We narrowed to 1,616 results for: luciferase reporter
-
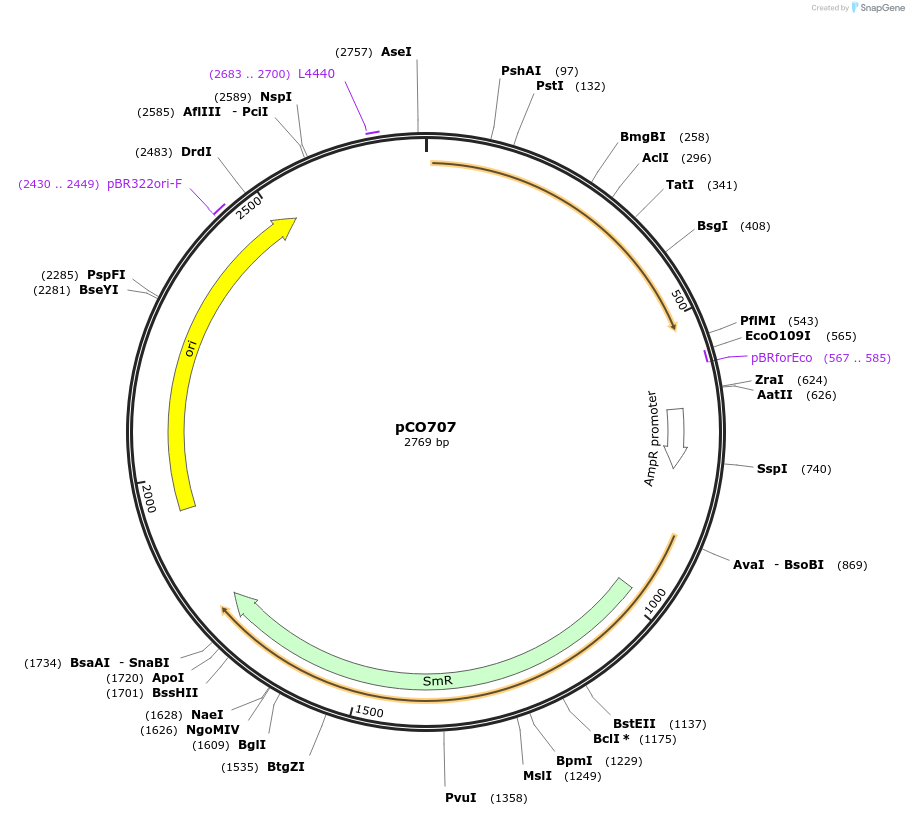
Plasmid#139744PurposeIndividual component of Caffeic Acid biosynthesis pathway, hpaCDepositorInserthpaC
UseMoclo level 0 componentPromoternoneAvailable SinceMay 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
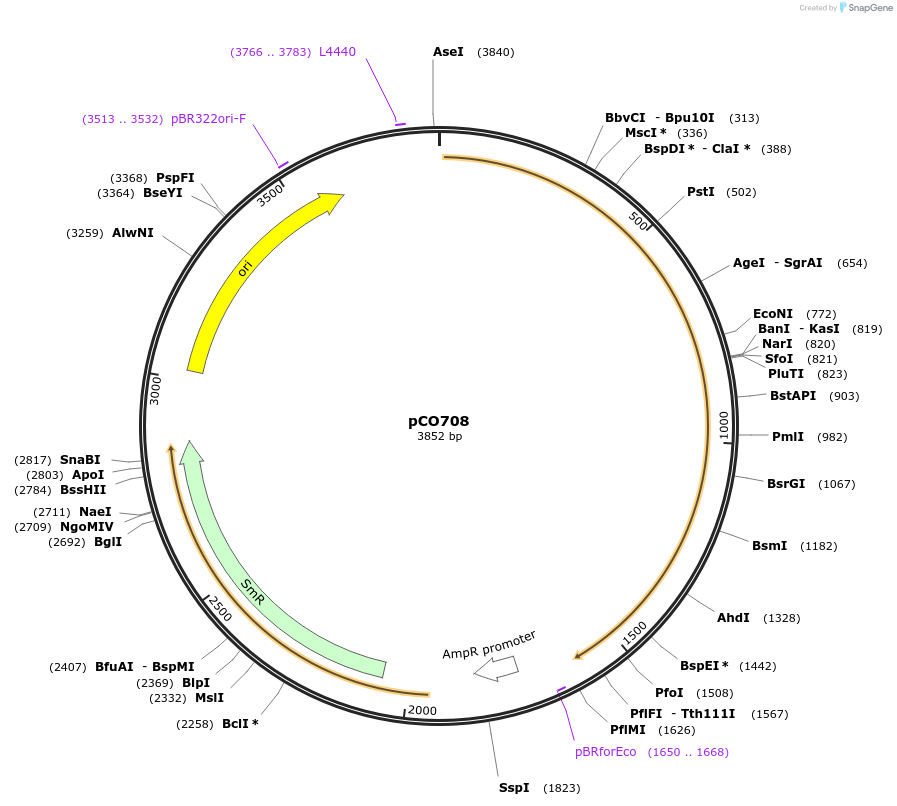
pCO708
Plasmid#139745PurposeIndividual component of Caffeic Acid biosynthesis pathway, hpaCDepositorInsertTAL
UseMoclo level 0 componentPromoternoneAvailable SinceMay 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
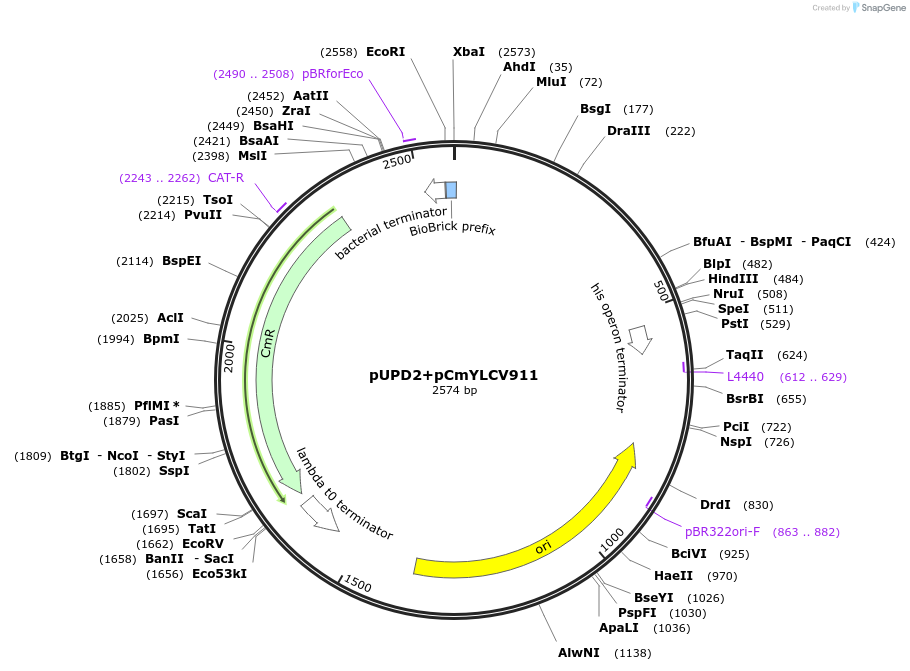
pUPD2+pCmYLCV911
Plasmid#170878PurposePhytobrick - pCmYLCV911DepositorInsertpCmYLCV911
ExpressionPlantAvailable SinceAug. 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
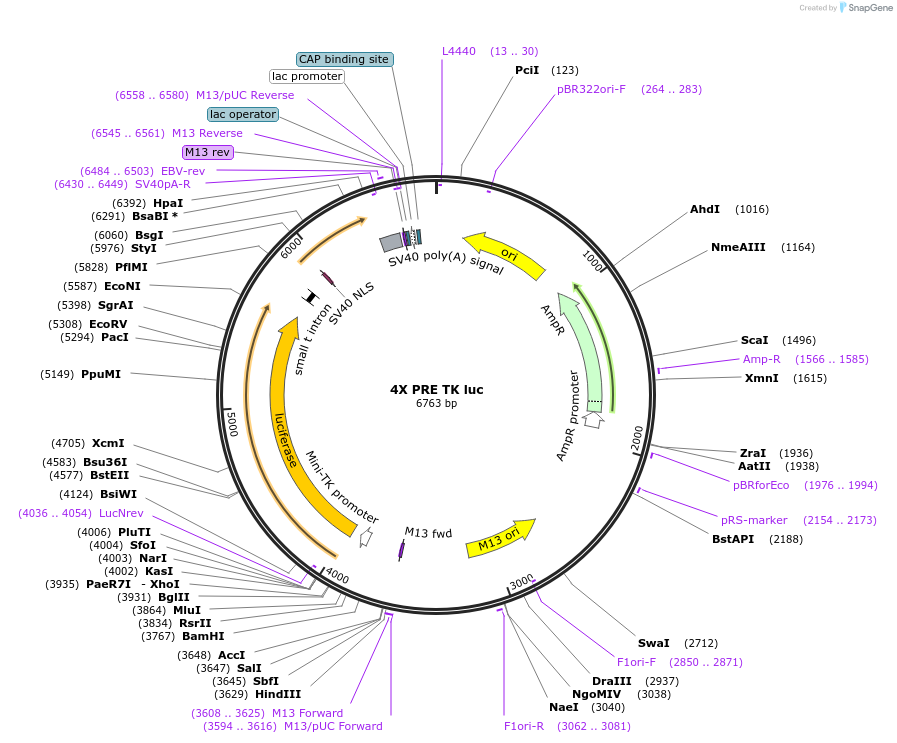
4X PRE TK luc
Plasmid#206159PurposeLuciferase reporter construct containing 4 consensus PGR binding sites upstream of a minimal TK promoter, derived from Addgene #11350DepositorInsert4X PRE TK luciferase
UseLuciferaseExpressionMammalianAvailable SinceNov. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
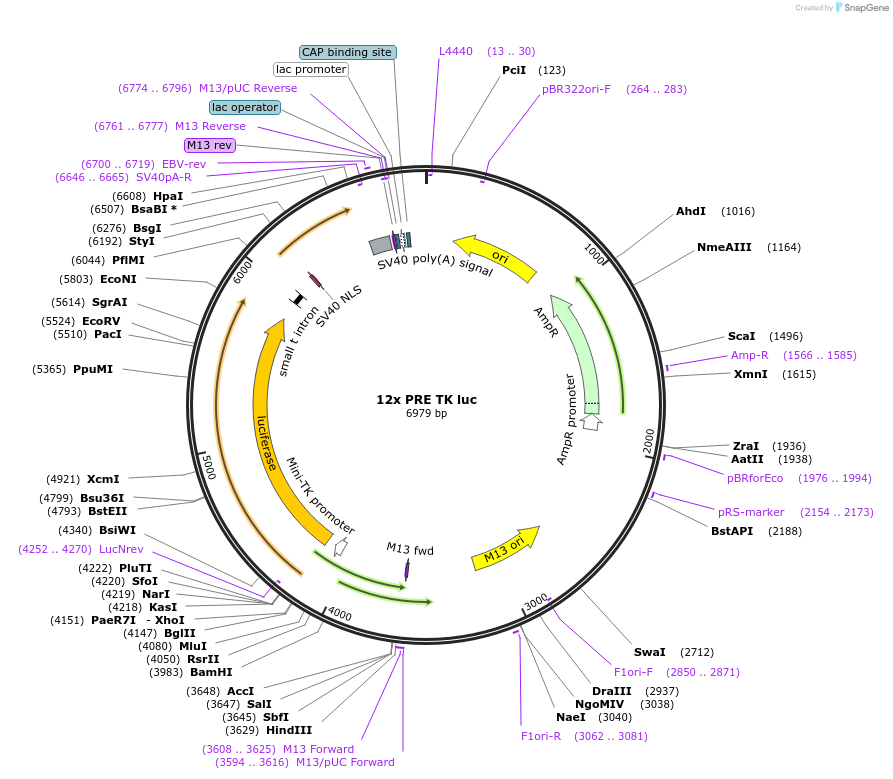
12x PRE TK luc
Plasmid#206163PurposeLuciferase reporter construct containing 12 consensus PGR binding sites upstream of a minimal TK promoter, derived from Addgene #11350DepositorInsert12X PRE TK luciferase
UseLuciferaseExpressionMammalianPromotermin TKAvailable SinceNov. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
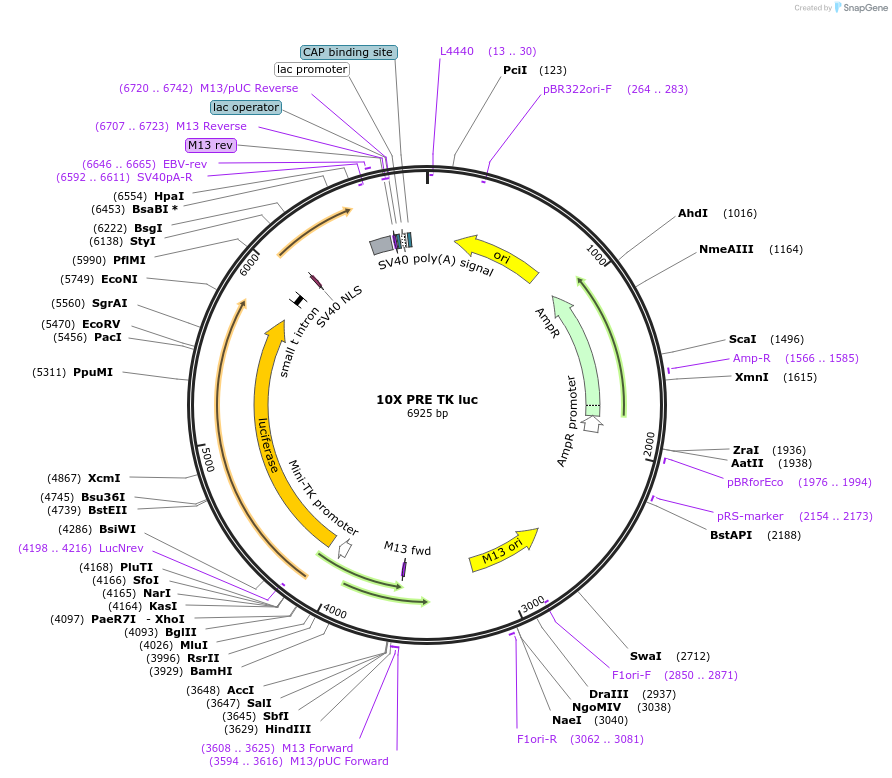
10X PRE TK luc
