We narrowed to 10,071 results for: EPO
-
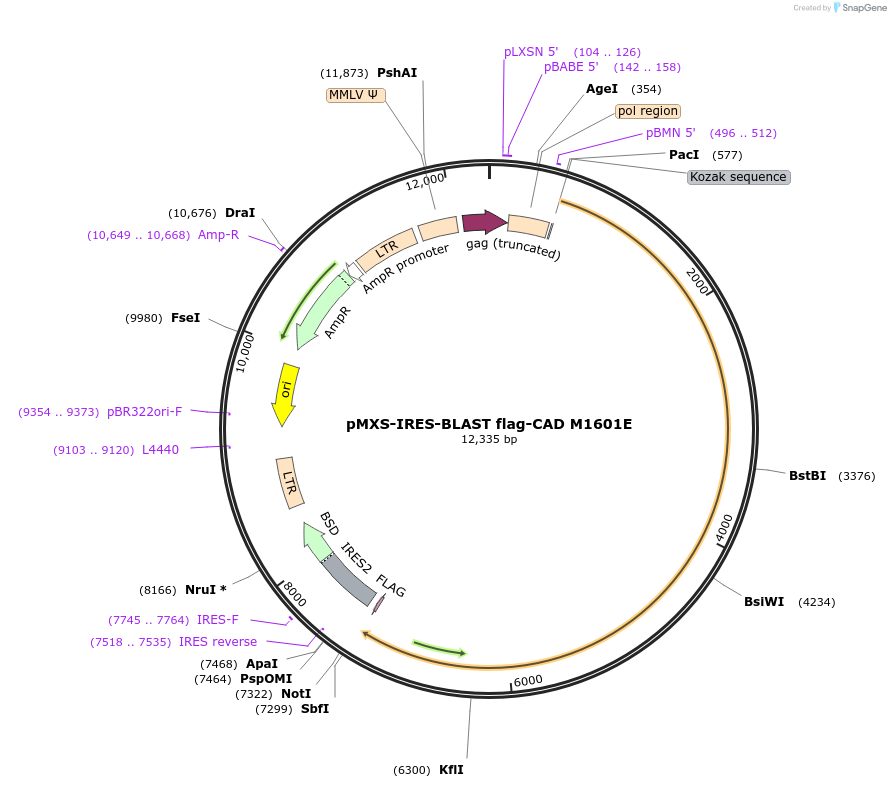
Plasmid#169828PurposeExpresses C-terminal flag-tagged CAD with mutation at reported dimerization interface of the DHOase domainDepositorInsertCAD (CAD Human)
UseRetroviralTagsFlag tagExpressionMammalianMutationM1601E; TCCC -> AGTC silent mutations at nt527…Available SinceJuly 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
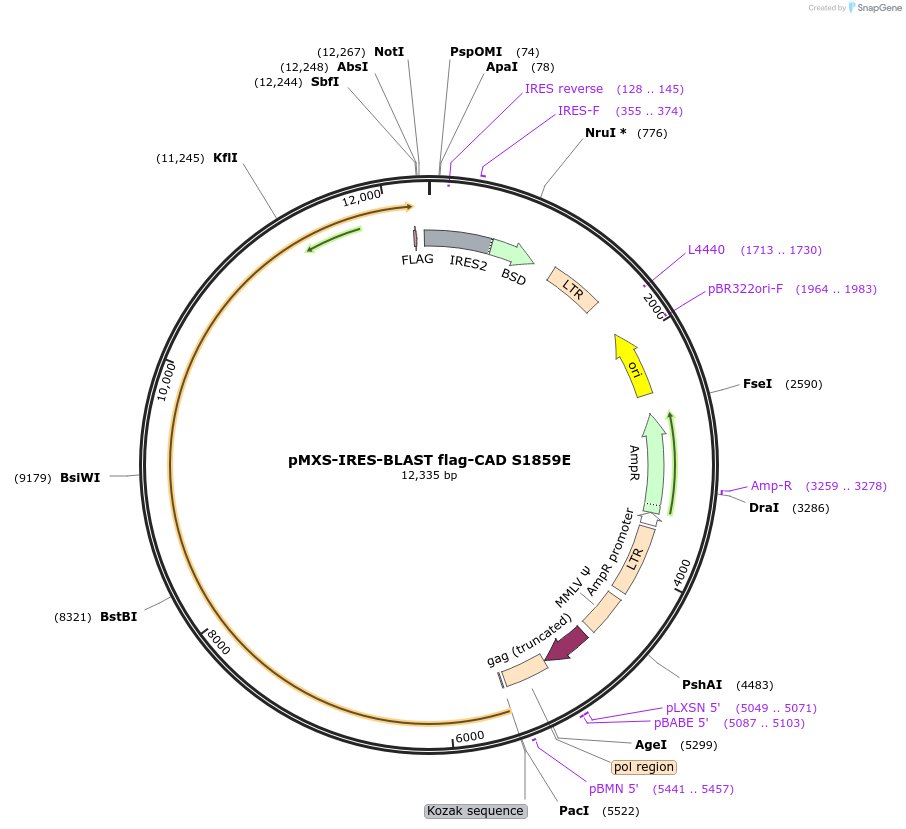
pMXS-IRES-BLAST flag-CAD S1859E
Plasmid#169822PurposeExpresses C-terminal flag-tagged CAD with mutation of reported S6K phosphorylation siteDepositorInsertCAD (CAD Human)
UseRetroviralTagsFlag tagExpressionMammalianMutationS1859E; TCCC -> AGTC silent mutations at nt527…Available SinceJuly 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
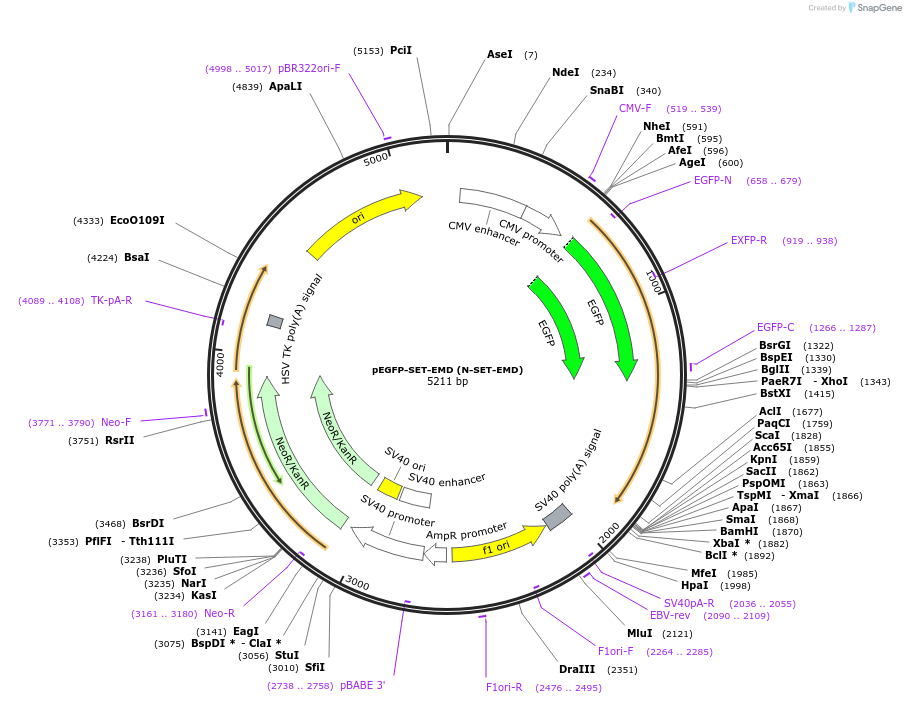
pEGFP-SET-EMD (N-SET-EMD)
Plasmid#217765PurposeFluorescent reporter for ceramide (putative, mammalian expression)DepositorAvailable SinceJune 12, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
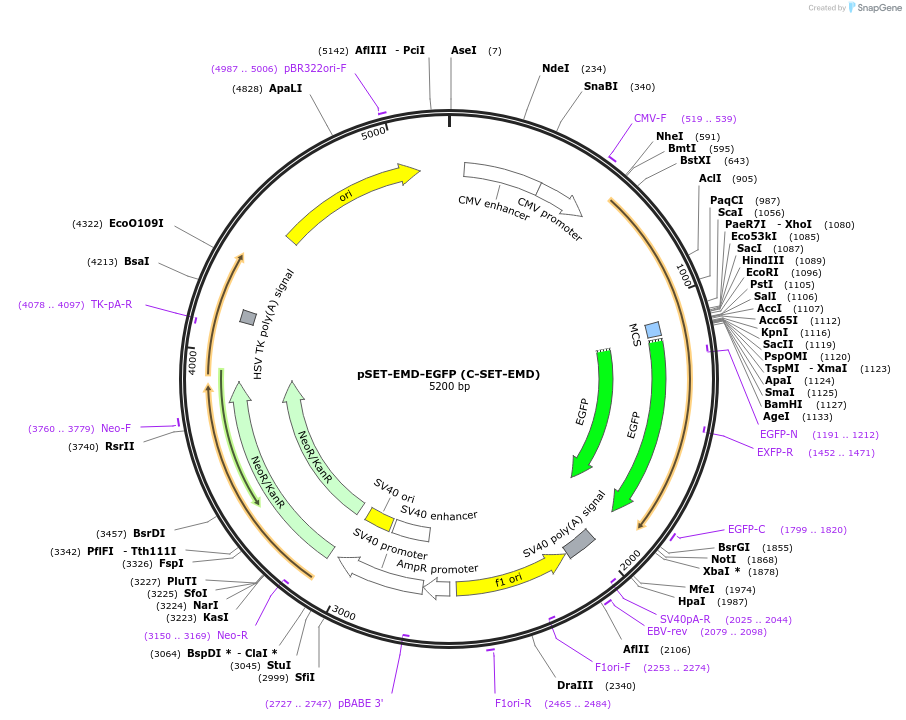
pSET-EMD-EGFP (C-SET-EMD)
Plasmid#217764PurposeFluorescent reporter for ceramide (putative, mammalian expression)DepositorAvailable SinceJune 12, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
