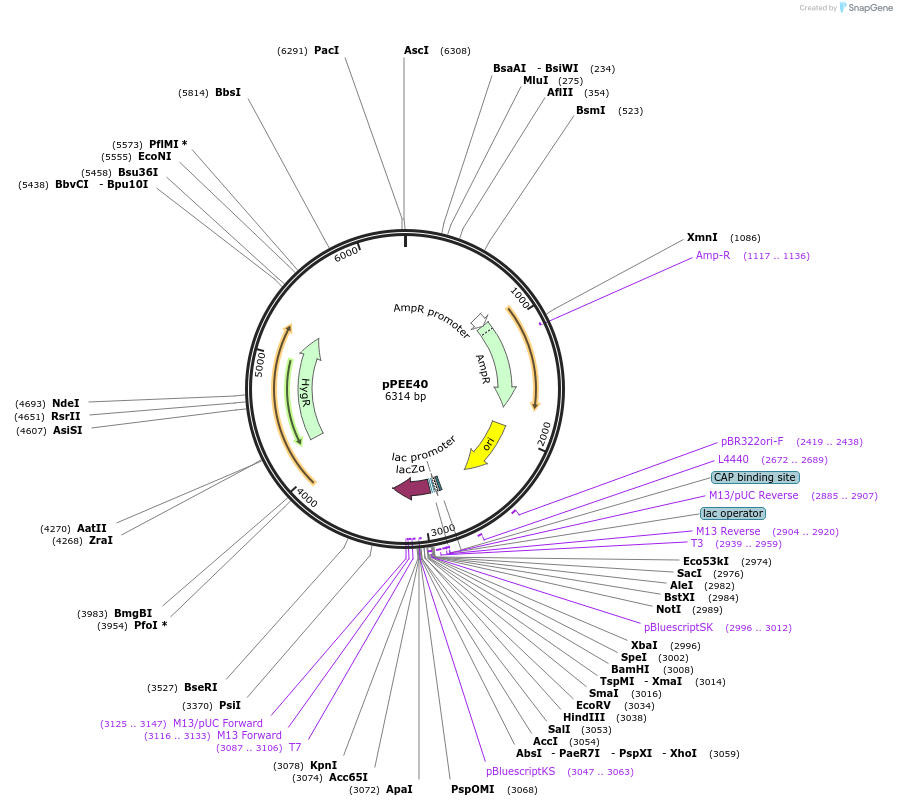
pPEE40
Plasmid#128380PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
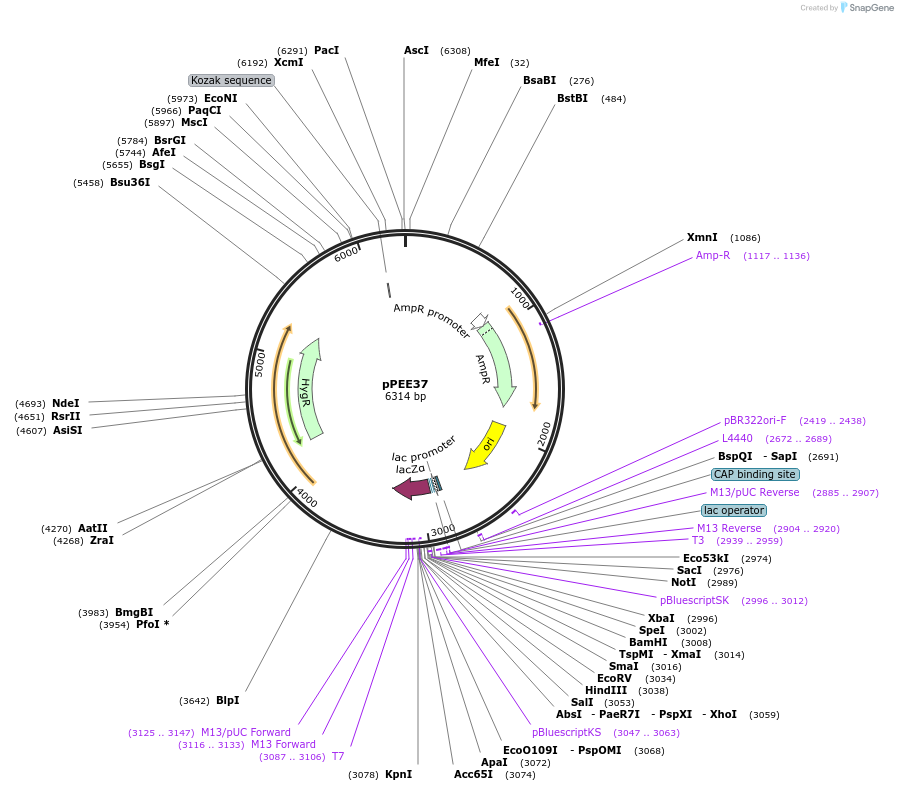
pPEE37
Plasmid#128377PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
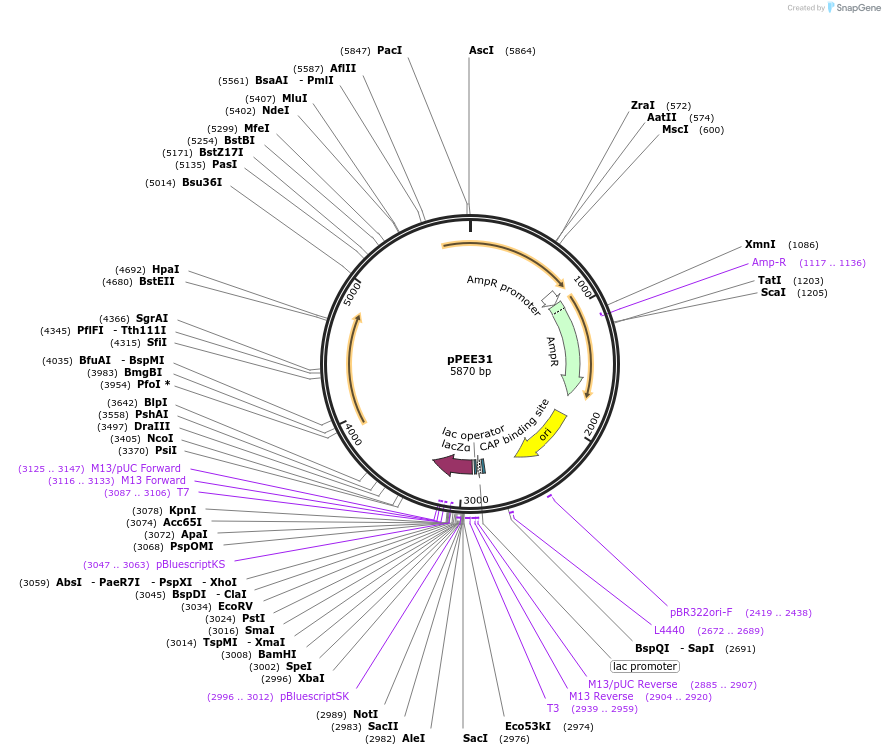
pPEE31
Plasmid#128371PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
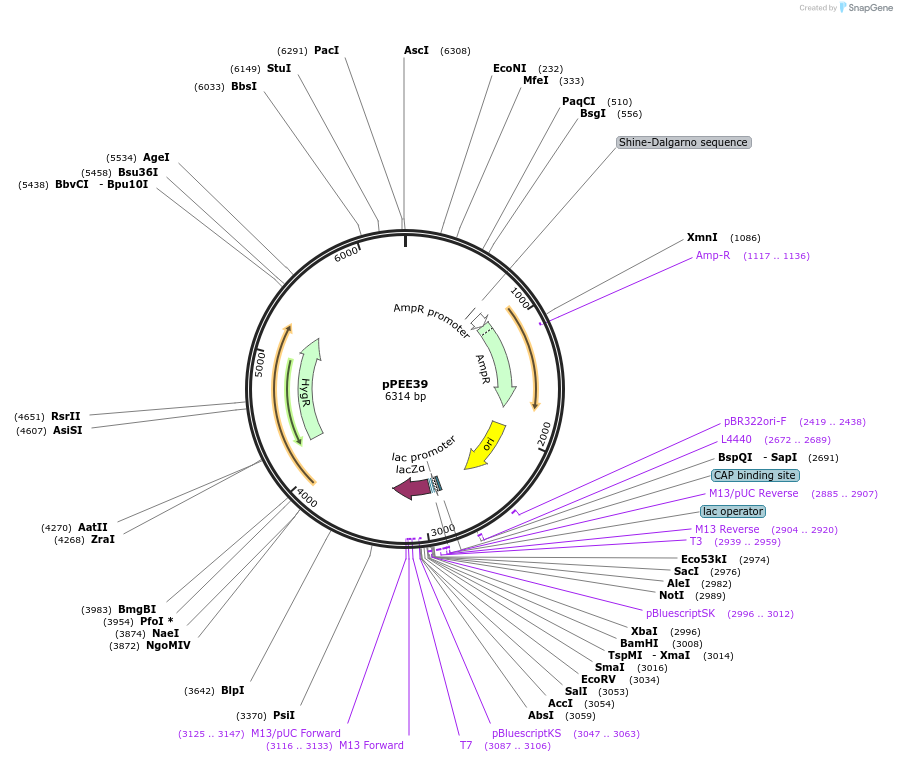
pPEE39
Plasmid#128379PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the HYG resistance marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
