We narrowed to 3,846 results for: 28
-
Plasmid#173978PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #6
UseLuciferaseMutationTiled deletion #6PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGL3-noSV40-humanEC1.45_p300peak1-2_del4
Plasmid#173976PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #4
UseLuciferaseMutationTiled deletion #4PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGL3-noSV40-humanEC1.45_p300peak1-2_del1
Plasmid#173973PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #1
UseLuciferaseMutationTiled deletion #1PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGL3-noSV40-humanEC1.45_p300peak1-2_del10
Plasmid#173982PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #10
UseLuciferaseMutationTiled deletion #10PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
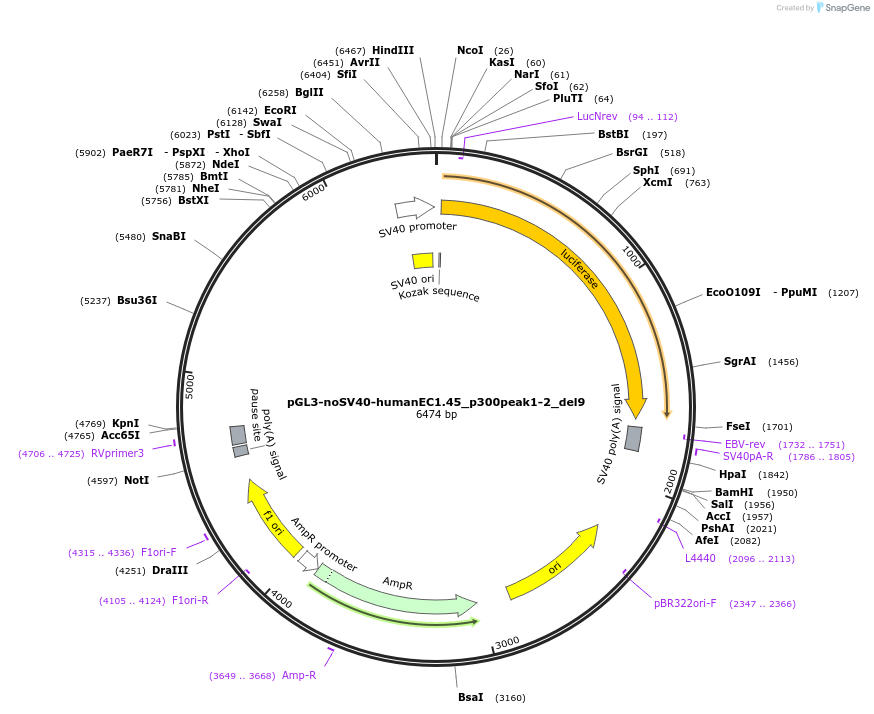
pGL3-noSV40-humanEC1.45_p300peak1-2_del9
Plasmid#173981PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #9
UseLuciferaseMutationTiled deletion #9PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
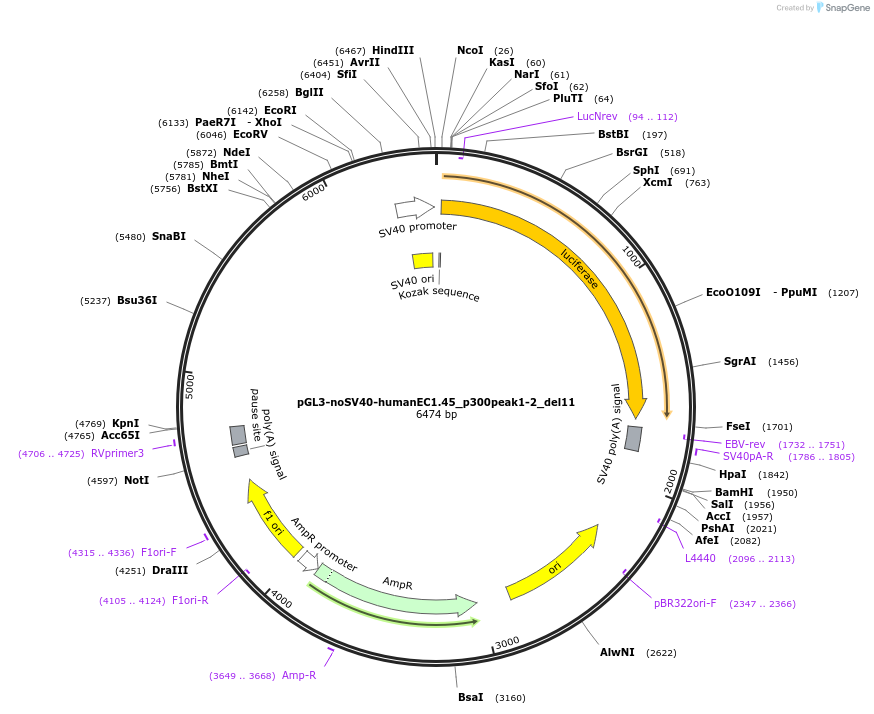
pGL3-noSV40-humanEC1.45_p300peak1-2_del11
Plasmid#173983PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #11
UseLuciferaseMutationTiled deletion #11PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
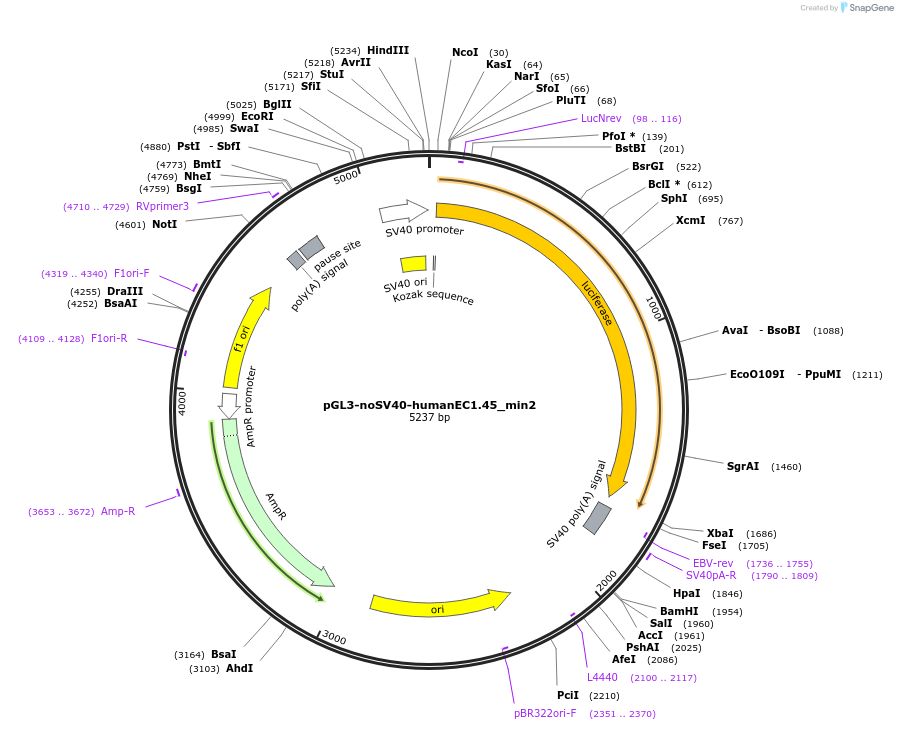
