We narrowed to 24,790 results for: Spr
-
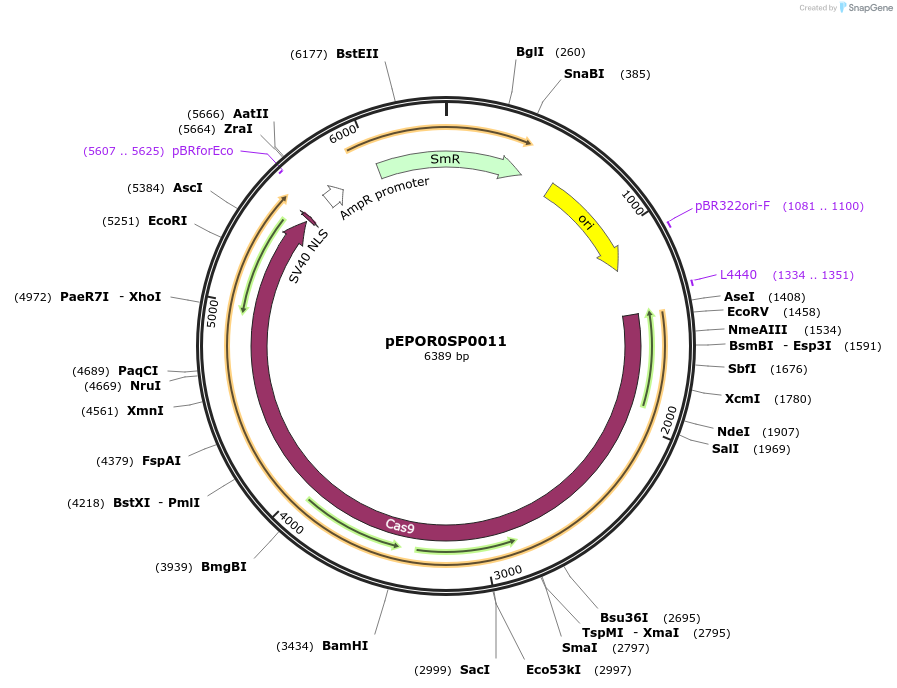
Plasmid#117530PurposeLevel0 Golden Gate part: CDS, Sp-xCas9 3.7 (no stop codon)DepositorInsertSp-xCas9 3.7 (no stop codon)
UseCRISPRMutationA262T, R324L, S409I, E480K, E543D, M694I, E1219VAvailable SinceNov. 7, 2018AvailabilityAcademic Institutions and Nonprofits only -
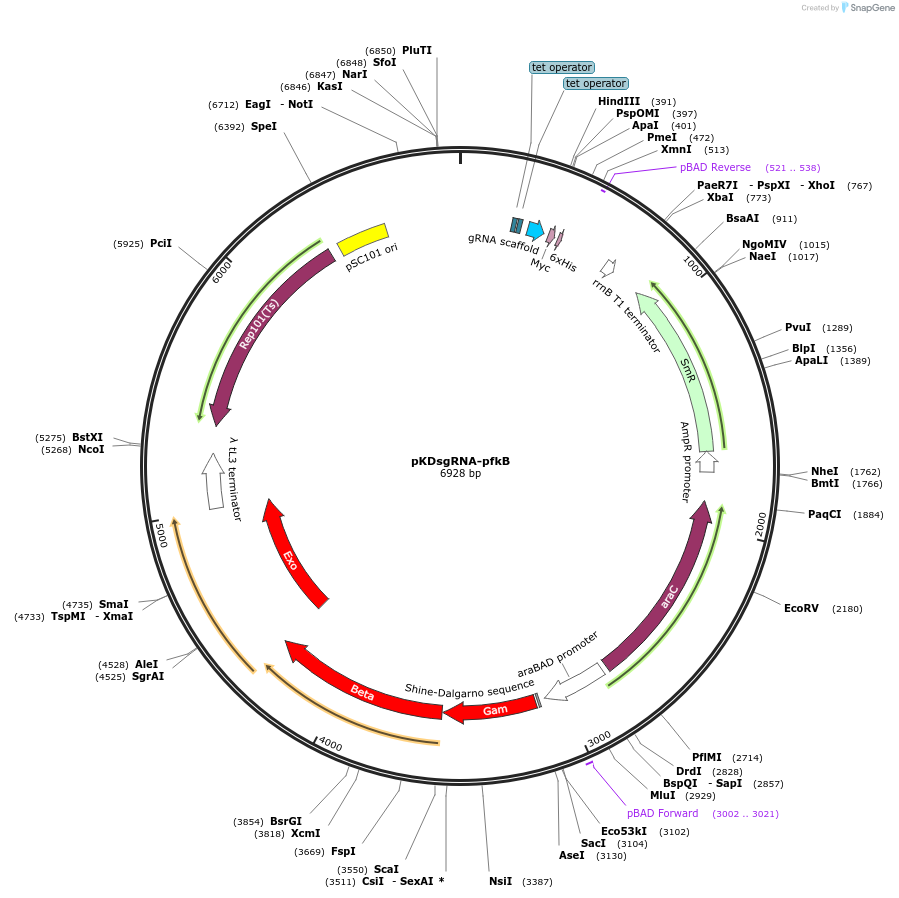
pKDsgRNA-pfkB
Plasmid#102284PurposeContains arabinose-induced lambda Red and a tet-inducible gRNA targeting pfkB.DepositorInsertpfkB gRNA
UseCRISPRExpressionBacterialPromoterpTetAvailable SinceApril 12, 2018AvailabilityAcademic Institutions and Nonprofits only -
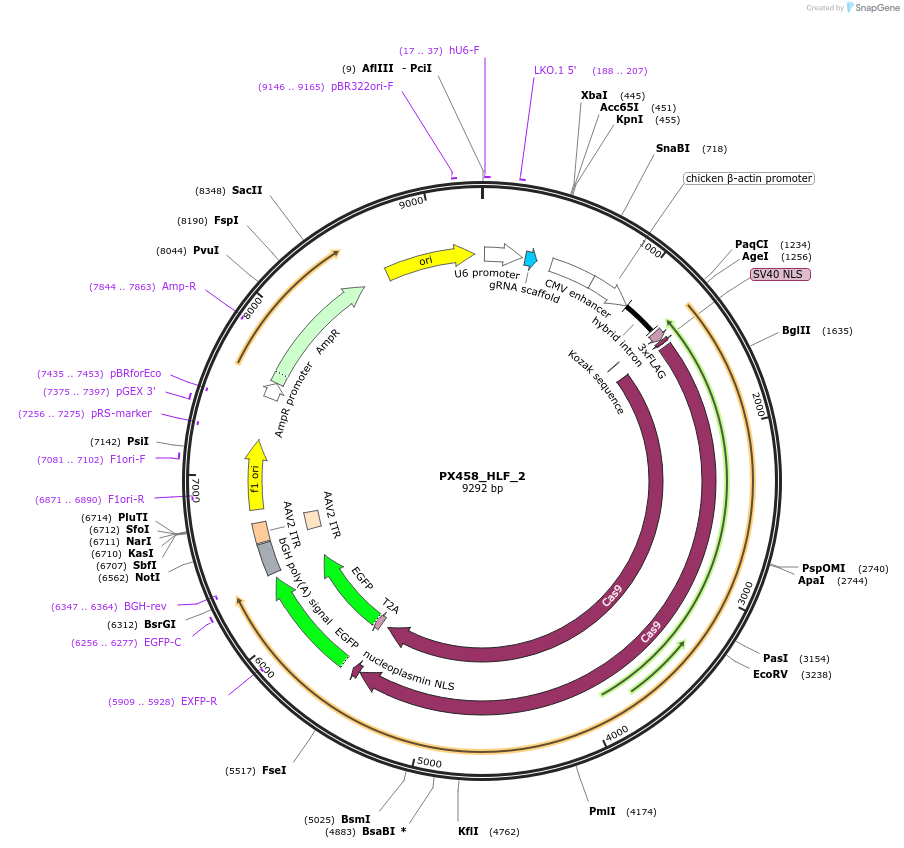
PX458_HLF_2
Plasmid#86302PurposeEncodes gRNA for 3' target of human HLFDepositorInsertgRNA against HLF (HLF Human)