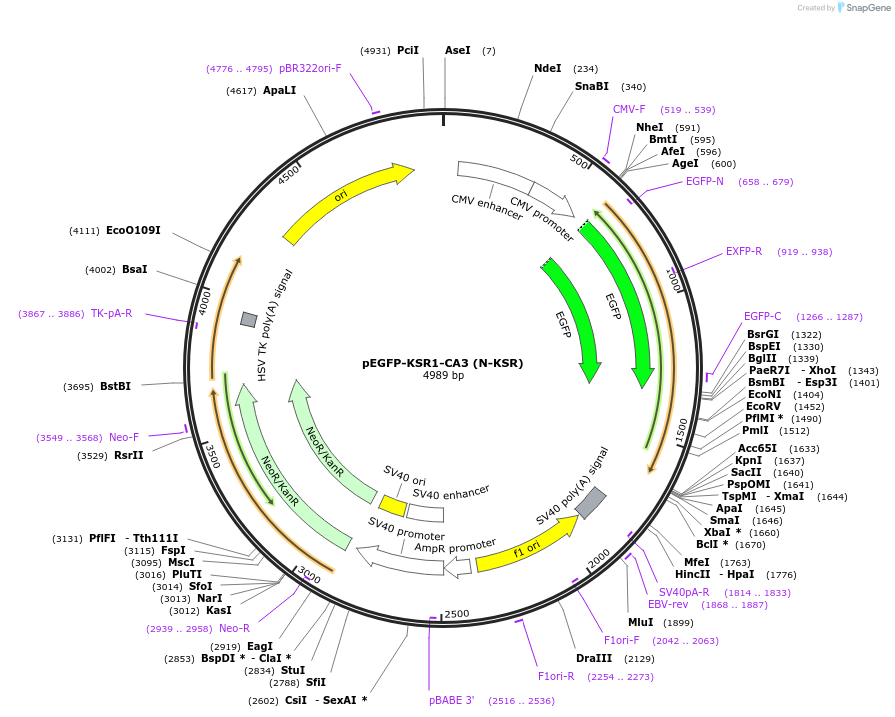
pEGFP-KSR1-CA3 (N-KSR)
Plasmid#217755PurposeFluorescent reporter for glucosyl-ceramide (putative, mammalian expression)DepositorInsertKinase suppressor of ras 1 CA3 domain (KSR1 Human)
TagsEGFPExpressionMammalianMutationCA3 domain aa 317-400PromoterCMVAvailable SinceJune 11, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
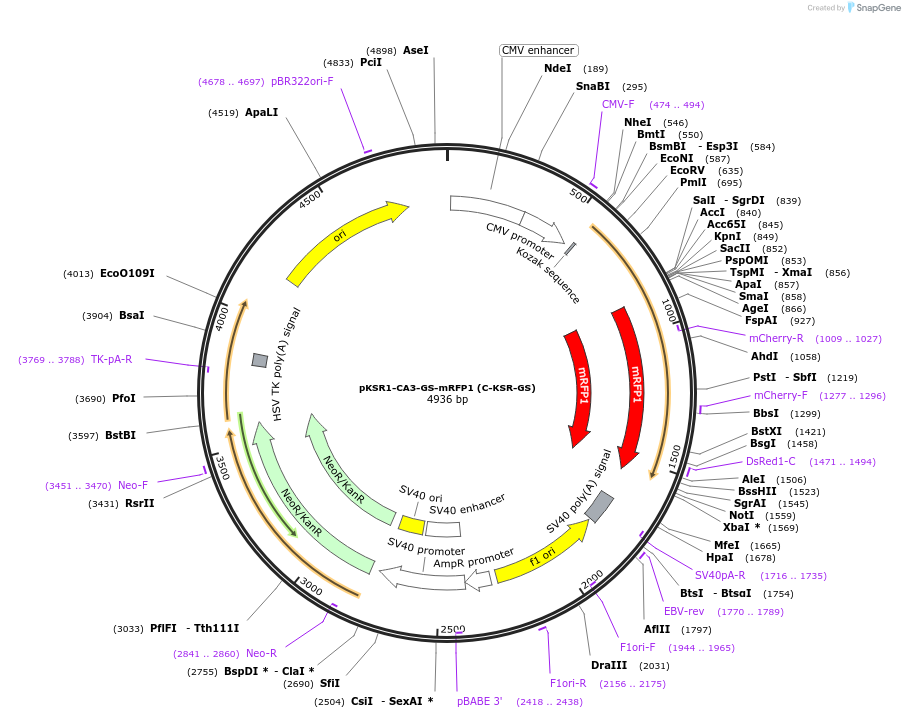
pKSR1-CA3-GS-mRFP1 (C-KSR-GS)
Plasmid#217758PurposeFluorescent reporter for ceramide (mammalian expression)DepositorInsertKinase suppressor of ras 1 CA3 domain (KSR1 Human)
TagsmRFPExpressionMammalianMutationCA3 domain aa 317-400 with GGSSGGGGA linkerPromoterCMVAvailable SinceJune 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
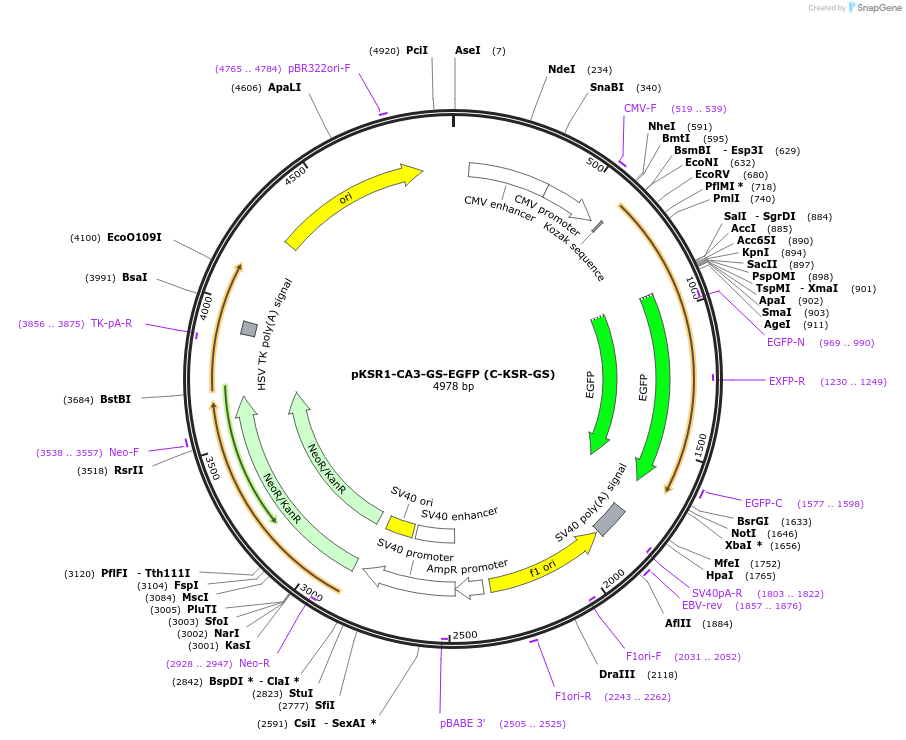
pKSR1-CA3-GS-EGFP (C-KSR-GS)
Plasmid#217757PurposeFluorescent reporter for ceramide (mammalian expression)DepositorInsertKinase suppressor of ras 1 CA3 domain (KSR1 Human)
TagsEGFPExpressionMammalianMutationCA3 domain aa 317-400 with GGSSGGGGA linkerPromoterCMVAvailable SinceJune 10, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
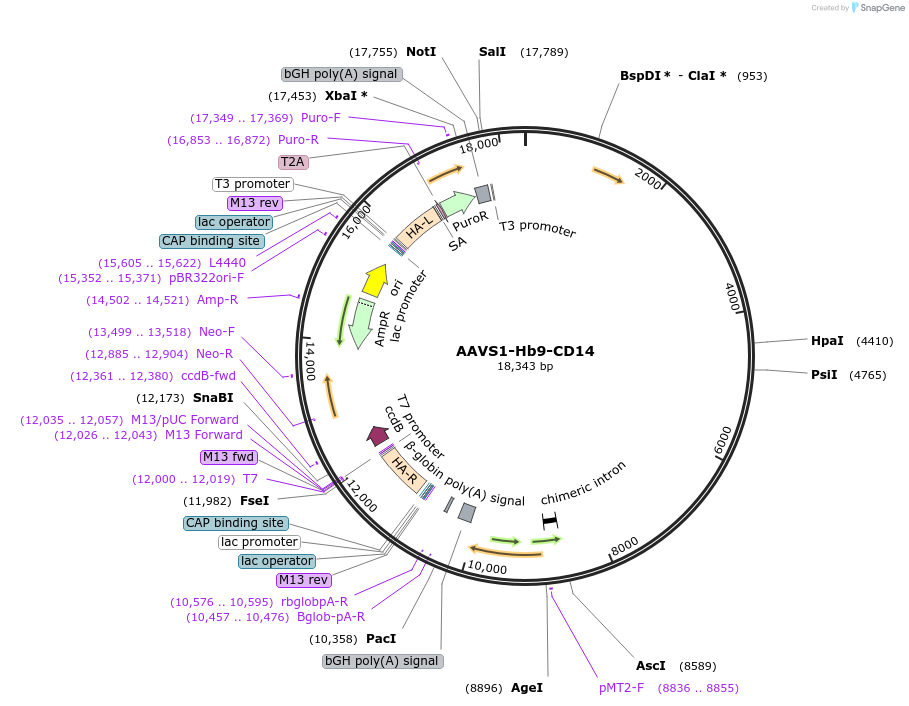
AAVS1-Hb9-CD14