pPEE33
Plasmid#128373PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
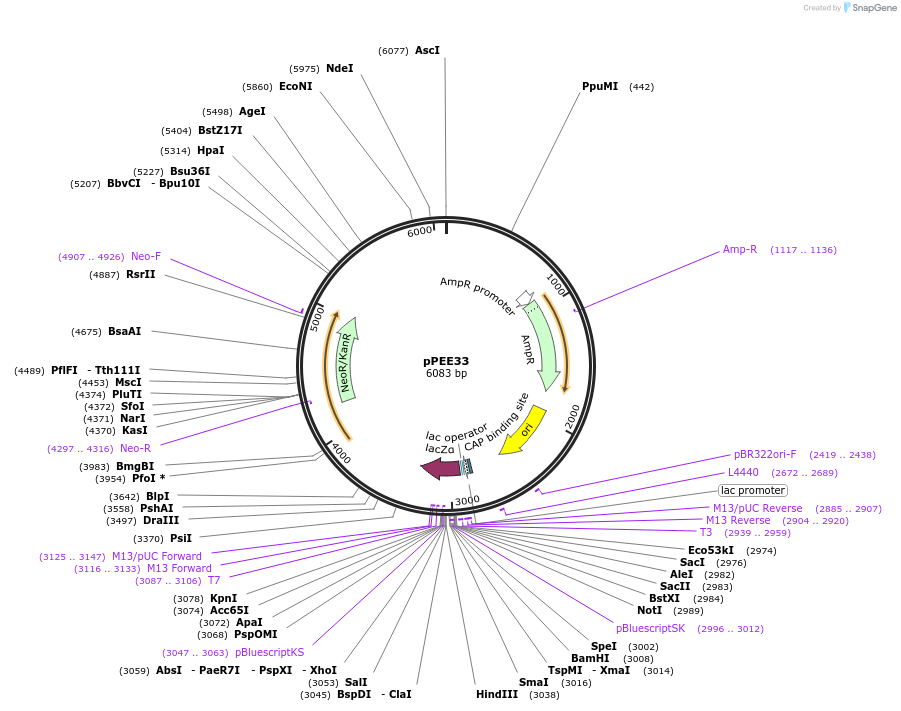
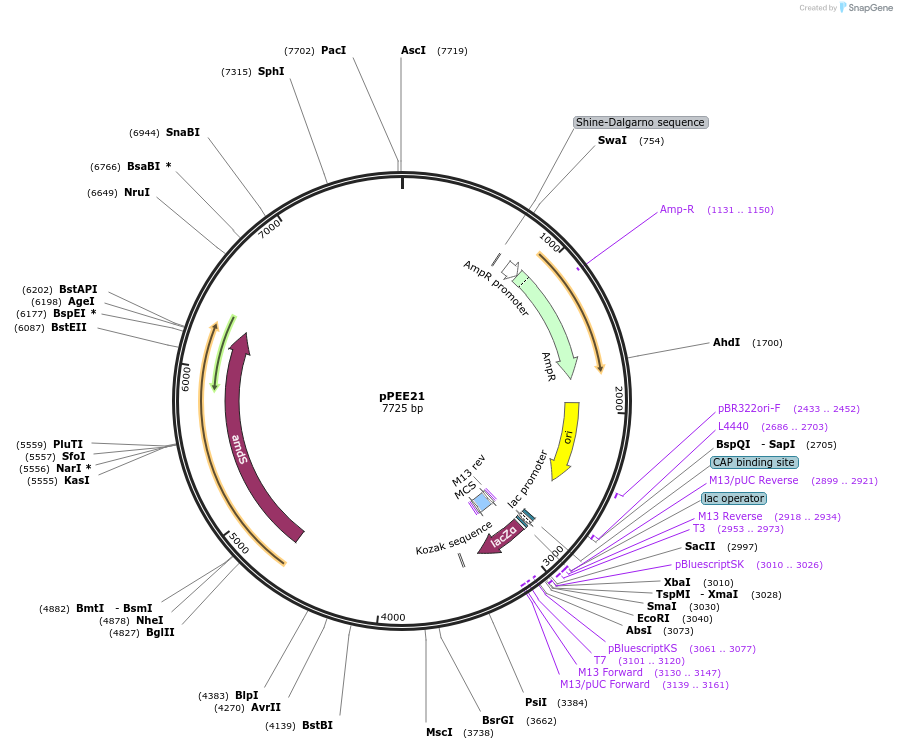
pPEE22
Plasmid#128365PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
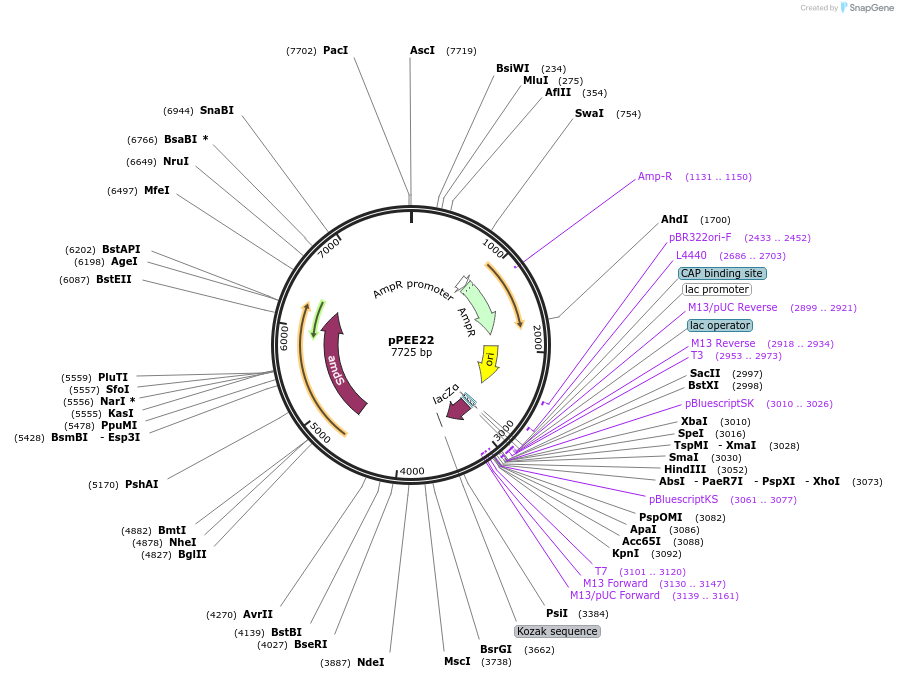
pPEE21
Plasmid#128364PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
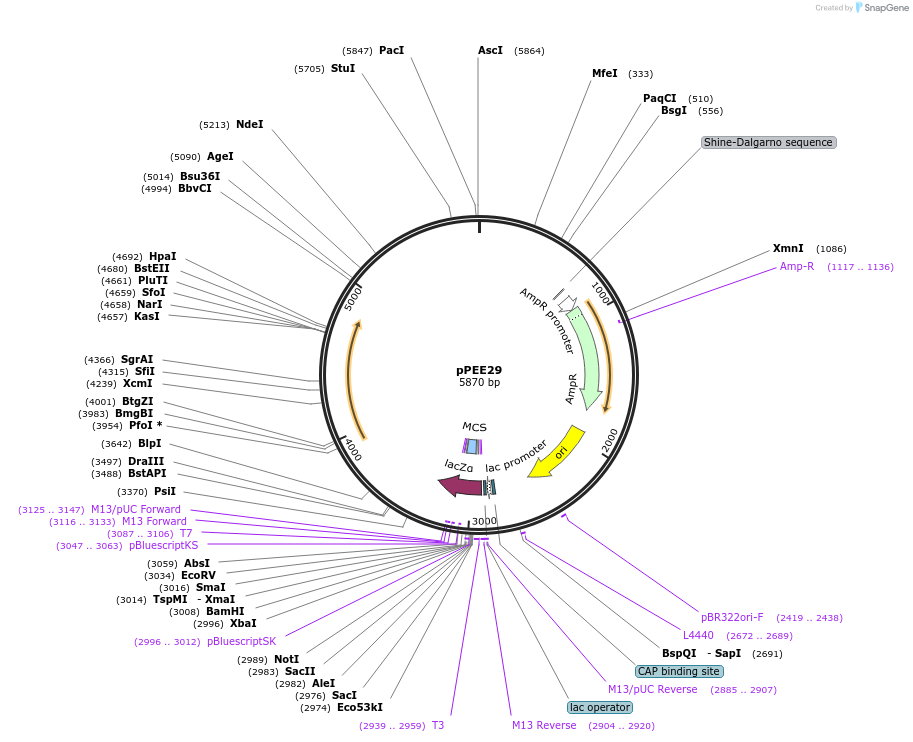
pPEE29
Plasmid#128369PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