UseCRISPRAvailable SinceFeb. 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
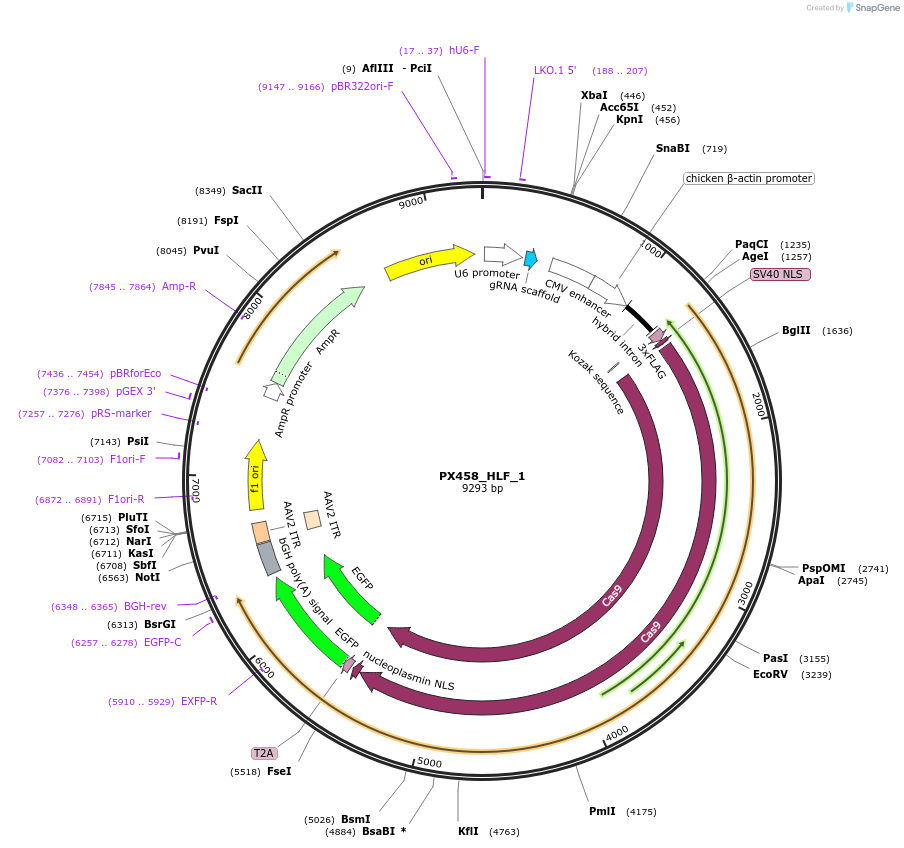
PX458_HLF_1
Plasmid#86301PurposeEncodes gRNA for 3' target of human HLFDepositorInsertgRNA against HLF (HLF Human)
UseCRISPRAvailable SinceJan. 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
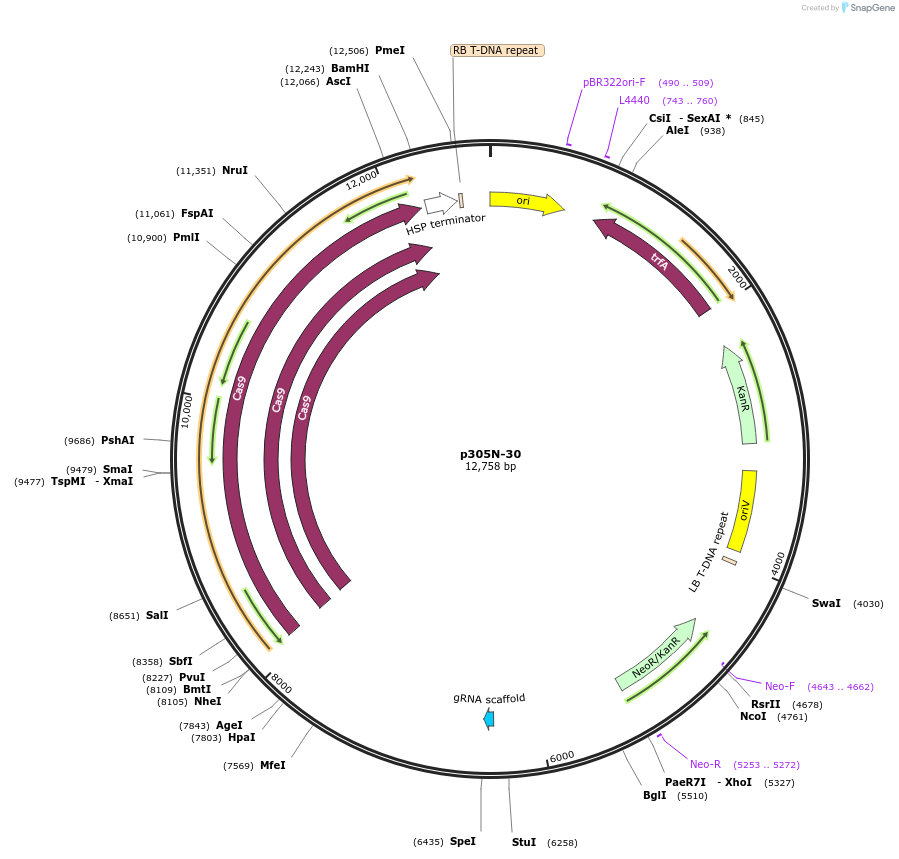
p305N-30
Plasmid#246313PurposeEvaluation of PtU6.2c3 promoter (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c3 promoter
UseCRISPRExpressionPlantMutationdeletion: -T (-29 from TSS) within TATAPromoterPtU6.2c3 promoterAvailable SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
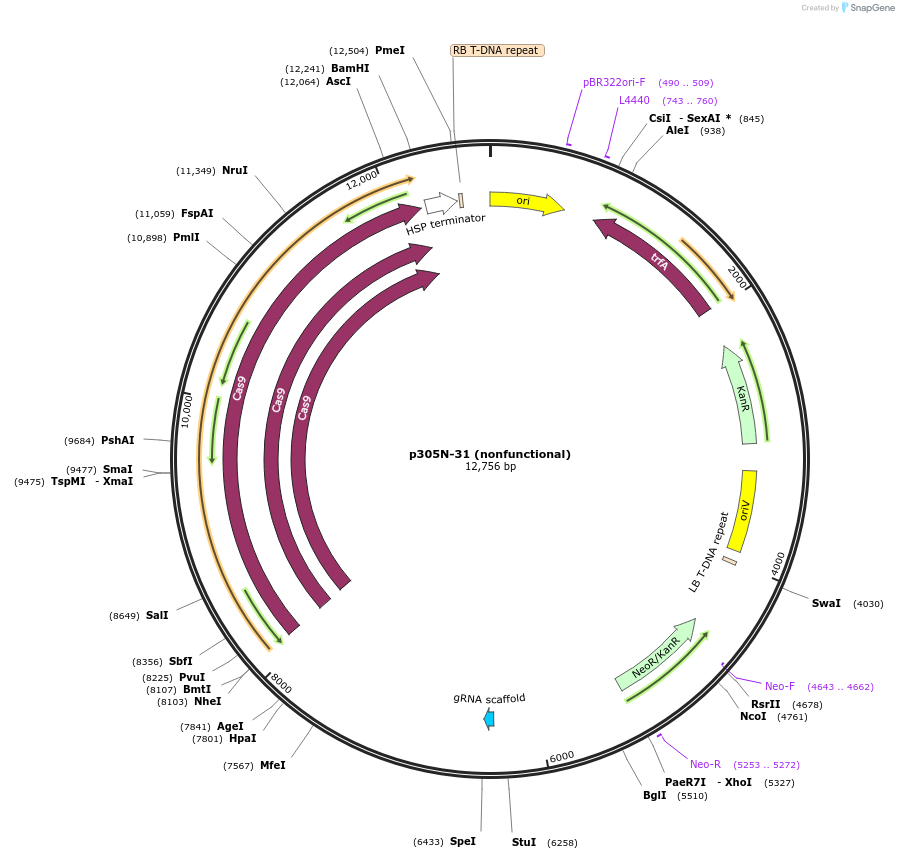
p305N-31 (nonfunctional)