Plasmid#206162PurposeLuciferase reporter construct containing 10 consensus PGR binding sites upstream of a minimal TK promoter, derived from Addgene #11350DepositorInsert10X PRE TK luciferase
UseLuciferaseExpressionMammalianPromotermin TKAvailable SinceNov. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
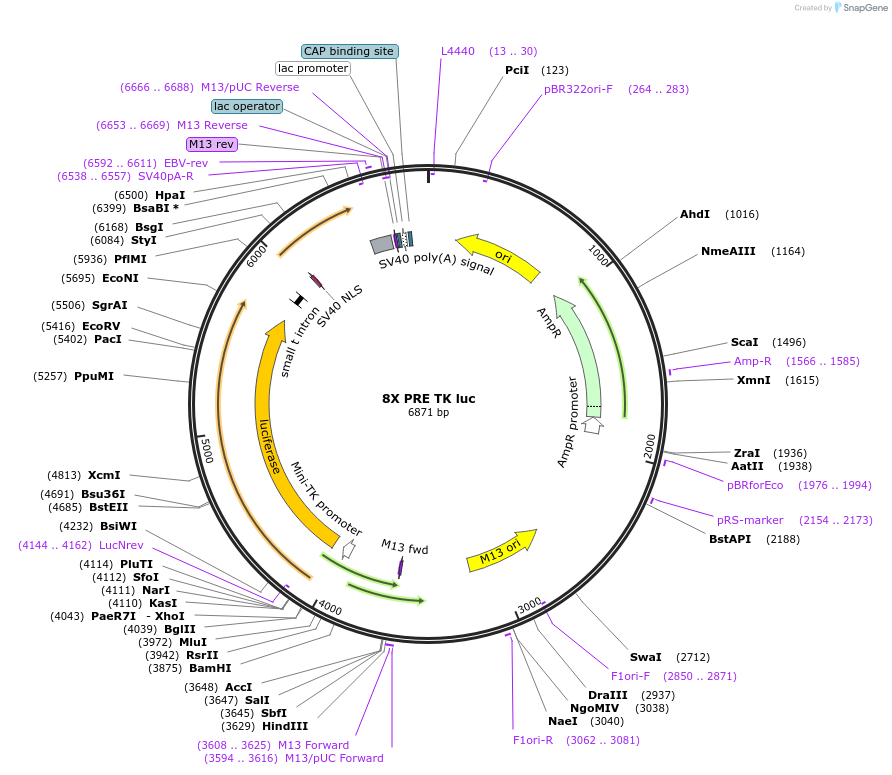
8X PRE TK luc
Plasmid#206161PurposeLuciferase reporter construct containing 8 consensus PGR binding sites upstream of a minimal TK promoter, derived from Addgene #11350DepositorInsert8X PRE TK luciferase
UseLuciferaseExpressionMammalianPromotermin TKAvailable SinceNov. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
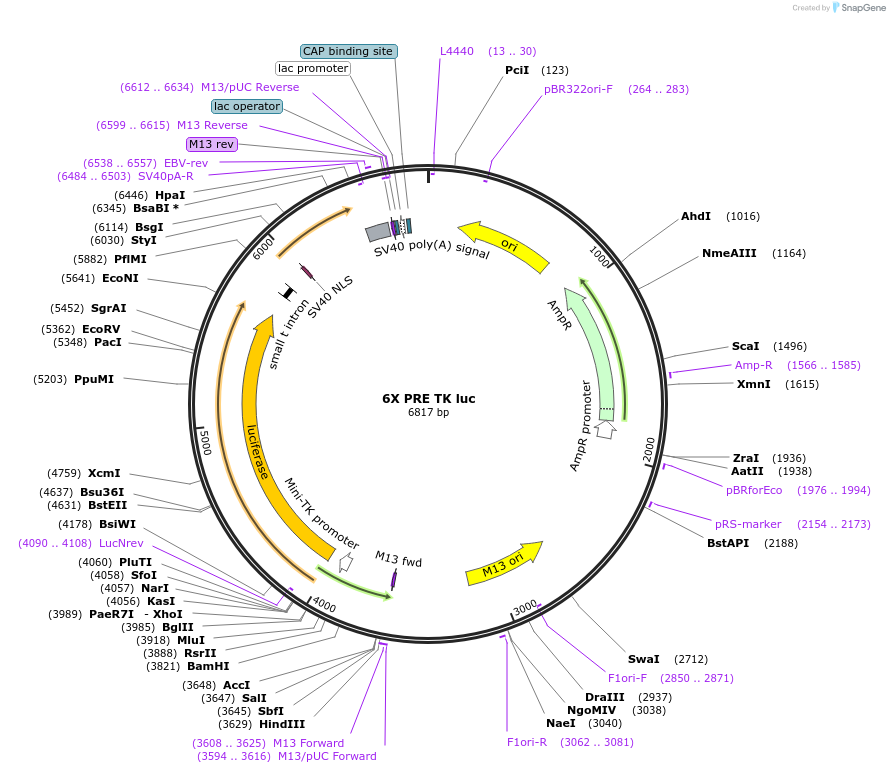
6X PRE TK luc
Plasmid#206160PurposeLuciferase reporter construct containing 6 consensus PGR binding sites upstream of a minimal TK promoter, derived from Addgene #11350DepositorInsert6X PRE TK luciferase
UseLuciferaseExpressionMammalianPromotermin TKAvailable SinceNov. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
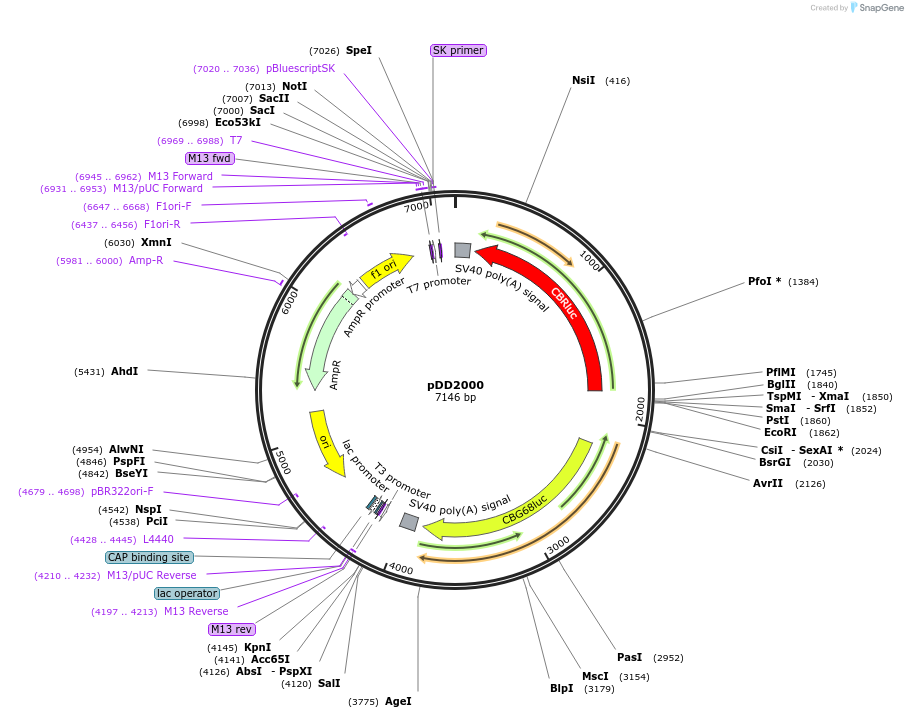
pDD2000
Plasmid#101197PurposeName: pBluescript-LANApi,K14. Bidirectional reporter driving LANApi side [green luciferase isoform from pCBG68] and K14 side [red luciferase isoform from pCBR] in pBluescript backbone.DepositorInsertLANApi side [green luciferase isoform of pCBG68], K14 side [red luciferase isoform of pCBR]
UseLuciferaseAvailabilityAcademic Institutions and Nonprofits only -
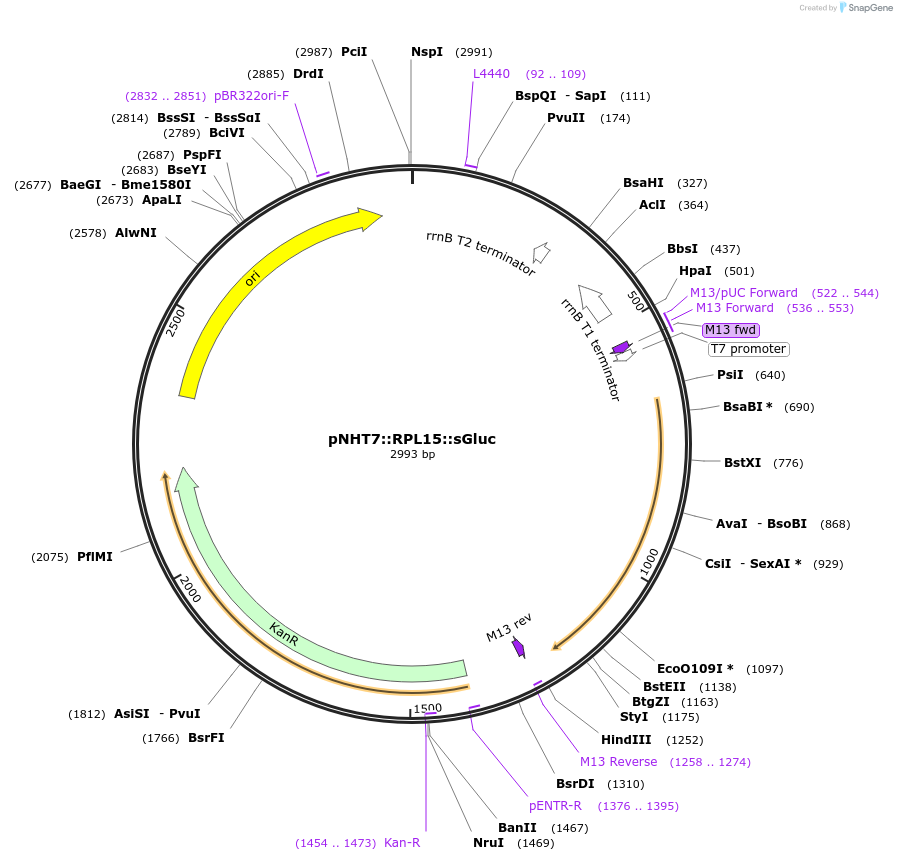
pNHT7::RPL15::sGluc
Plasmid#186769PurposeAn mRNA production vector encoding planarian codon-optimized gaussia luciferase (sGluc) with RPL15 5' and 3' UTRs.DepositorInsertGaussia luciferase