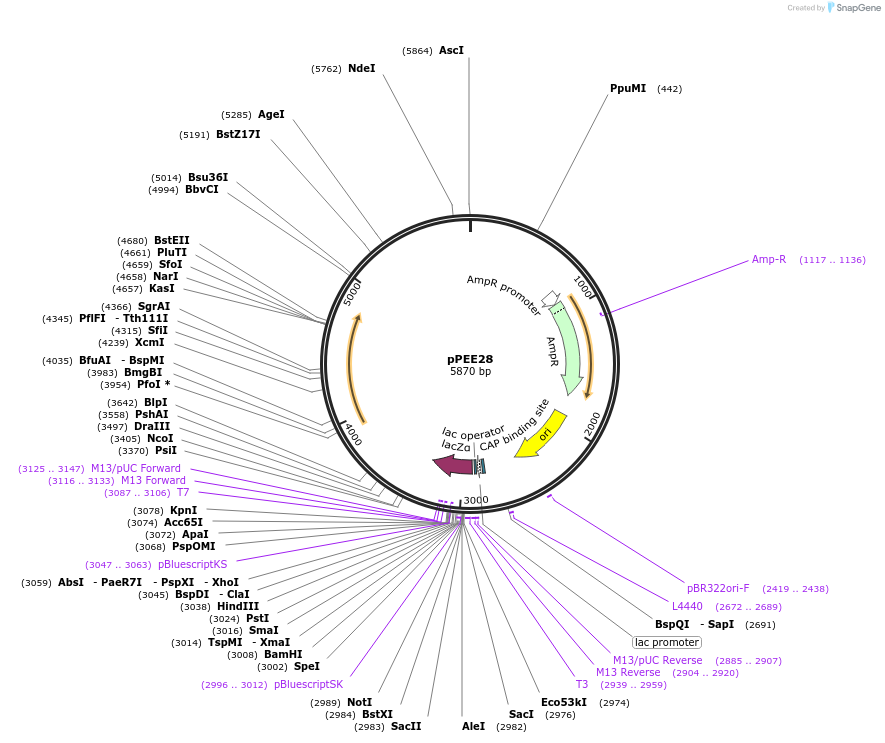
pPEE28
Plasmid#128368PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 4, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
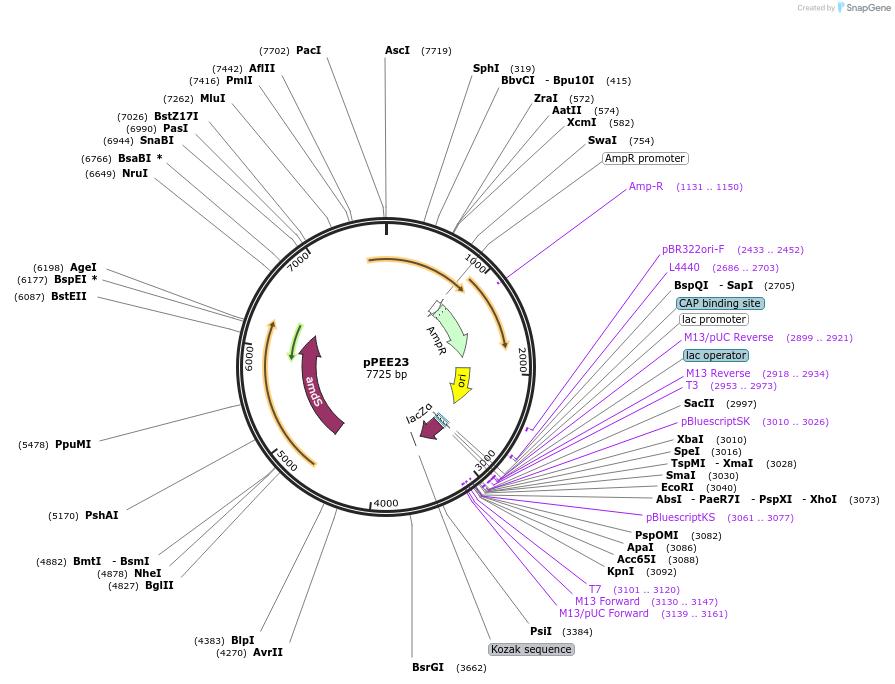
pPEE23
Plasmid#128366PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 7, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
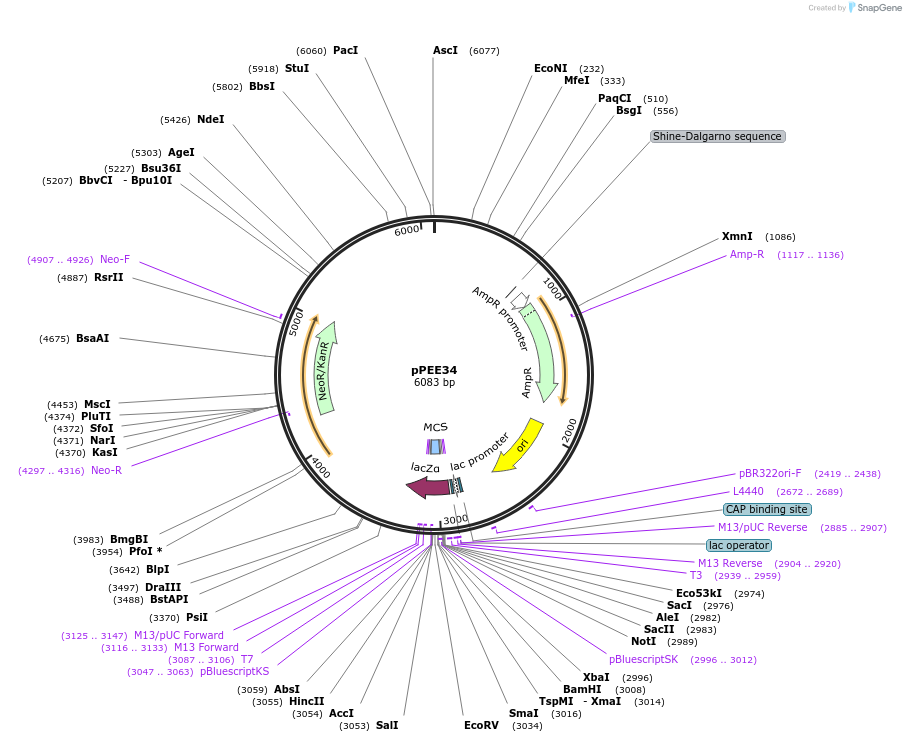
pPEE34
Plasmid#128374PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 5, a small