Plasmid#204344PurposeKnock-in vector to insert a motor neuron-specific MACS-sortable genetic reporter into the AAVS1 locus in human pluripotent stem cells. Allows isolation of human iPSC-derived motor neurons.DepositorAvailable SinceSept. 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
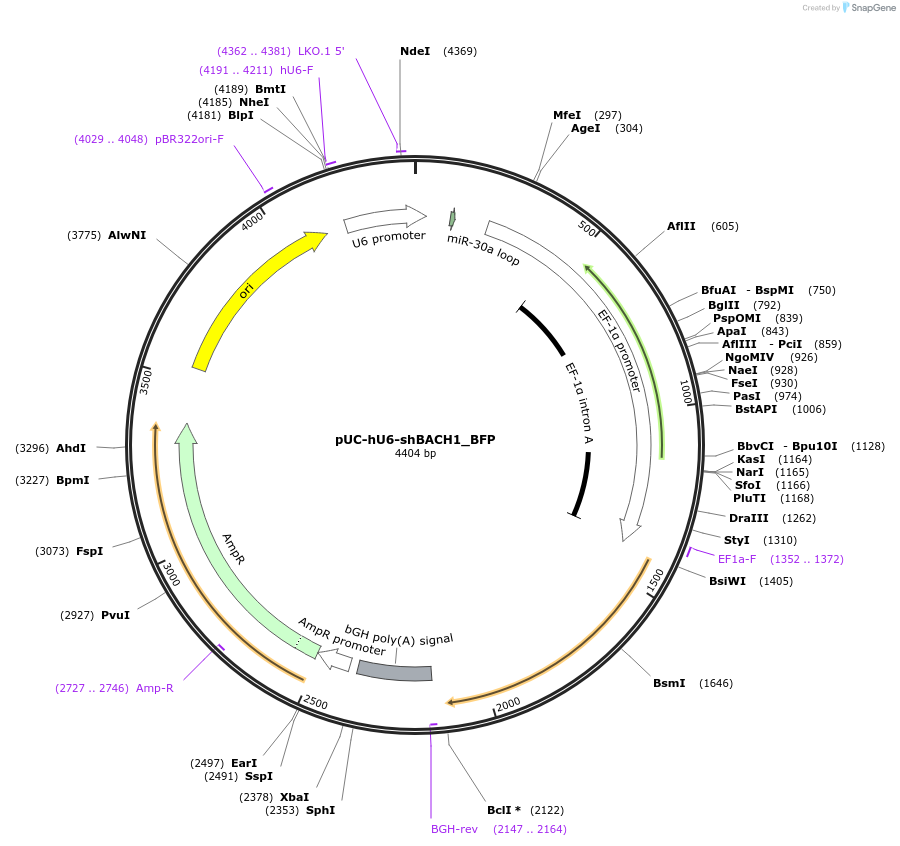
pUC-hU6-shBACH1_BFP
Plasmid#199217PurposeExpression plasmid with shRNAmiR targeting BACH1, with a BFP reporter on the backbone.DepositorAvailable SinceAug. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
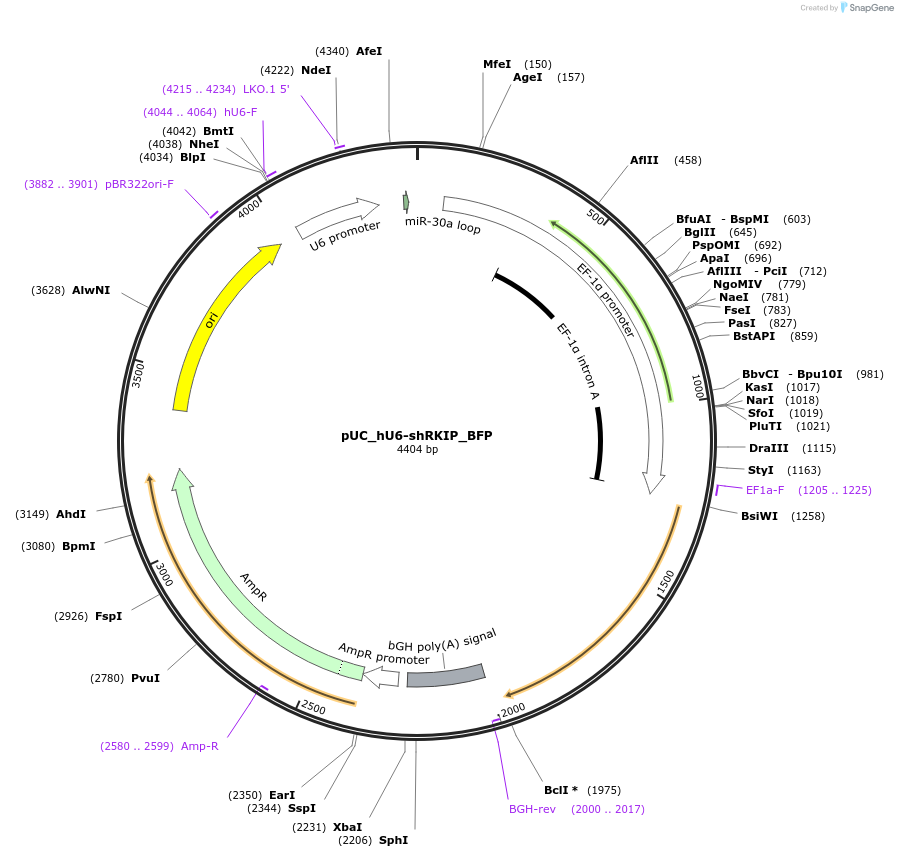
pUC_hU6-shRKIP_BFP
Plasmid#199216PurposeExpression plasmid with shRNAmiR targeting RKIP (PEBP1), with a BFP reporter on the backbone.DepositorAvailable SinceJune 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
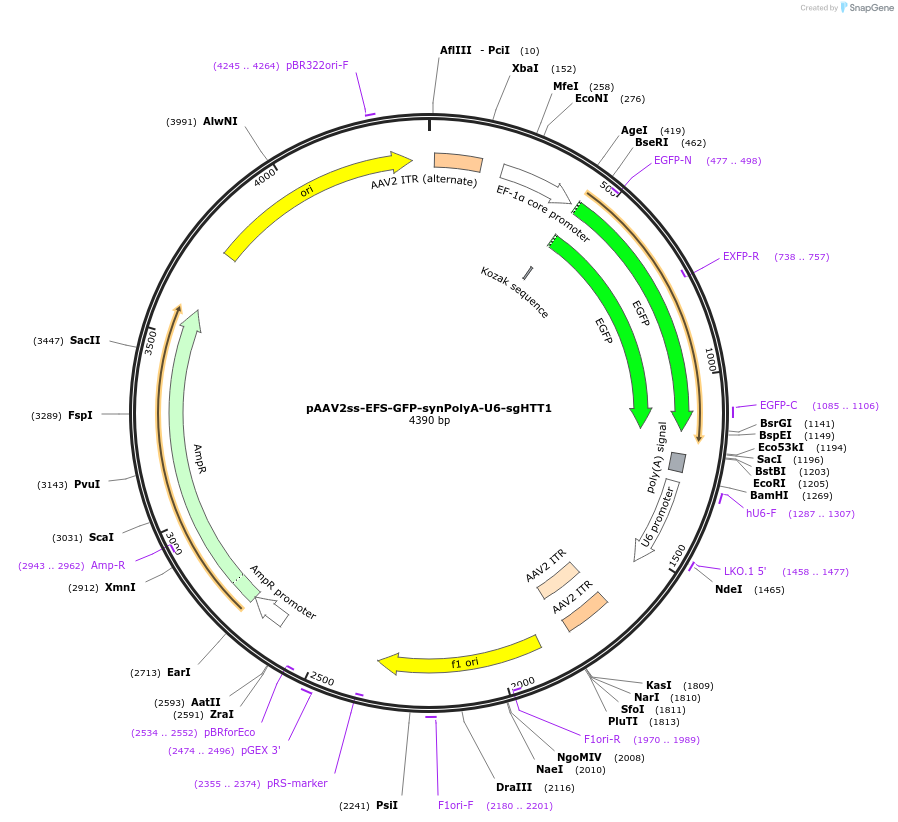
pAAV2ss-EFS-GFP-synPolyA-U6-sgHTT1
Plasmid#190902PurposeAAV vector expressing thew GFP reporter gene and human sgHTTDepositorAvailable SinceMarch 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
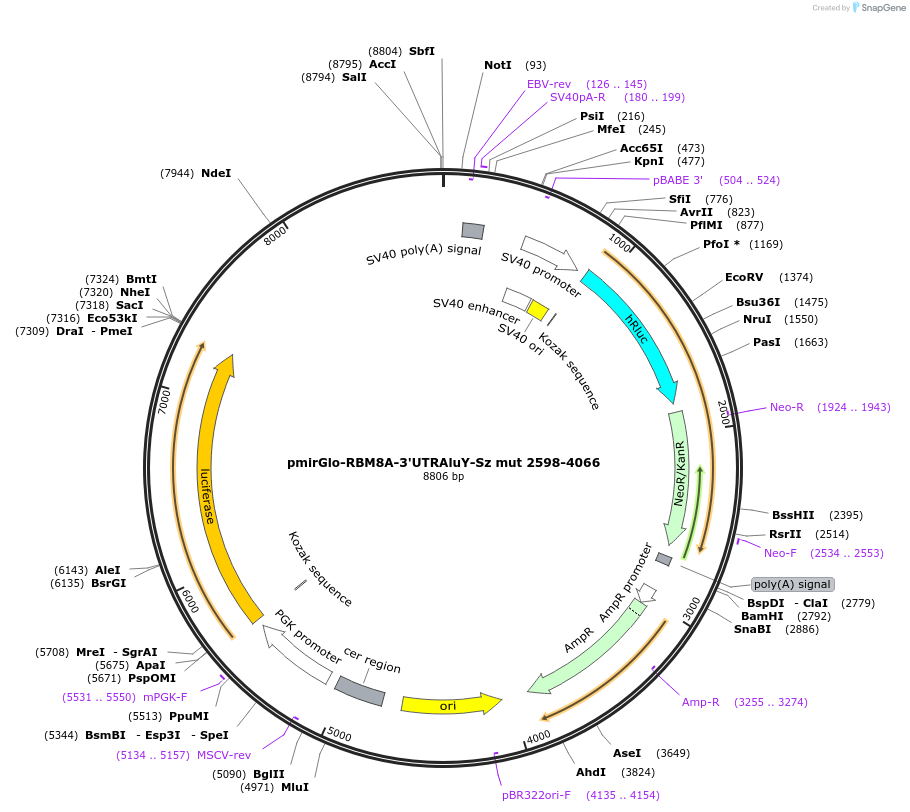
pmirGlo-RBM8A-3'UTRAluY-Sz mut 2598-4066
Plasmid#171135PurposereporterDepositorInsertRBM8A 3'UTR (RBM8A Human)
ExpressionMammalianMutationA3302G, A3356G, A3373G, A3389G,A3451G, T3729C, A3…Available SinceDec. 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