PromoterT7Available SinceOct. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
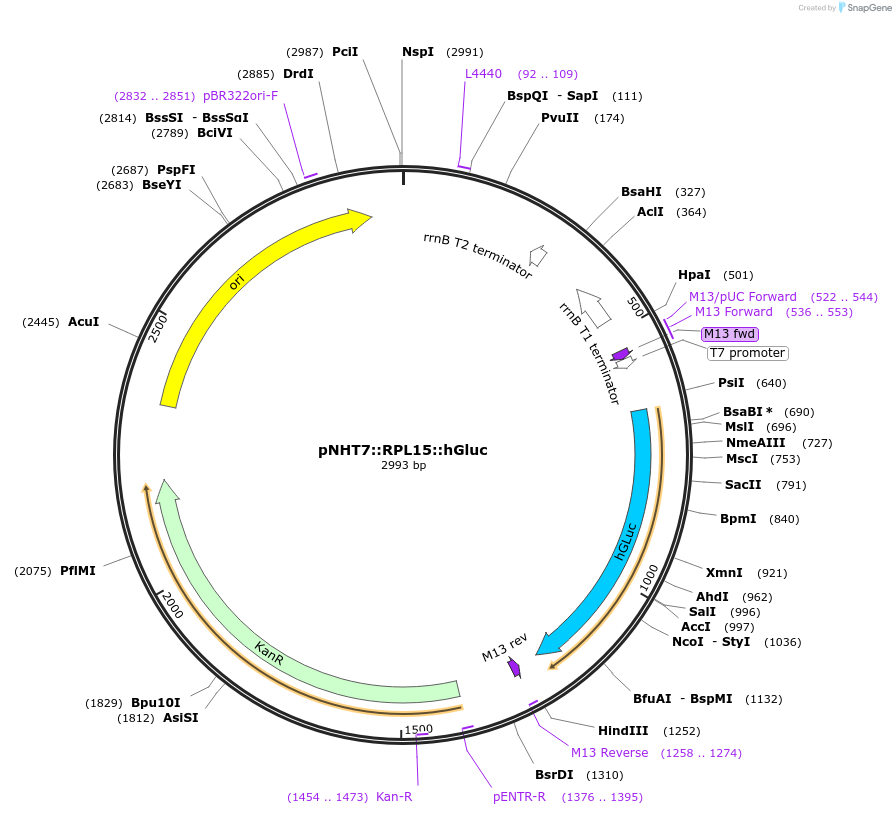
pNHT7::RPL15::hGluc
Plasmid#186768PurposeAn mRNA production vector encoding human codon-optimized gaussia luciferase (hGluc) with RPL15 5' and 3' UTRs.DepositorInsertGaussia luciferase
PromoterT7Available SinceOct. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
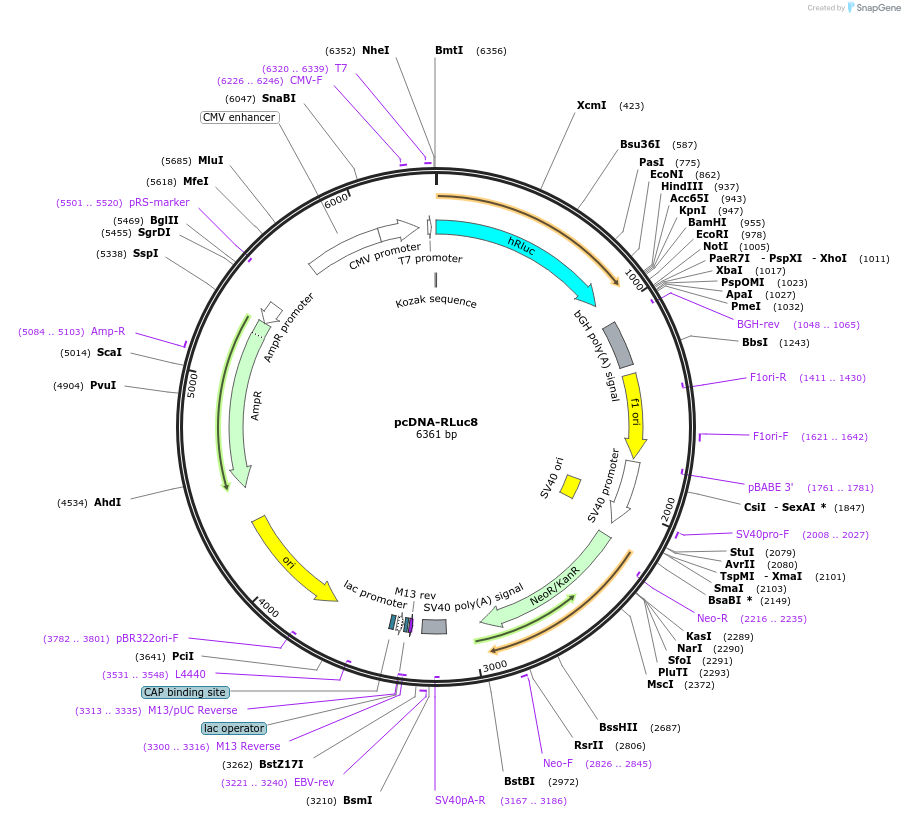
pcDNA-RLuc8
Plasmid#87121PurposeMammalian expression vector expressing mutant version of the reporter gene-Renilla Luciferase (Rluc8), which is more stable in mouse serum and has more light output.DepositorInsertRenilla Luciferase
UseLuciferaseExpressionMammalianMutationA55T, C124A, S130A, K136R, A143M, M185V, M253L, a…PromoterCMVAvailable SinceApril 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
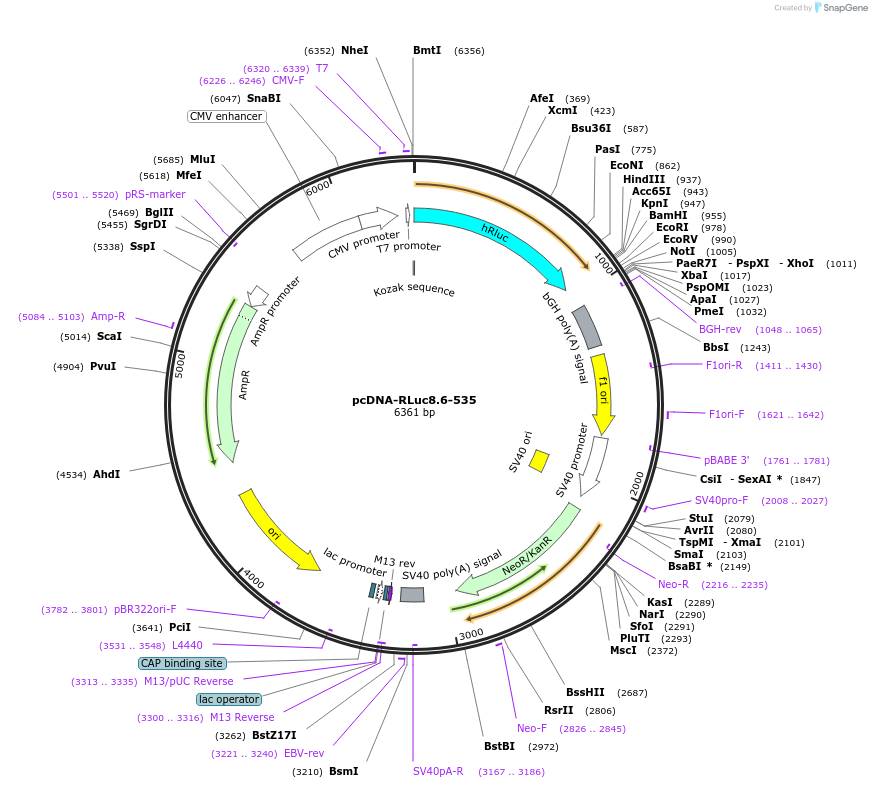
pcDNA-RLuc8.6-535
Plasmid#87125PurposeMammalian expression vector expressing red shifted variant of the reporter gene-Renilla luciferase (Rluc8.6)DepositorInsertRenilla Luciferase
ExpressionMammalianMutationRLuc8/ A123S/D154M/E155G/D162E/I163L/V185L.PromoterCMVAvailable SinceApril 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
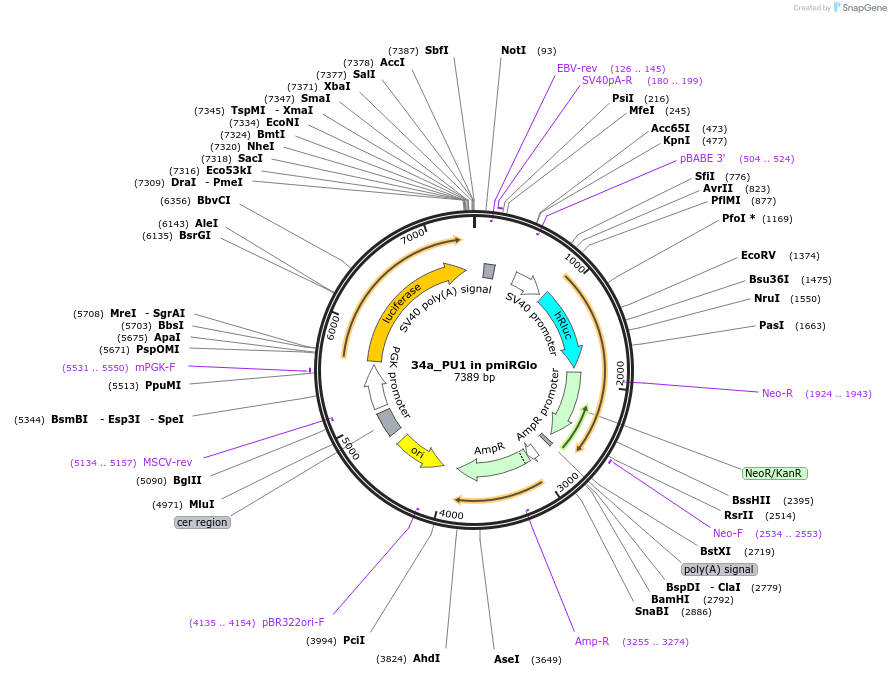
34a_PU1 in pmiRGlo
Plasmid#78131Purposeluciferase reporter for miRNA activityDepositorInsertPU1
UseLuciferaseTagsluciferaseExpressionMammalianMutation3'UTR containing miRNA binding sitesPromoterPGKAvailable SinceJuly 5, 2016AvailabilityAcademic Institutions and Nonprofits only -
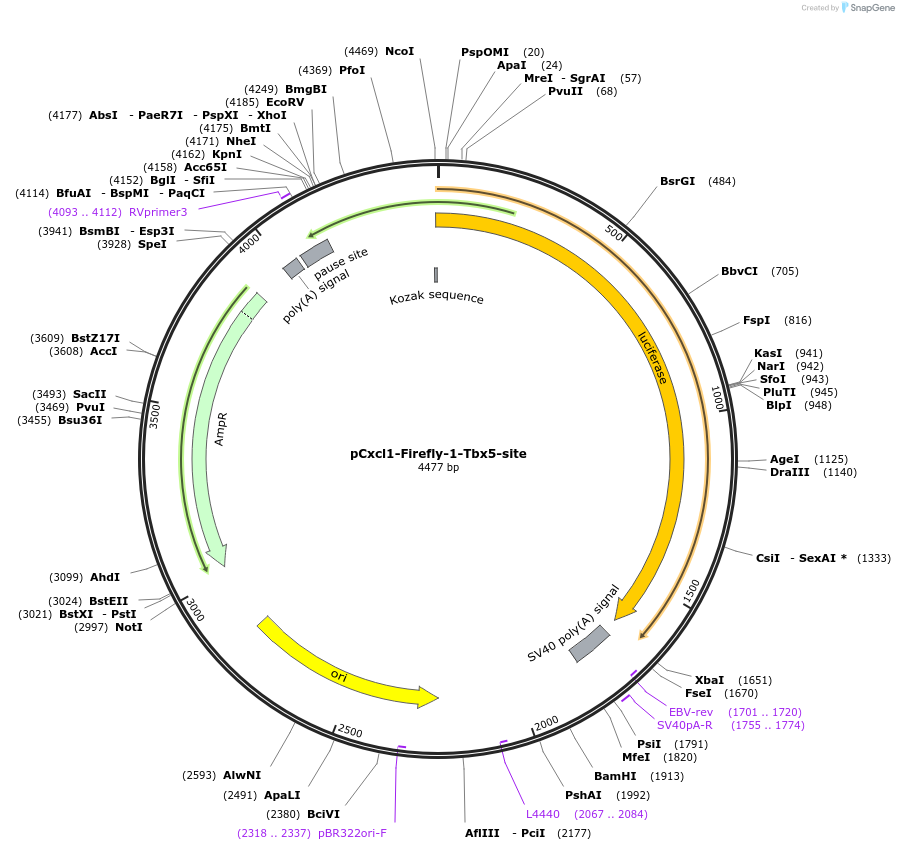
pCxcl1-Firefly-1-Tbx5-site
Plasmid#177832PurposeLuciferase reporter for mouse Cxcl1 with one Tbx5 consensus binding siteDepositorInsertLuciferase