Plasmid#246314PurposeEvaluation of PtU6.2c4 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertPtU6.2c4 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationdeletions: -A (-41 from TSS), -T (-30), -T (-29);…PromoterPtU6.2c4 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
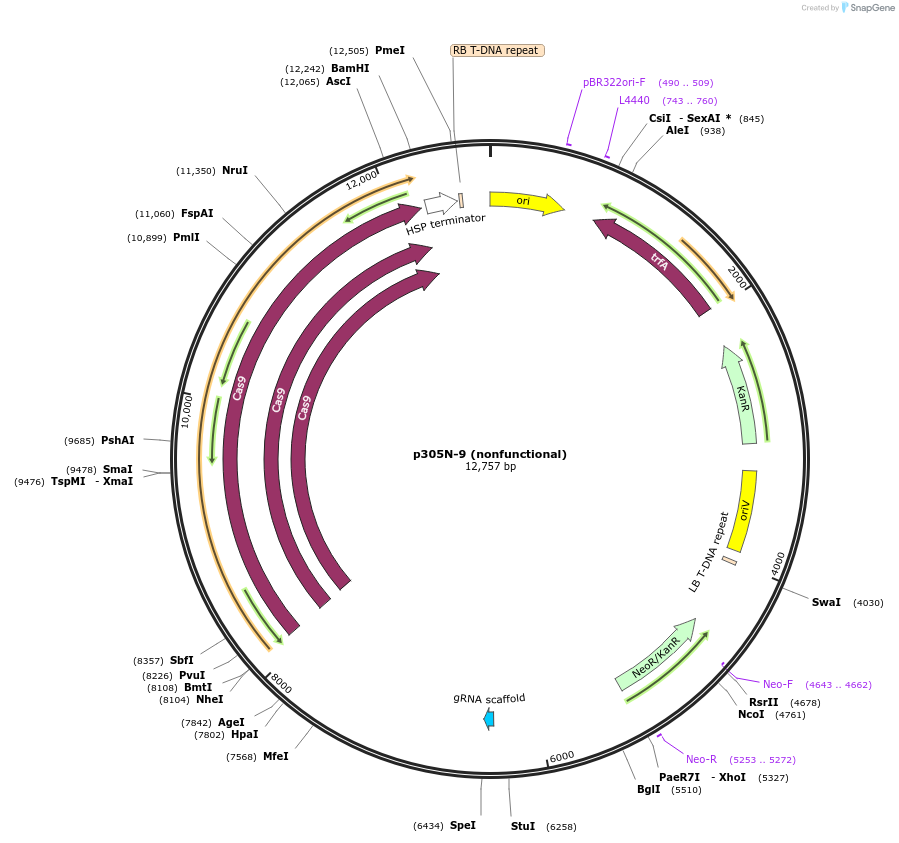
p305N-9 (nonfunctional)
Plasmid#246292PurposeEvaluation of AtU6.29c13 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.29c13 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationdeletions: -A (-58 from TSS), -T (-57) within USEPromoterAtU6.29c13 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
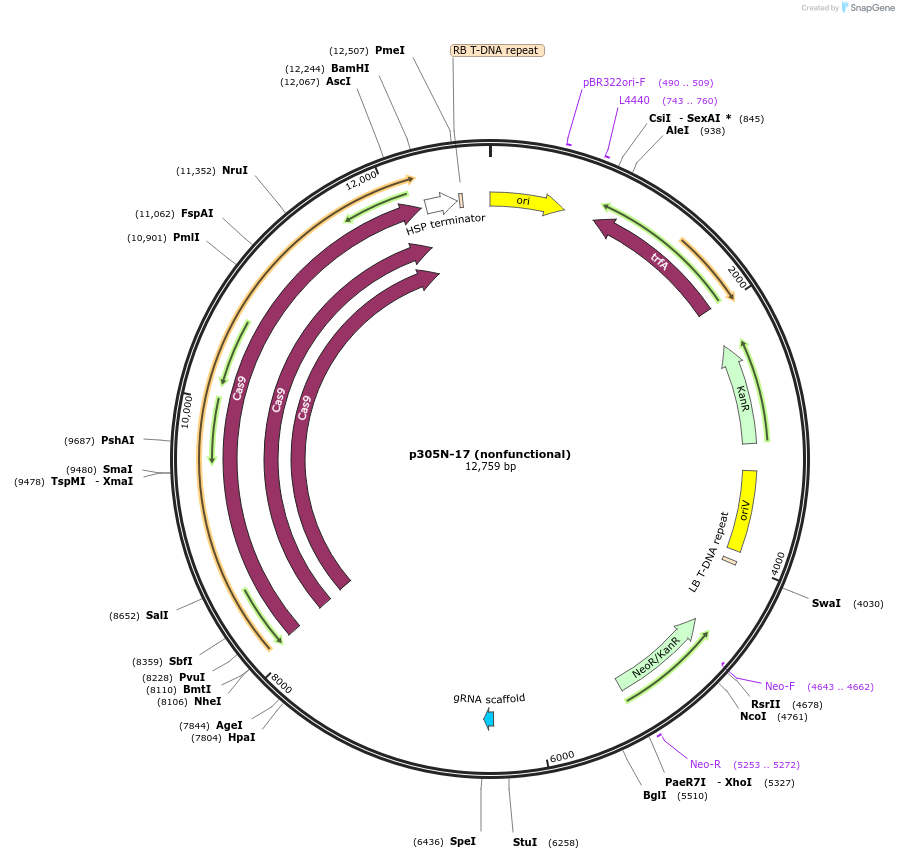
p305N-17 (nonfunctional)