gene-free region. Contains the NEO resistance markerDepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
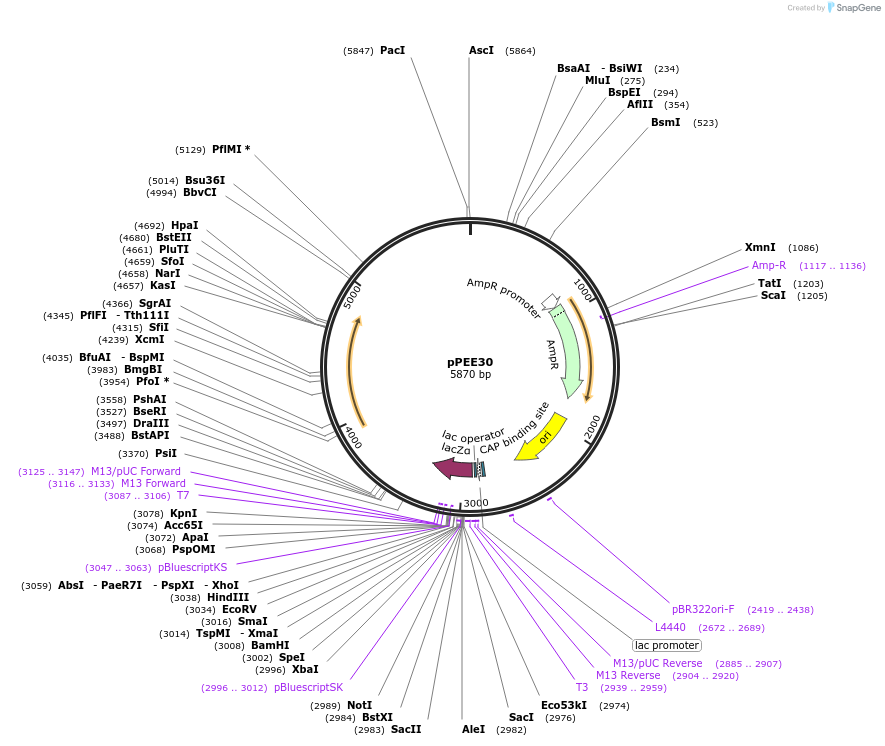
pPEE30
Plasmid#128370PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
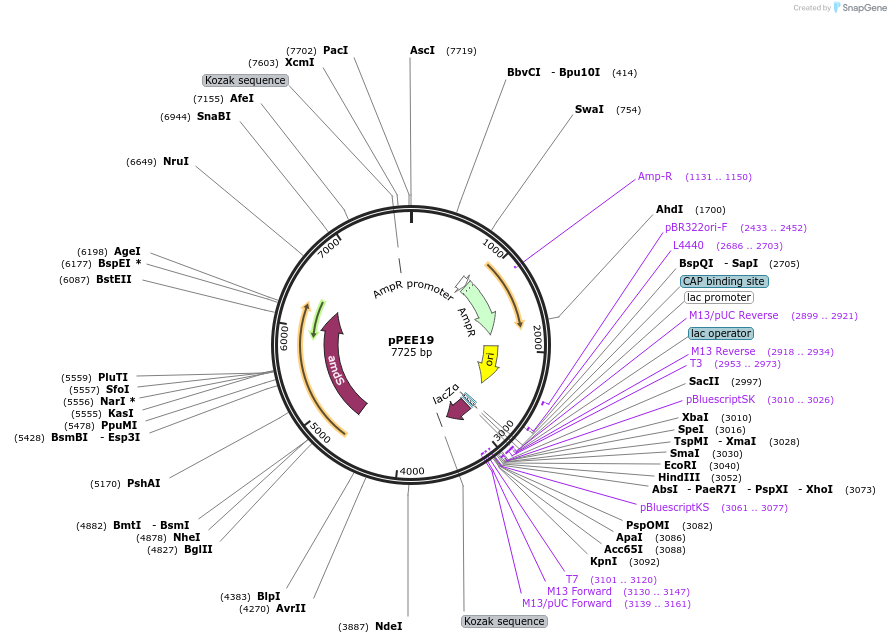
pPEE19
Plasmid#128362PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
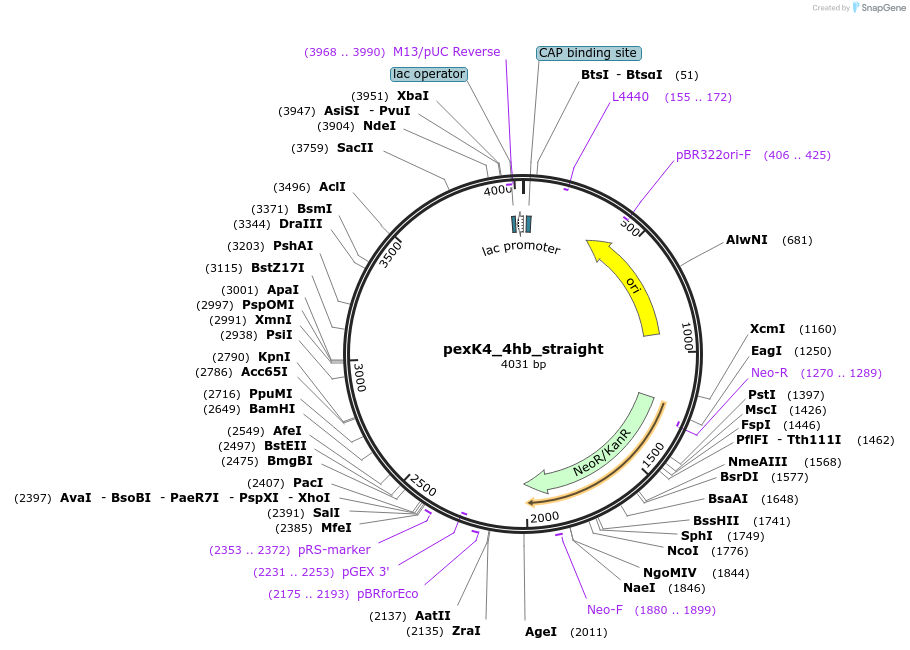
pexK4_4hb_straight
Plasmid#89678PurposePlasmid for the construction of DNA-protein hybrid nanostructures. Template