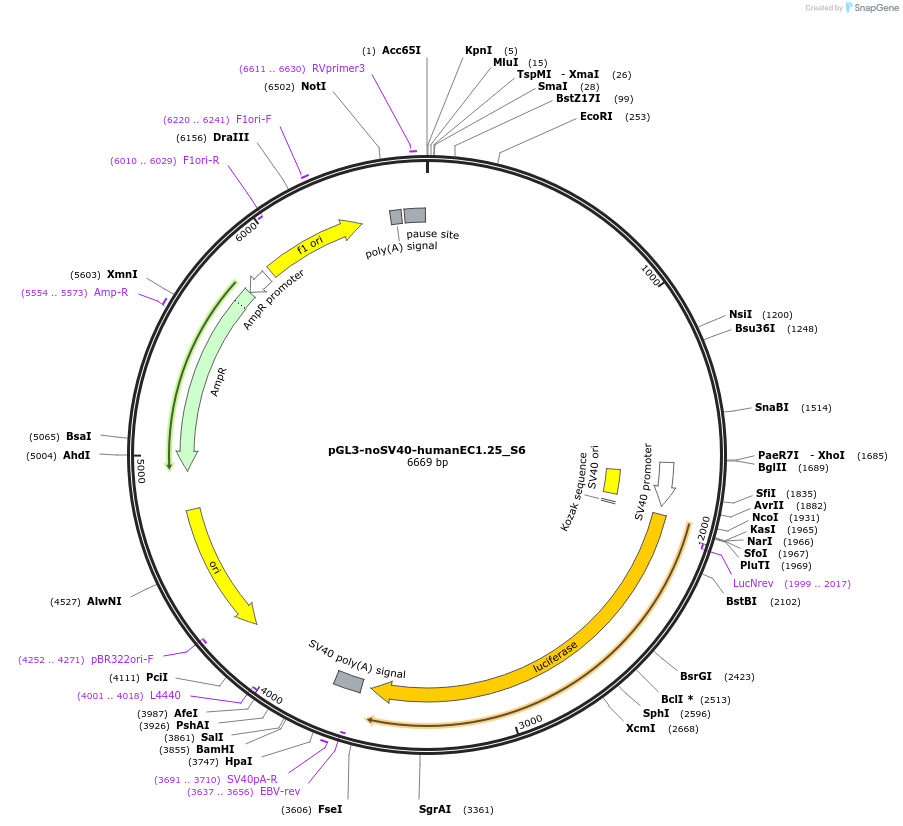
pGL3-noSV40-humanEC1.25_S6
Plasmid#173960PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertEnhancer region 6 (S6) from SOX9 enhancer cluster EC1.25
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
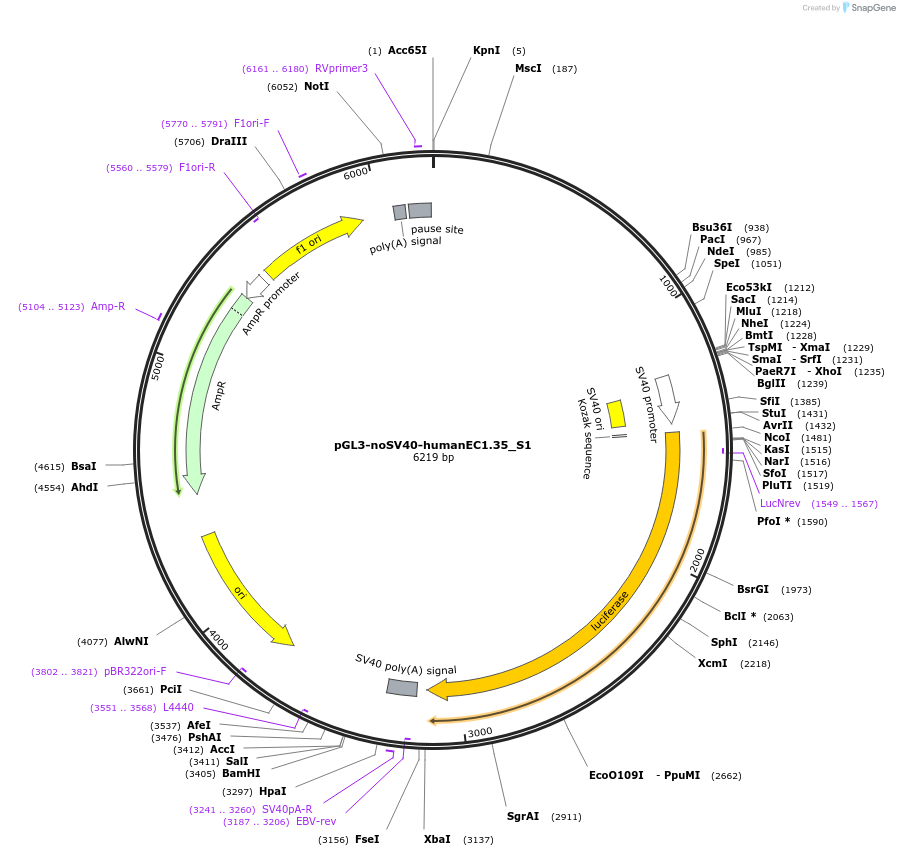
pGL3-noSV40-humanEC1.35_S1
Plasmid#173954PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertEnhancer region 1 (S1) from SOX9 enhancer cluster EC1.35
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
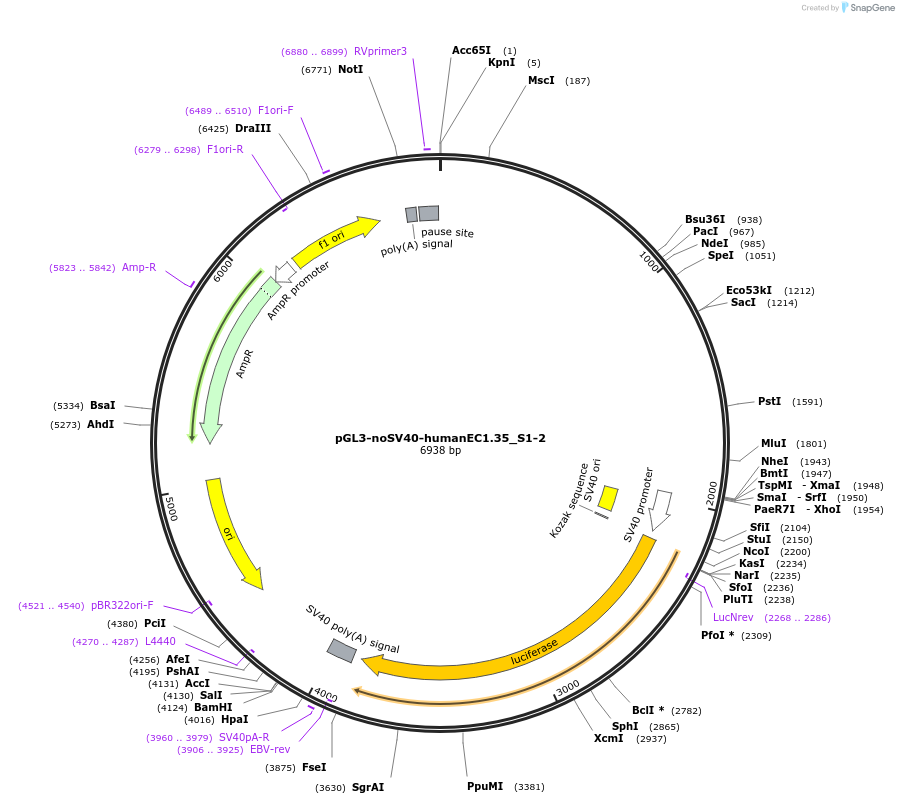
pGL3-noSV40-humanEC1.35_S1-2
Plasmid#173953PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertEnhancer regions 1 and 2 (S1 and S2) from SOX9 enhancer cluster EC1.35
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
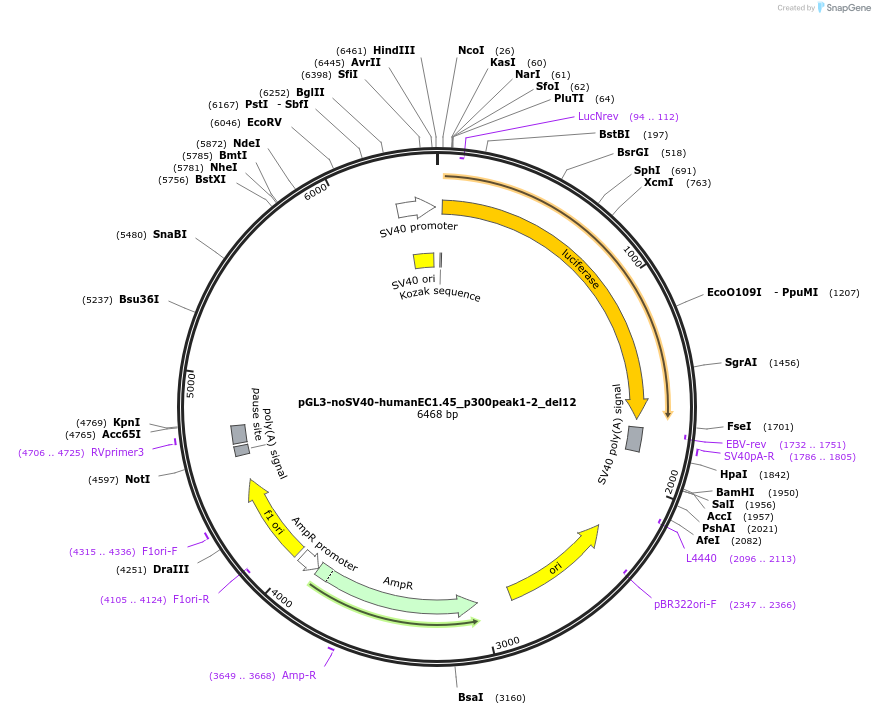
pGL3-noSV40-humanEC1.45_p300peak1-2_del12
Plasmid#173984PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #12
UseLuciferaseMutationTiled deletion #12PromoterSV40 promoterAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
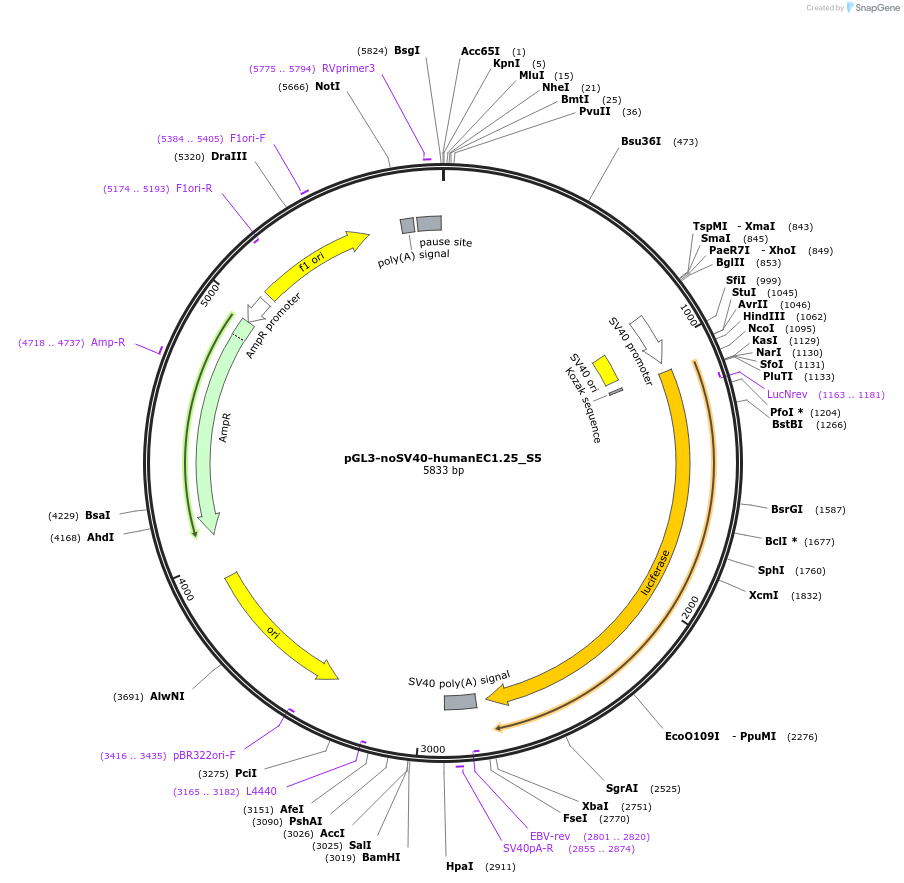
pGL3-noSV40-humanEC1.25_S5
Plasmid#173959PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertEnhancer region 5 (S5) from SOX9 enhancer cluster EC1.25