Plasmid#246300PurposeEvaluation of HbU6.2m3 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertHbU6.2m3 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-T (-29 from TSS), G-to-A (-24)PromoterHbU6.2m3 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
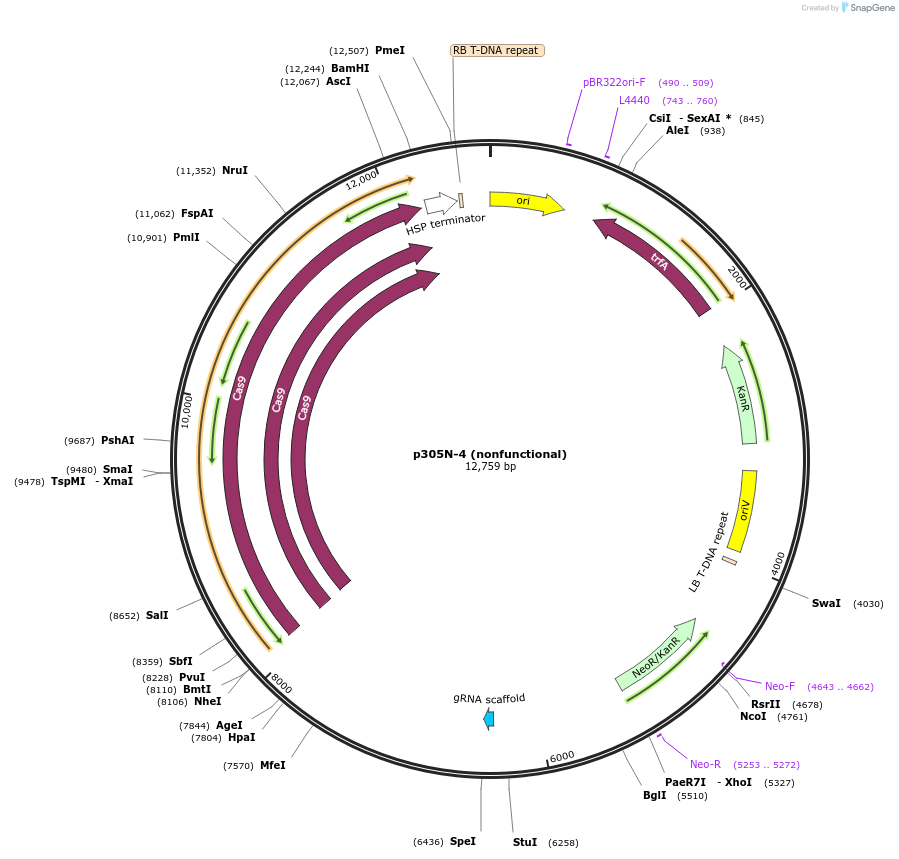
p305N-4 (nonfunctional)
Plasmid#246287PurposeEvaluation of AtU6.1c1 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertAtU6.1c1 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationG-to-C at -15 from TSSPromoterAtU6.1c1 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
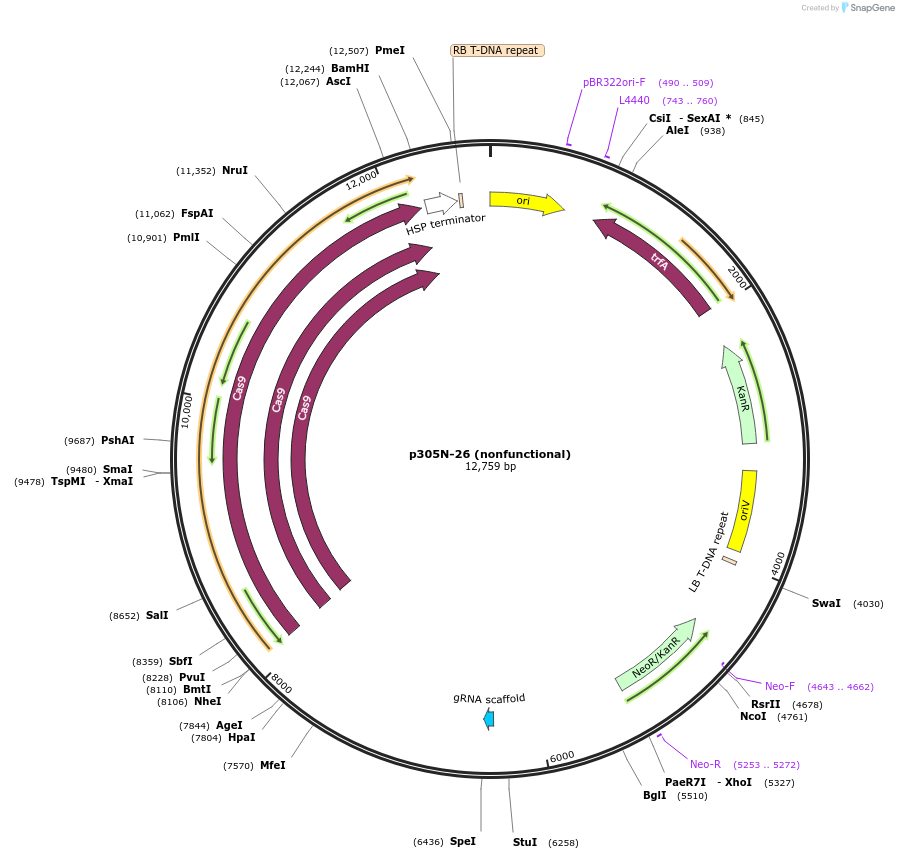
p305N-26 (nonfunctional)
Plasmid#246309PurposeEvaluation of MtU6.6m7 promoter (nonfunctional) (Pol III promoter) sequence variation for CRISPR-Cas9 editing of mEGFP in a Nicotiana benthamiana reporter lineDepositorInsertMtU6.6m7 promoter (nonfunctional)