for the straight four-helix-bundle shown in figure 4A of associated article.DepositorInsert4hb_straight
UseUnspecifiedAvailable SinceAug. 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
pexK4_4hb_bent
Plasmid#89679PurposePlasmid for the construction of DNA-protein hybrid nanostructures. Template for the bent four-helix-bundle shown in figure 4E of associated article.DepositorInsert4hb_bent
UseUnspecifiedAvailable SinceAug. 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
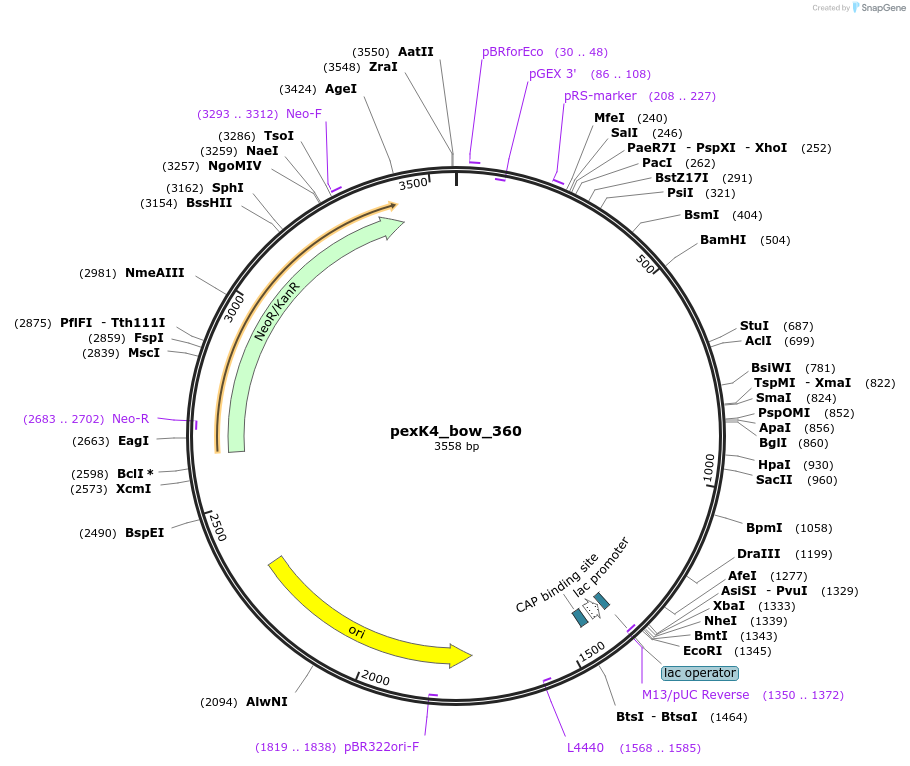
pexK4_bow_360
Plasmid#89673PurposePlasmid for the construction of DNA-protein hybrid nanostructures. Template for the bent two-helix-bundle shown in figure 3B of associated article.DepositorInsertbow_360
UseUnspecifiedAvailable SinceAug. 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
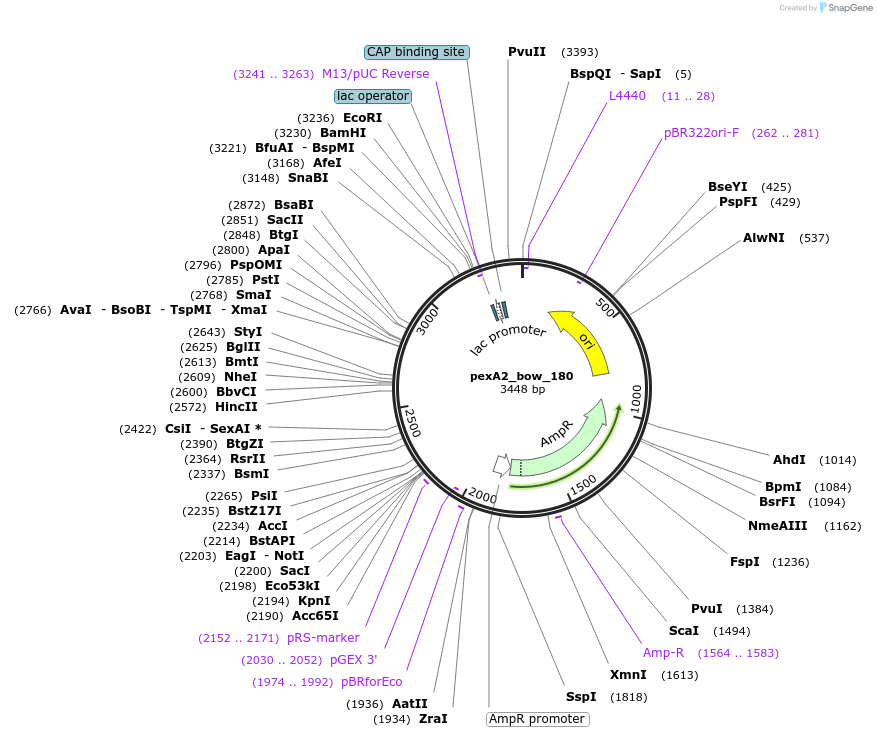
pexA2_bow_180
Plasmid#89671PurposePlasmid for the construction of DNA-protein hybrid nanostructures. Template for the bent