UseLuciferasePromoterCxcl1Available SinceAug. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
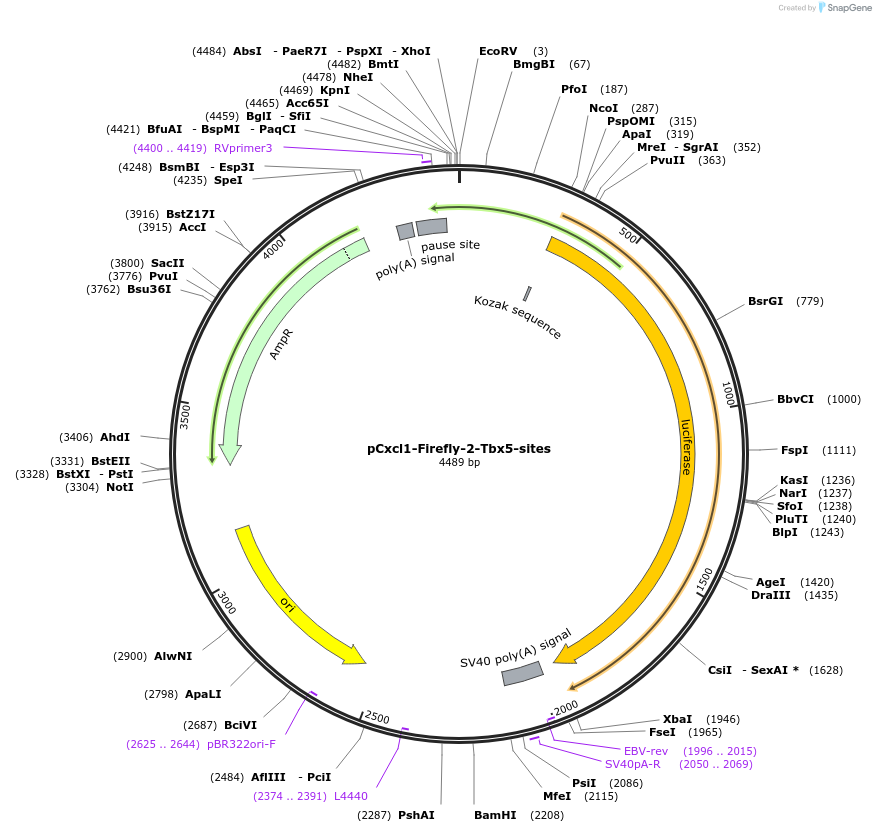
pCxcl1-Firefly-2-Tbx5-sites
Plasmid#177833PurposeLuciferase reporter for mouse Cxcl1 with two Tbx5 consensus binding sitesDepositorInsertLuciferase
UseLuciferasePromoterCxcl1Available SinceAug. 29, 2022AvailabilityAcademic Institutions and Nonprofits only -
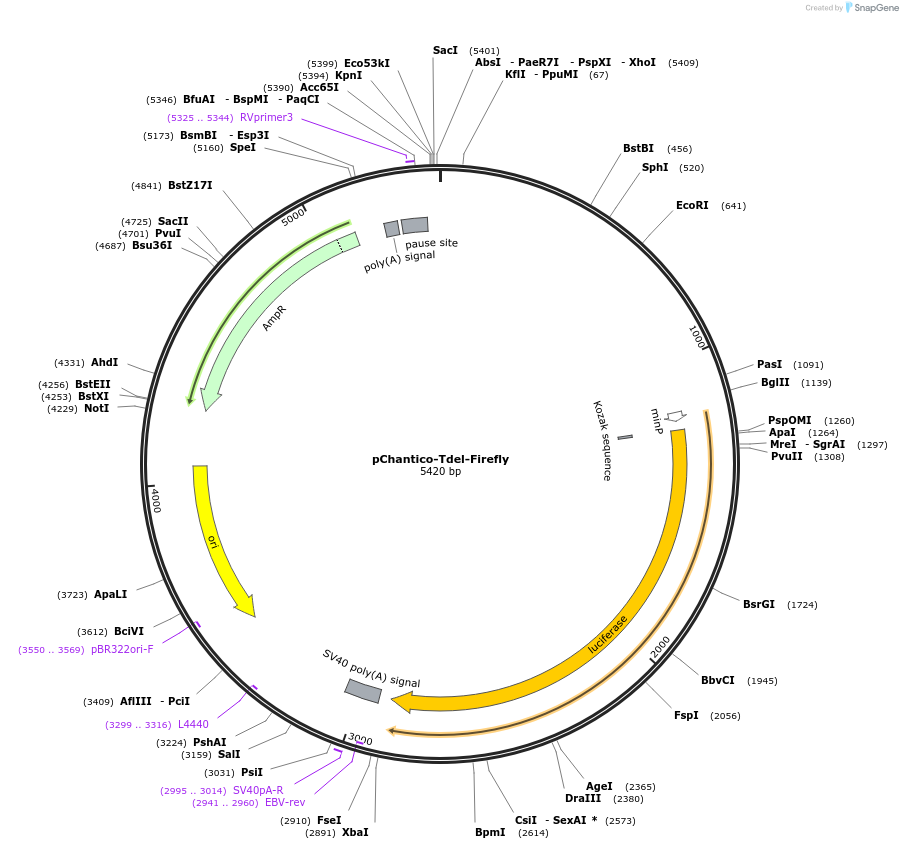
pChantico-Tdel-Firefly
Plasmid#163902PurposeLuciferase reporter for Chantico lncRNA with Tbx5 site deletionDepositorInsertLuciferase
UseLuciferasePromoterChantico with Tbx5 consensus binding site deletionAvailable SinceJan. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
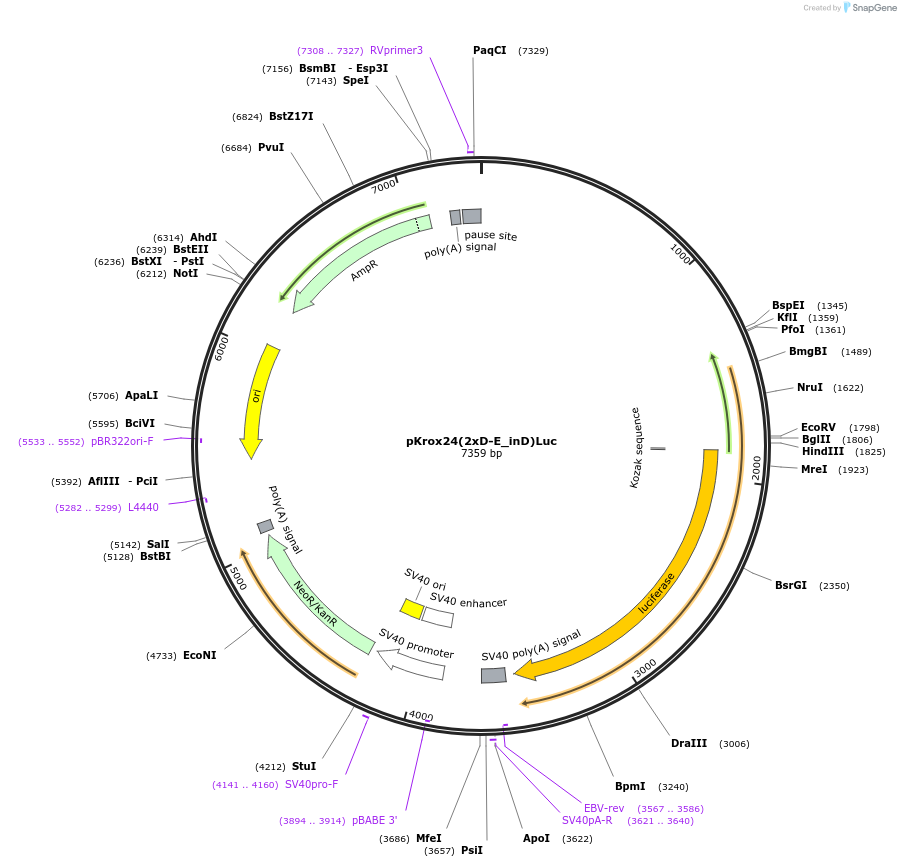
pKrox24(2xD-E_inD)Luc
Plasmid#200109PurposeLuciferase reporter for in cell monitoring of receptor tyrosine kinase-MAP ERK pathway activityDepositorInsertmodified EGR1 promoter
ExpressionMammalianMutationhighly modified sequencePromoter2 copies of D-E element in front of EGR1 promoter…Available SinceNov. 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
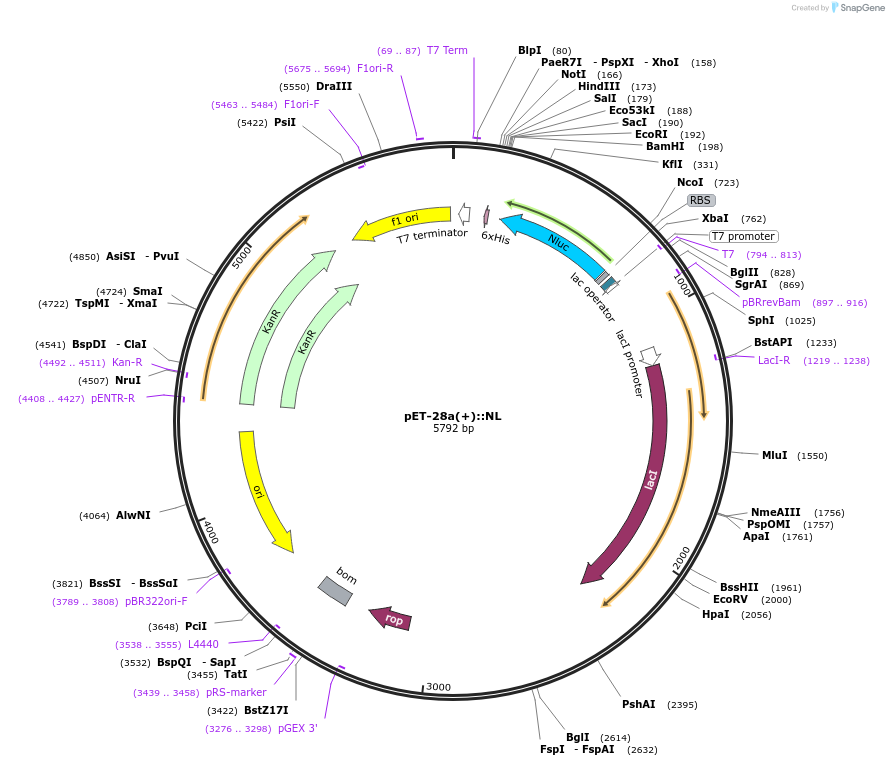
pET-28a(+)::NL
Plasmid#141289PurposeNLUC in pET-28 a (+) backbone for bacterial IPTG inducible expression (KmR)DepositorInsertNLUC
UseLuciferaseExpressionBacterialMutationEngineered for high stability (t1/2 = 11.5 days a…Promoterpromoter for bacteriophage T7 RNA polymeraseAvailable SinceFeb. 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
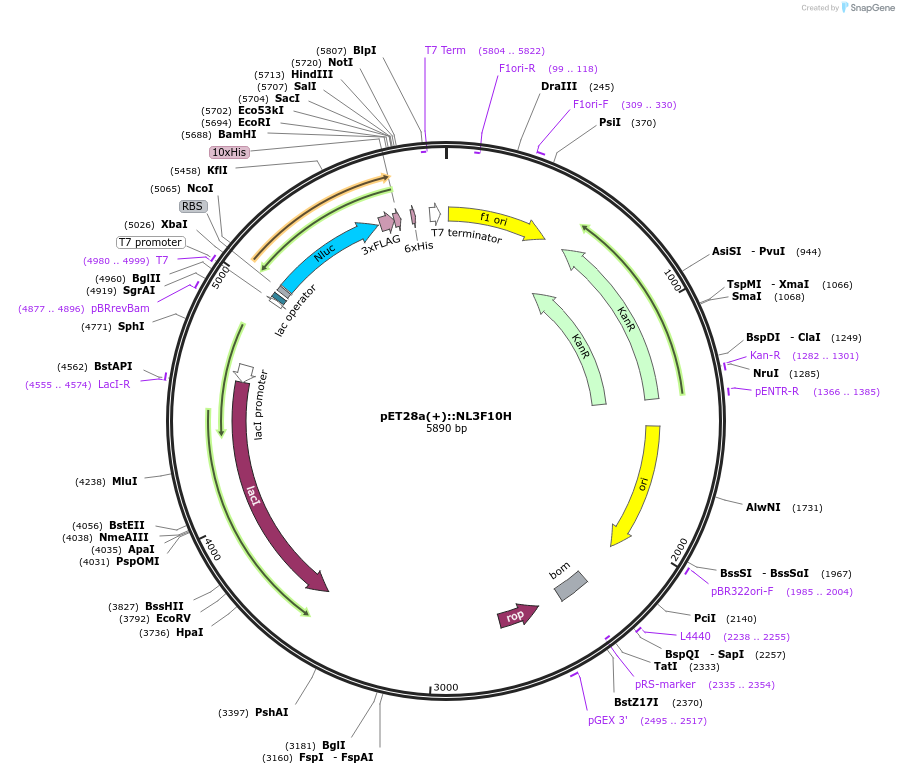
pET28a(+)::NL3F10H
