We narrowed to 10,182 results for: yeast
-
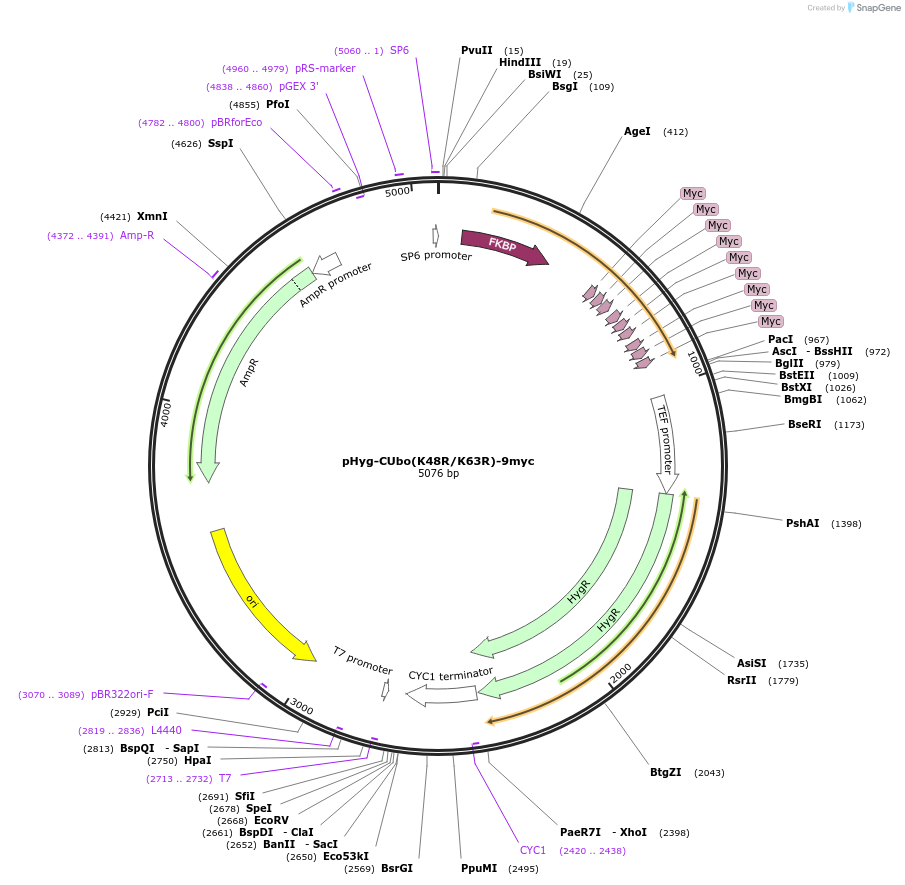
Plasmid#212789PurposeC-terminal PCR-based tagging with CUbo(K48R/K63R)-9myc in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 29, 2024AvailabilityAcademic Institutions and Nonprofits only
-
pHyg-CUbo-AID*-GFP
Plasmid#212770PurposeC-terminal PCR-based tagging with CUbo-AID*-yeGFP in budding yeast with HygR selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
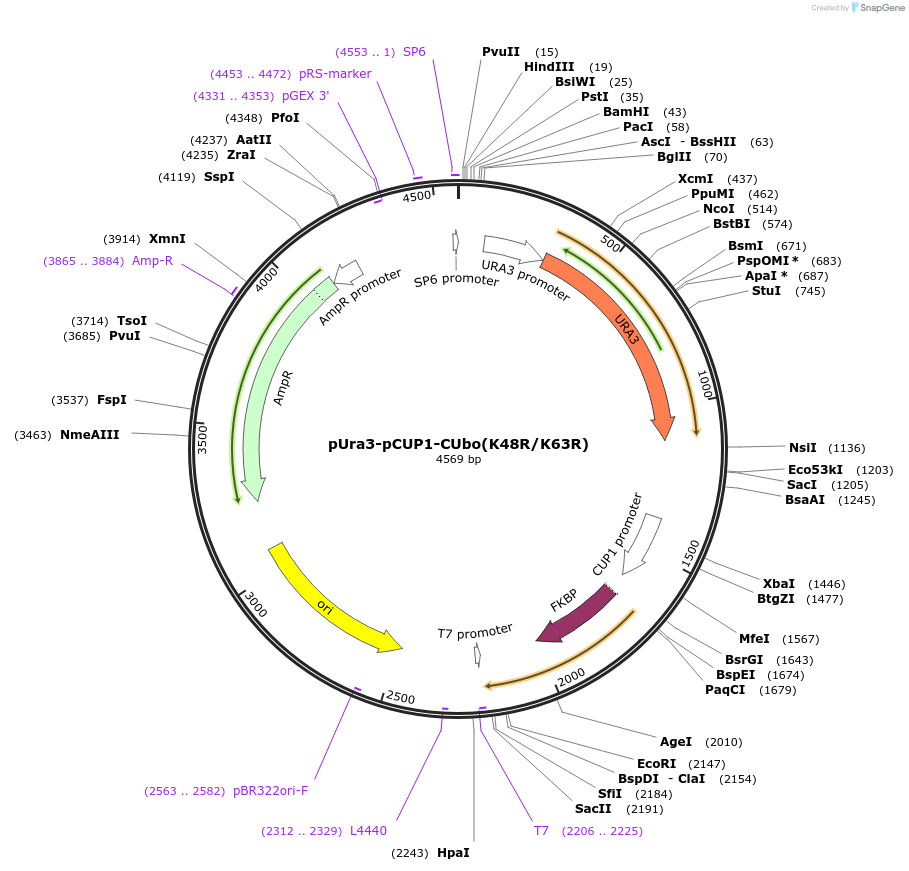
pUra3-pCUP1-CUbo(K48R/K63R)
Plasmid#212765PurposeN-terminal PCR-based tagging with pCUP1-CUbo(K48R/K63R) in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
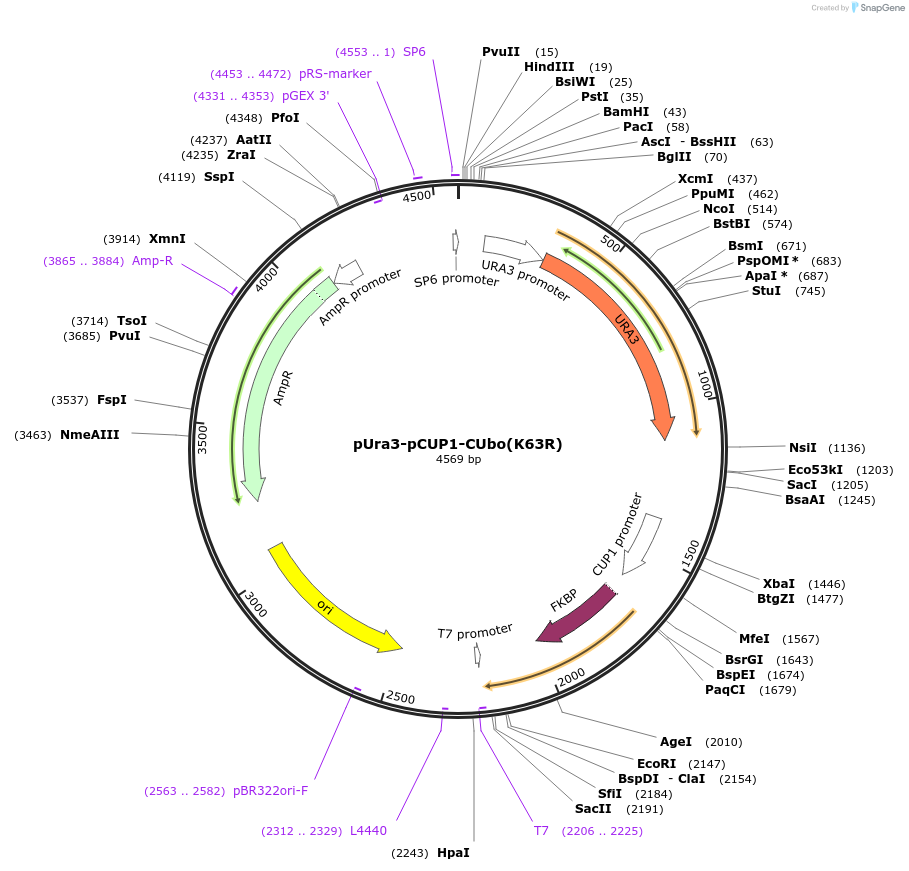
pUra3-pCUP1-CUbo(K63R)
Plasmid#212764PurposeN-terminal PCR-based tagging with pCUP1-CUbo(K63R) in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
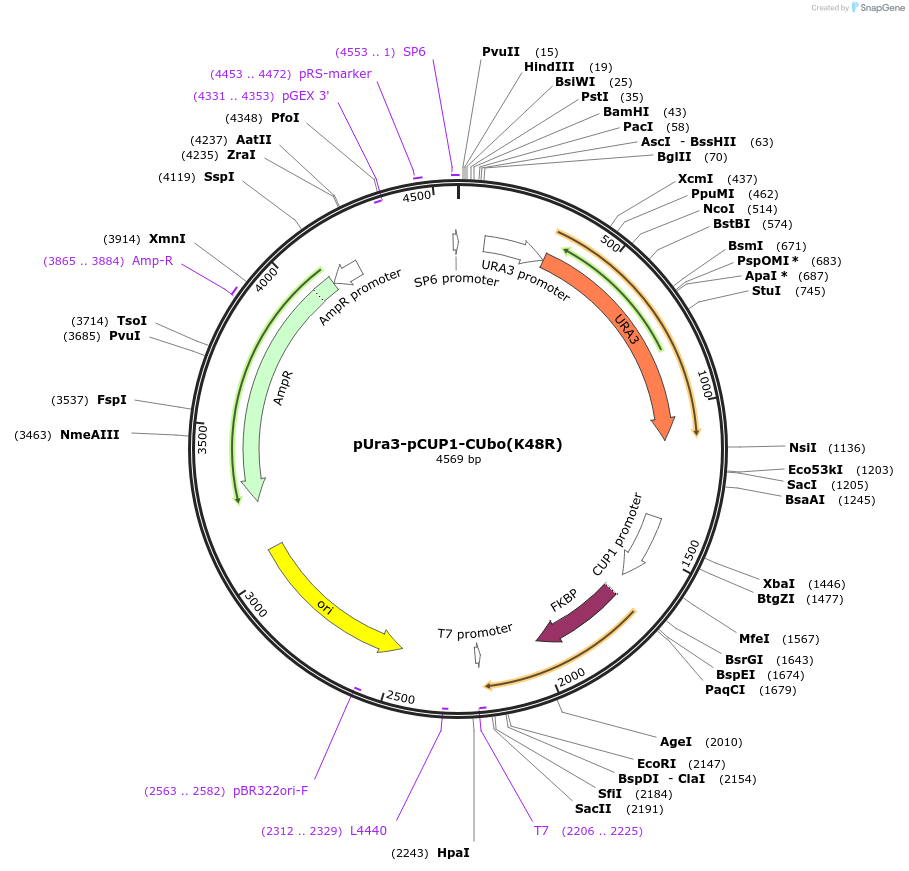
pUra3-pCUP1-CUbo(K48R)
Plasmid#212763PurposeN-terminal PCR-based tagging with pCUP1-CUbo(K48R) in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
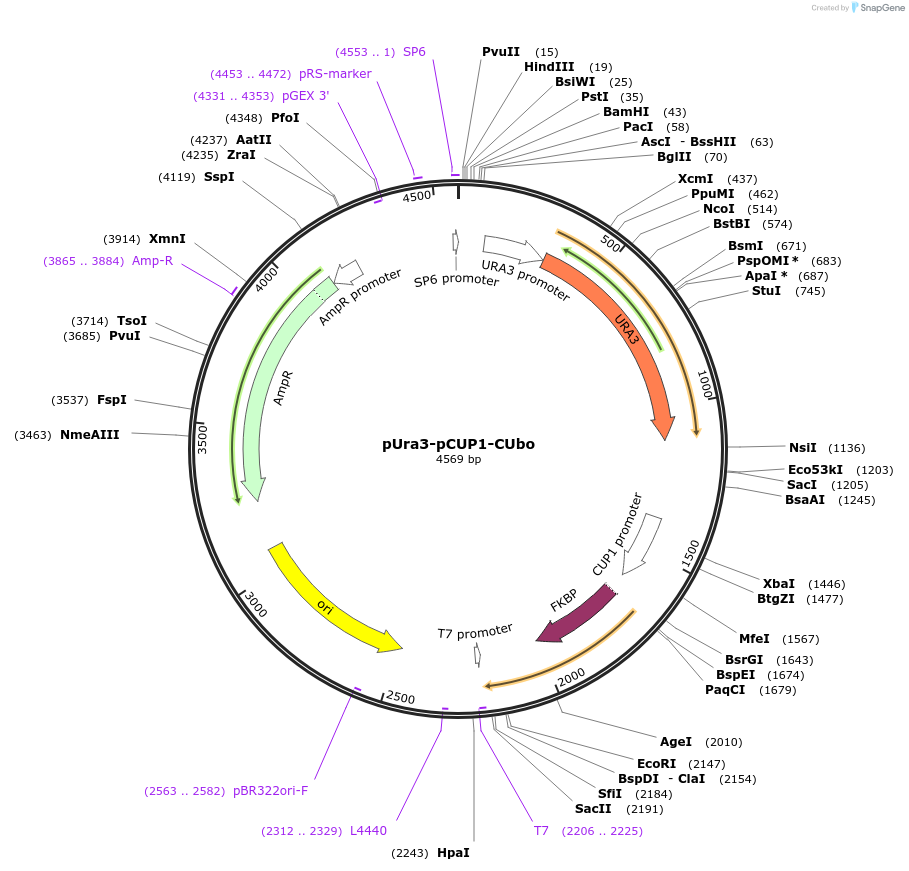
pUra3-pCUP1-CUbo
Plasmid#212762PurposeN-terminal PCR-based tagging with pCUP1-CUbo in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
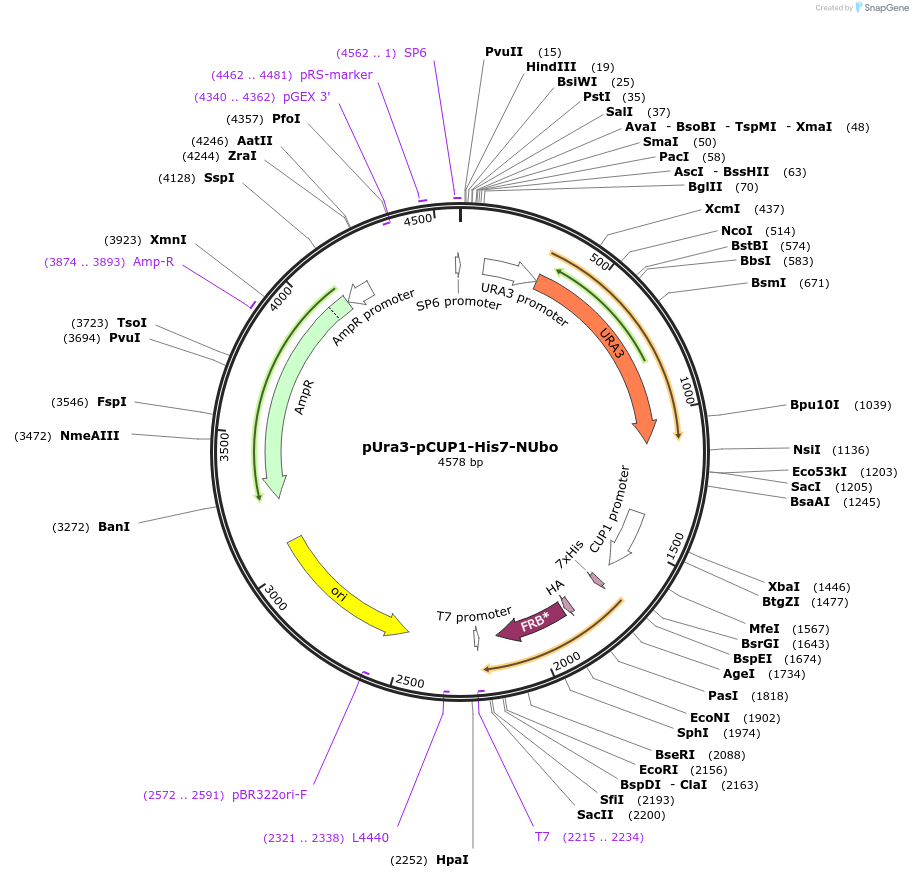
pUra3-pCUP1-His7-NUbo
Plasmid#212761PurposeN-terminal PCR-based tagging with pCUP1-His7-NUbo in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
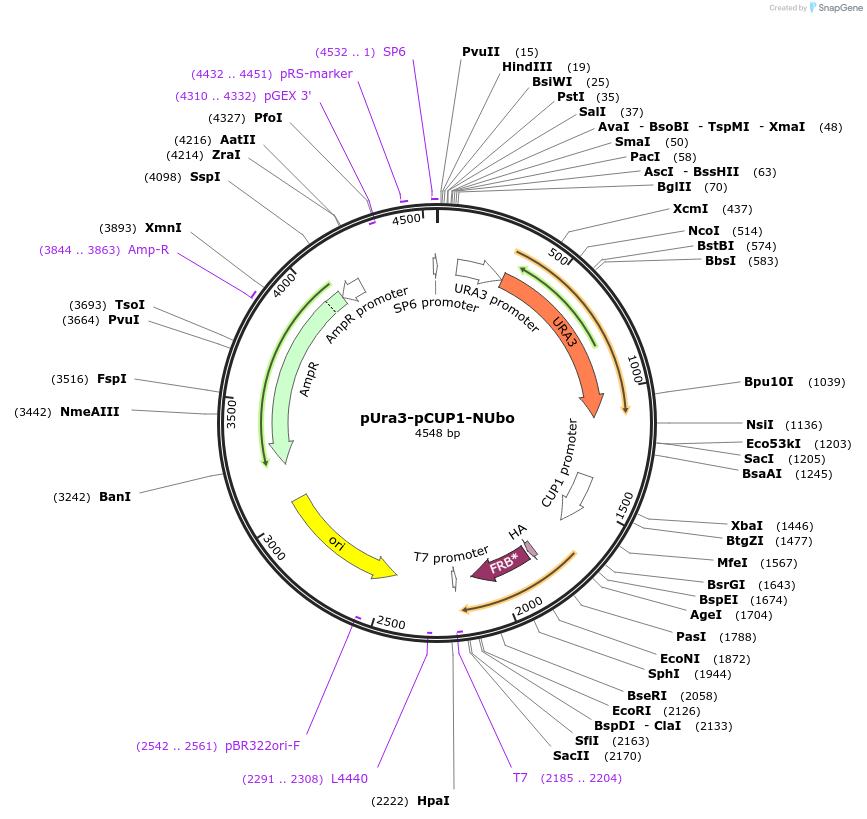
pUra3-pCUP1-NUbo
Plasmid#212760PurposeN-terminal PCR-based tagging with pCUP1-NUbo in budding yeast with URA3 selectionDepositorTypeEmpty backboneUsePcr-based taggingAvailable SinceJan. 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
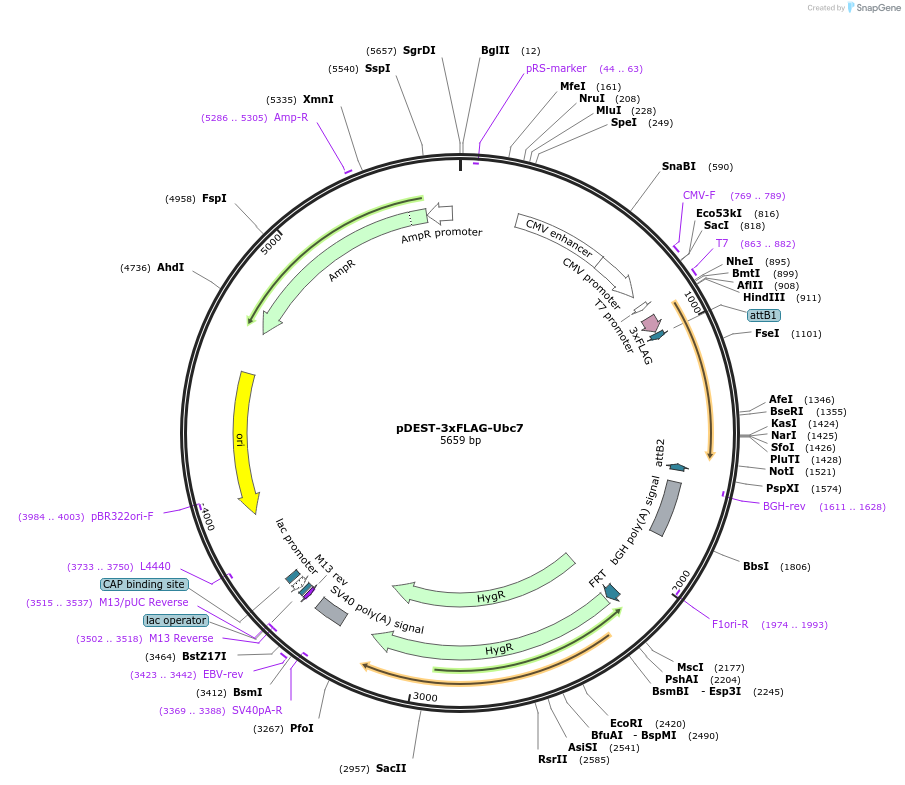
pDEST-3xFLAG-Ubc7