pGL3-noSV40-humanEC1.45_min2
Plasmid#173951PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertMinimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
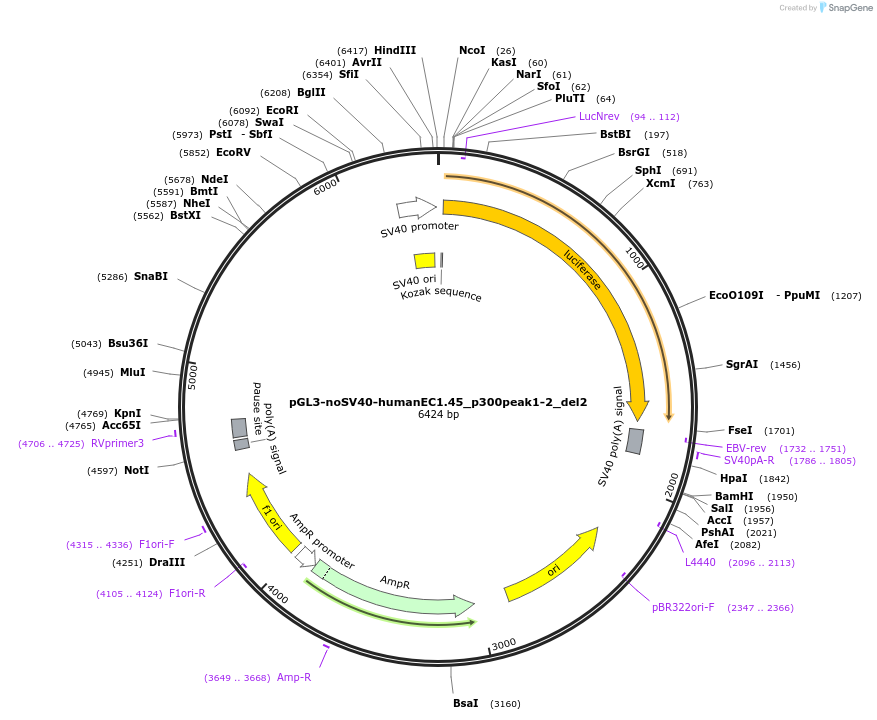
pGL3-noSV40-humanEC1.45_p300peak1-2_del2
Plasmid#173974PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #2
UseLuciferaseMutationTiled deletion #2PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
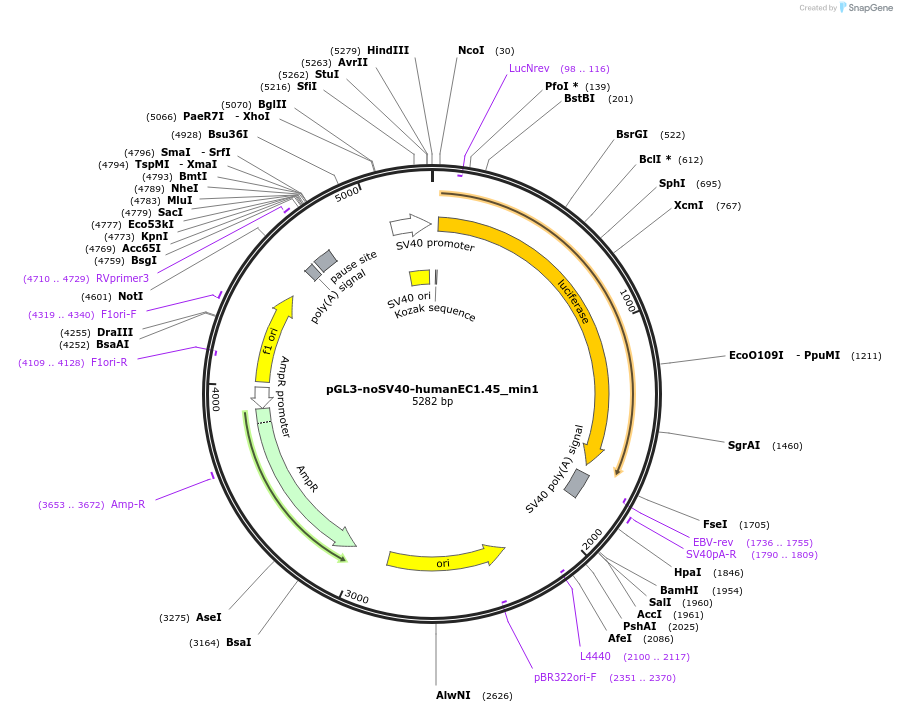
pGL3-noSV40-humanEC1.45_min1
Plasmid#173950PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertMinimal enhancer region 1 (min1) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
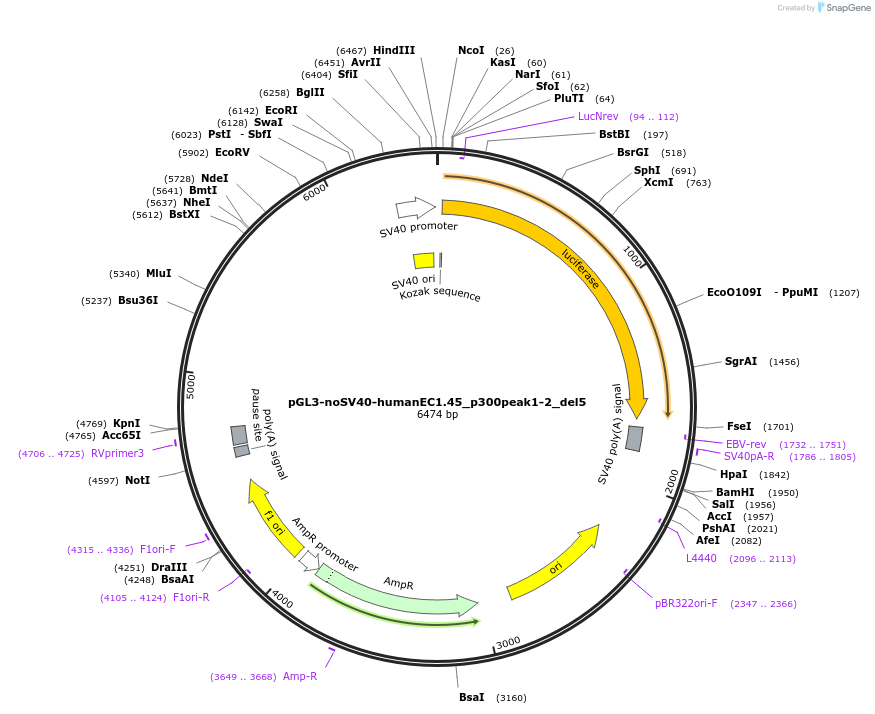