Plasmid#141290PurposeNLUC:3xFlag:10xHis in pET-28 a (+) backbone for bacterial IPTG inducible expression (KmR)DepositorInsertNLUC3F10H
UseLuciferaseTagsFlag (x3) and His (x10)ExpressionBacterialMutationEngineered for high stability (t1/2 = 11.5 days a…Promoterpromoter for bacteriophage T7 RNA polymeraseAvailable SinceFeb. 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
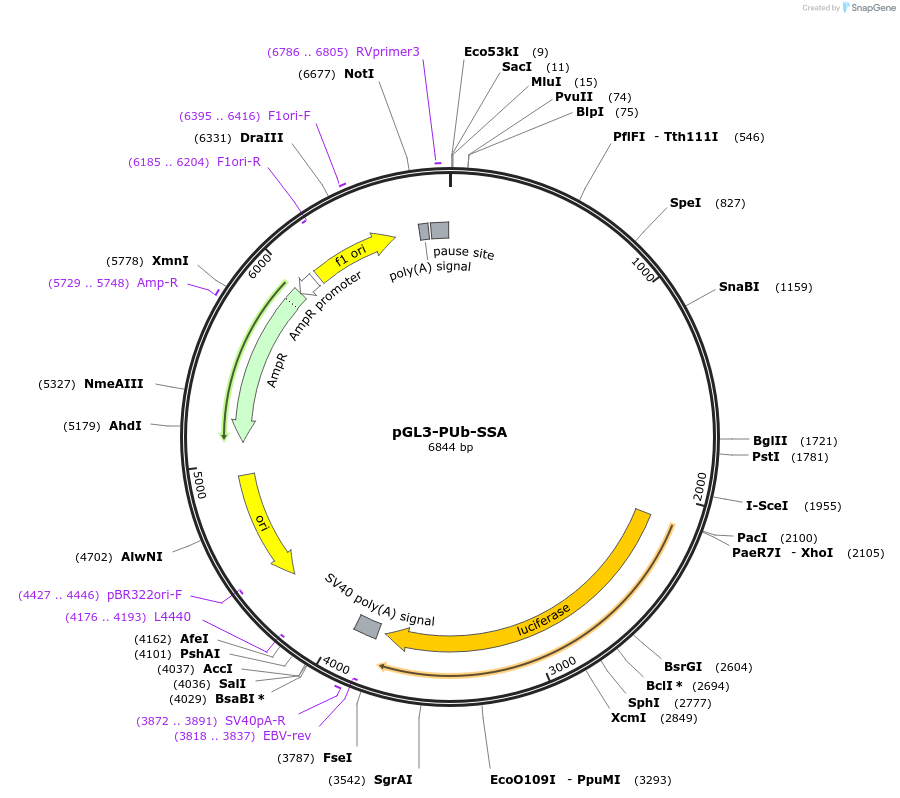
pGL3-PUb-SSA
Plasmid#52910PurposeAe aegypti PUb promoter Single Stranded Annealing reporterDepositorInsertLuciferase duplication-TALEN recognition sites
UseLuciferasePromoterAe aegypti polyubiquitinAvailable SinceMay 27, 2014AvailabilityAcademic Institutions and Nonprofits only -
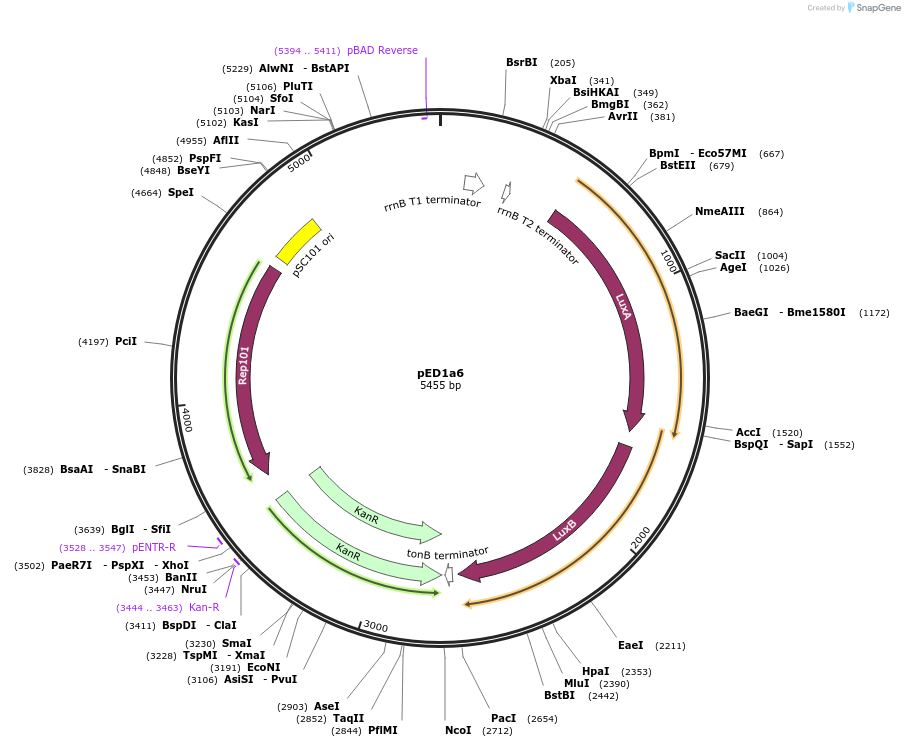
pED1a6
Plasmid#134787PurposeLuciferase reporter for quadruplet codon TAGADepositorInsertluxAB-357-TAGA
UseLuciferase and Synthetic BiologyExpressionBacterialMutation357-TAGAAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
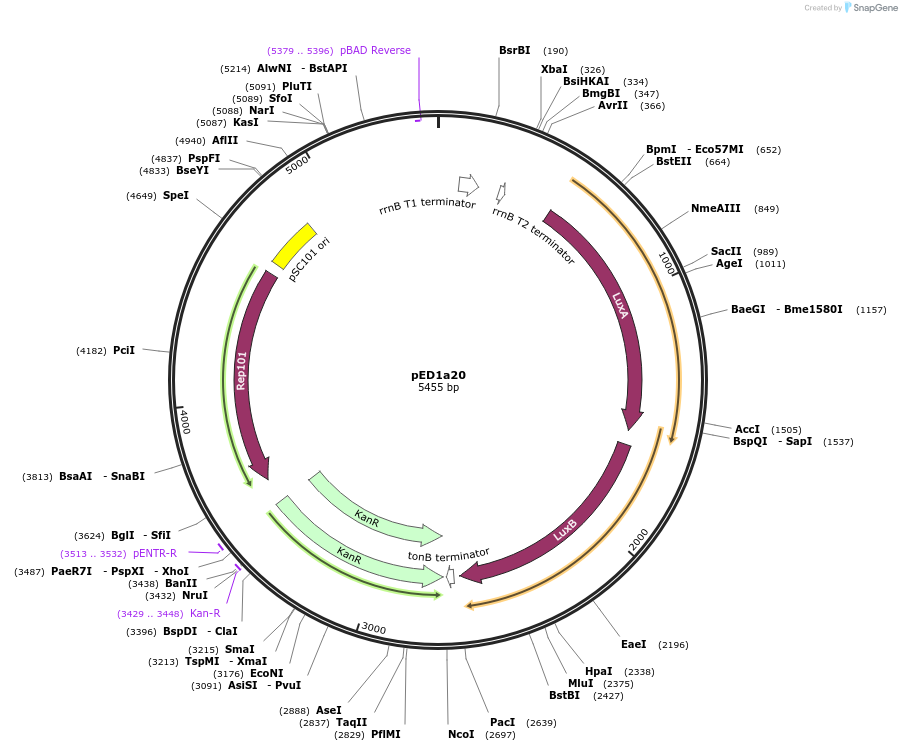
pED1a20
Plasmid#134788PurposeLuciferase reporter for quadruplet codon CAGGDepositorInsertluxAB-357-CAGG
UseLuciferase and Synthetic BiologyExpressionBacterialMutation357-CAGGAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
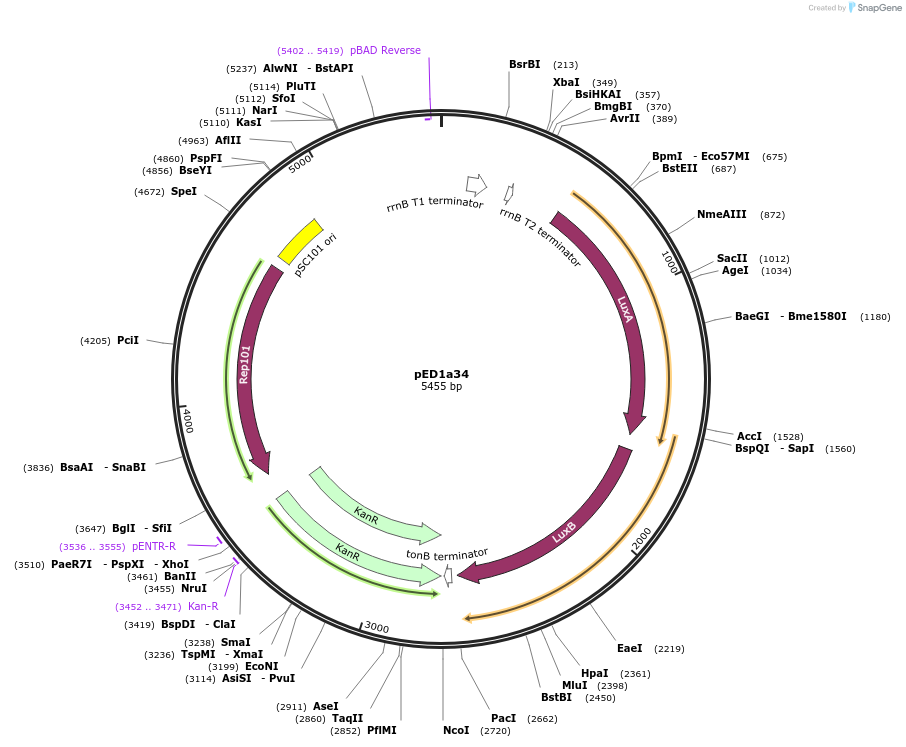
pED1a34
Plasmid#134789PurposeLuciferase reporter for quadruplet codon AGGGDepositorInsertluxAB-357-AGGG
UseLuciferase and Synthetic BiologyExpressionBacterialMutation357-AGGGAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
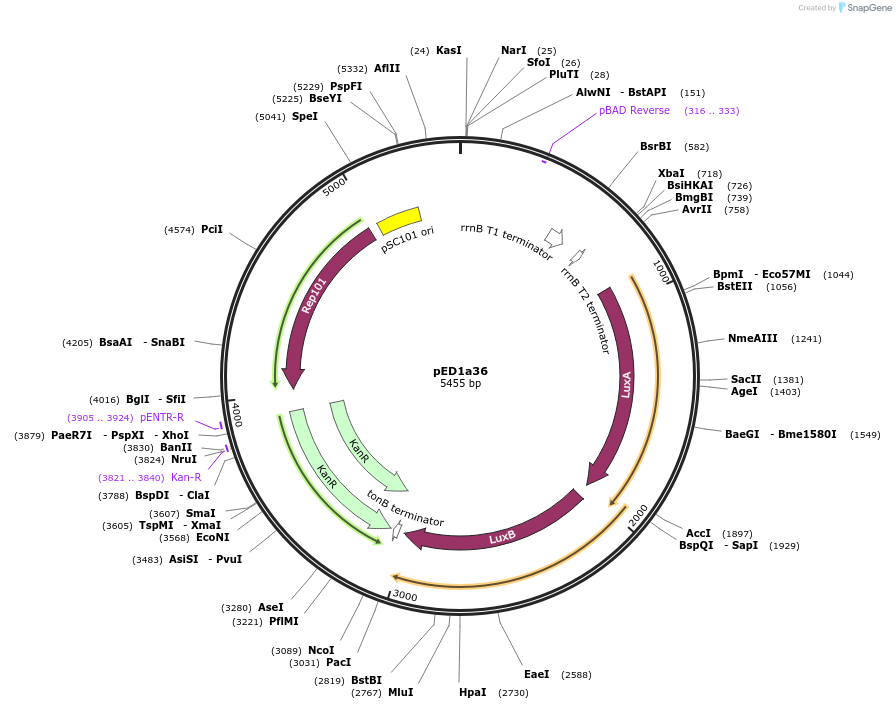
pED1a36
Plasmid#134790PurposeLuciferase reporter for quadruplet codon CGGTDepositorInsertluxAB-357-CGGT