Plasmid#212801PurposeConstitutive expression of 3xFLAG-tagged budding yeast Ubc7 in mammalian cellsDepositorInsertUbc7
Tags3xFLAGExpressionMammalianAvailable SinceJan. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
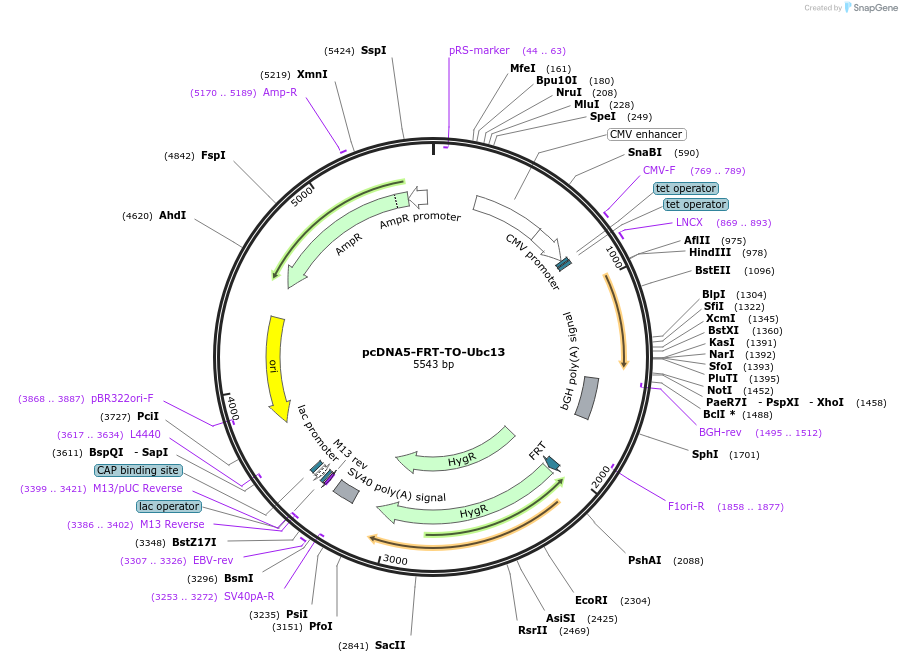
pcDNA5-FRT-TO-Ubc13
Plasmid#212806PurposeConstitutive or doxycycline-inducible expression of budding yeast Ubc13 in mammalian cellsDepositorInsertUbc13
ExpressionMammalianAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
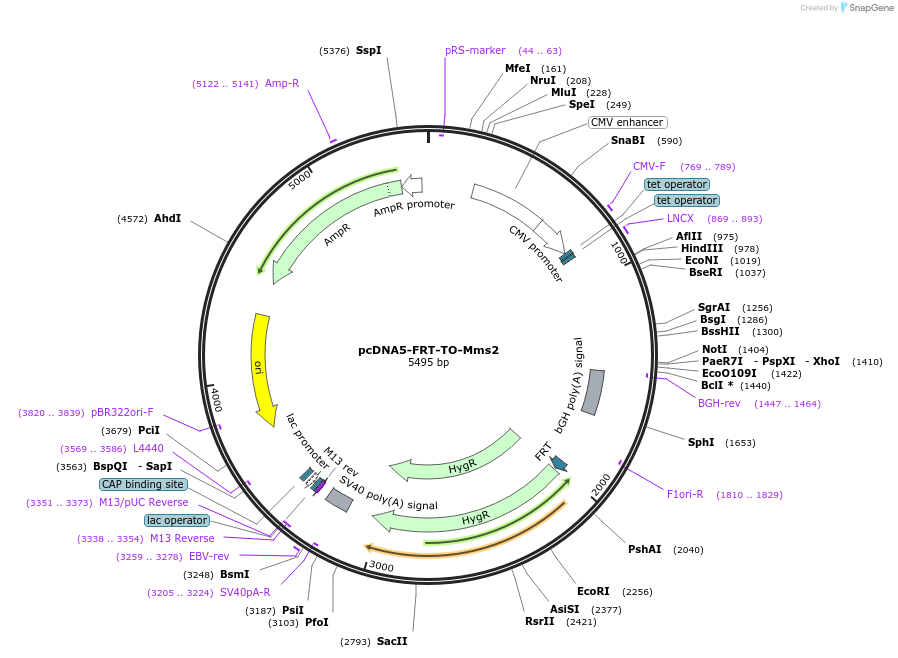
pcDNA5-FRT-TO-Mms2
Plasmid#212807PurposeConstitutive or doxycycline-inducible expression of budding yeast Mms2 in mammalian cellsDepositorInsertMms2
ExpressionMammalianAvailable SinceJan. 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
pDN0617 (pDEST-mEGFP C-term, NatR)
Plasmid#210484PurposepDEST vector to tag gene of interest with mEGFP on the C-terminus to perform localization assays in S. cerevisiae.DepositorTypeEmpty backboneUseSynthetic Biology; Destination vector for gateway…TagsmEGFPExpressionYeastPromoterADH1Available SinceDec. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
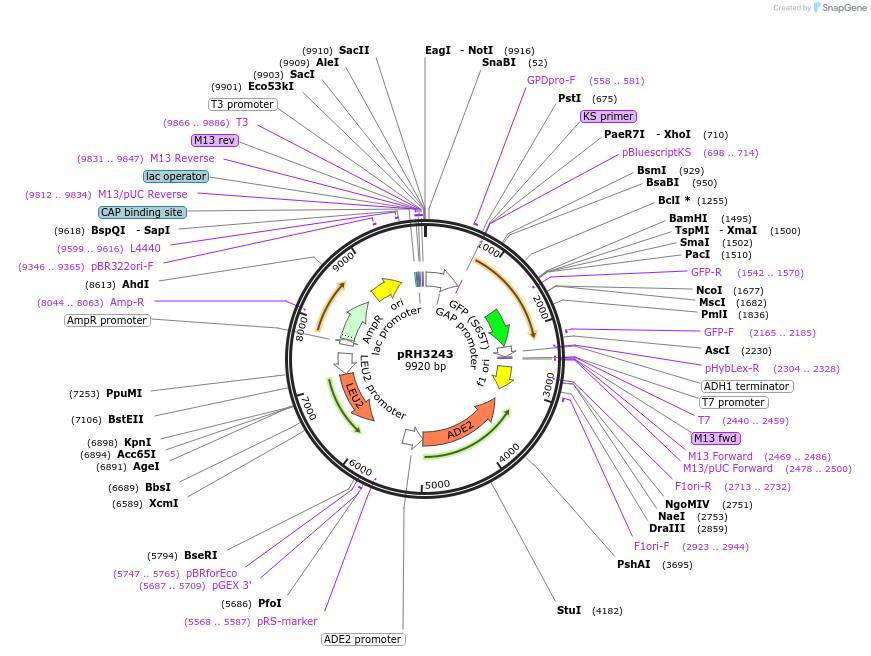
pRH3243
Plasmid#210187PurposeProtein quality control substrate and associated control YIp ADE2-LEU2 pTDH3::aro7-R33P-GFP::tADH1DepositorInsertYIp ADE2-LEU2 pTDH3::aro7-R33P-GFP::tADH1 (ARO7 Budding Yeast)
ExpressionYeastMutationR33PAvailable SinceNov. 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
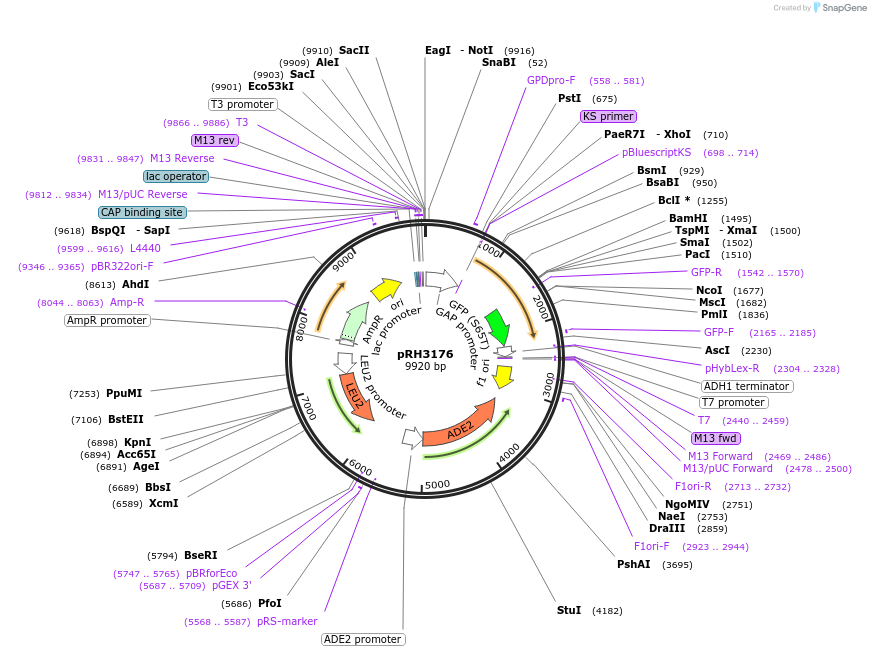
pRH3176
Plasmid#210178PurposeProtein quality control substrate and associated control YIp ADE2-LEU2 pTDH3::aro7-G141S-GFP::tADH1DepositorInsertYIp ADE2-LEU2 pTDH3::aro7-G141S-GFP::tADH1 (ARO7 Budding Yeast)
ExpressionYeastMutationG141SAvailable SinceNov. 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
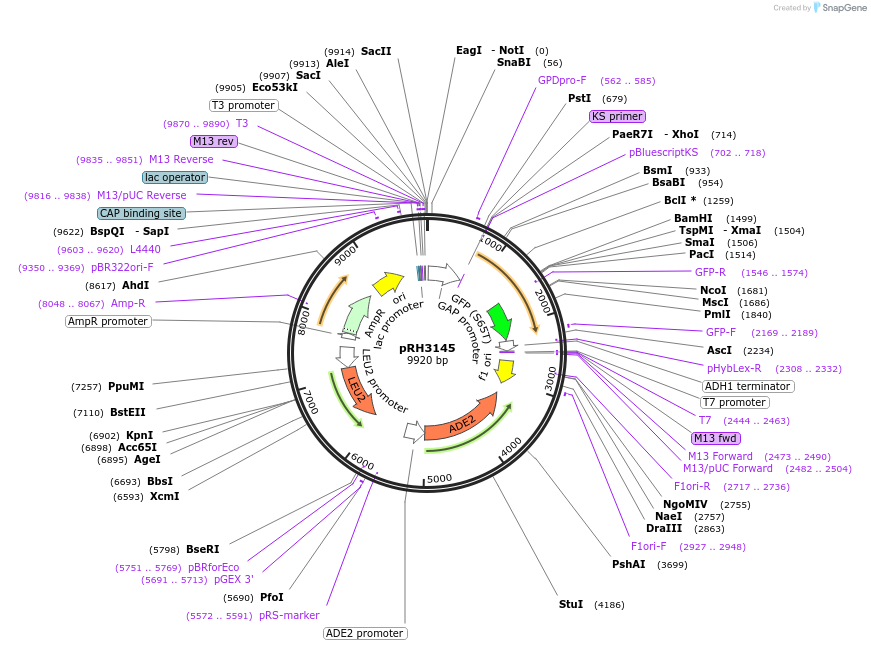