UseCRISPRExpressionPlantMutationC-to-A (-63 from TSS), T-to-G (-57)PromoterMtU6.6m7 promoter (nonfunctional)Available SinceOct. 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
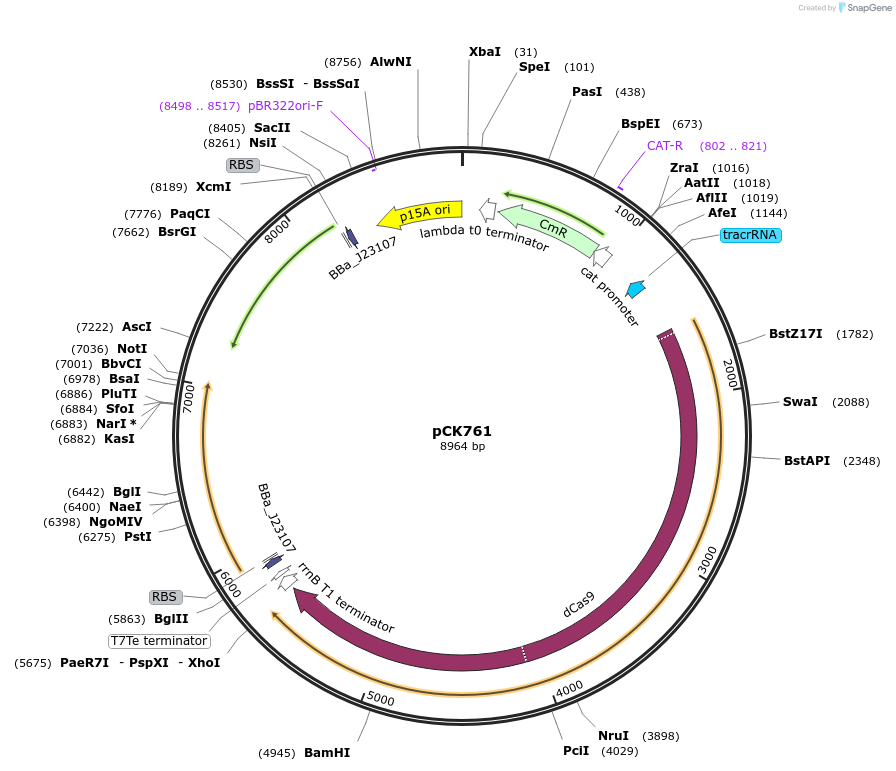
pCK761
Plasmid#242916PurposeConstitutive expression of dCas9, αNTD-MCP, and PspF-λN22DepositorAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
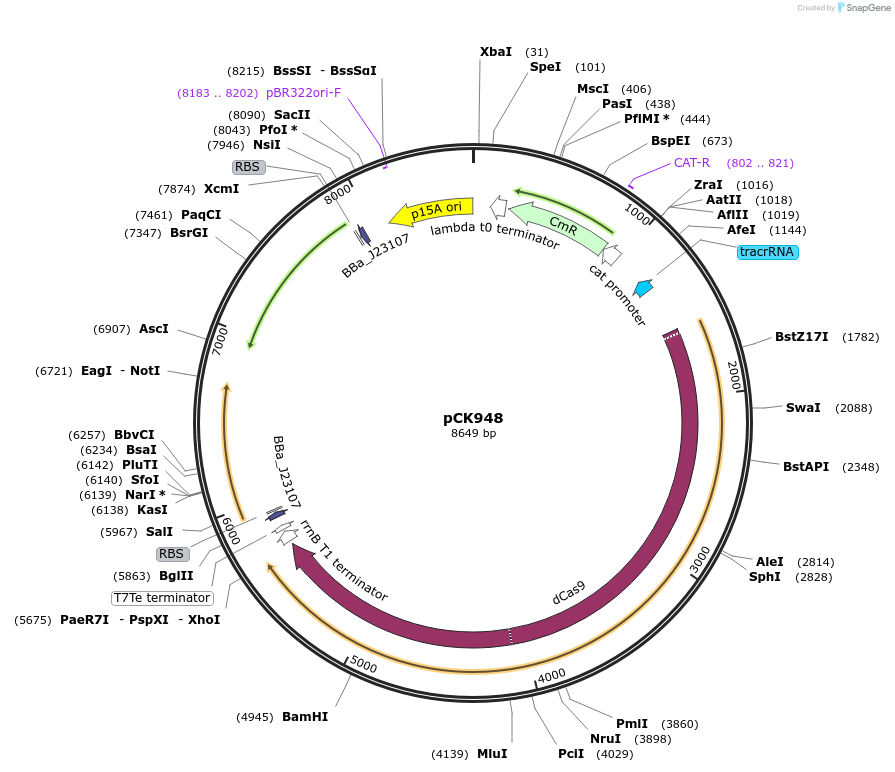
pCK948
Plasmid#242917PurposeConstitutive expression of dCas9, MCP-TetD, and PspF-λN22DepositorAvailable SinceOct. 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
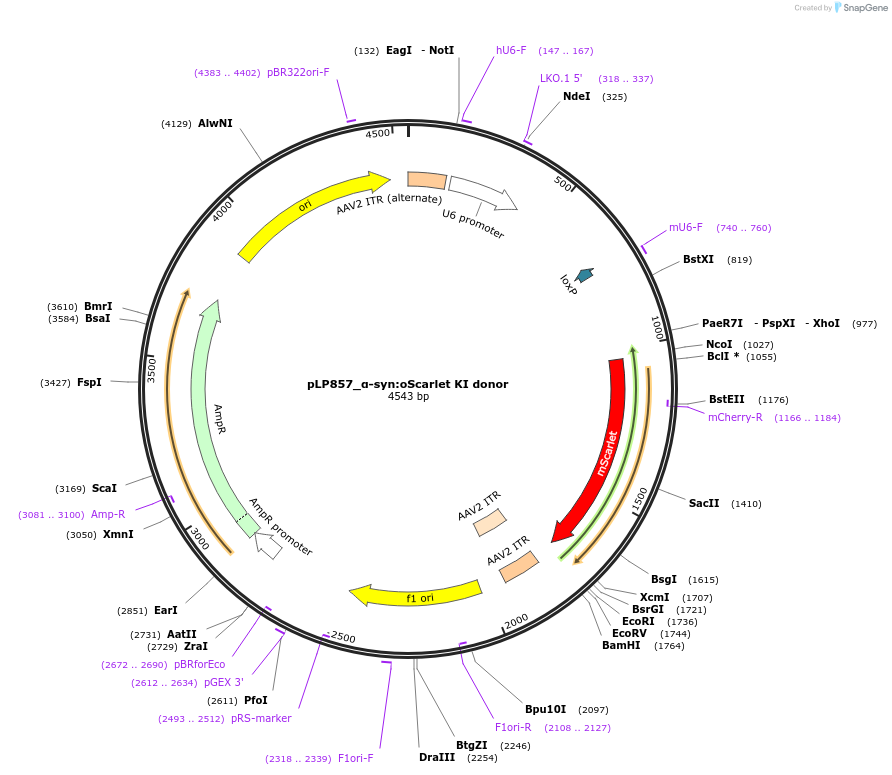
pLP857_α-syn:oScarlet KI donor