UseLuciferase and Synthetic BiologyExpressionBacterialMutation357-CGGTAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
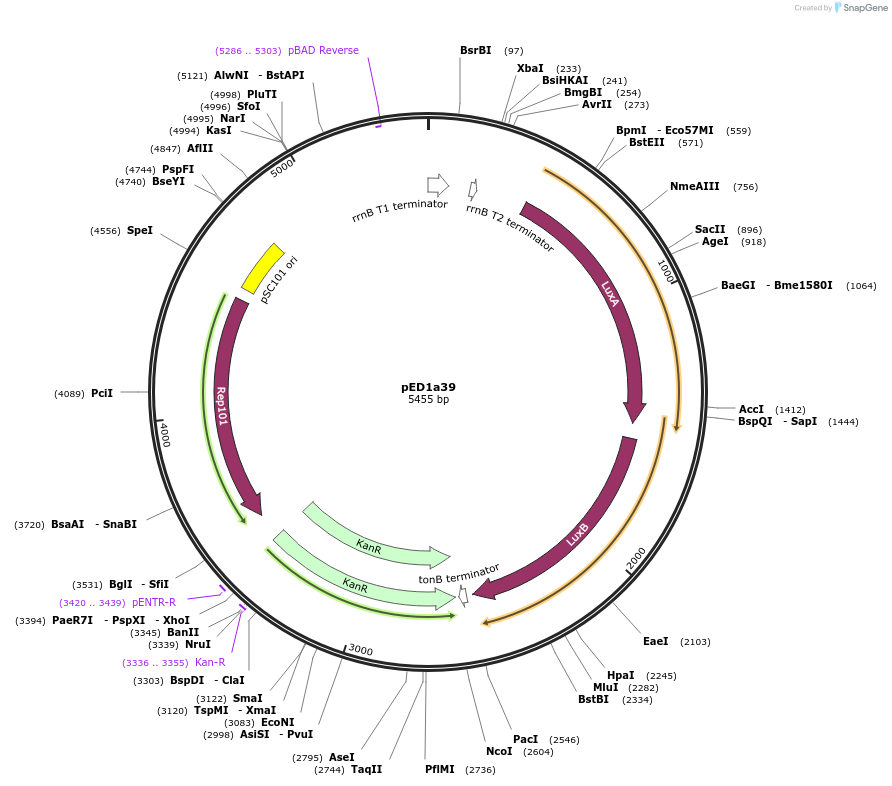
pED1a39
Plasmid#134791PurposeLuciferase reporter for quadruplet codon TACADepositorInsertluxAB-357-TACA
UseLuciferase and Synthetic BiologyExpressionBacterialMutation357-TACAAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
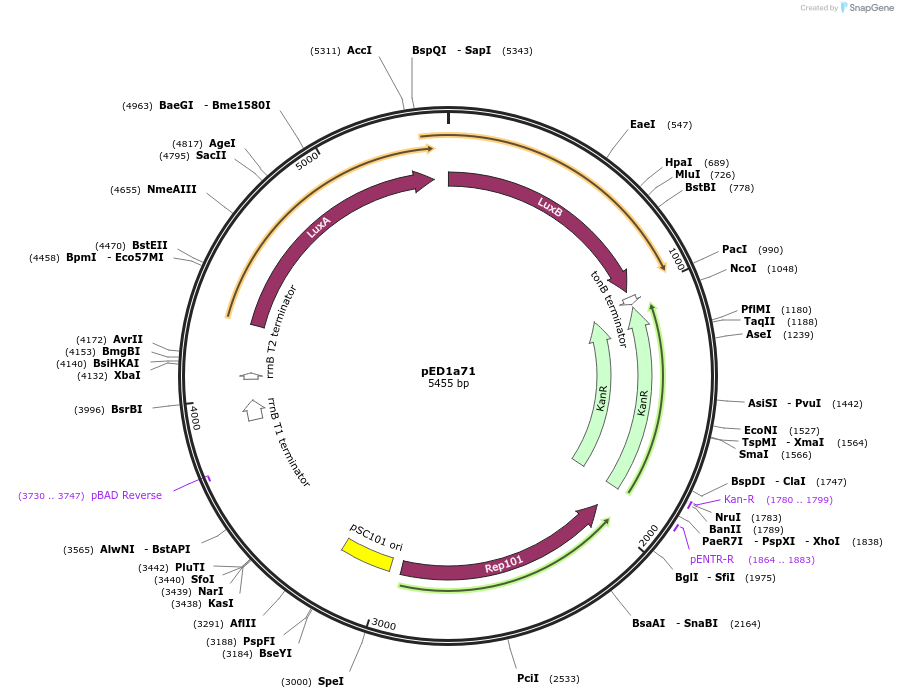
pED1a71
Plasmid#134792PurposeLuciferase reporter for quadruplet codon AGGADepositorInsertluxAB-357-AGGA
UseLuciferase and Synthetic BiologyExpressionBacterialMutation357-AGGAAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
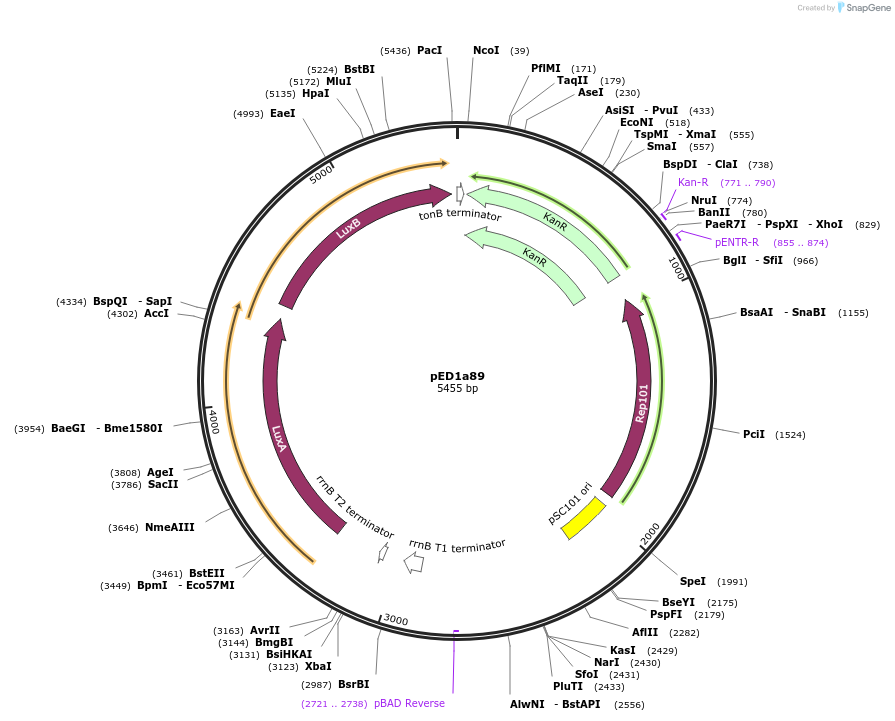
pED1a89
Plasmid#134793PurposeLuciferase reporter for quadruplet codon CGGCDepositorInsertluxAB-357-CGGC
UseLuciferase and Synthetic BiologyExpressionBacterialMutation357-CGGCAvailable SinceSept. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
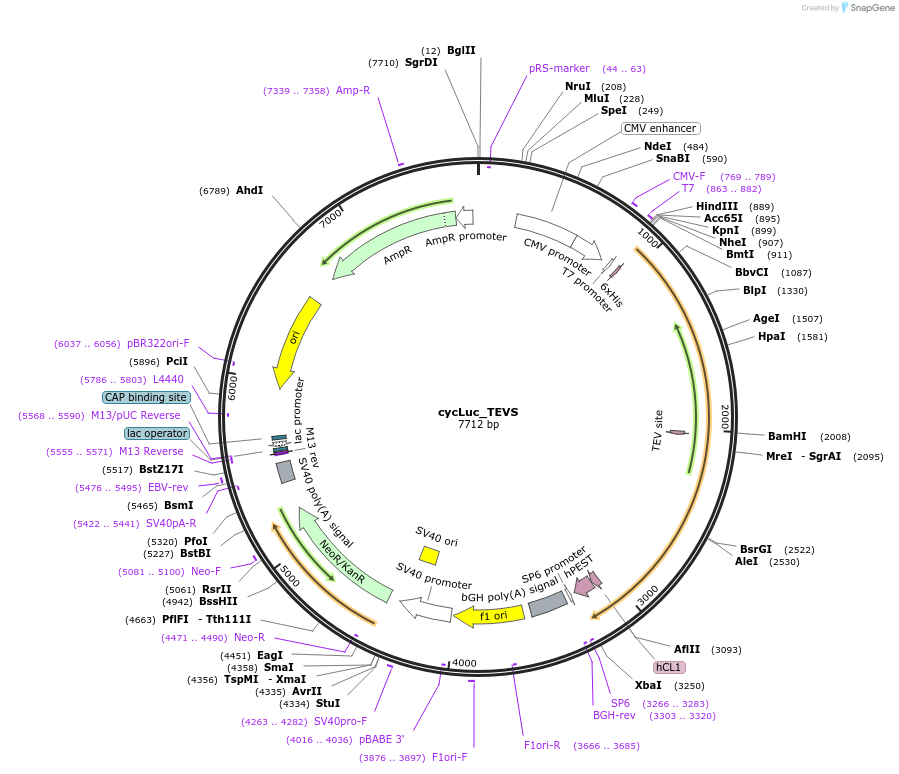
cycLuc_TEVS
Plasmid#119207PurposeCircularly permutated firefly luciferase with TEV cleavage site (Luciferase reporter for detection of proteolytic activity)DepositorInsertcyclic luciferase with TEVs cleavage site
UseLuciferaseTags6xHis and AU1 tagExpressionMammalianPromoterCMVAvailable SinceApril 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
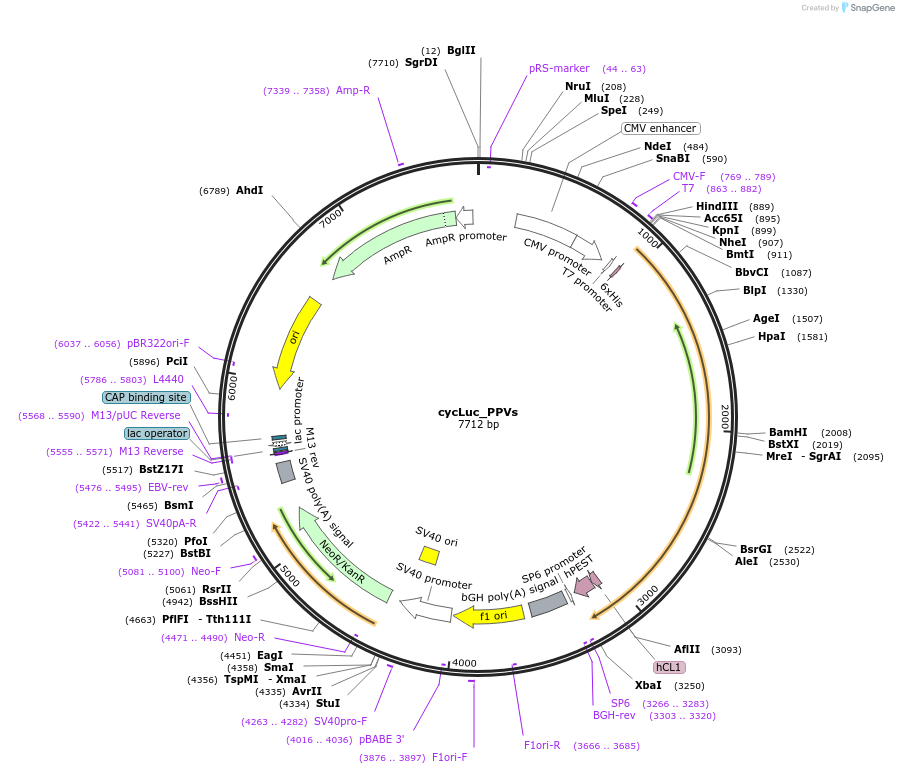
cycLuc_PPVs
Plasmid#119208PurposeCircularly permutated firefly luciferase with PPVs cleavage site (Luciferase reporter for detection of proteolytic activity)DepositorInsertcyclic luciferase with PPVp cleavage site
UseLuciferaseTagsAU1 tagPromoterCMVAvailable SinceJune 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