pRH3145
Plasmid#210169PurposeProtein quality control substrate and associated control YIp ADE2-LEU2 pTDH3::aro7-D147V-GFP::tADH1DepositorInsertYIp ADE2-LEU2 pTDH3::aro7-D147V-GFP::tADH1 (ARO7 Budding Yeast)
ExpressionYeastMutationD147VAvailable SinceNov. 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
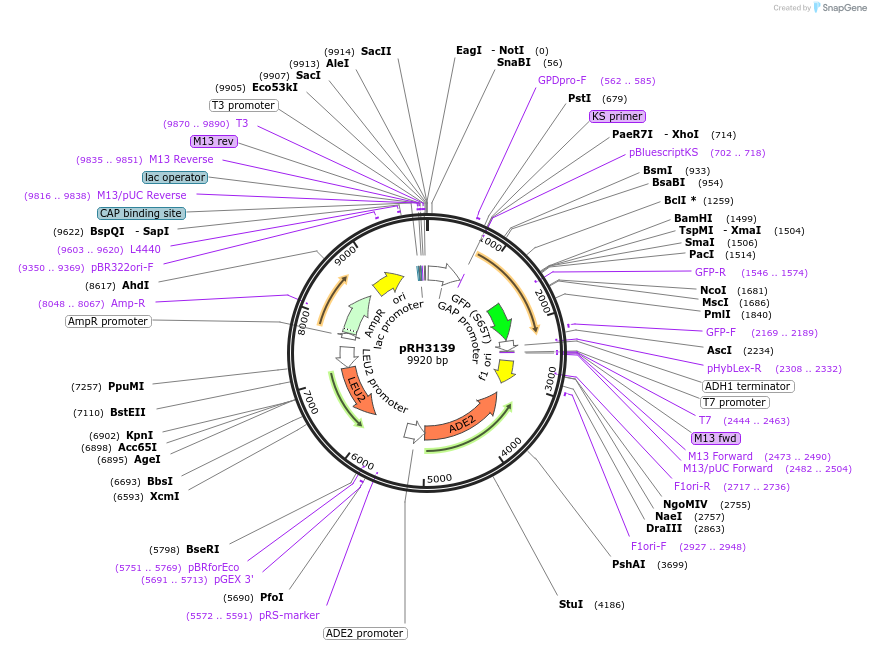
pRH3139
Plasmid#210167PurposeProtein quality control substrate and associated control YIp ADE2-LEU2 pTDH3::aro7-R33G-GFP::tADH1DepositorInsertYIp ADE2-LEU2 pTDH3::aro7-R33G-GFP::tADH1 (ARO7 Budding Yeast)
ExpressionYeastMutationR33GAvailable SinceNov. 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
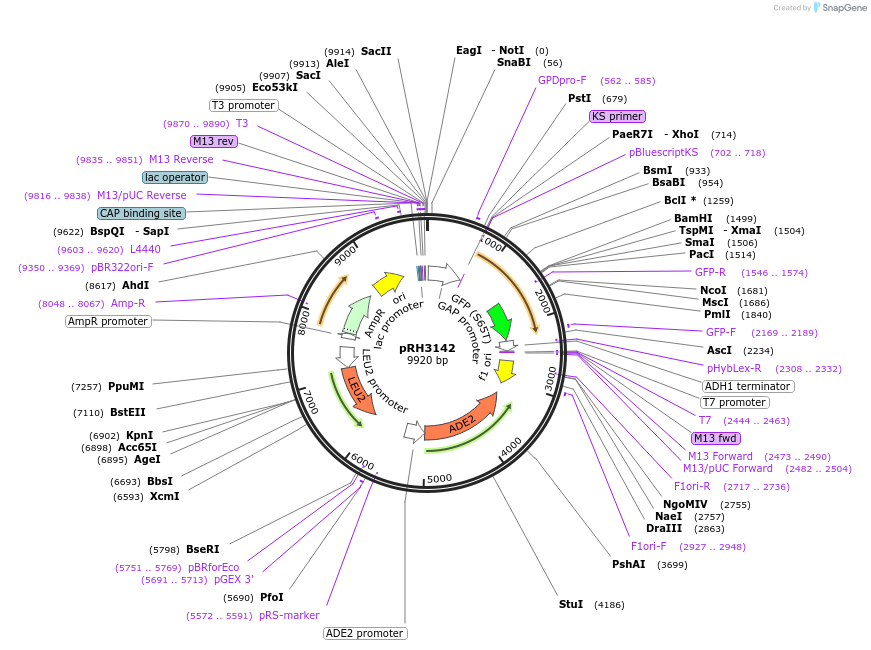
pRH3142
Plasmid#210168PurposeProtein quality control substrate and associated control YIp ADE2-LEU2 pTDH3::aro7-D147G-GFP::tADH1DepositorInsertYIp ADE2-LEU2 pTDH3::aro7-D147G-GFP::tADH1 (ARO7 Budding Yeast)
ExpressionYeastMutationD147GAvailable SinceNov. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
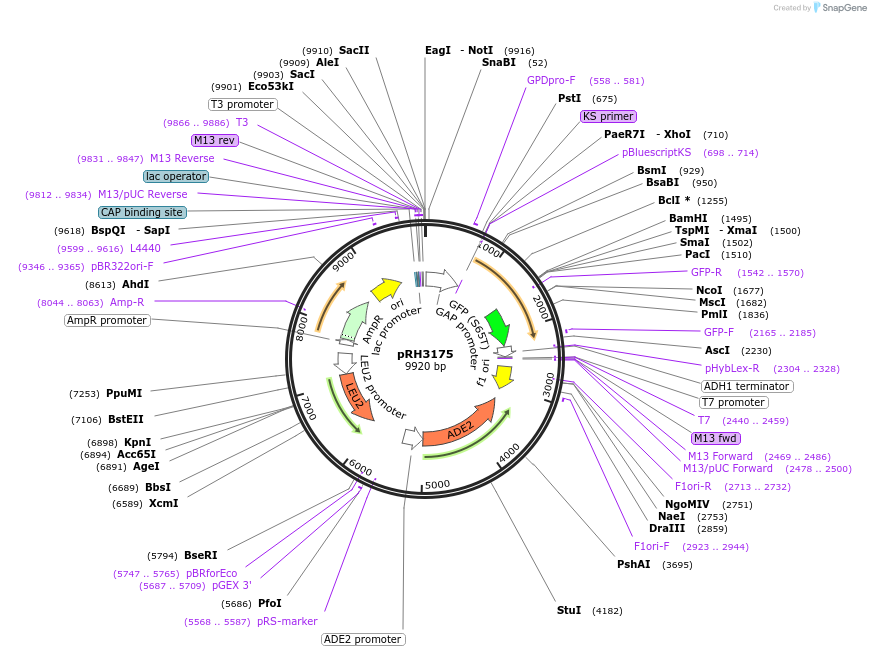
pRH3175
Plasmid#210177PurposeProtein quality control substrate and associated control YIp ADE2-LEU2 pTDH3::aro7-R33G,G141S-GFP::tADH1DepositorInsertYIp ADE2-LEU2 pTDH3::aro7-R33G,G141S-GFP::tADH1 (ARO7 Budding Yeast)
ExpressionYeastMutationR33G,G141SAvailable SinceNov. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
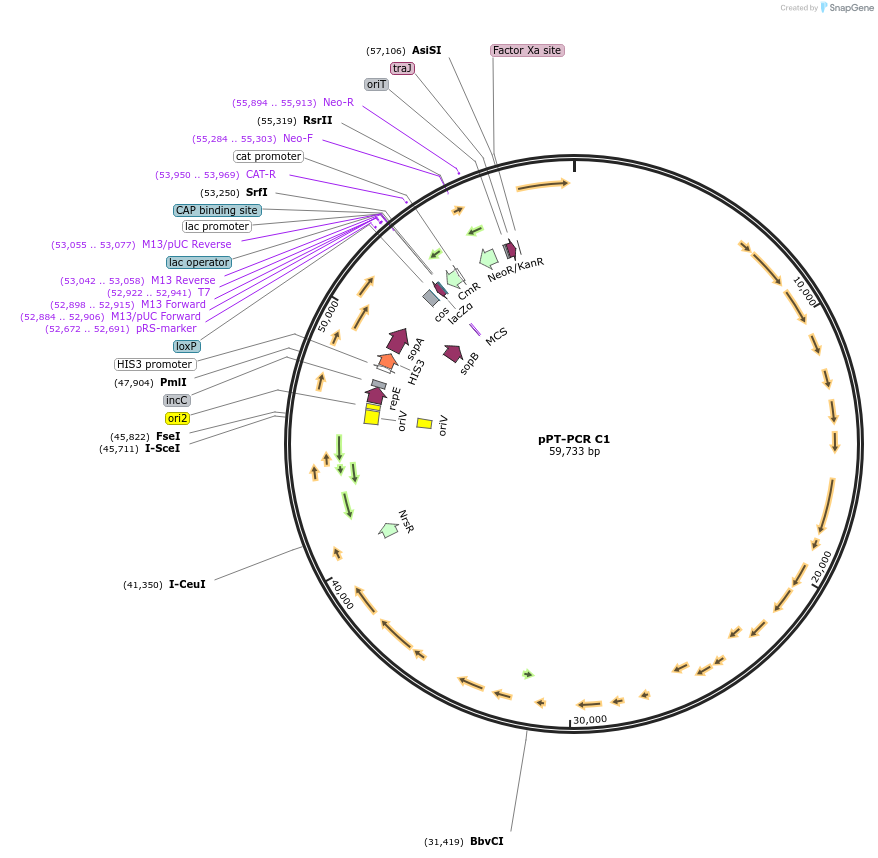
pPT-PCR C1
Plasmid#141248PurposePCR cloned Phaeodactylum tricornutum Mitochondrial Genome - Clone 1DepositorInsertReduced Mitochondrial Genome of Phaeodactylum tricornutum
UseSynthetic Biology; Diatom expressionExpressionBacterial and YeastMutationRepeat region removed, additional minor mutations…Available SinceMay 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
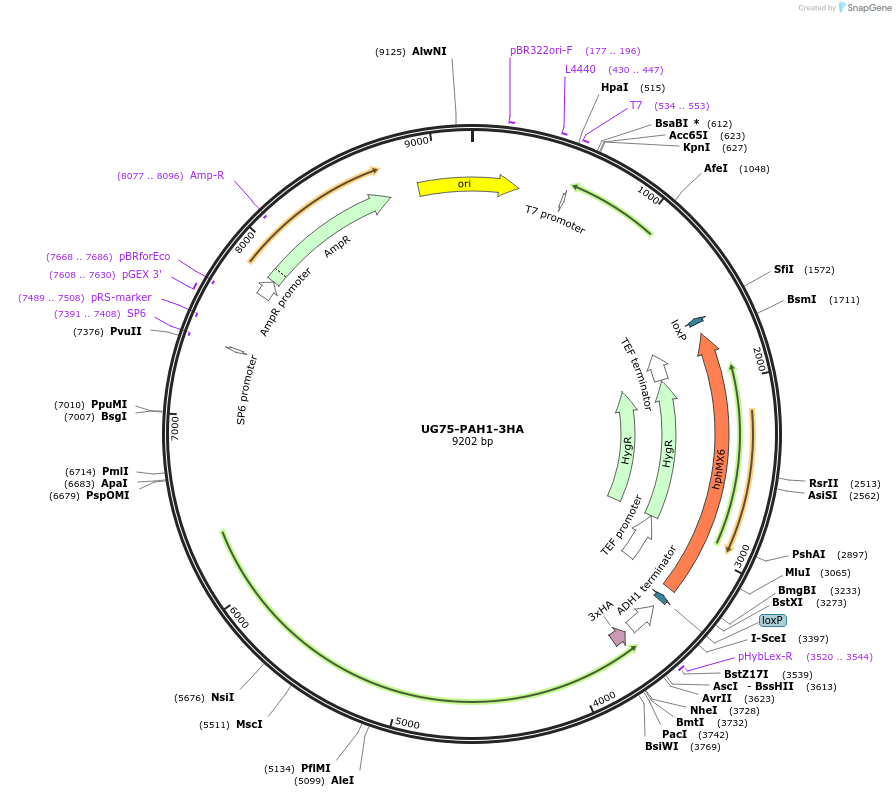