pGL3-noSV40-humanEC1.45_p300peak1-2_del5
Plasmid#173977PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #5
UseLuciferaseMutationTiled deletion #5PromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
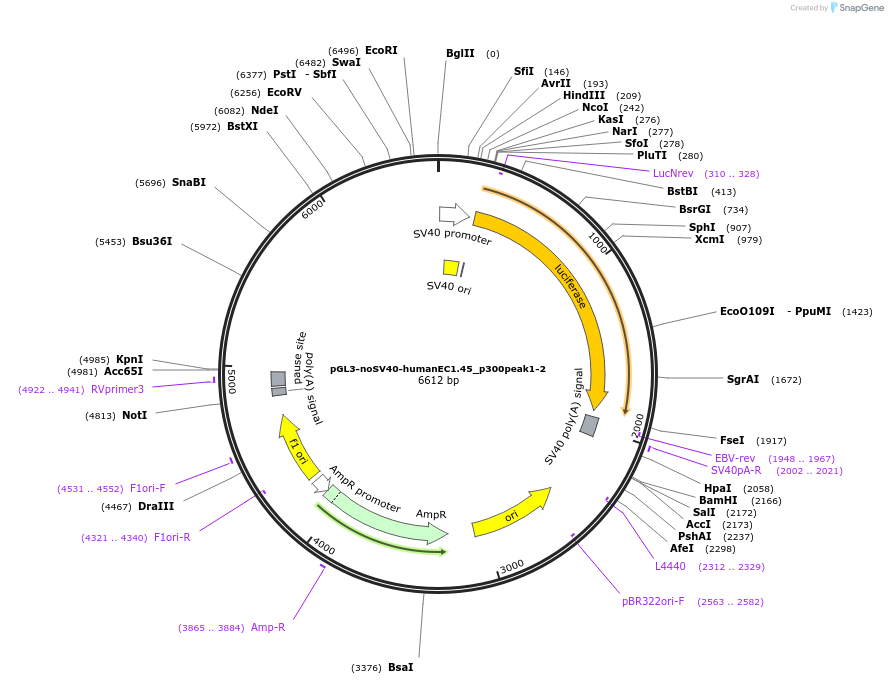
pGL3-noSV40-humanEC1.45_p300peak1-2
Plasmid#173949PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
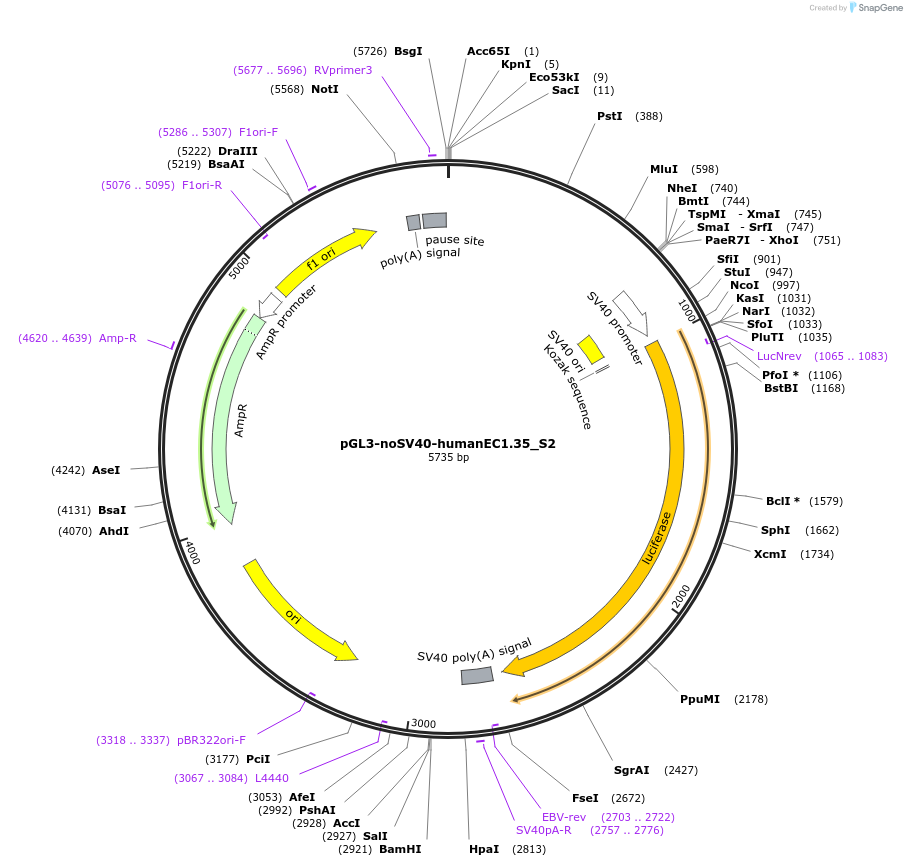
pGL3-noSV40-humanEC1.35_S2
Plasmid#173955PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertEnhancer region 2 (S2) from EC1.35 SOX9 enhancer cluster EC1.35
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
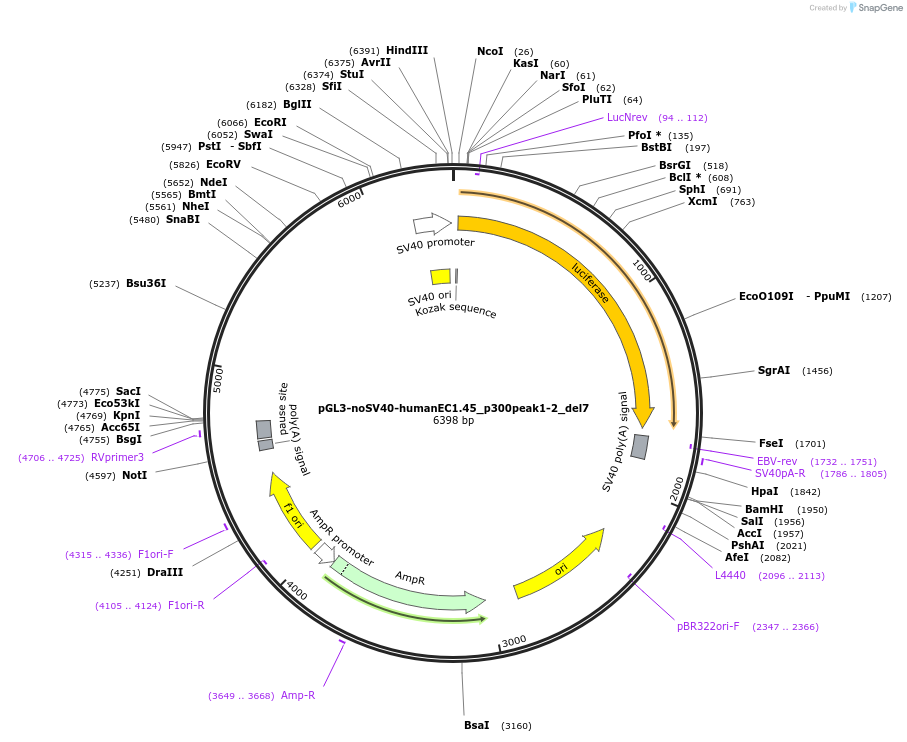
pGL3-noSV40-humanEC1.45_p300peak1-2_del7