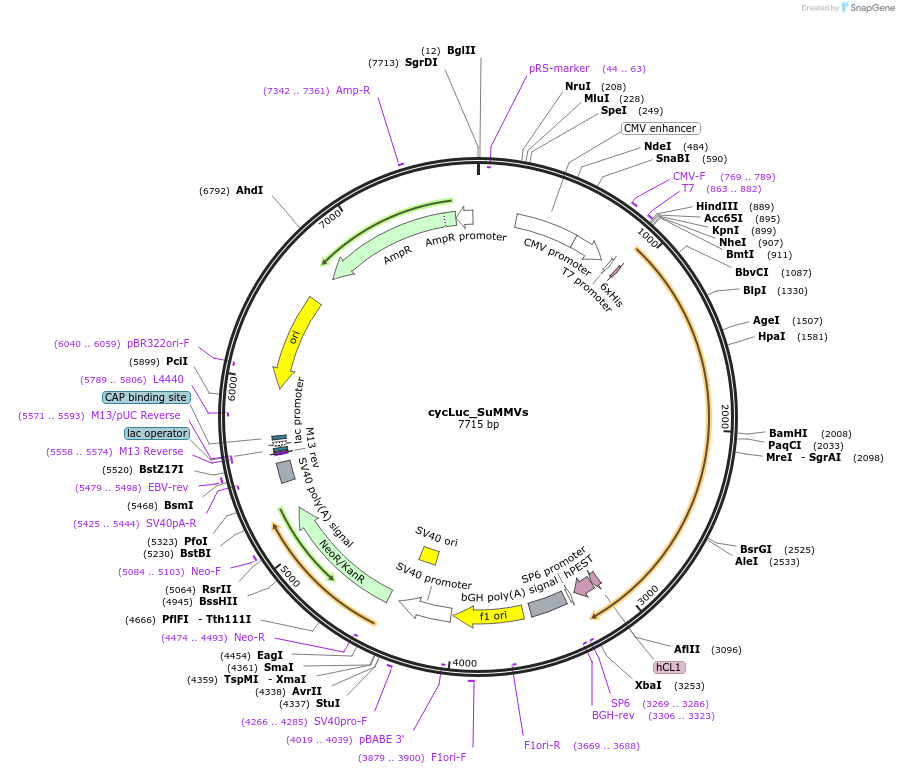
cycLuc_SuMMVs
Plasmid#119210PurposeCircularly permutated firefly luciferase with SuMMVs cleavage site (Luciferase reporter for detection of proteolytic activity)DepositorInsertcyclic luciferase with SuMMVp cleavage site
UseLuciferaseTagsAU1 tagExpressionMammalianPromoterCMVAvailable SinceFeb. 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
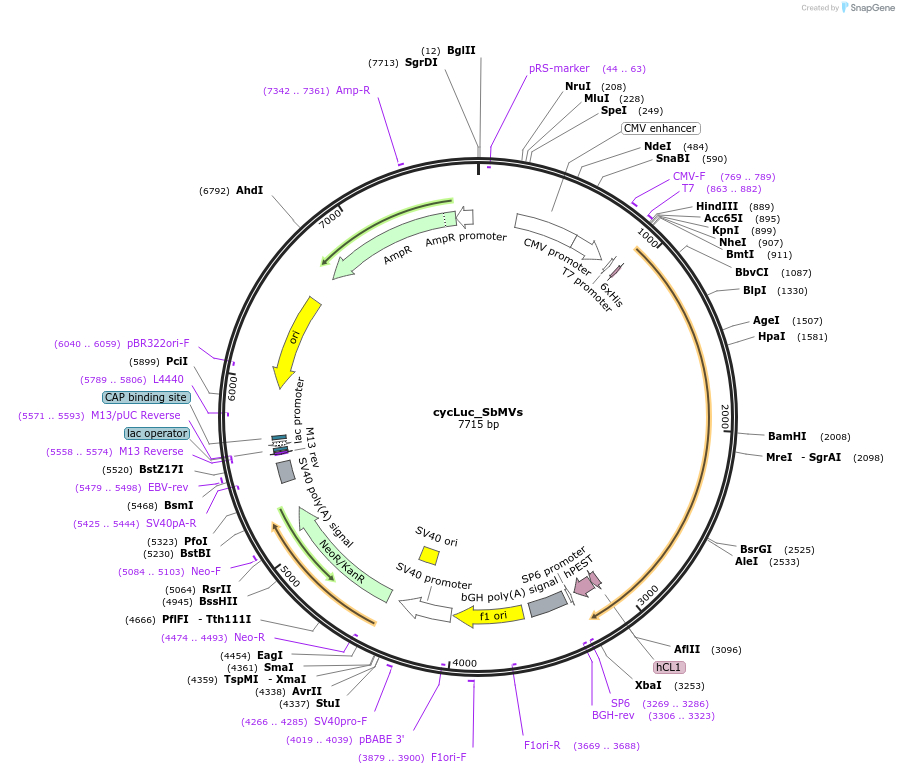
cycLuc_SbMVs
Plasmid#119209PurposeCircularly permutated firefly luciferase with SbMV cleavage site (Luciferase reporter for detection of proteolytic activity)DepositorInsertcyclic luciferase with SbMVp cleavage site
UseLuciferaseTagsAU1 tagExpressionMammalianPromoterCMVAvailable SinceFeb. 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
RAB14 3'UTR D1D2
Plasmid#61492Purposeluciferase reporter containing RAB14 3' UTR with both predicted MiR144 binding site deletedDepositorInsertRAB14 3' UTR (RAB14 Human)
UseLuciferaseTagsluciferaseExpressionMammalianMutationcontains position 837-1637 (numbering relative to…PromoterPGKAvailable SinceSept. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
RAB14 3'UTR D1
Plasmid#61490Purposeluciferase reporter containing RAB14 3' UTR with the first predicted MiR144 binding site deletedDepositorInsertRAB14 3' UTR (RAB14 Human)
UseLuciferaseTagsluciferaseExpressionMammalianMutationcontains position 837-1637 (numbering relative to…PromoterPGKAvailable SinceJan. 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
RAB14 3'UTR D2
Plasmid#61491Purposeluciferase reporter containing RAB14 3' UTR with the second predicted MiR144 binding site deletedDepositorInsertRAB14 3' UTR (RAB14 Human)