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
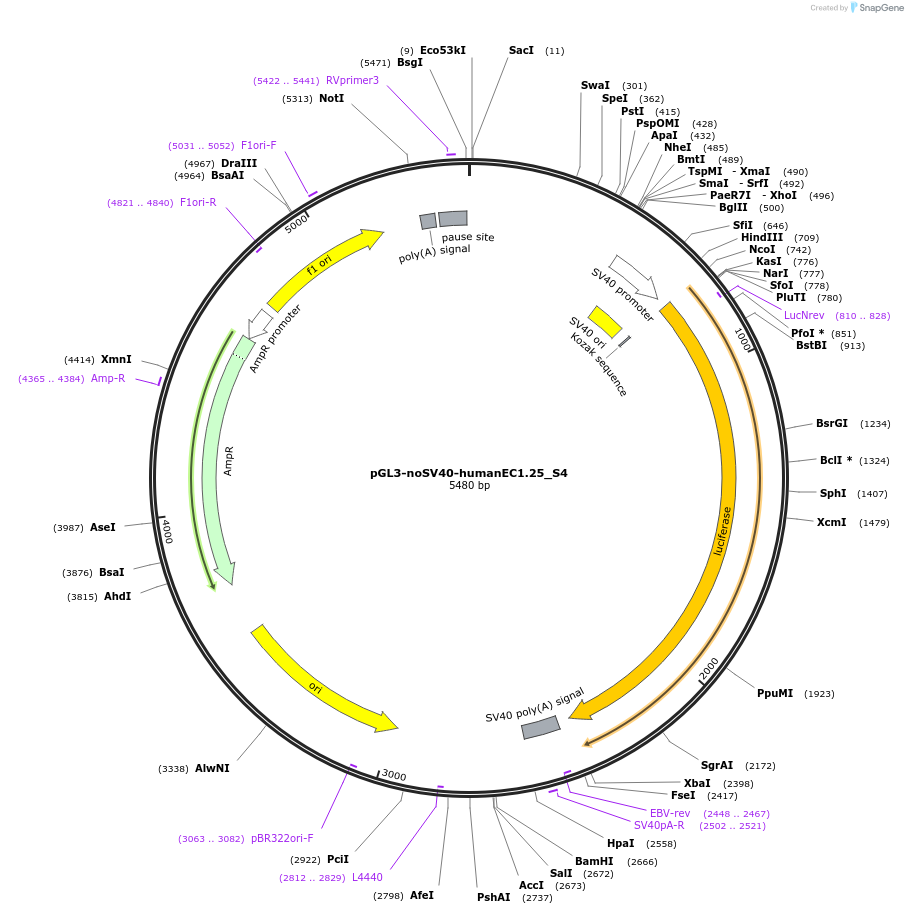
pGL3-noSV40-humanEC1.25_S4
Plasmid#173958PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertEnhancer region 4 (S4) from SOX9 enhancer cluster EC1.25
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
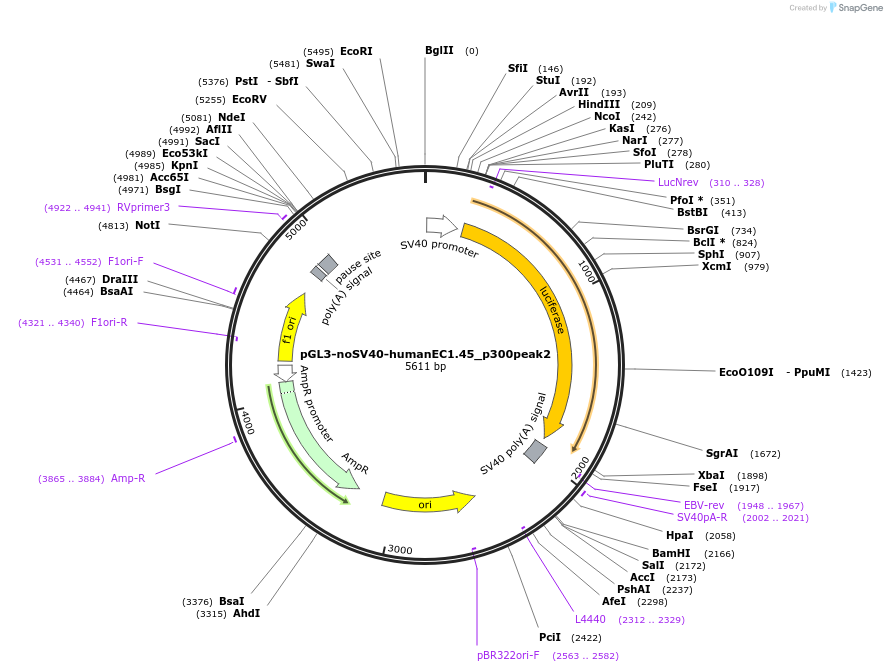
pGL3-noSV40-humanEC1.45_p300peak2
Plasmid#173948PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 2 from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
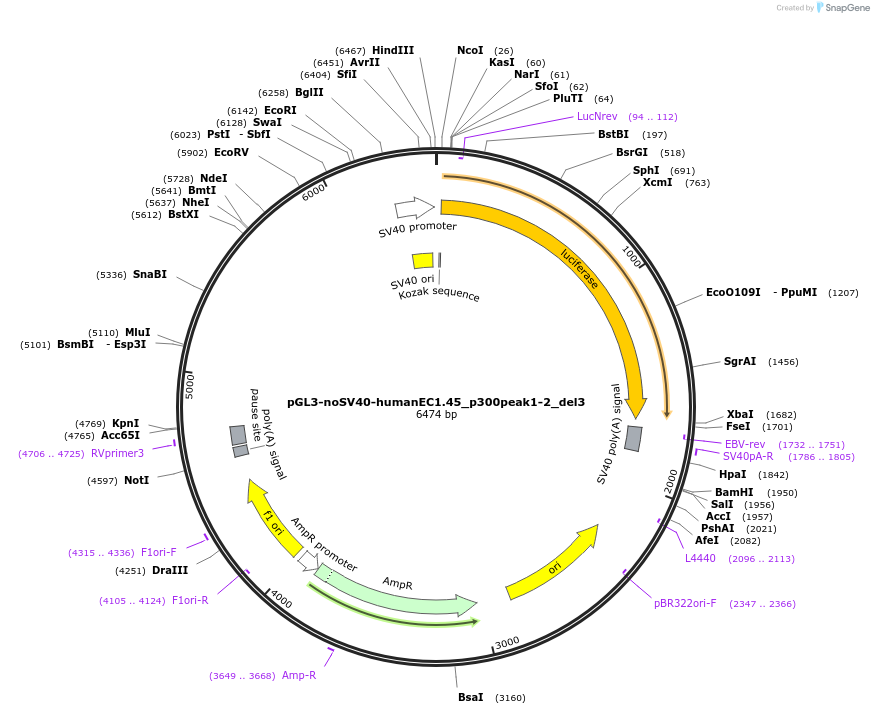
pGL3-noSV40-humanEC1.45_p300peak1-2_del3
Plasmid#173975PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #3
UseLuciferaseMutationTiled deletion #3PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
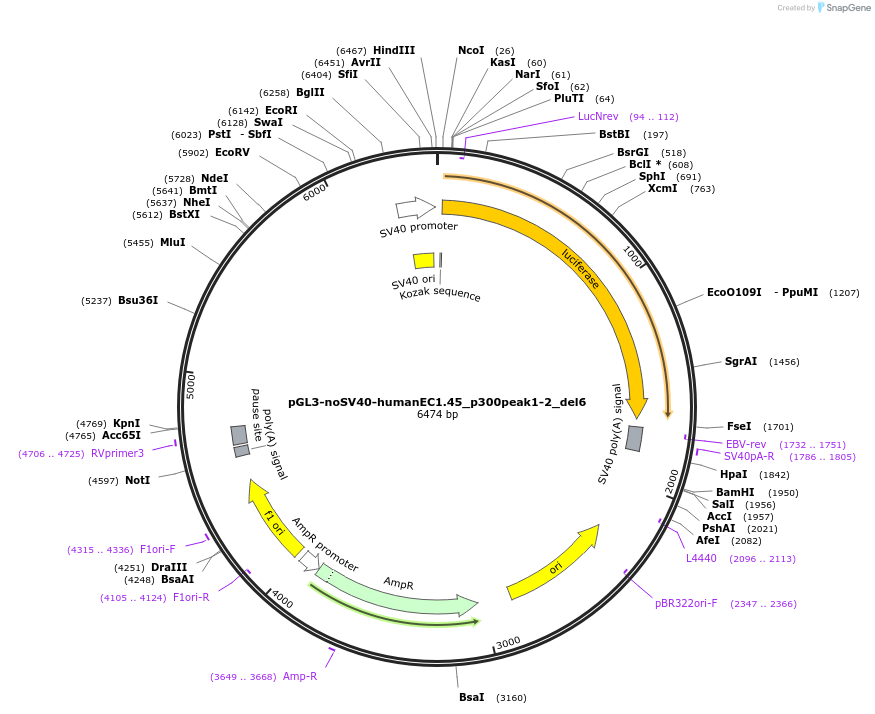
pGL3-noSV40-humanEC1.45_p300peak1-2_del6
Plasmid#173978PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #6
UseLuciferaseMutationTiled deletion #6PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
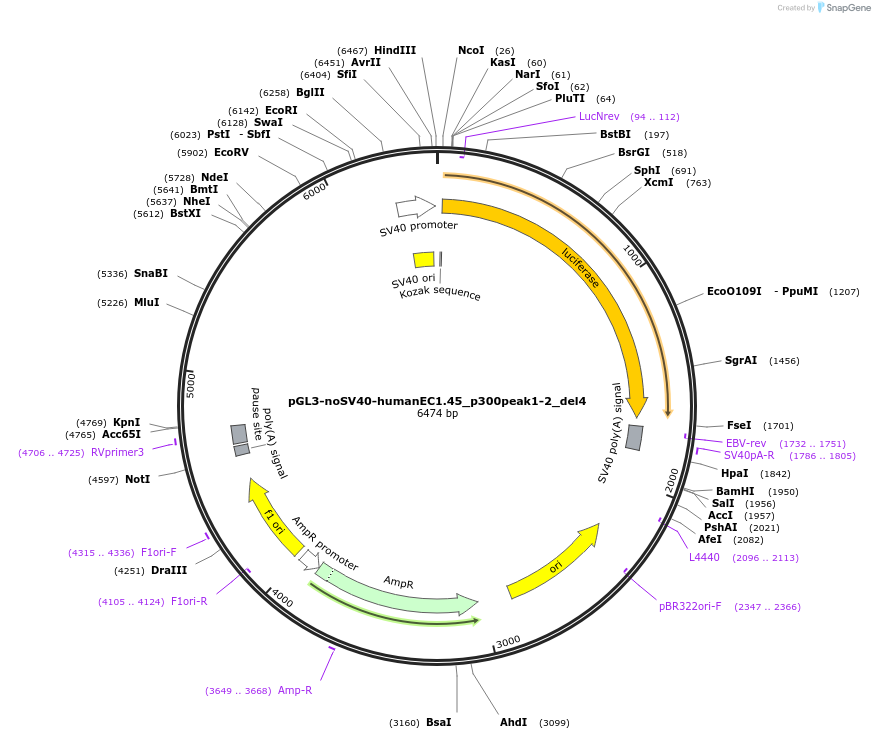
pGL3-noSV40-humanEC1.45_p300peak1-2_del4
Plasmid#173976PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #4
UseLuciferaseMutationTiled deletion #4PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