Plasmid#173979PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #7
UseLuciferaseMutationTiled deletion #7PromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
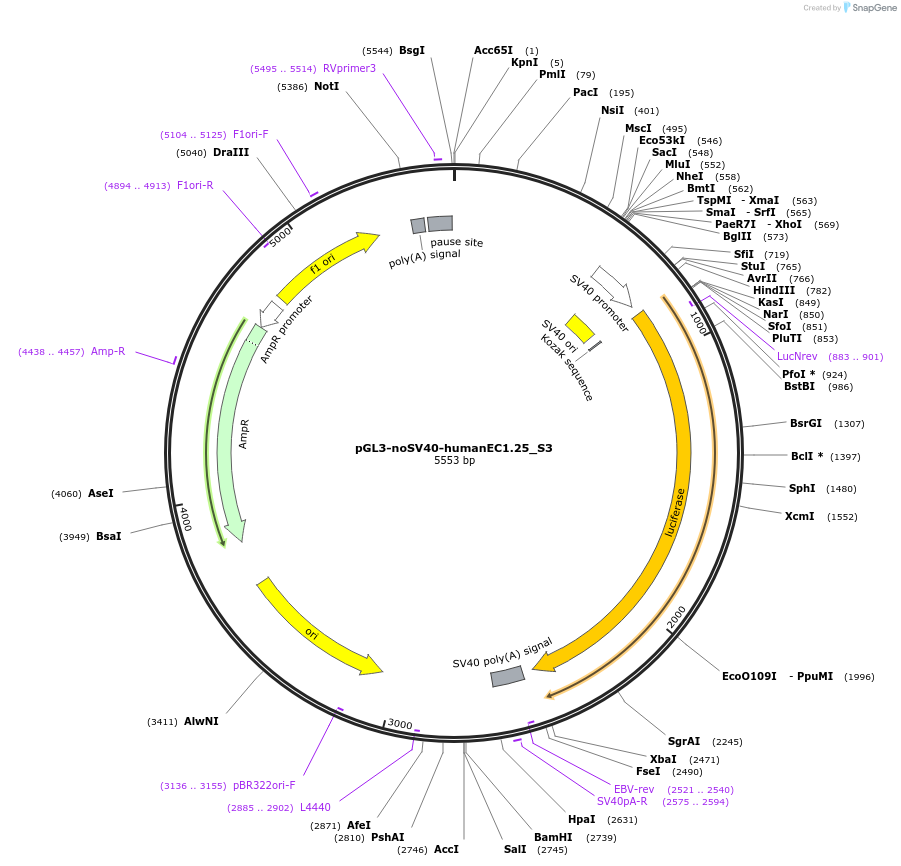
pGL3-noSV40-humanEC1.25_S3
Plasmid#173957PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertEnhancer region 3 (S3) from SOX9 enhancer cluster EC1.25
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
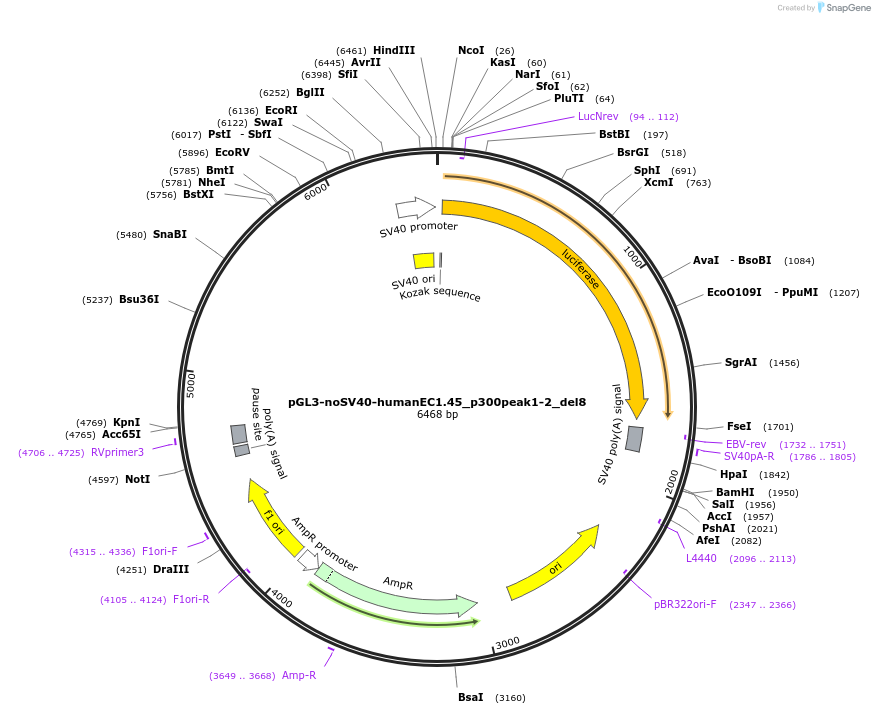
pGL3-noSV40-humanEC1.45_p300peak1-2_del8
Plasmid#173980PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #8
UseLuciferaseMutationTiled deletion #8PromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