UseLuciferaseTagsluciferaseExpressionMammalianMutationcontains position 837-1637 (numbering relative to…PromoterPGKAvailable SinceSept. 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
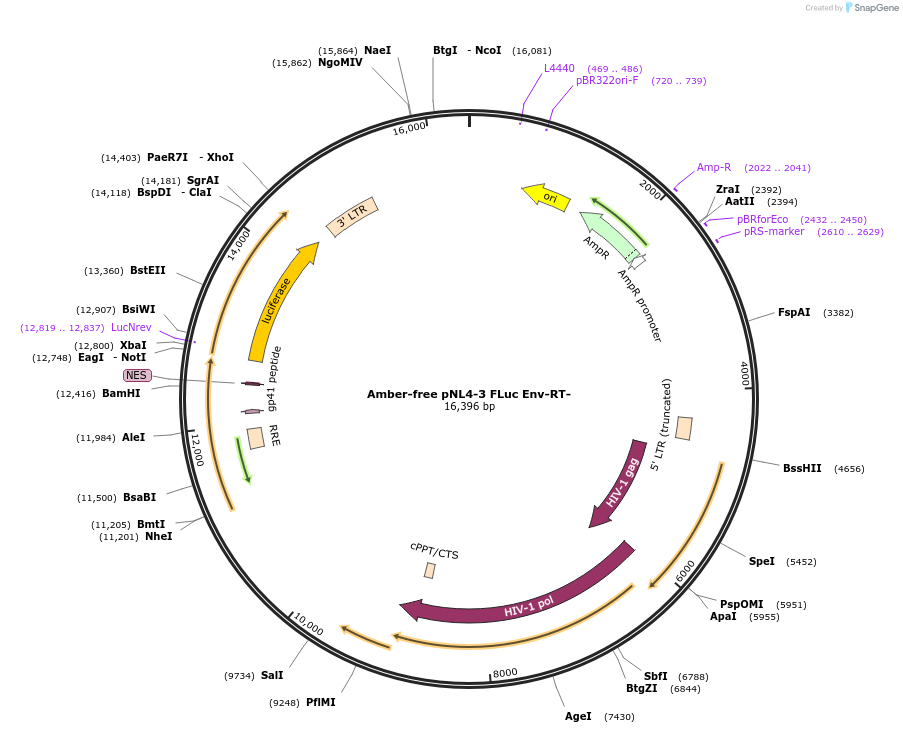
Amber-free pNL4-3 FLuc Env-RT-
Plasmid#238415PurposeAn amber-free HIV NL4-3 carrying a Firefly luciferase reporter gene, RT and Env impaired.DepositorInsertFirefly luciferase (LOC116160065 Synthetic)
UseLentiviral and LuciferaseExpressionMammalianMutationAlternation of amber stop codons "TAG" …Available SinceAug. 25, 2025AvailabilityAcademic Institutions and Nonprofits only -
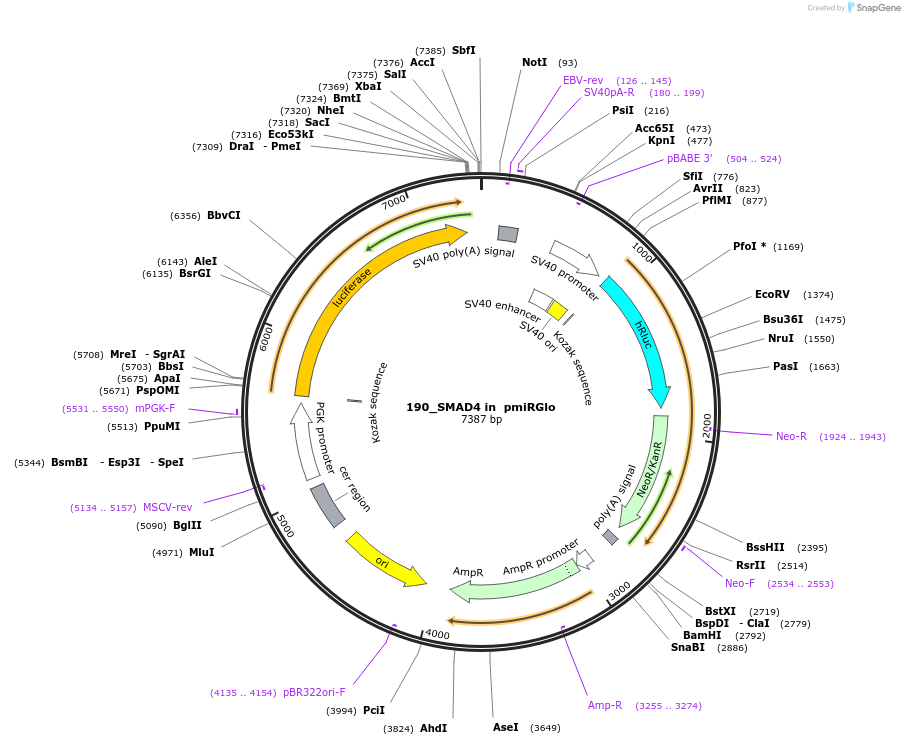
190_SMAD4 in pmiRGlo
Plasmid#78128Purposeluciferase reporter for miRNA activityDepositorInsertSMAD4 (SMAD4 Human)
UseLuciferaseTagsluciferaseExpressionMammalianMutation3'UTR containing miRNA binding sitesPromoterPGKAvailable SinceMay 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
pGL3 GATA Luc
Plasmid#85695PurposeLuciferase reporter containing three repeats of the GATA consensus siteDepositorInsert3xGATA and minimal MT promoter
UseLuciferaseTagsluciferaseExpressionMammalianPromoter3xGATA and minimal MT promoterAvailable SinceMarch 21, 2017AvailabilityAcademic Institutions and Nonprofits only -
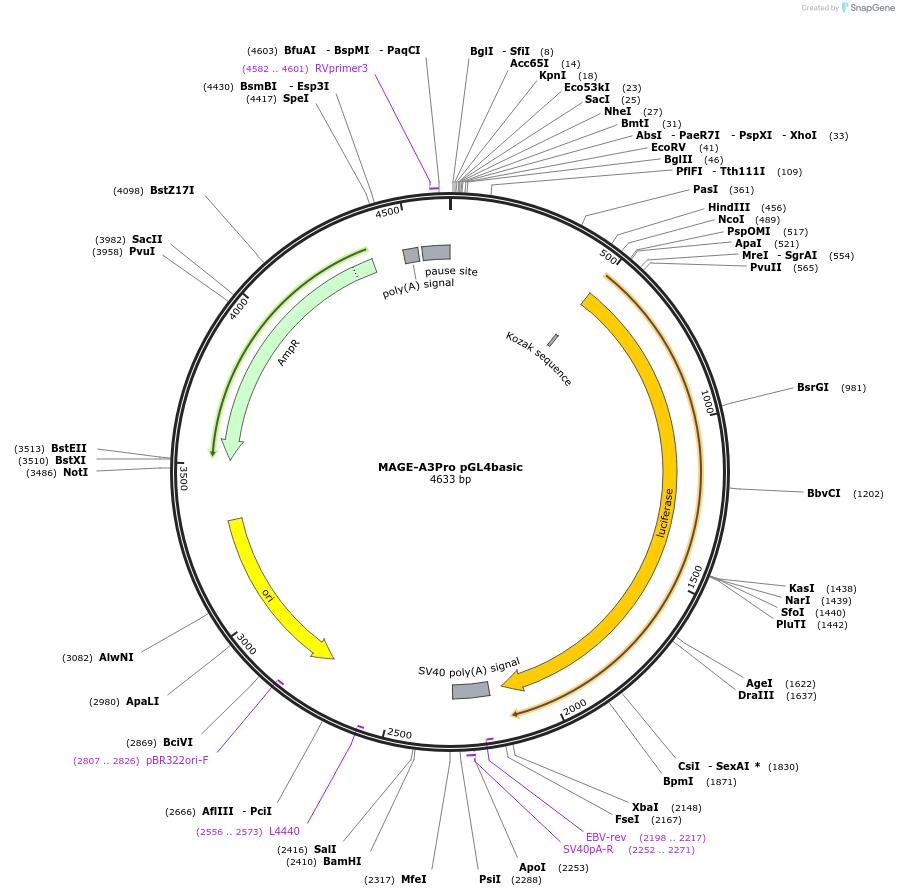
MAGE-A3Pro pGL4basic
Plasmid#78138Purposeluciferase reporter plasmidDepositorInsertMAGE-A3 (MAGEA3 Human)
UseLuciferaseTagsluciferaseExpressionMammalianMutationcontains -418/+12 of promoterPromoterMAGE-A3Available SinceJuly 5, 2016AvailabilityAcademic Institutions and Nonprofits only -