UG75-PAH1-3HA
Plasmid#184761PurposePAH1 overexpression. Promotes the formation of diacylglycerol from phosphatidate. Uses antibiotic resistance marker hphMX6.DepositorAvailable SinceFeb. 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
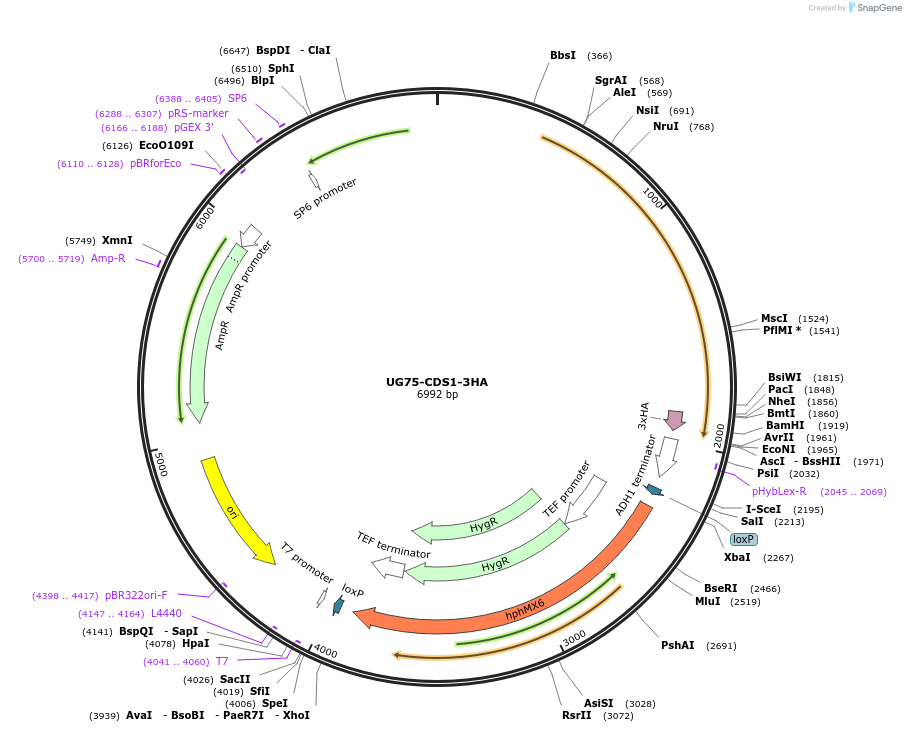
UG75-CDS1-3HA
Plasmid#184760PurposeCDS1 overexpression. Diverts glycerolipid synthesis pathway towards phospholipids. Uses antibiotic resistance marker hphMX6.DepositorAvailable SinceFeb. 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
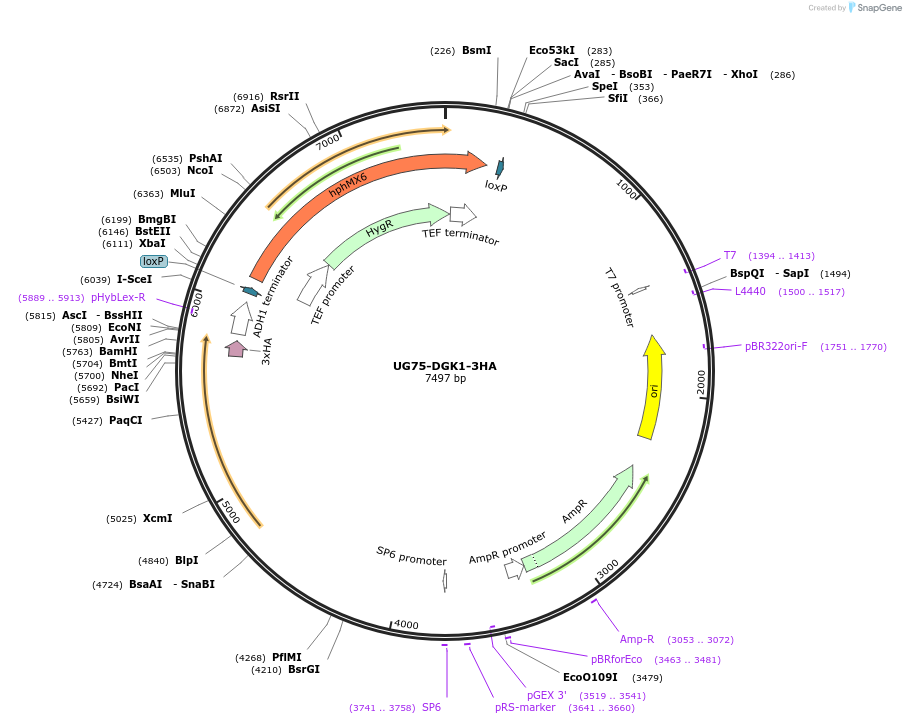
UG75-DGK1-3HA
Plasmid#184759PurposeDGK1 overexpression. Promotes the formation of phosphatidate from diacylglycerol. Uses antibiotic resistance marker hphMX6.DepositorAvailable SinceFeb. 17, 2023AvailabilityAcademic Institutions and Nonprofits only -
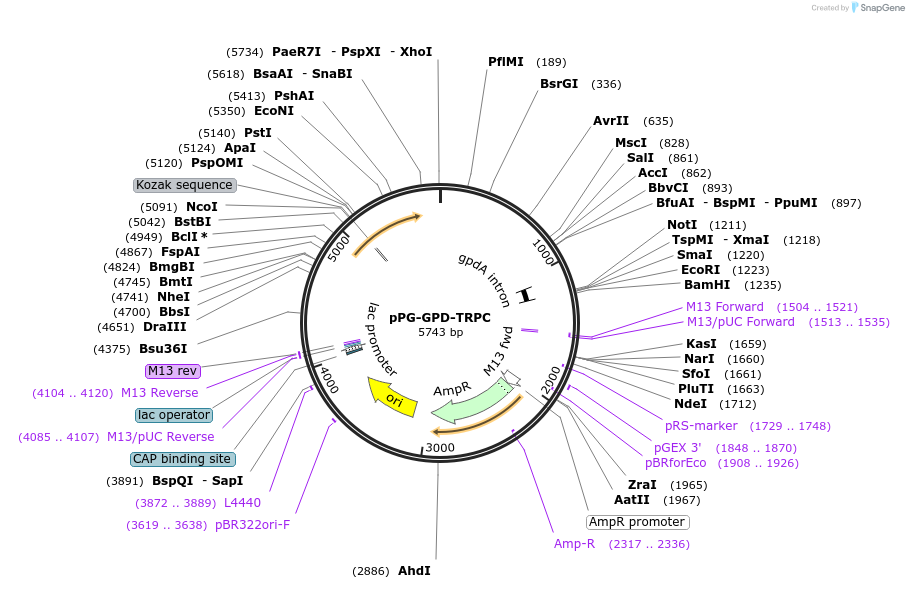
pPG-GPD-TRPC
Plasmid#190113PurposeExpression vector for fungal (Aspergillus flavus) genes based on Aspergillus parasiticus pyrG gene selectionDepositorTypeEmpty backboneExpressionYeastAvailable SinceJan. 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
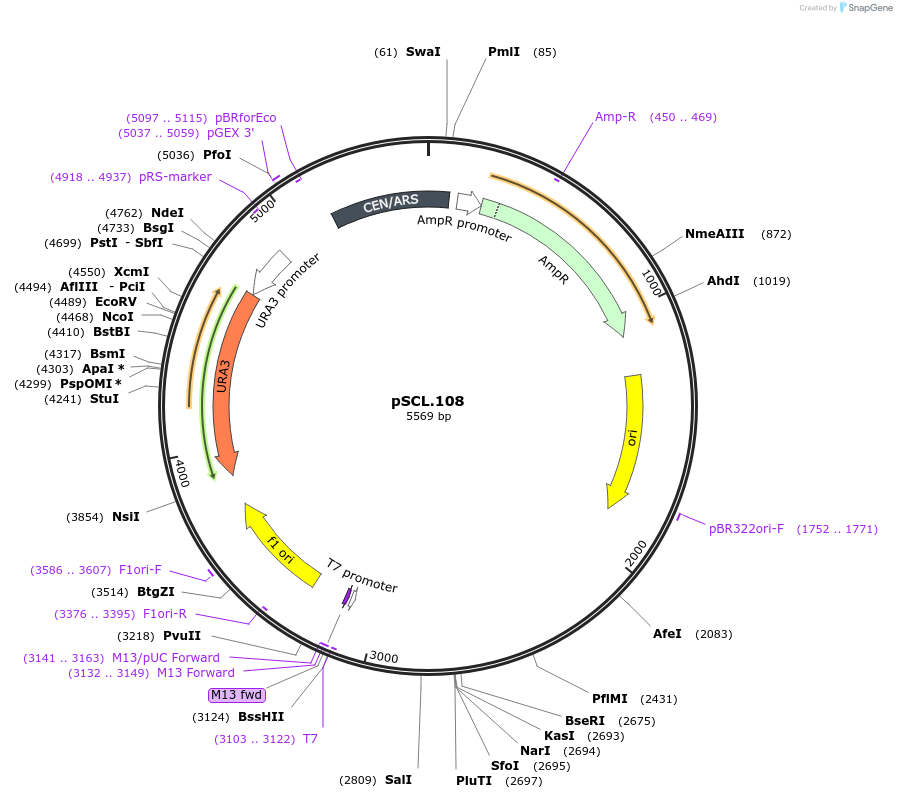
pSCL.108
Plasmid#184977PurposeExpress -Eco1 LYP1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 12
ExpressionYeastMutationLYP1 donor E27stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
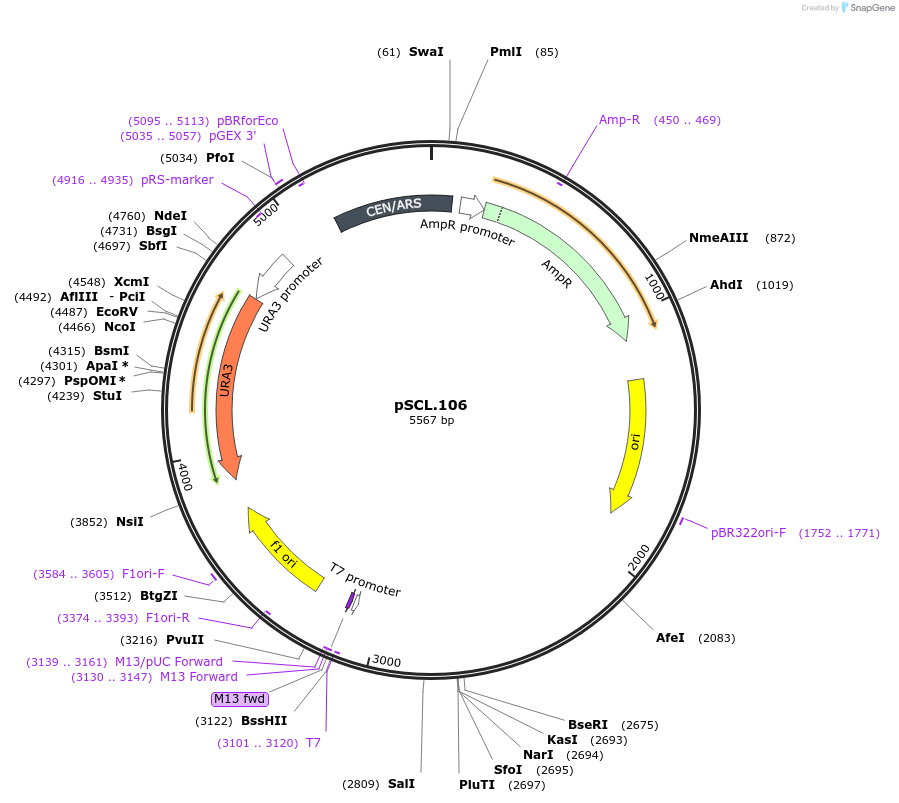
pSCL.106
Plasmid#184975PurposeExpress -Eco1 CAN1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, CAN1 G444X, a1/a2 length: 12
ExpressionYeastMutationCAN1 donor G444stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
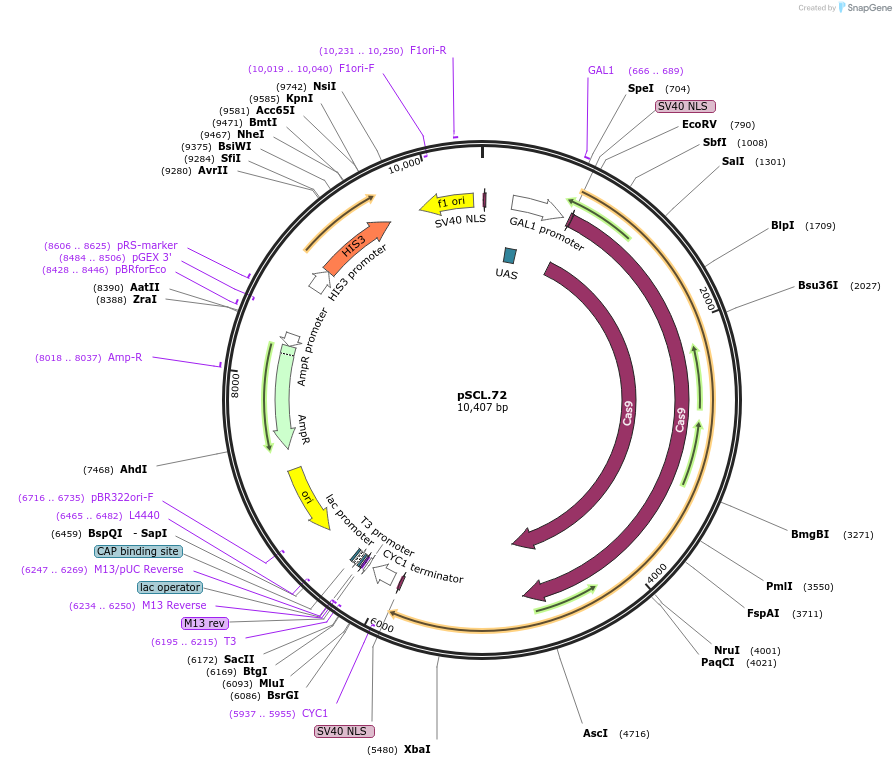
pSCL.72
Plasmid#184984PurposeExpress Cas9/-Eco1 RT fusion, using linker1DepositorInsertCas9-fusion_linker1-Eco1RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