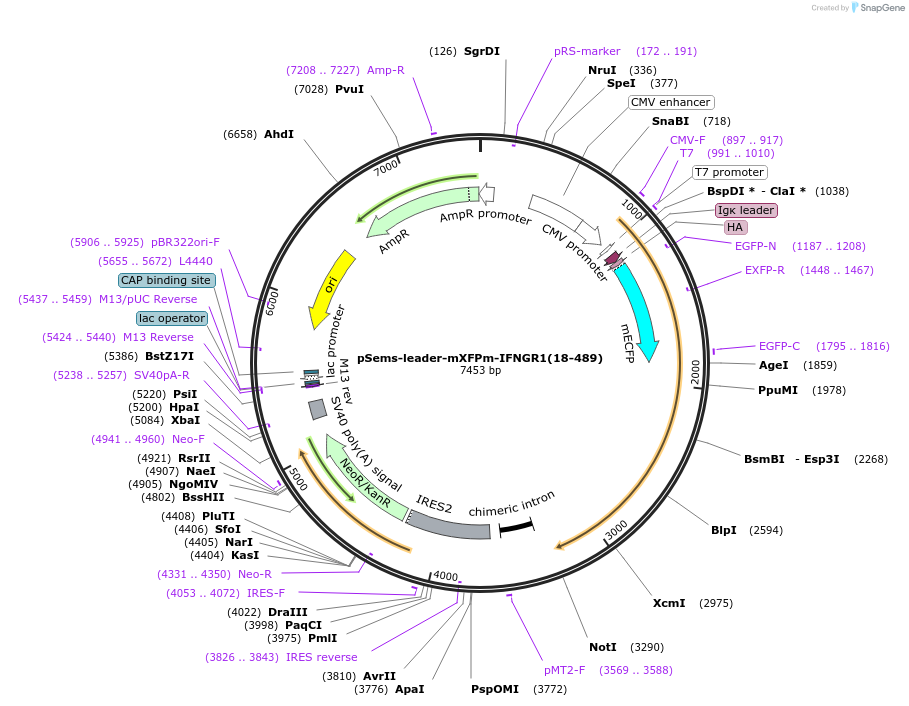
pSems-leader-mXFPm-IFNGR1(18-489)
Plasmid#192786PurposeExpression of mXFPm-tagged IFNGR1 at the plasma membrane, e.g. for single molecule fluorescence microscopyDepositorInsertIFNGR1 (IFNGR1 Human)
TagsIg k-chain leader sequence and mXFPmExpressionMammalianMutationmXFPm: tryptophan 66 to phenylalanine, gluatmic a…PromoterCMVAvailable SinceDec. 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
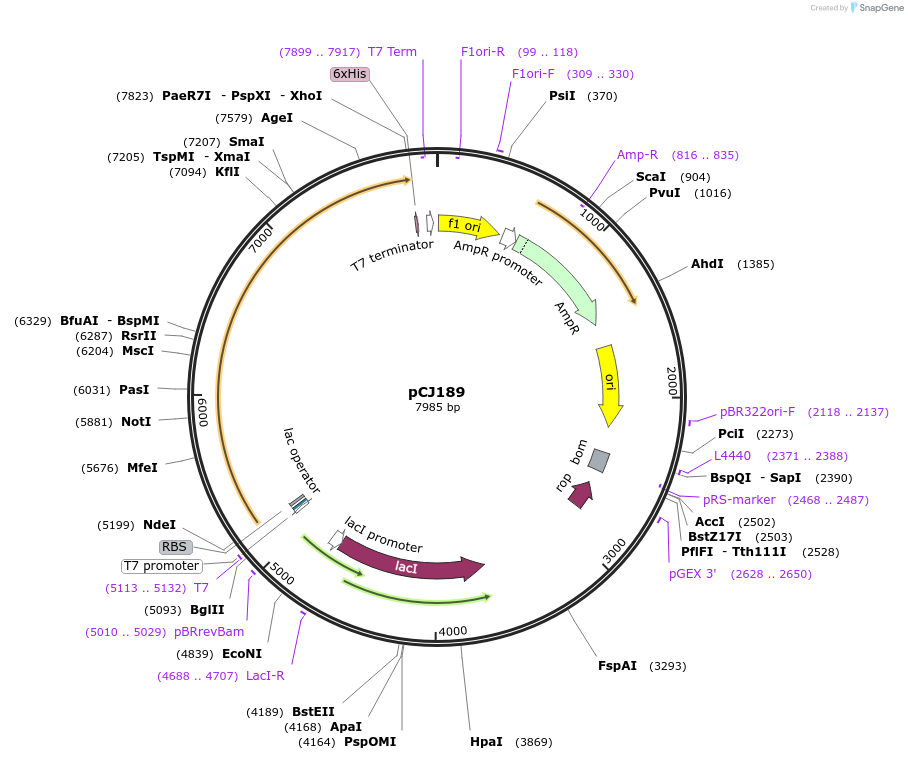
pCJ189
Plasmid#162666PurposepET-21b(+) based plasmid for expresion of MHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by an 8 residue Gly/Ser