Plasmid#239403PurposeAAV vector for SpCas9-mediated CRISPR KI of oScarlet at the C-terminus of mouse α-synDepositorInsertoScarlet
UseAAV and CRISPRTagsoScarletExpressionMammalianAvailable SinceSept. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
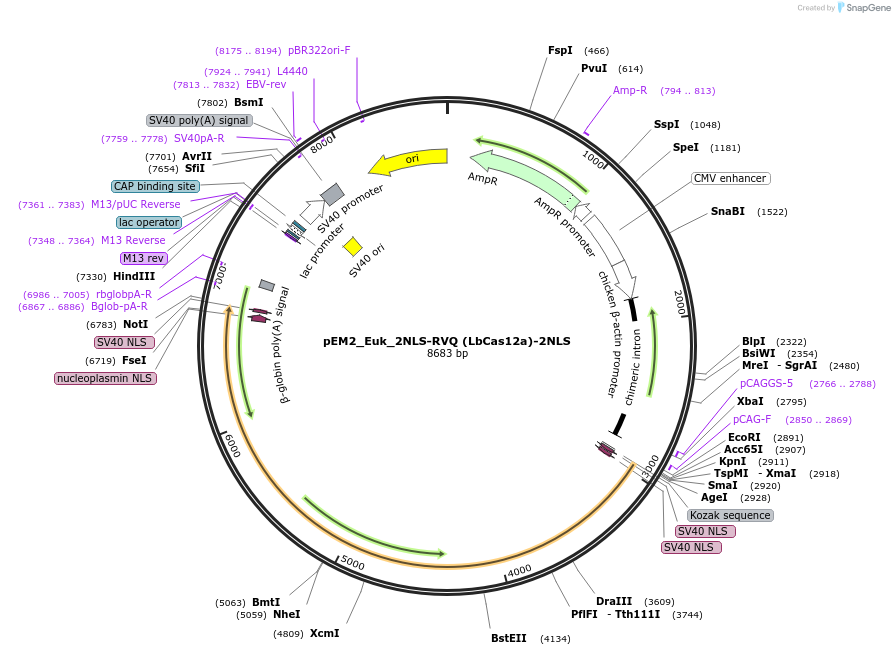
pEM2_Euk_2NLS-RVQ (LbCas12a)-2NLS
Plasmid#244837PurposeMammalian expression of LbCas12a-RVQ with 2xSV40 NLS at N-terminus and 2xSV40 at C-terminusDepositorInsertLbCas12a-RVQ
UseCRISPRTags2xSV40 NLSExpressionMammalianMutationG146R/R182V/E795QPromoterT7Available SinceSept. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
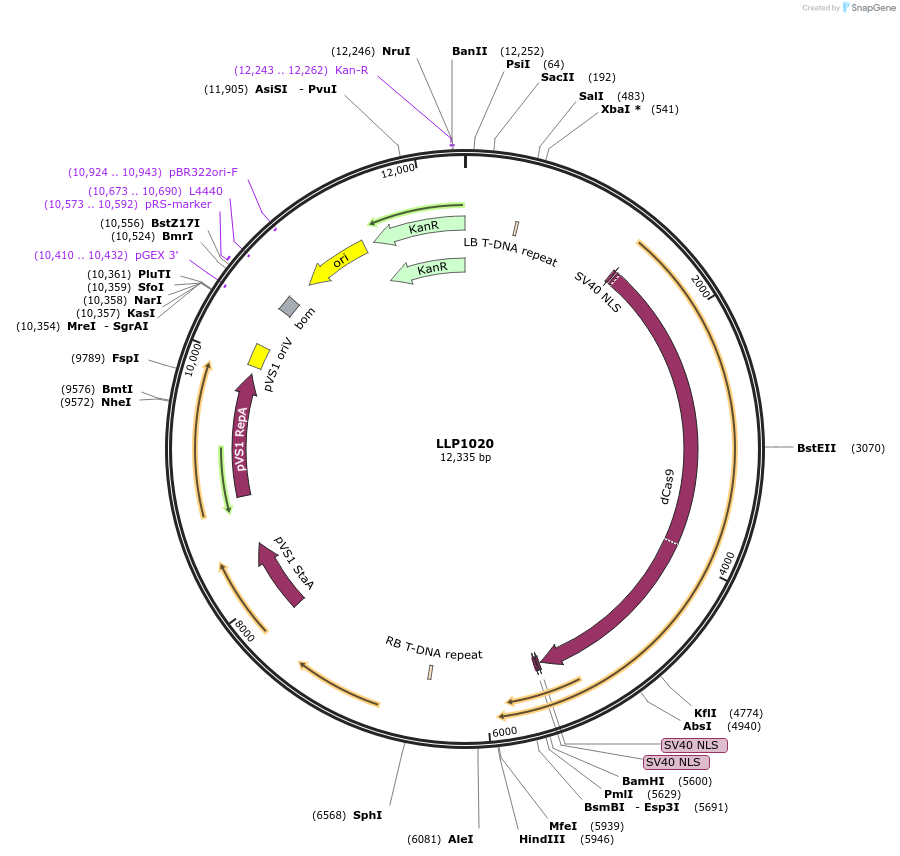
LLP1020
Plasmid#239942PurposeAtACT2 promoter driving the expression of dCas9-ZAT10(2x)DepositorInsertAtACT2::dCas9-ZAT10(2x)