two-helix-bundle shown in figure 3A of associated article.DepositorInsertbow_180
UseUnspecifiedAvailable SinceAug. 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
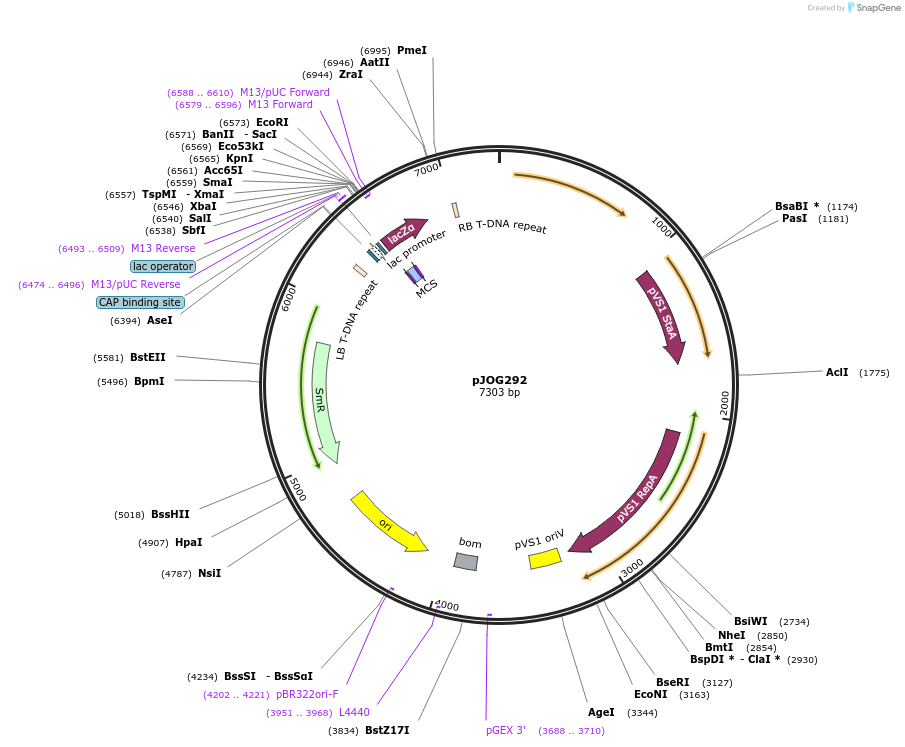
pJOG292
Plasmid#80586Purposeempty vector (lacZ) for 2nd generation recipient assemblyDepositorTypeEmpty backboneExpressionPlantAvailable SinceSept. 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
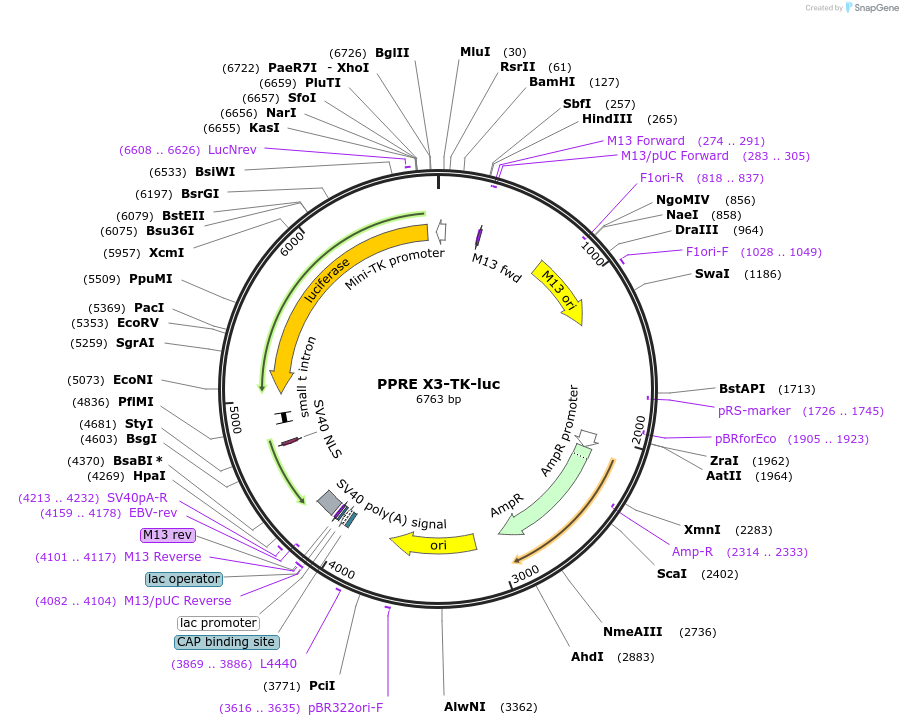
PPRE X3-TK-luc