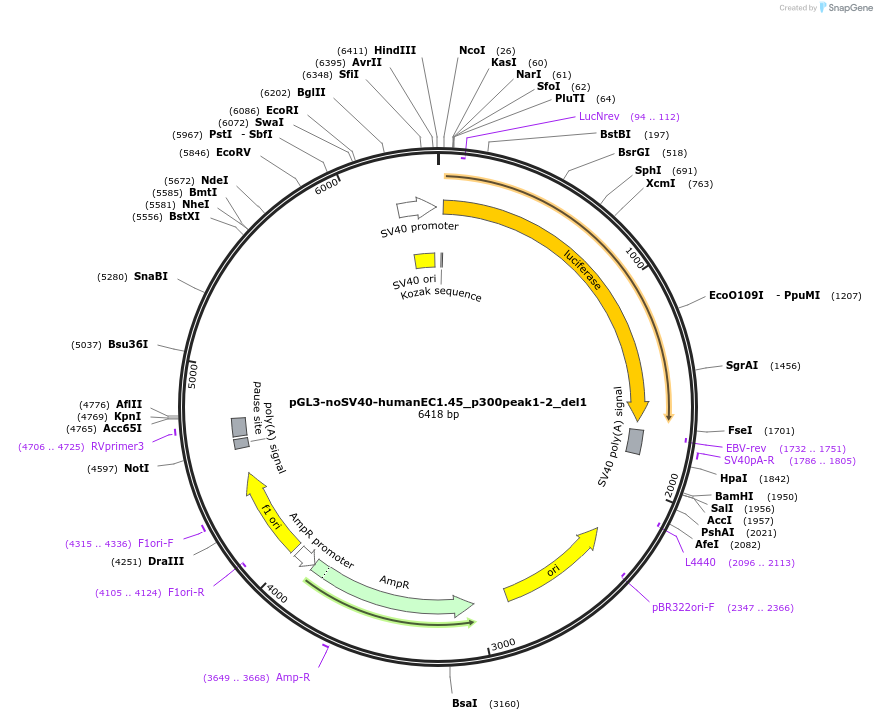
pGL3-noSV40-humanEC1.45_p300peak1-2_del1
Plasmid#173973PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #1
UseLuciferaseMutationTiled deletion #1PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
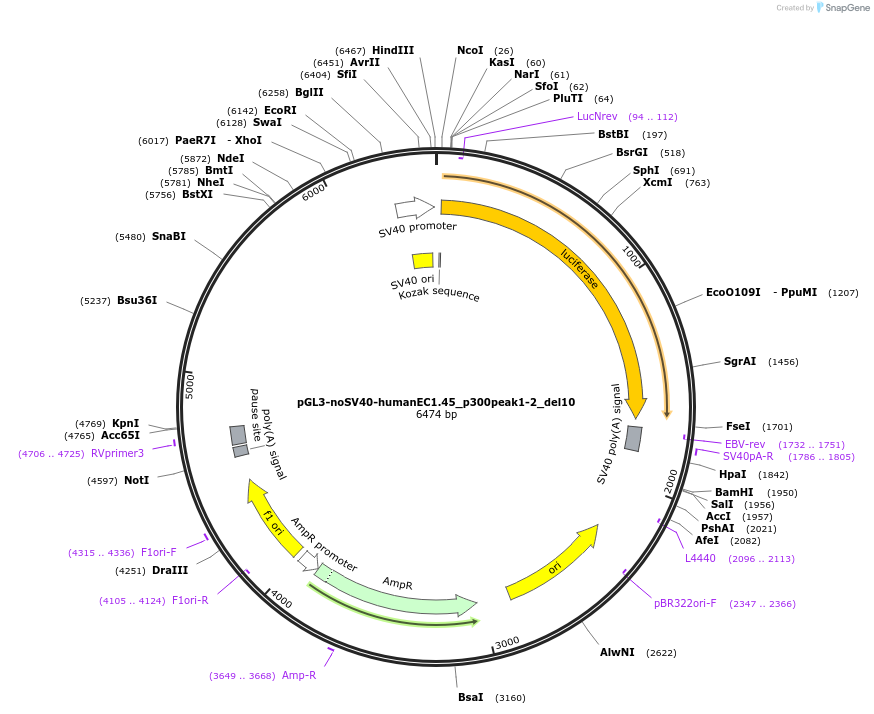
pGL3-noSV40-humanEC1.45_p300peak1-2_del10
Plasmid#173982PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #10
UseLuciferaseMutationTiled deletion #10PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
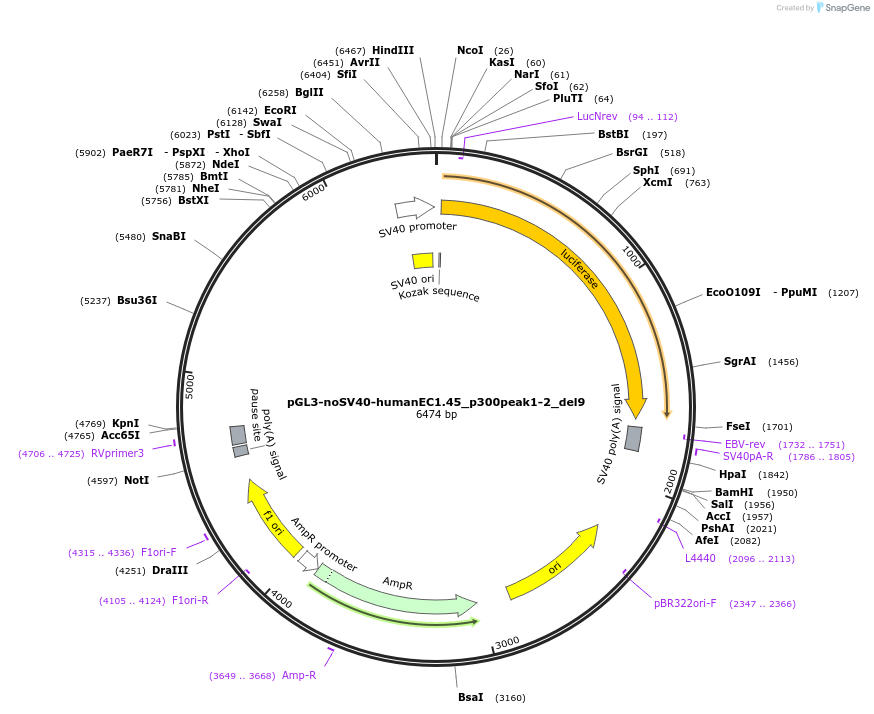
pGL3-noSV40-humanEC1.45_p300peak1-2_del9
Plasmid#173981PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #9
UseLuciferaseMutationTiled deletion #9PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
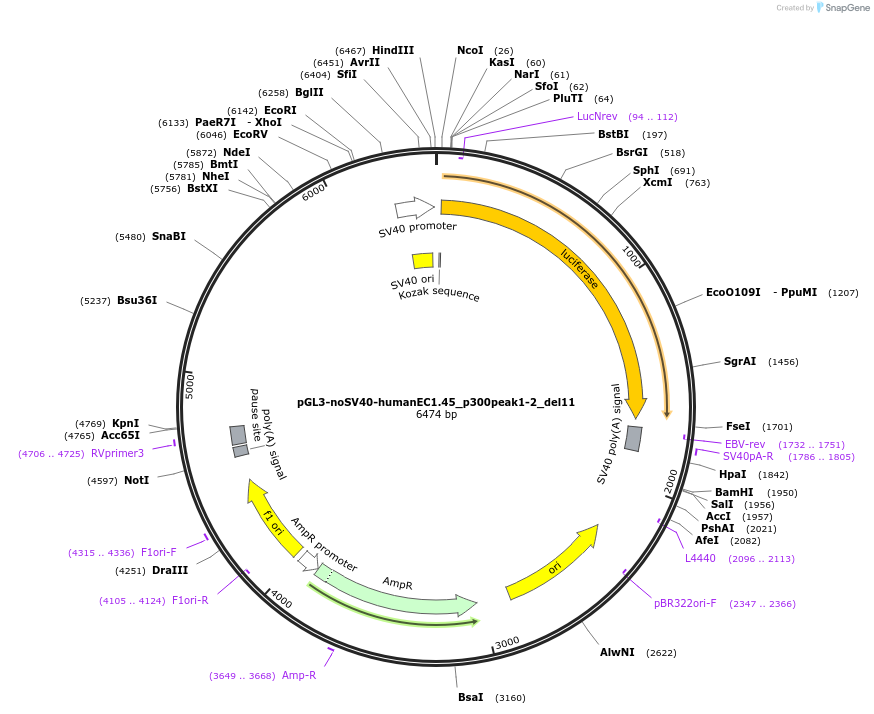
pGL3-noSV40-humanEC1.45_p300peak1-2_del11
Plasmid#173983PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #11
UseLuciferaseMutationTiled deletion #11PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
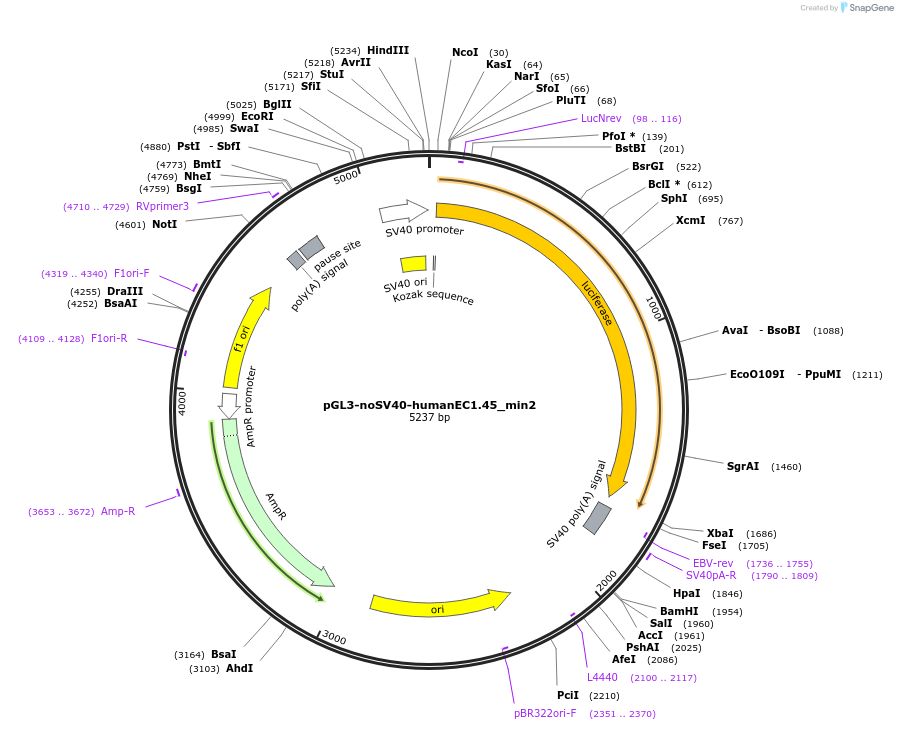
pGL3-noSV40-humanEC1.45_min2
Plasmid#173951PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertMinimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
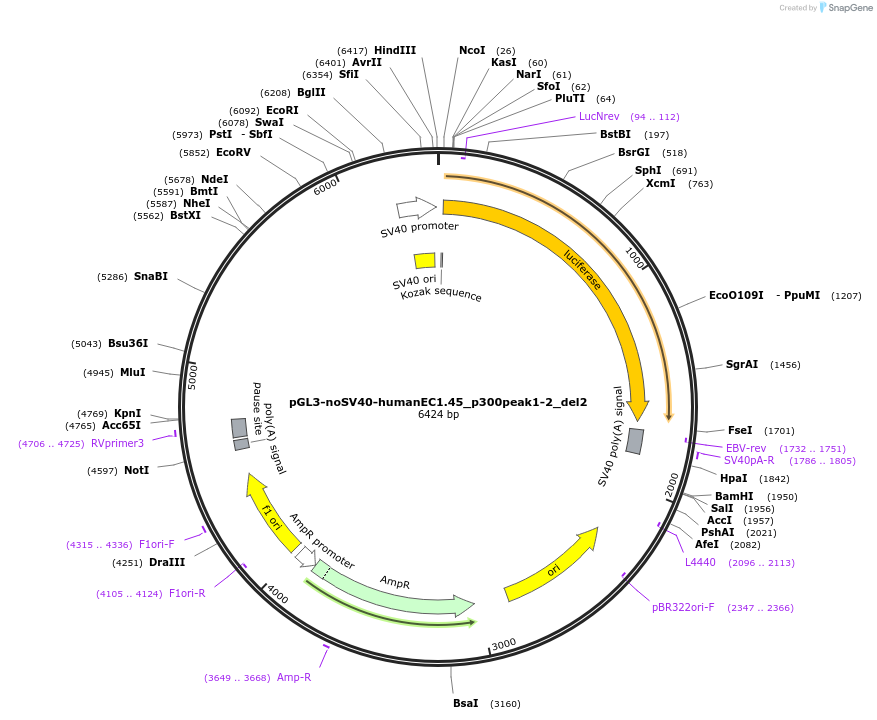