linker; codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertMHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by an 8 residue Gly/Ser linker
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceFeb. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
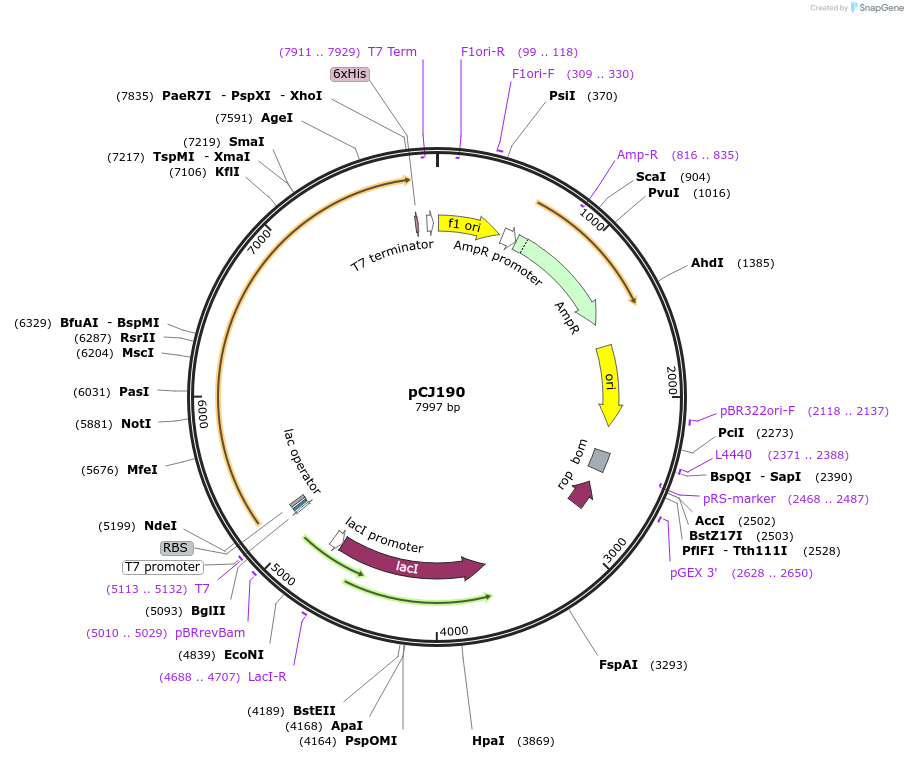
pCJ190
Plasmid#162667PurposepET-21b(+) based plasmid for expresion of MHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by a 12 residue Gly/Ser linker; codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertMHETase (Genbank GAP38911.1) linked to PETase (Genbank GAP38373.1) from Ideonella sakaiensis by a 12 residue Gly/Ser linker
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
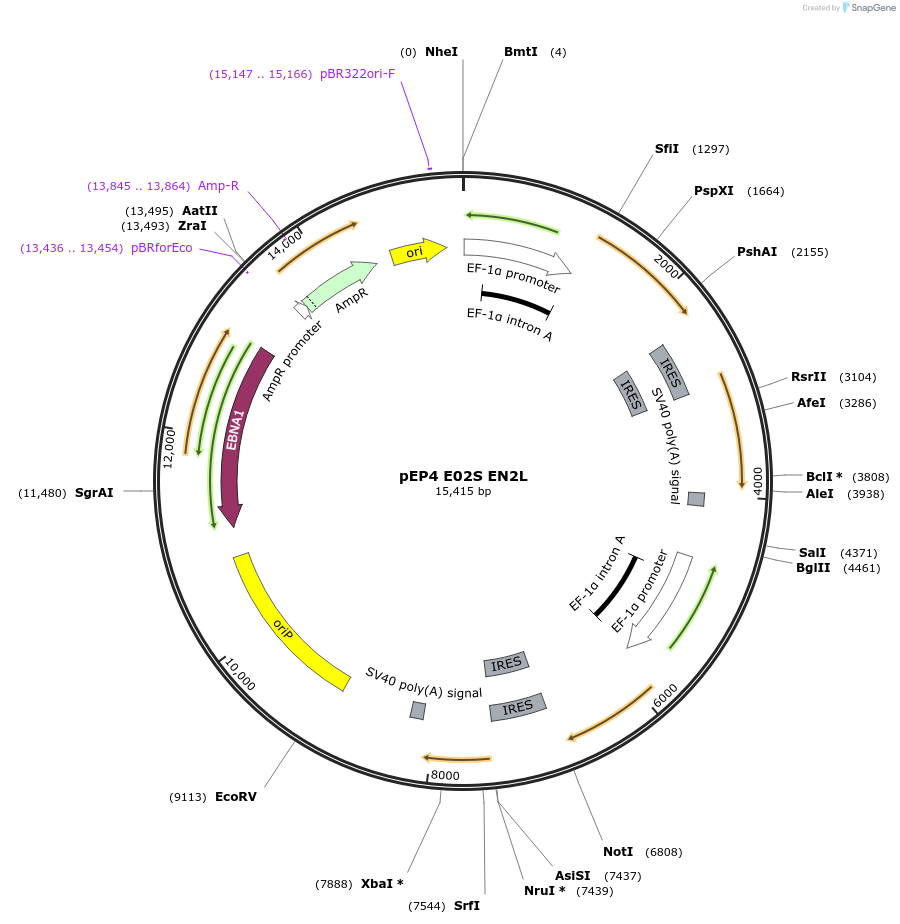
pEP4 E02S EN2L
Plasmid#20922Purposeused in the derivation of human iPS cells using non-integrating episomal vectors; expresses Oct4 and Sox2; Nanog and Lin28DepositorArticleAvailable SinceMay 20, 2009AvailabilityAcademic Institutions and Nonprofits only -
pSems-leader-mXFPe-IFNGR2(30-337)
Plasmid#192787PurposeExpression of mXFPe-tagged IFNGR2 at the plasma membrane, e.g. for single molecule fluorescence microscopyDepositorInsertIFNGR2 (IFNGR2 Human)
TagsIg k-chain leader sequence and mXFPeExpressionMammalianMutationmXFPe: tyrosine 66 to phenylalanine, asparagine 1…PromoterCMVAvailable SinceDec. 21, 2022AvailabilityAcademic Institutions and Nonprofits only