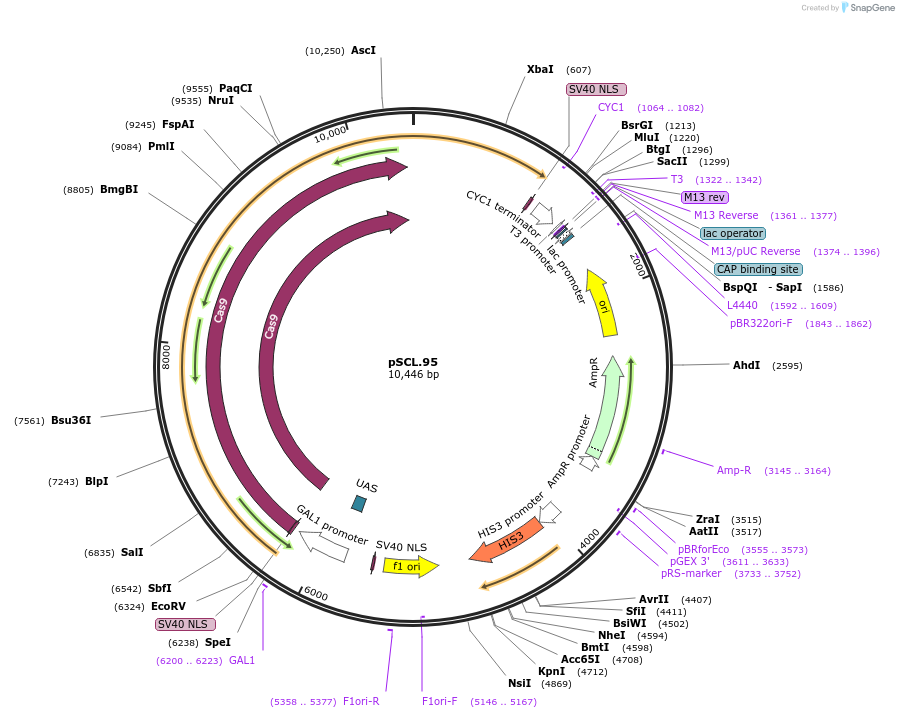
pSCL.95
Plasmid#184986PurposeExpress Cas9/-Eco1 RT fusion, using linker2DepositorInsertCas9-fusion_linker2-Eco1RT
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
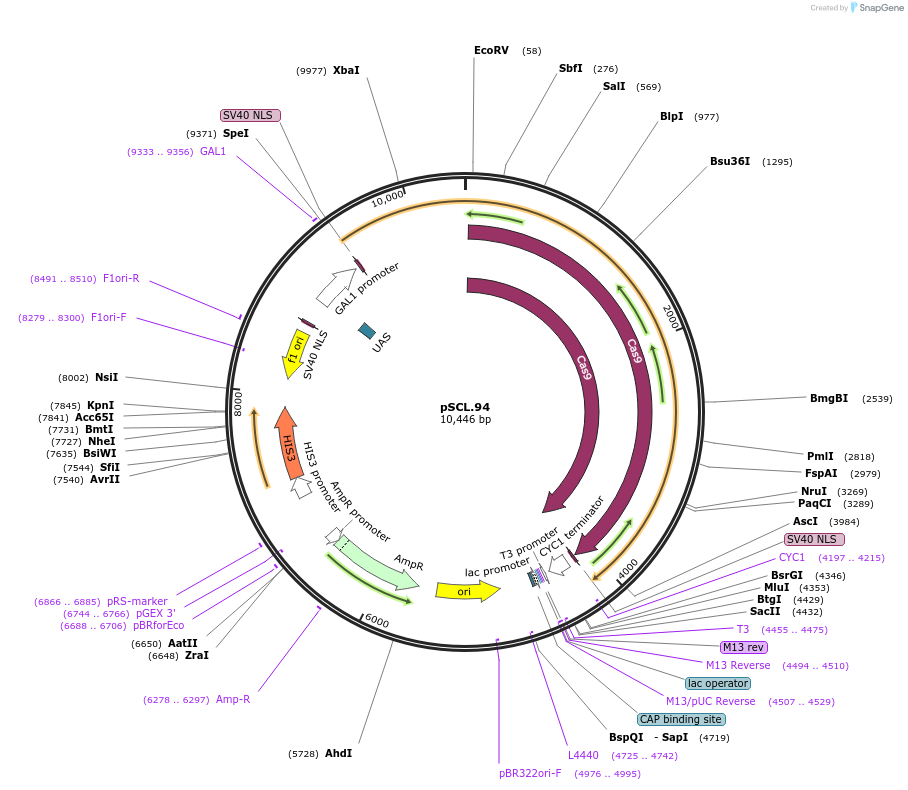
pSCL.94
Plasmid#184985PurposeExpress -Eco1 RT/Cas9 fusion, using linker2DepositorInsertEco1RT-fusion_linker2-Cas9
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
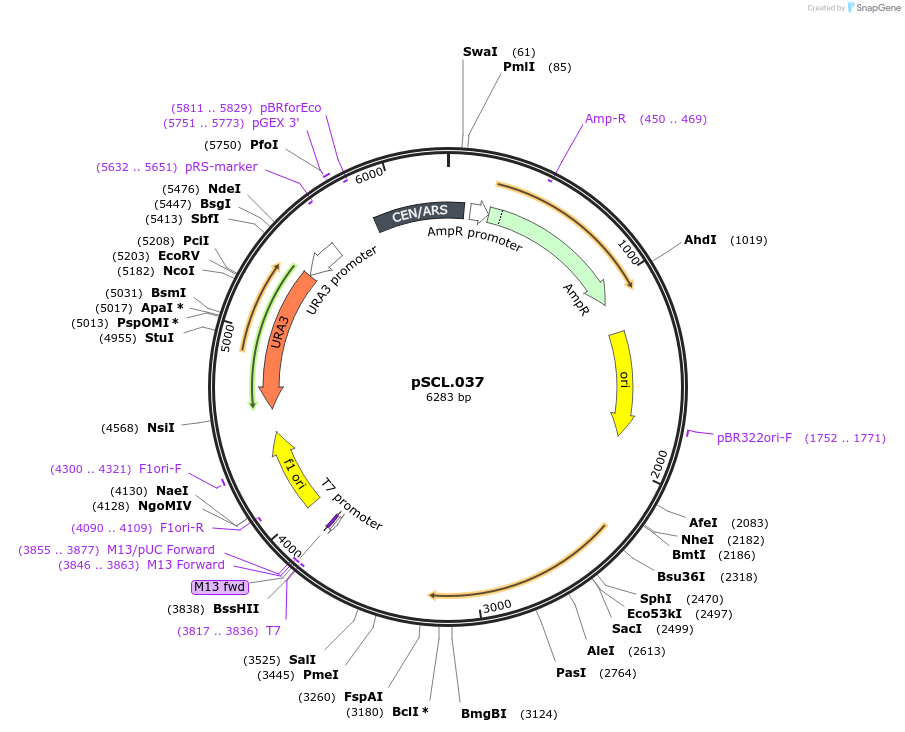
pSCL.037
Plasmid#184966PurposeExpress -Eco1 dead RT and ncRNA in yeasstDepositorInsertEco1: RT and ncRNA(wt), a1/a2 length: 12, dead RT
ExpressionYeastMutationHuman codon optimized RT, YXDD catalytic region m…PromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
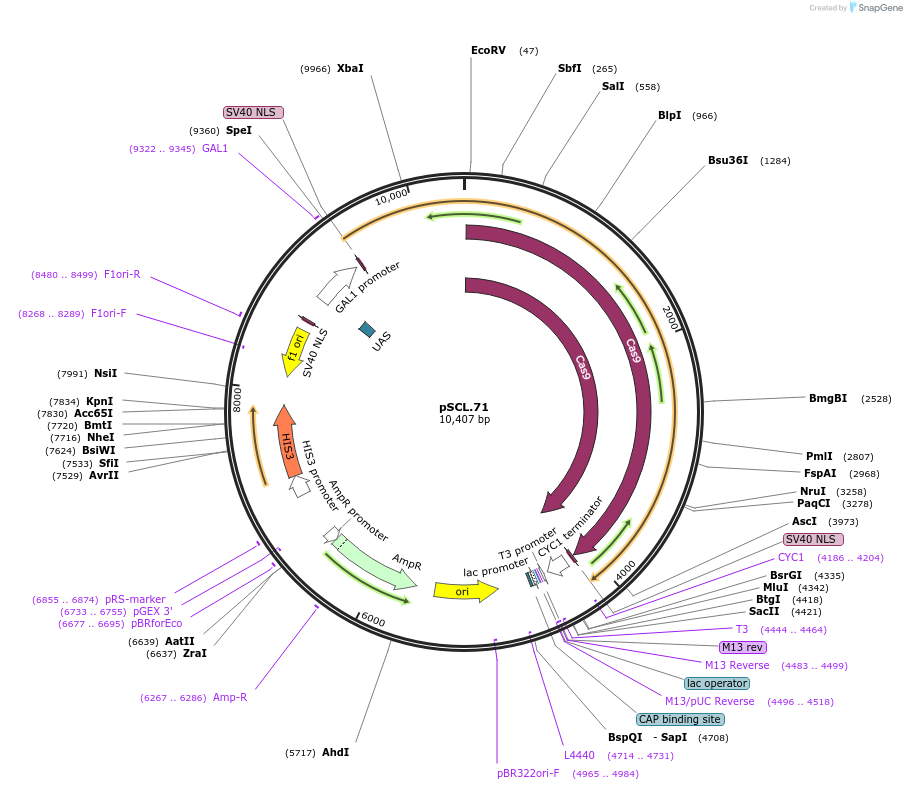
pSCL.71
Plasmid#184983PurposeExpress -Eco1 RT/Cas9 fusion, using linker1DepositorInsertEco1RT-fusion_linker1-Cas9
TagsSV40NLSExpressionYeastMutationhuman codon optimized RTPromoterGal1-10Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
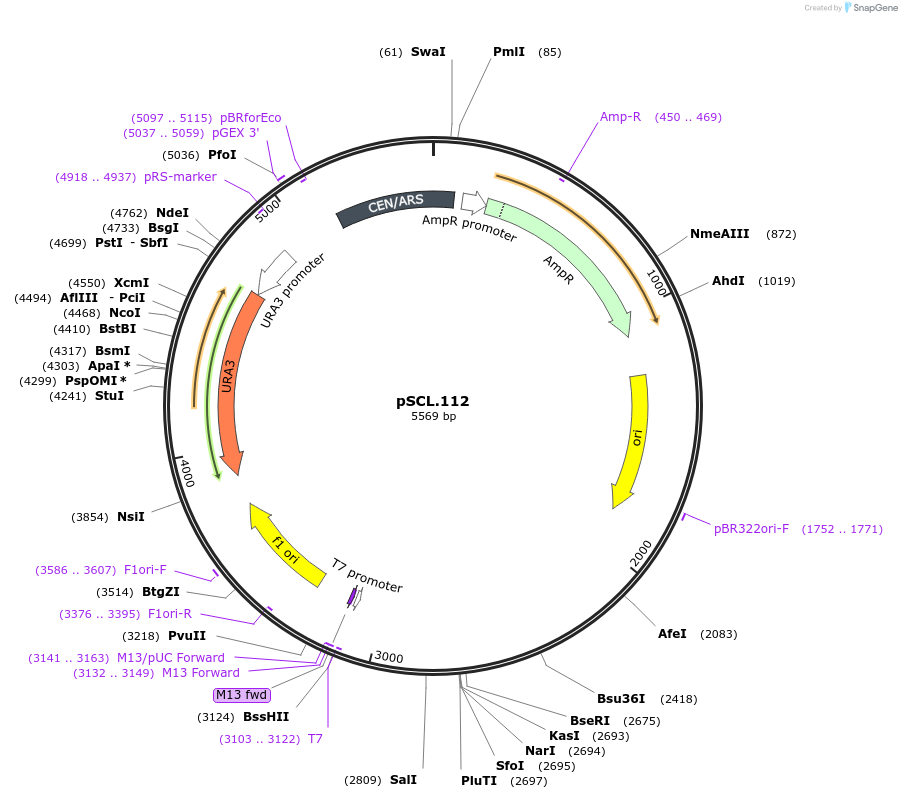
pSCL.112
Plasmid#184981PurposeExpress -Eco1 FAA1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, FAA1 P233X, a1/a2 length: 12
ExpressionYeastMutationFAA1 donor P233stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
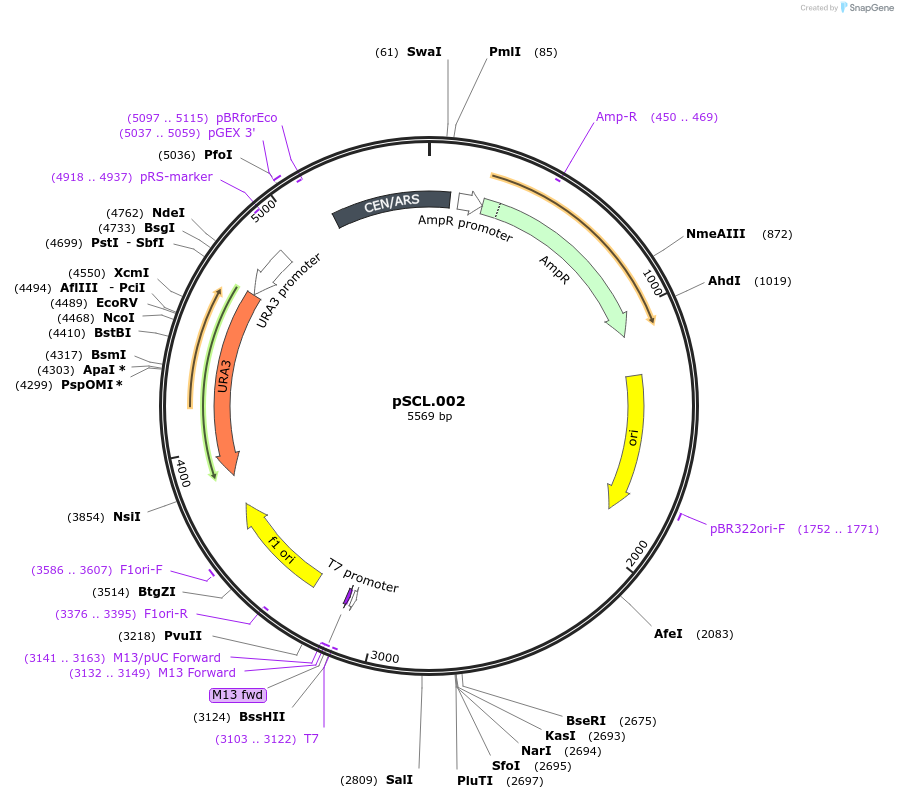
pSCL.002
Plasmid#184972PurposeExpress -Eco1 ADE2 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 12
ExpressionYeastMutationADE2 donor P272stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
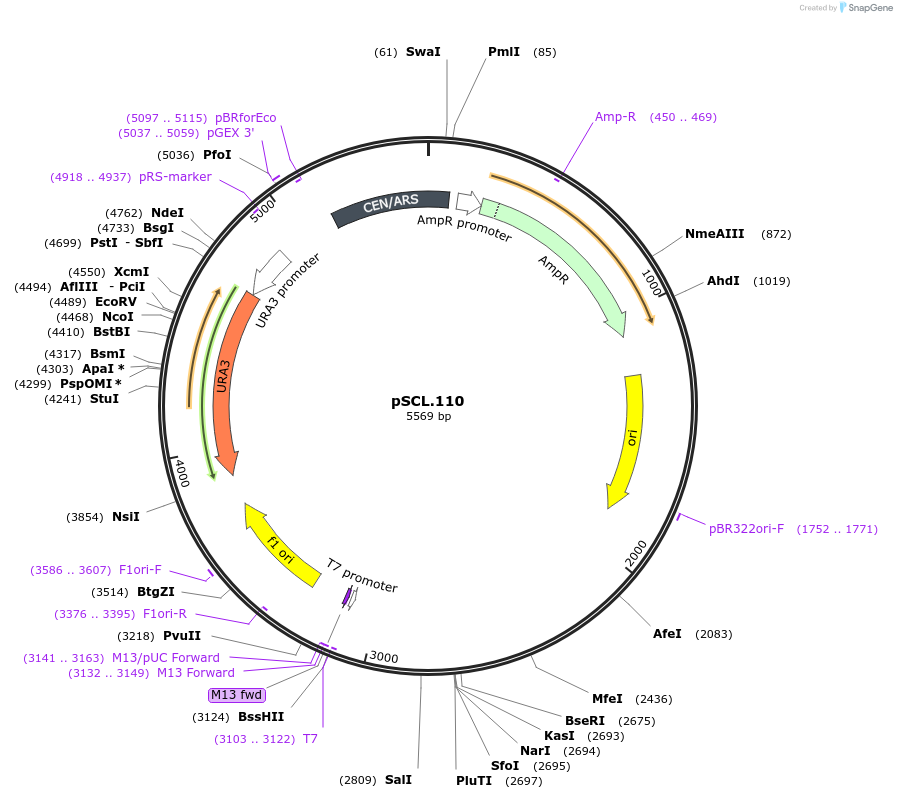
pSCL.110
Plasmid#184979PurposeExpress -Eco1 TRP2 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, TRP2 E64X, a1/a2 length: 12
ExpressionYeastMutationTRP2 donor E64stopPromoterGal7Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