pGL3-noSV40-humanEC1.45_p300peak1-2_del2
Plasmid#173974PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #2
UseLuciferaseMutationTiled deletion #2PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
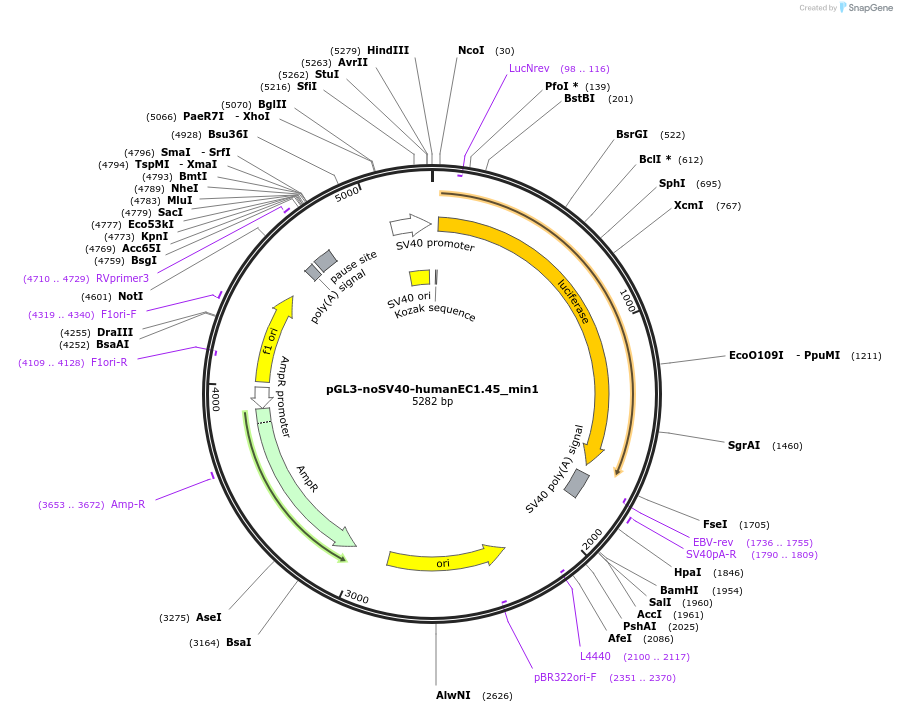
pGL3-noSV40-humanEC1.45_min1
Plasmid#173950PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertMinimal enhancer region 1 (min1) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
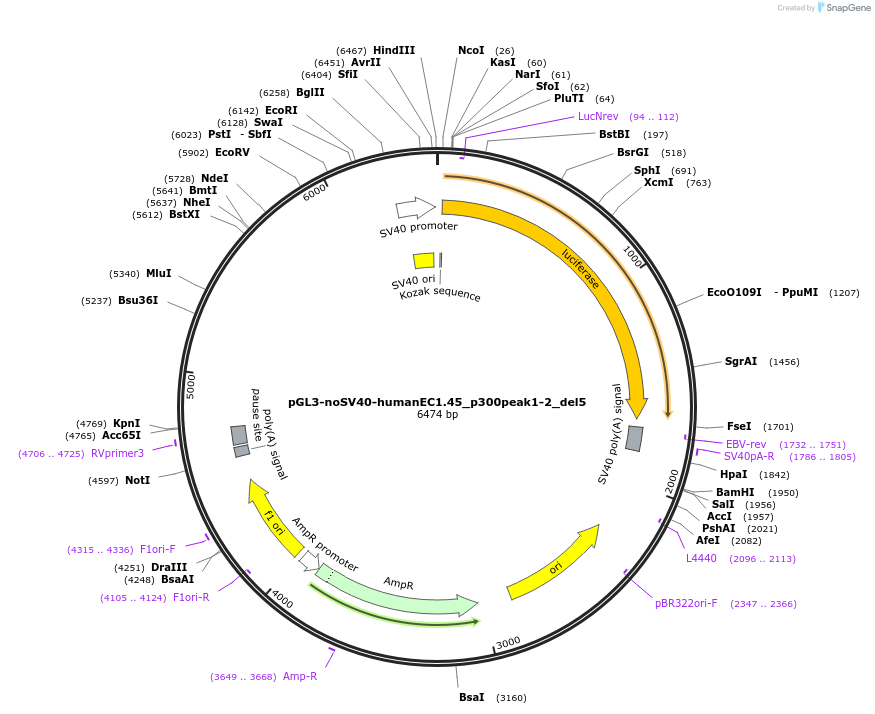
pGL3-noSV40-humanEC1.45_p300peak1-2_del5
Plasmid#173977PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #5
UseLuciferaseMutationTiled deletion #5PromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
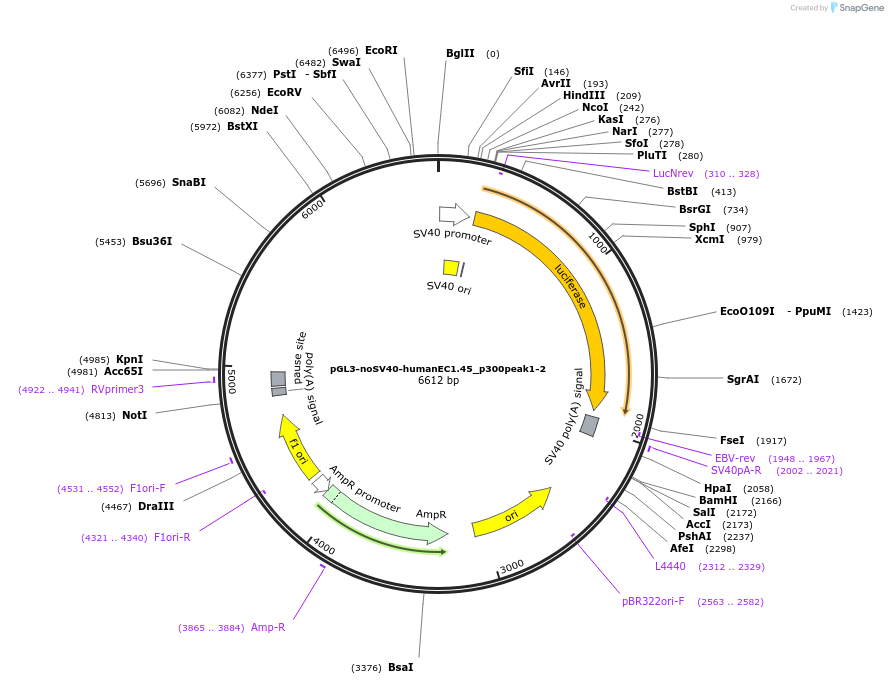
pGL3-noSV40-humanEC1.45_p300peak1-2
Plasmid#173949PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
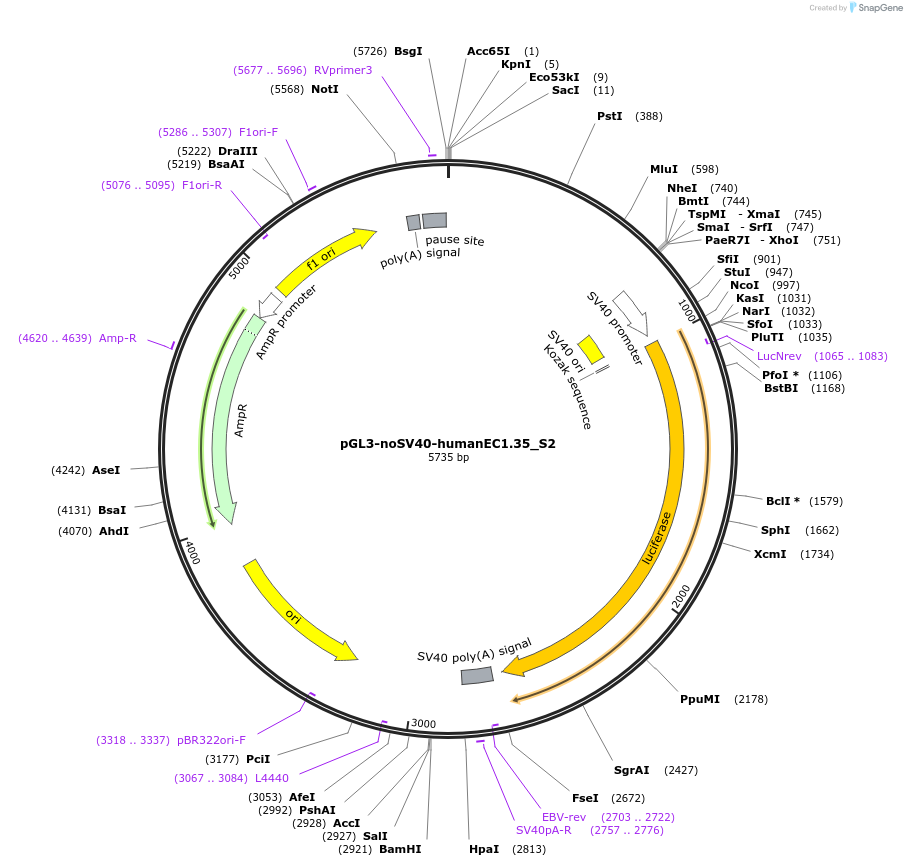
pGL3-noSV40-humanEC1.35_S2
Plasmid#173955PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertEnhancer region 2 (S2) from EC1.35 SOX9 enhancer cluster EC1.35
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
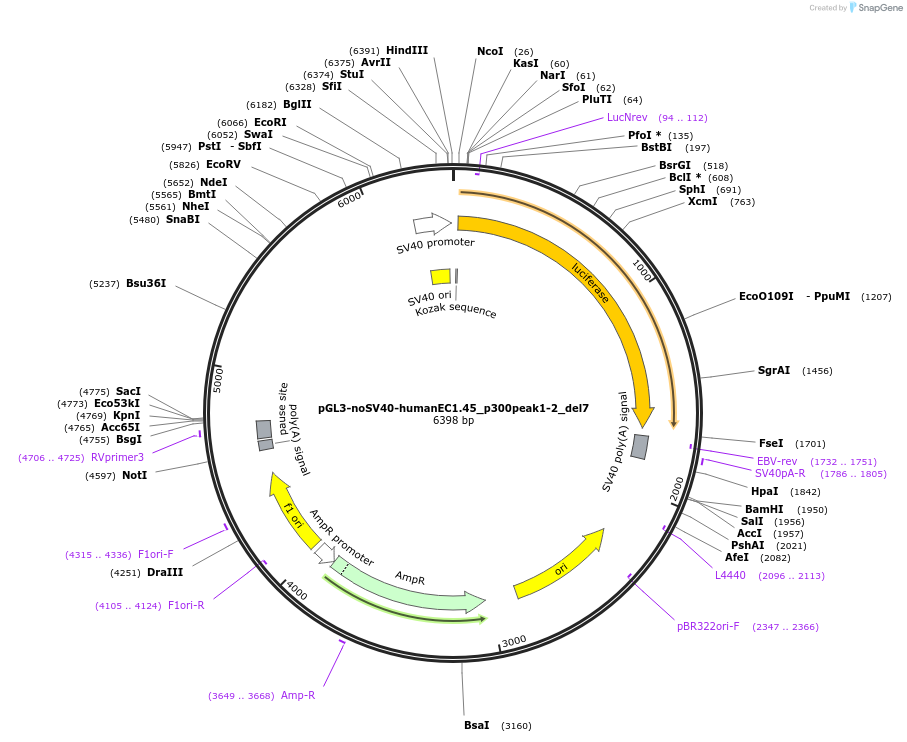
pGL3-noSV40-humanEC1.45_p300peak1-2_del7
Plasmid#173979PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #7