UseCRISPR and Synthetic BiologyExpressionPlantMutationN/AAvailable SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
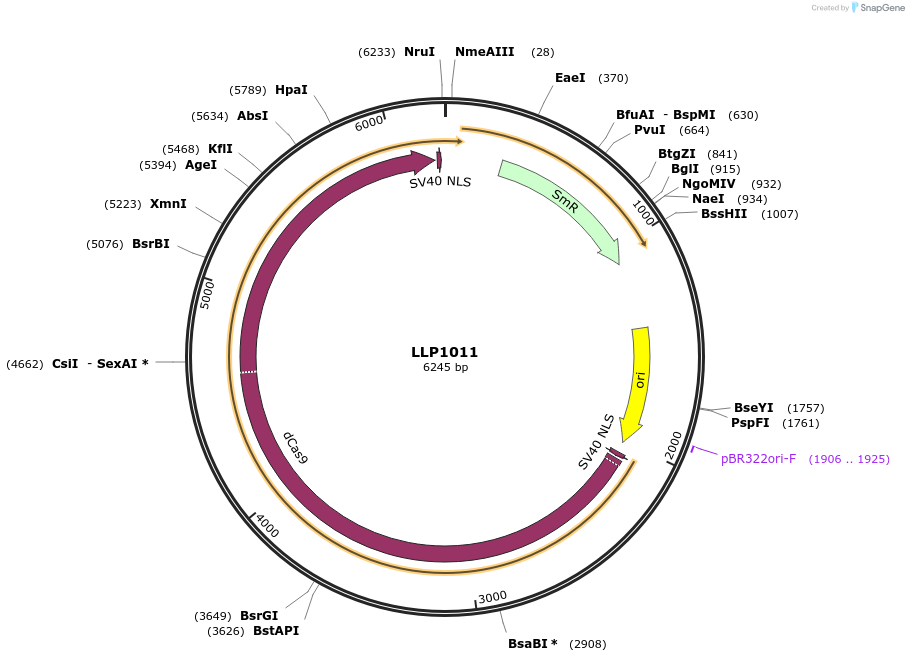
LLP1011
Plasmid#239933PurposedCas9 without Stop codon as Level-0 part (Golden Gate cloning)DepositorInsertdCas9
UseCRISPR and Synthetic BiologyMutationN/AAvailable SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
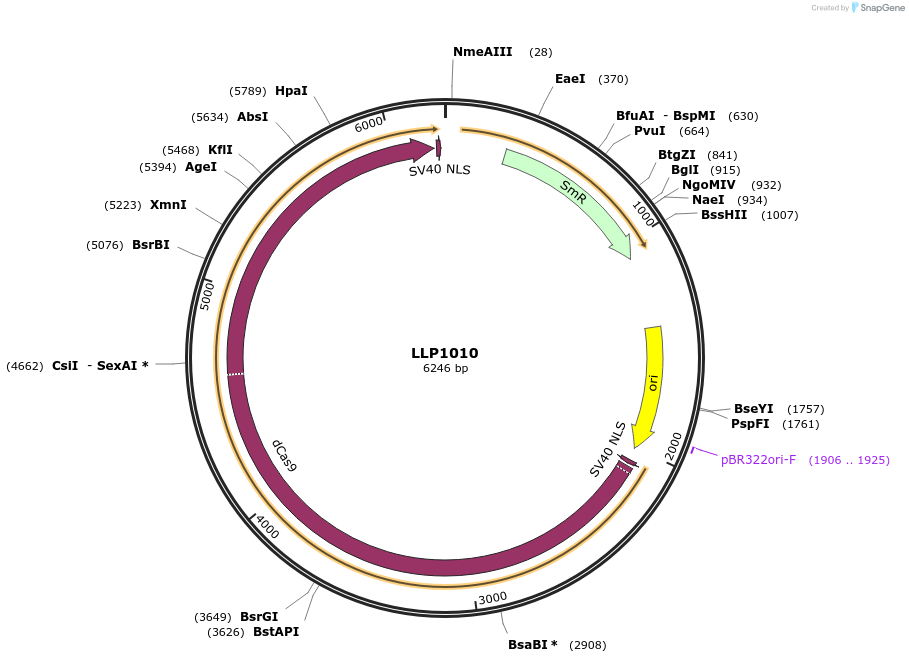
LLP1010
Plasmid#239932PurposedCas9 with Stop codon as Level-0 part (Golden Gate cloning)DepositorInsertdCas9
UseCRISPR and Synthetic BiologyMutationN/AAvailable SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
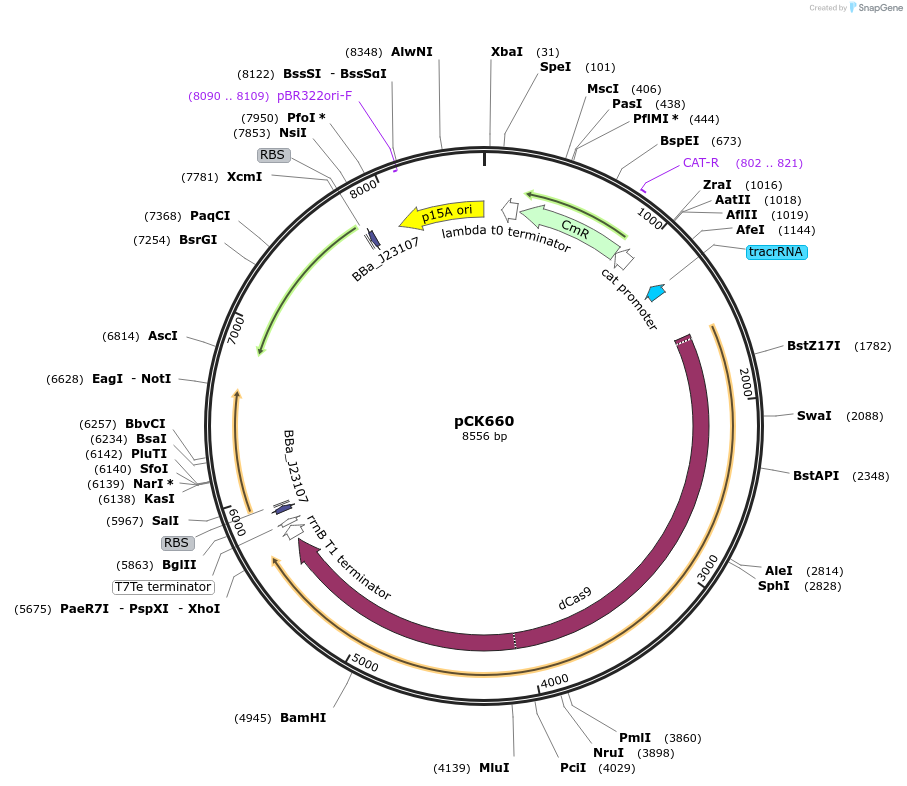
pCK660
Plasmid#242914PurposeConstitutive expression of dCas9, MCP-SoxS, and PspF-λN22DepositorAvailable SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