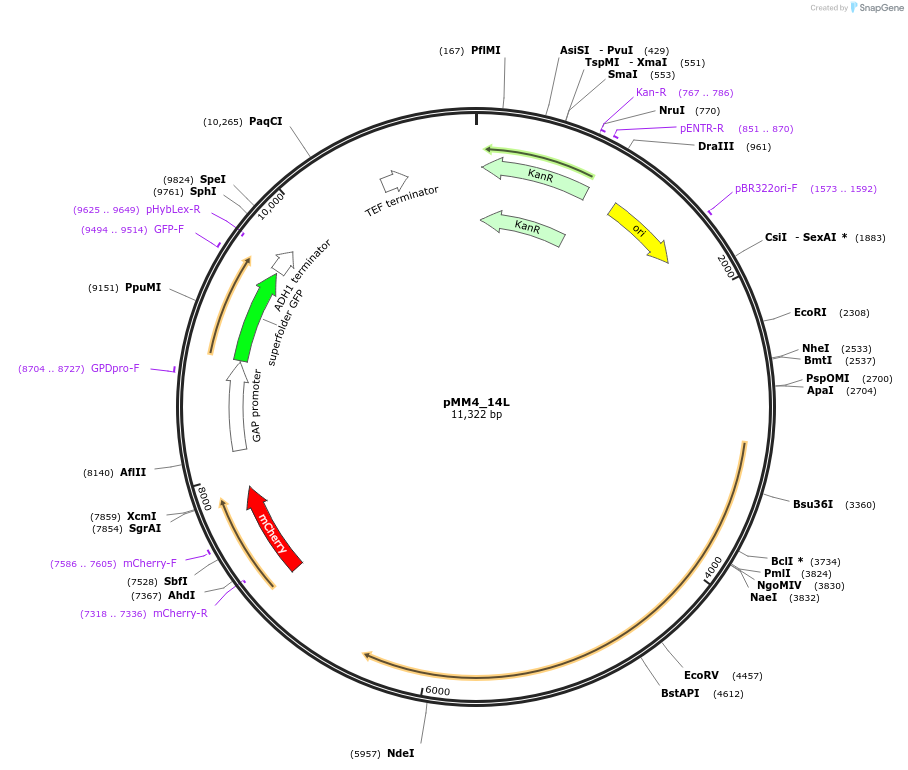
pMM4_14L
Plasmid#183063PurposePlasmid expressing a biosensor for acetic acid and pHluorin: BM3R1-Haa1-mTurquoise2 under RET2p, mCherry under HREsYGP1p and sfpHluorin under TDH3p with flanks for genome integration into HO locusDepositorInsertsmCherry
sfpHluorin
BM3R1-HAA1-mTurquoise2
UseSynthetic BiologyTagsmCherry and mTurquoise2ExpressionYeastPromoterRET2, Synthetic promoter, and TDH3Available SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
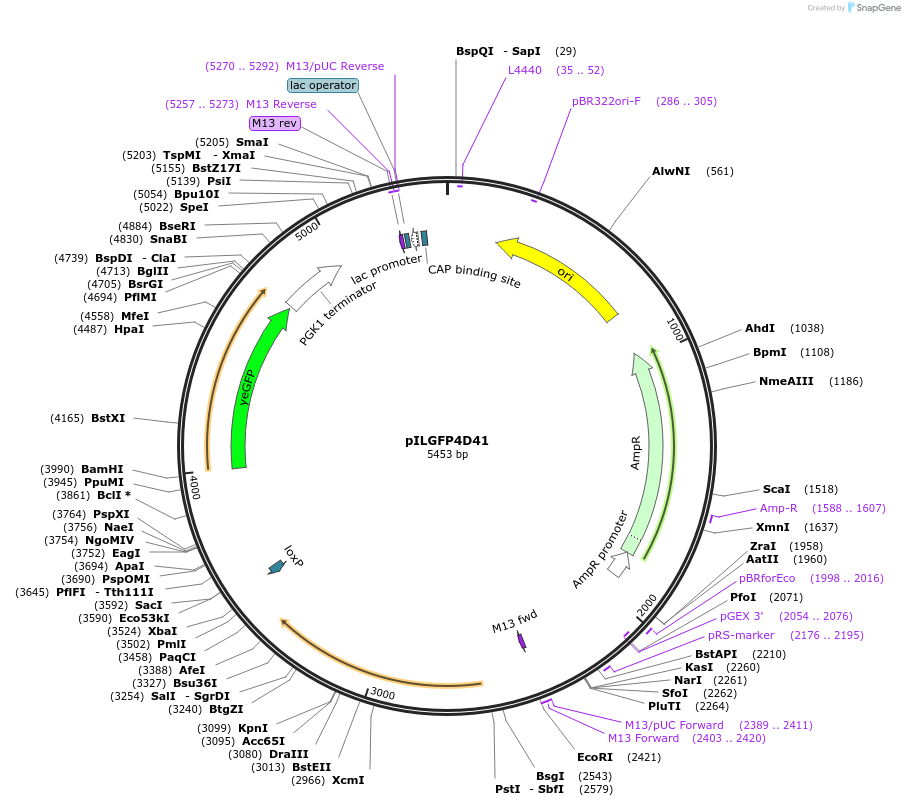
pILGFP4D41
Plasmid#185841PurposeTesting the CYC1 core promoter inserted with 4 Z268 elements using a green fluorescence protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1-PCYC1+[Z268]>yEGFP>TPGK1-TURA3
ExpressionYeastMutationNoneAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
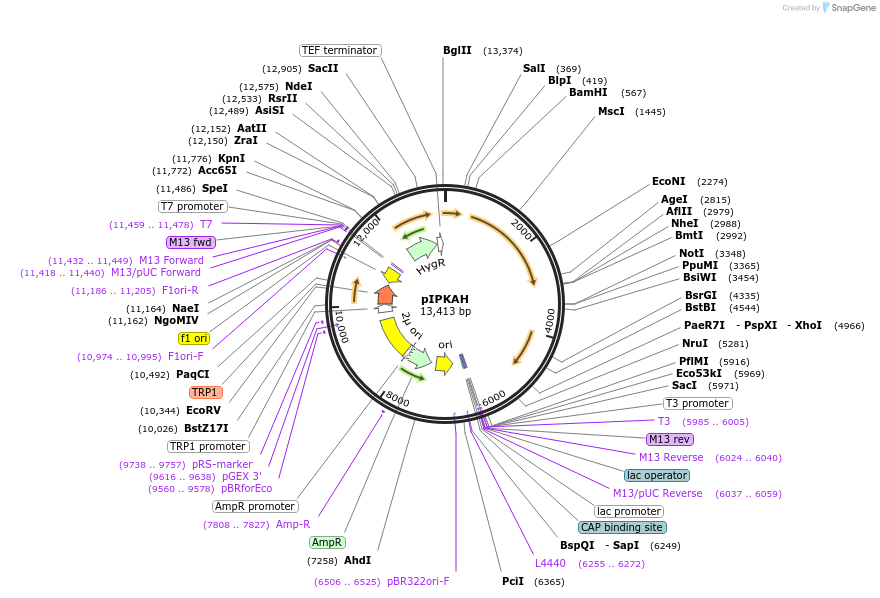
pIPKAH
Plasmid#185869PurposeKnocking out GPP1 and expressing a phosphoketolase pathway (Lactobacillus reuteri xfp and Methanosarcina barkeri pta)DepositorInsertGPP1(-220,80)> PAg.TEF1>hphMX6>TAg.TEF1-PENO2> Lr.xfp>T_TPI1-PTDH3>Mb.pta>TFBA1-GPP1(754, 1400)
ExpressionYeastMutationNoneAvailable SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
pILGFP10CF72 (B11)
Plasmid#185859PurposeTesting the SkGAL2 promoter inserted with 2 PZ4 element using a green fluorescent protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1- PSkGAL2M3(2*PZ4)>(BamHI)yEGFP>TURA3
ExpressionYeastMutationNoneAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
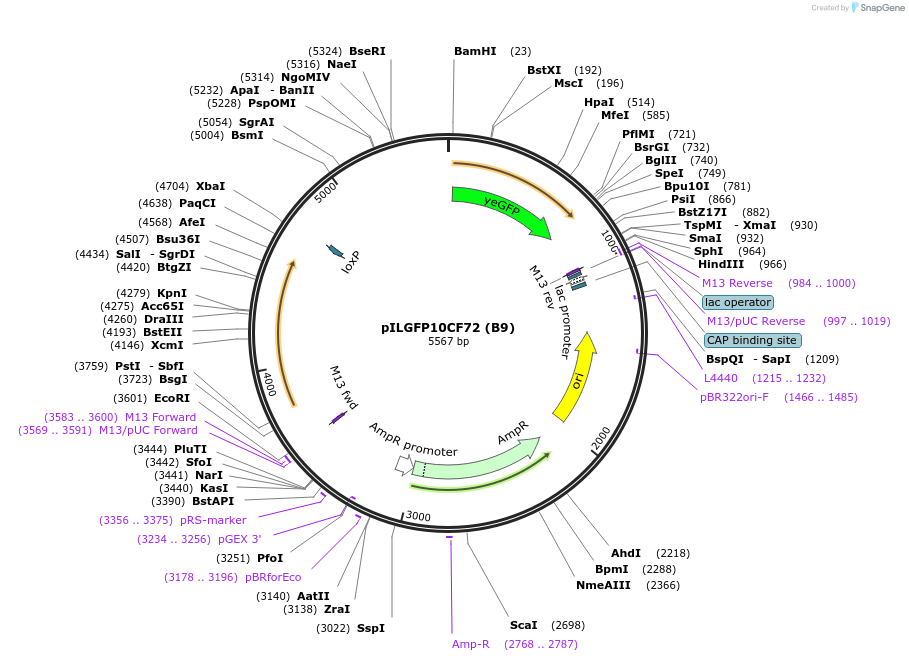
pILGFP10CF72 (B9)
Plasmid#185858PurposeTesting the SkGAL2 promoter inserted with 2 PZ4 element using a green fluorescent protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1- PSkGAL2M5(5*PZ4)>(BamHI)yEGFP>TURA3
ExpressionYeastMutationNoneAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
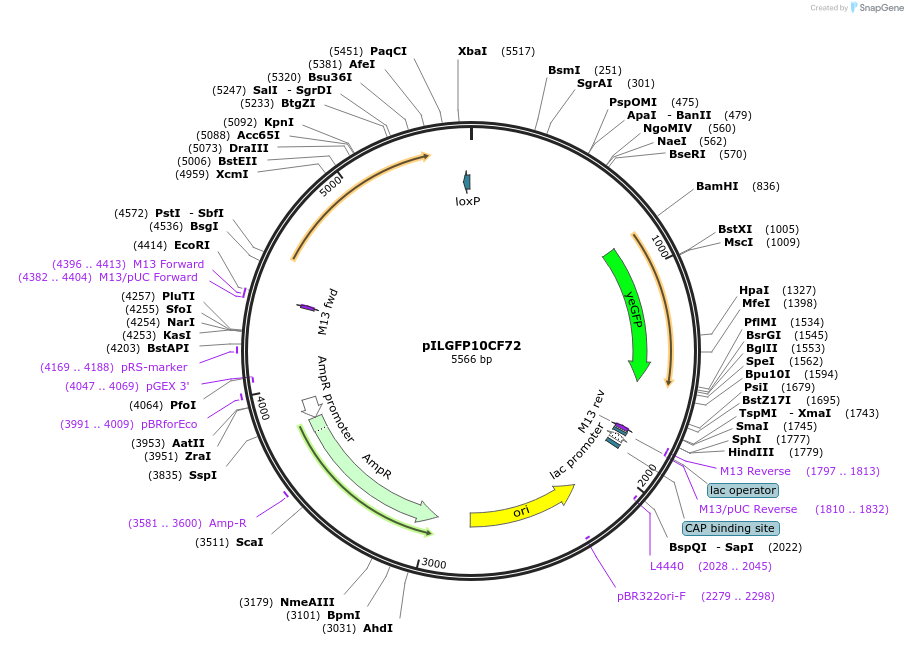
pILGFP10CF72
Plasmid#185857PurposeTesting the SkGAL2 promoter inserted with 4 PZ4 elements using a green fluorescent protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1- PSkGAL2M4(4*PZ4)>(BamHI)yEGFP>TURA3
ExpressionYeastMutationNoneAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
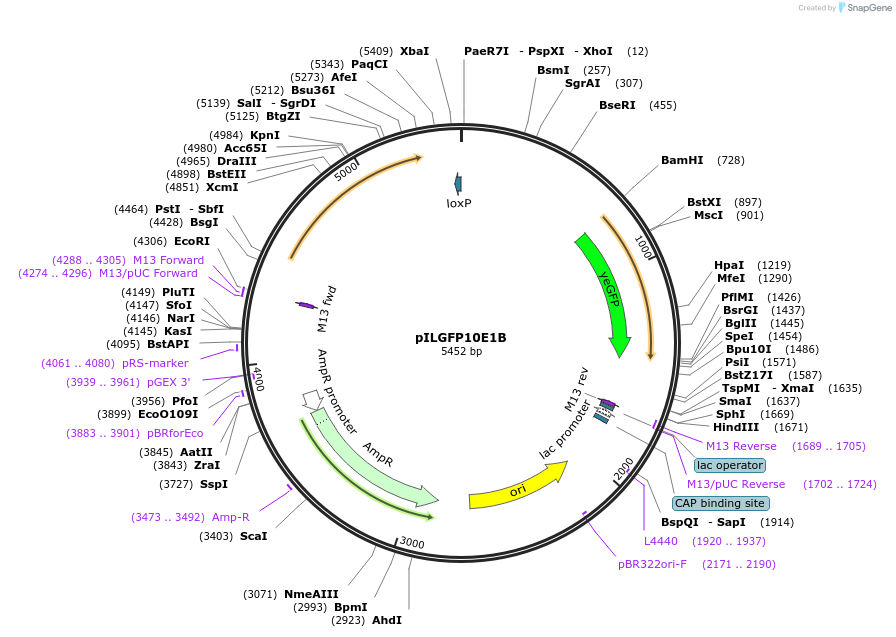
pILGFP10E1B
Plasmid#185856PurposeTesting the SkGAL2 promoter inserted with 3 PZ4 elements using a green fluorescent protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1- PSkGAL2M2(3*PZ4)>(BamHI)yEGFP>TURA3
ExpressionYeastMutationNoneAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
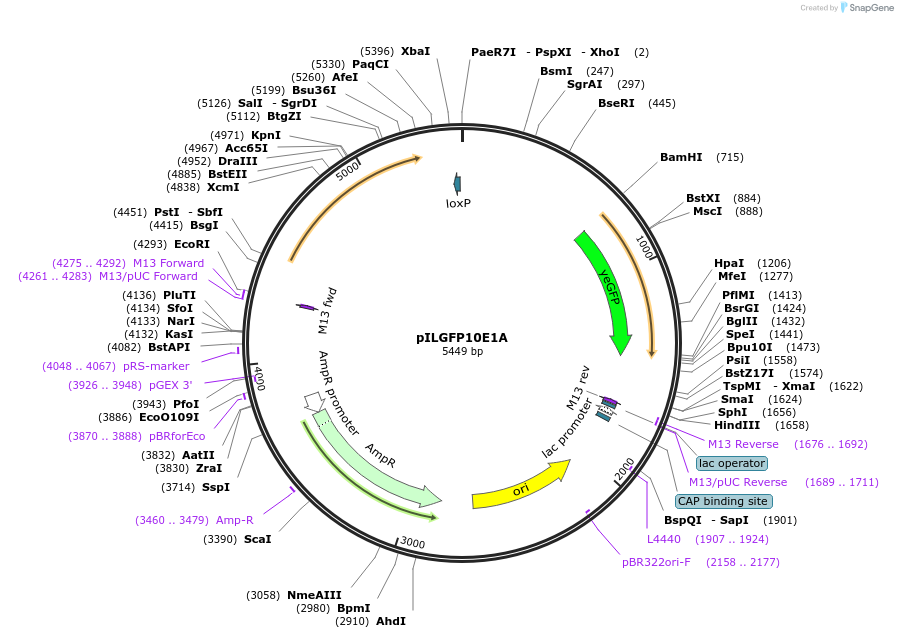
pILGFP10E1A
Plasmid#185855PurposeTesting the SkGAL2 promoter inserted with 2 PZ4 elements using a green fluorescent protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1- PSkGAL2M1(2*PZ4)> (BamHI)yEGFP>TURA3
ExpressionYeastMutationNoneAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
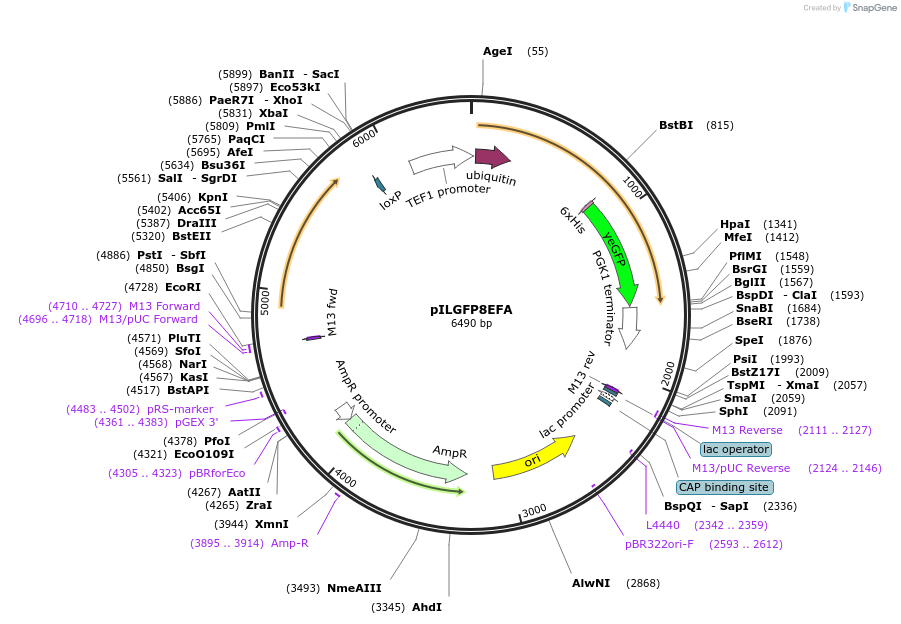
pILGFP8EFA
Plasmid#185854PurposeTesting a heat inducible protein degradation tag -DHFR P66L using a green fluorescent protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1-PTEF1>UBI4>RHGSGTMV>DHFRP66L>yEGFP>TPGK1-TURA3
ExpressionYeastMutationDHFR P66LAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
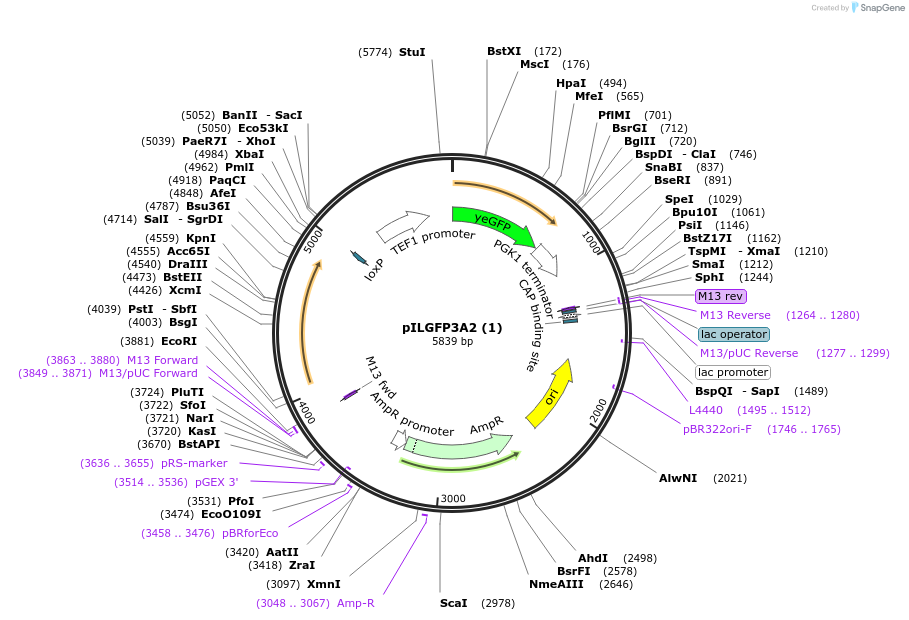
pILGFP3A2 (1)
Plasmid#185851PurposeTesting the TEF1 promoter appended with 2 tetracycline riboswitch using a green fluorescence protein as the reporterDepositorInsertPURA3>KlURA3>TAgTEF1-PTEF1>2*TcRb>yEGFP>TPGK1-TURA3
ExpressionYeastMutationNoneAvailable SinceAug. 31, 2022AvailabilityAcademic Institutions and Nonprofits only -