Plasmid#1015PurposePPAR reporter. 3xDR1 sites upstream of a luciferase reporter.DepositorInsertPPRE
UseLuciferaseExpressionMammalianAvailable SinceNov. 3, 2005AvailabilityAcademic Institutions and Nonprofits only -
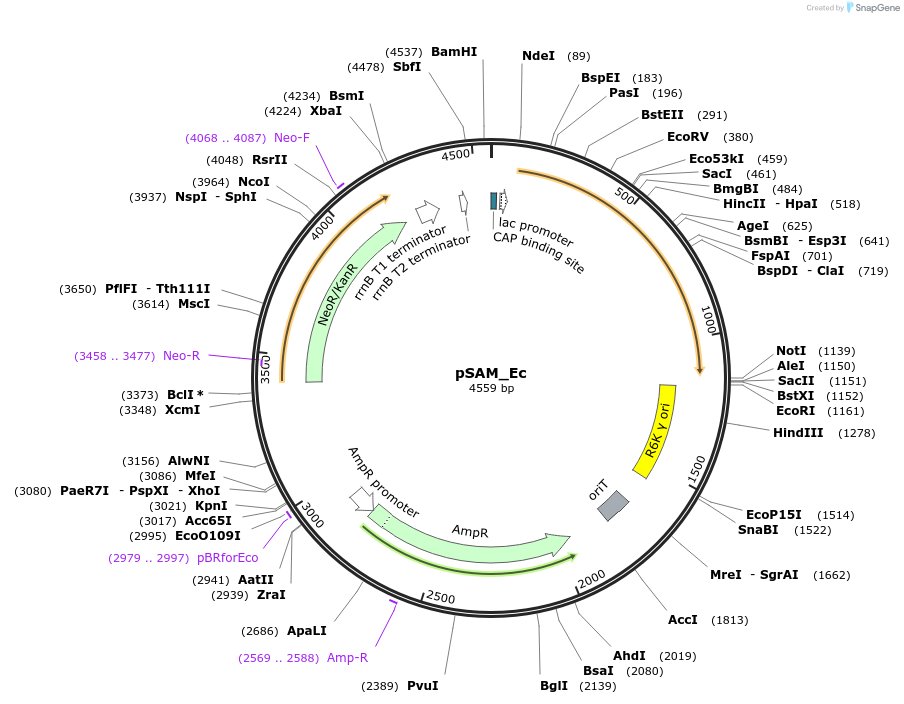
pSAM_Ec
Plasmid#102939PurposeTransposon mutagenesis and Tn-seqDepositorInsertmariner transposon flanked by MmeI modified inverted repeats and the himar1C9 transposase
ExpressionBacterialAvailable SinceApril 19, 2018AvailabilityAcademic Institutions and Nonprofits only