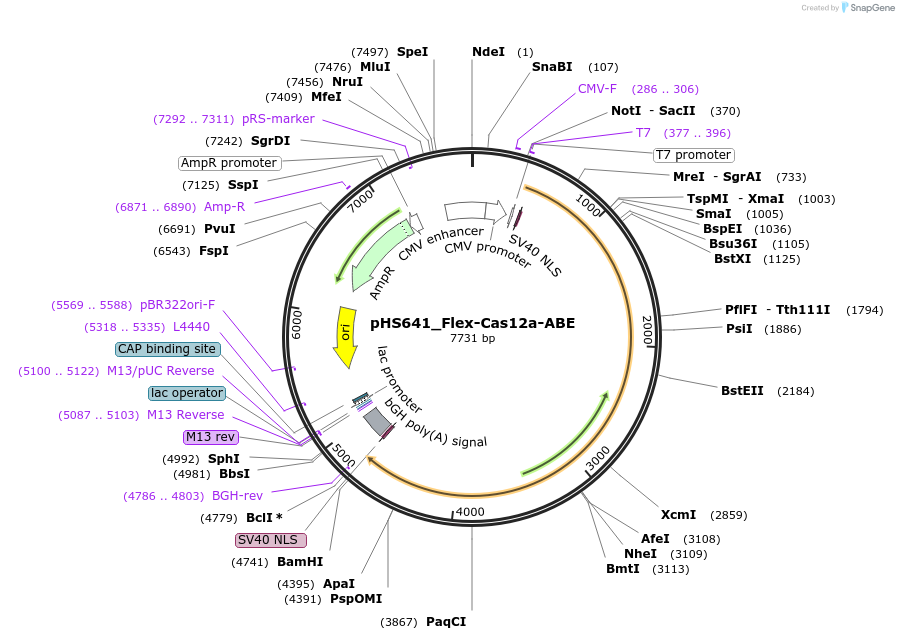
pHS641_Flex-Cas12a-ABE
Plasmid#244846PurposeMammalian expression of Flex-Cas12a adenine base editorDepositorInsertFlex-dCas12a-ABE
UseCRISPRTagsSV40 NLS-ABE8eExpressionMammalianMutationG146R/R182V/D535G/S551F/D665N/E795Q/D832APromoterCMVAvailable SinceSept. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
pMKR07
Plasmid#240097PurposeA CRISPR‑Cas9 system for knock‑out and knock‑in of high molecular weight DNA enables module‑swapping of the pikromycin synthase in its native hostDepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionBacterialPromoterermE* promoterAvailable SinceJuly 14, 2025AvailabilityAcademic Institutions and Nonprofits only