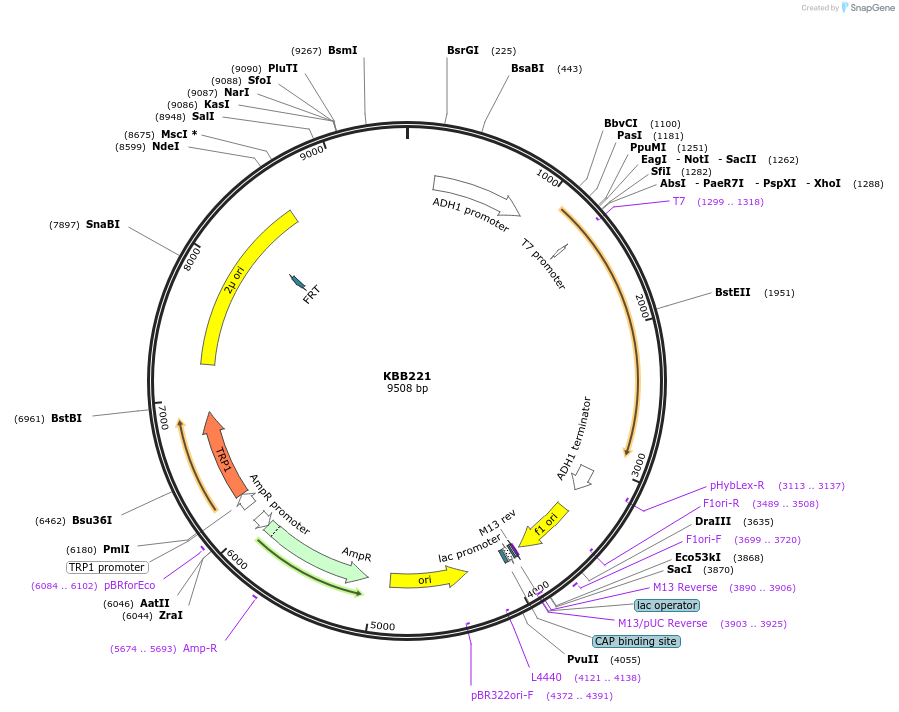
KBB221
Plasmid#185071PurposeSST::ACT2 "SA236"DepositorInsertACT2
ExpressionYeastMutationACT2-SA236Available SinceAug. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
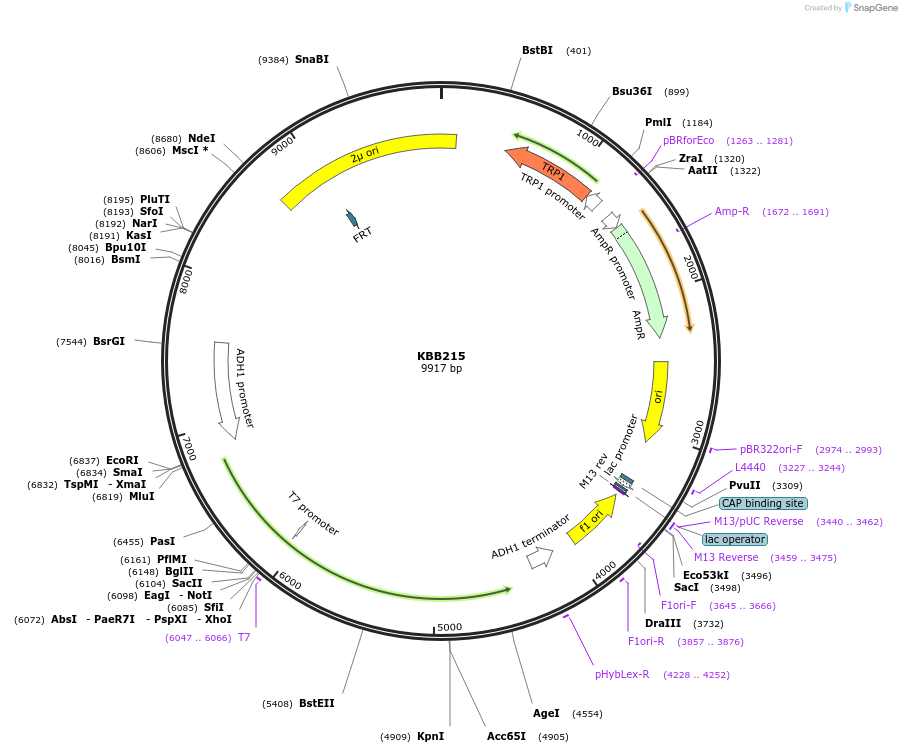
KBB215
Plasmid#185068PurposeSST::WAK2 EcoRI/ScaI fragment (secretion confirmed)DepositorInsertWAK2
ExpressionYeastAvailable SinceAug. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
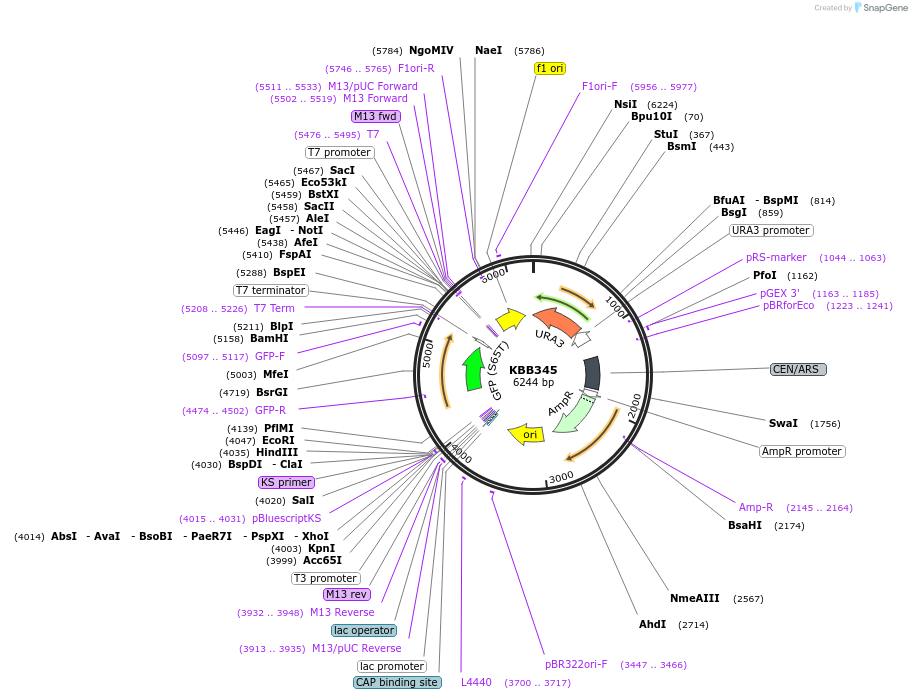
KBB345
Plasmid#185078Purposerfa3D46 truncation fused to GFPDepositorInsertRFA3
TagsGFPExpressionYeastMutationRfa3 truncation aa 1-46Available SinceJuly 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
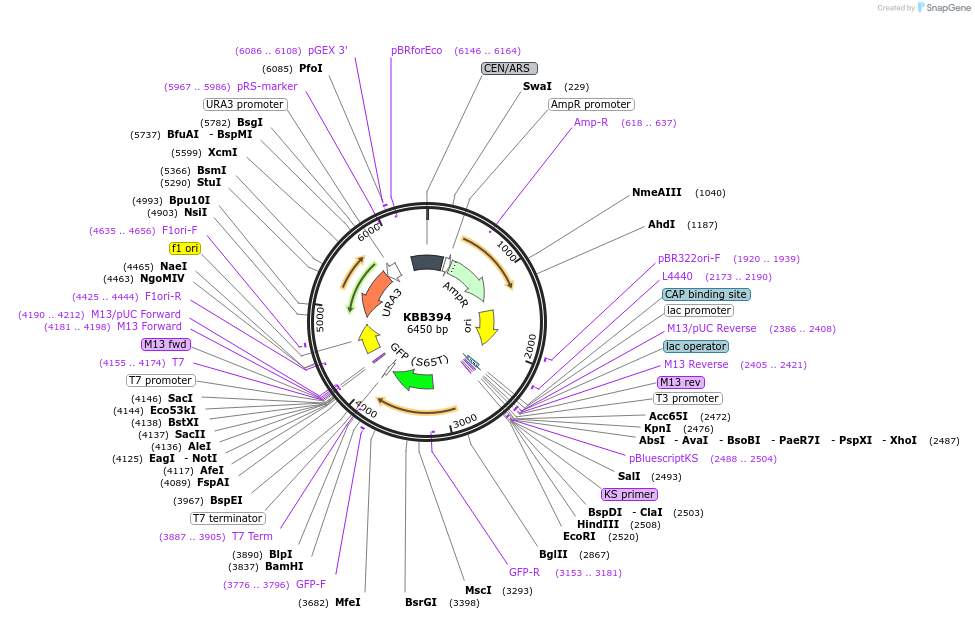
KBB394
Plasmid#185086PurposeNMA111 with both nuclear localization signals mutated to alanines fused to GFP (mutagenesis of KBB280)DepositorInsertNMA111
TagsGFPExpressionYeastMutationNma111 KKR 9-11 AAA; KRK 28-30 AAAAvailable SinceJuly 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
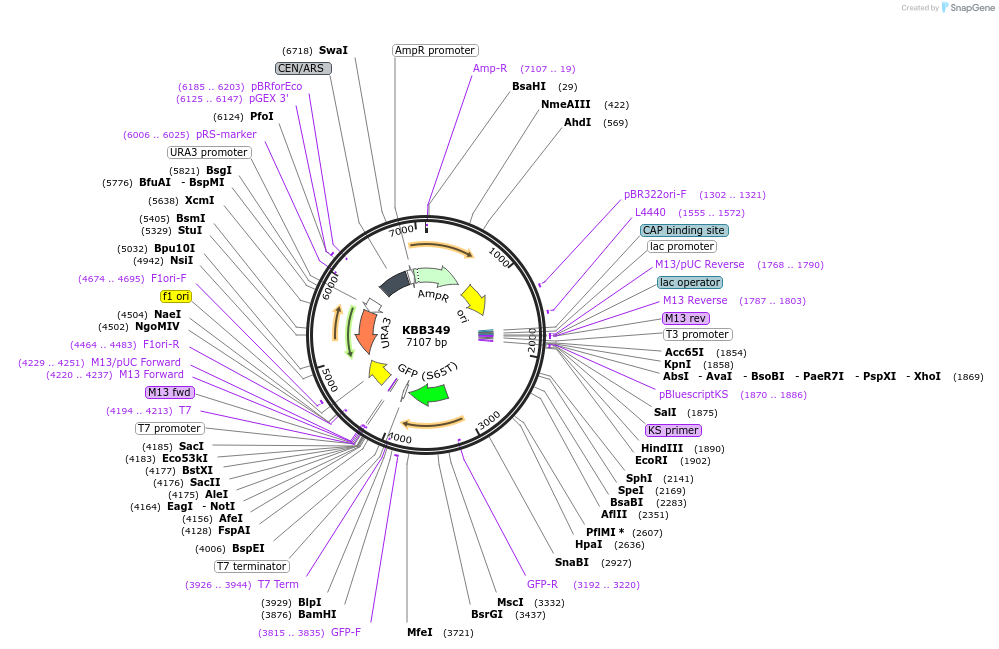
KBB349
Plasmid#185080Purposerfa2D248 truncation fused to GFPDepositorInsertRFA2
TagsGFPExpressionYeastMutationRfa2 truncation aa 1-247Available SinceJuly 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
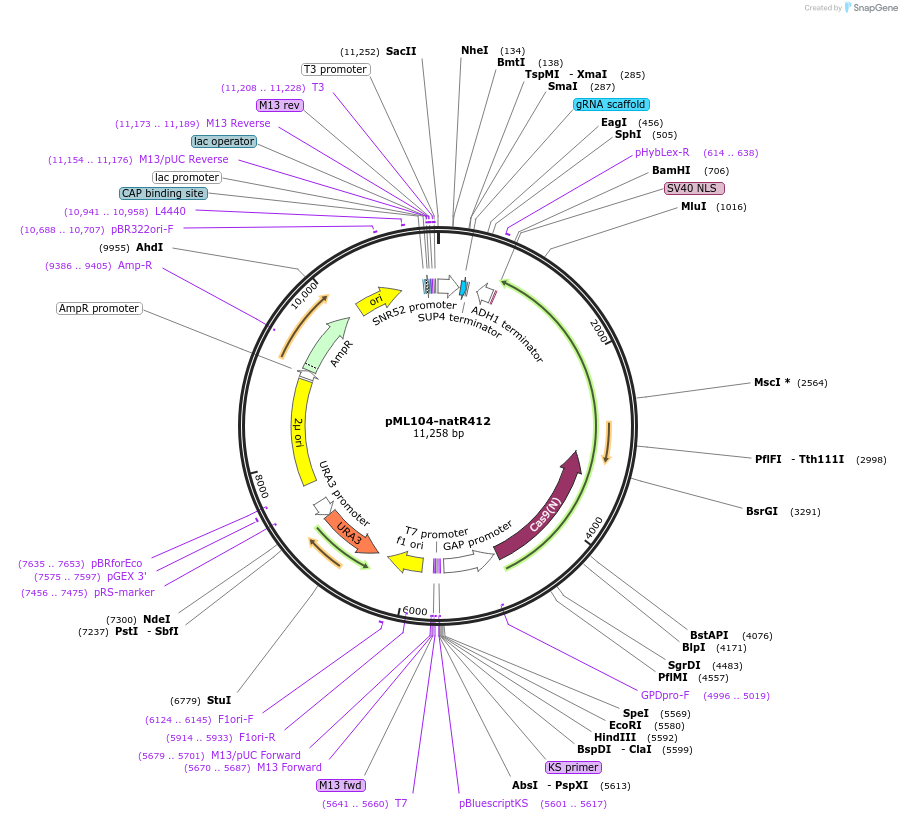
pML104-natR412
Plasmid#162042PurposeExpresses Cas9 and contains natR412 guide which targets the natR geneDepositorInsertnatR412 guide
UseCRISPRExpressionYeastAvailable SinceFeb. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
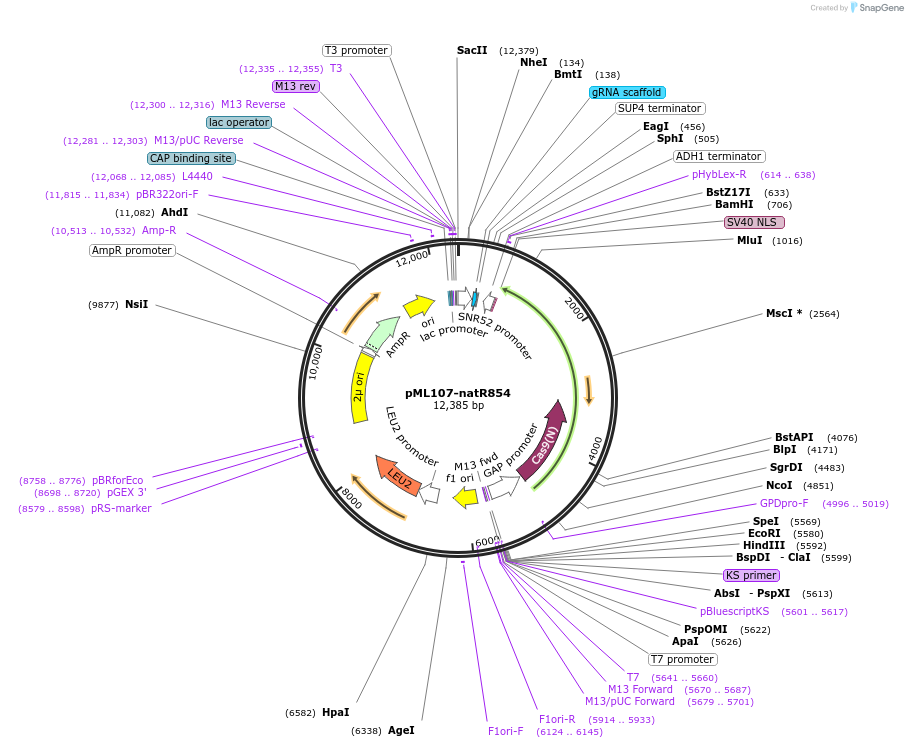
pML107-natR854
Plasmid#162043PurposeExpresses Cas9 and contains natR854 guide which targets the natR geneDepositorInsertnatR854 guide
UseCRISPRExpressionYeastAvailable SinceFeb. 11, 2022AvailabilityAcademic Institutions and Nonprofits only