UseLuciferaseMutationTiled deletion #7PromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
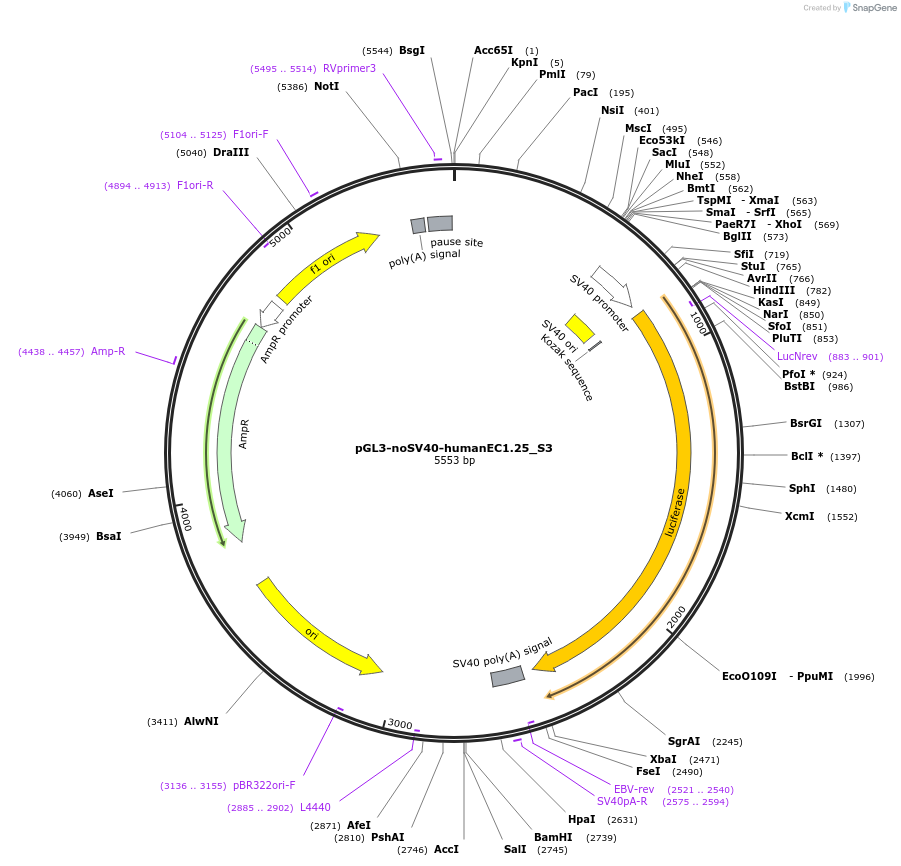
pGL3-noSV40-humanEC1.25_S3
Plasmid#173957PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertEnhancer region 3 (S3) from SOX9 enhancer cluster EC1.25
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
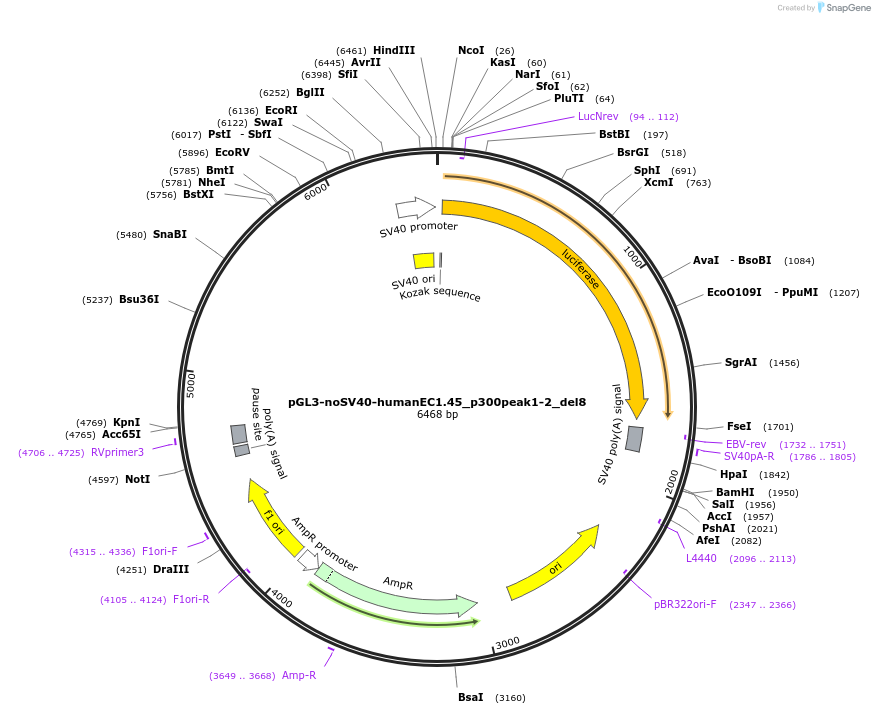
pGL3-noSV40-humanEC1.45_p300peak1-2_del8
Plasmid#173980PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with tiled deletion #8
UseLuciferaseMutationTiled deletion #8PromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
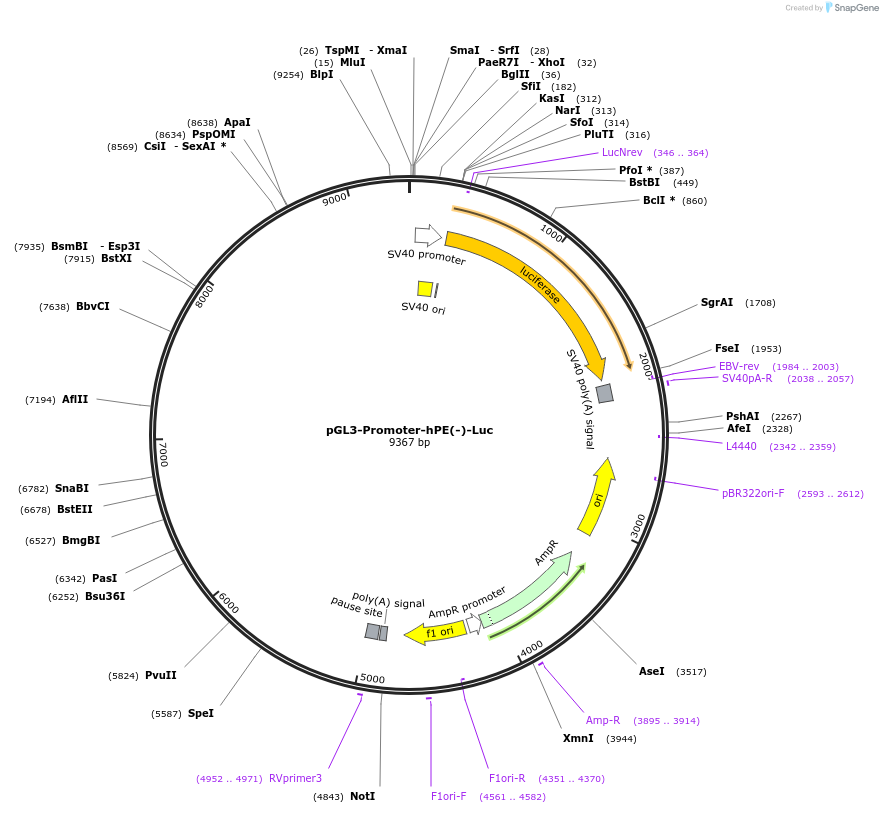
pGL3-Promoter-hPE(-)-Luc
Plasmid#172004PurposeLuciferase Reporter for Human PTEN Enhancer PE - Reverse OrientationDepositorInsertHuman PE sequence (hPE)
UseLuciferaseExpressionBacterial and MammalianPromoterSV40 PromoterAvailable SinceJuly 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGL3-Promoter-mPE(-)-Luc
Plasmid#172006PurposeLuciferase Reporter for Mouse PTEN Enhancer PE - Reverse OrientationDepositorInsertMouse PE sequence (mPE)
UseLuciferaseExpressionBacterial and MammalianPromoterSV40 PromoterAvailable SinceJuly 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
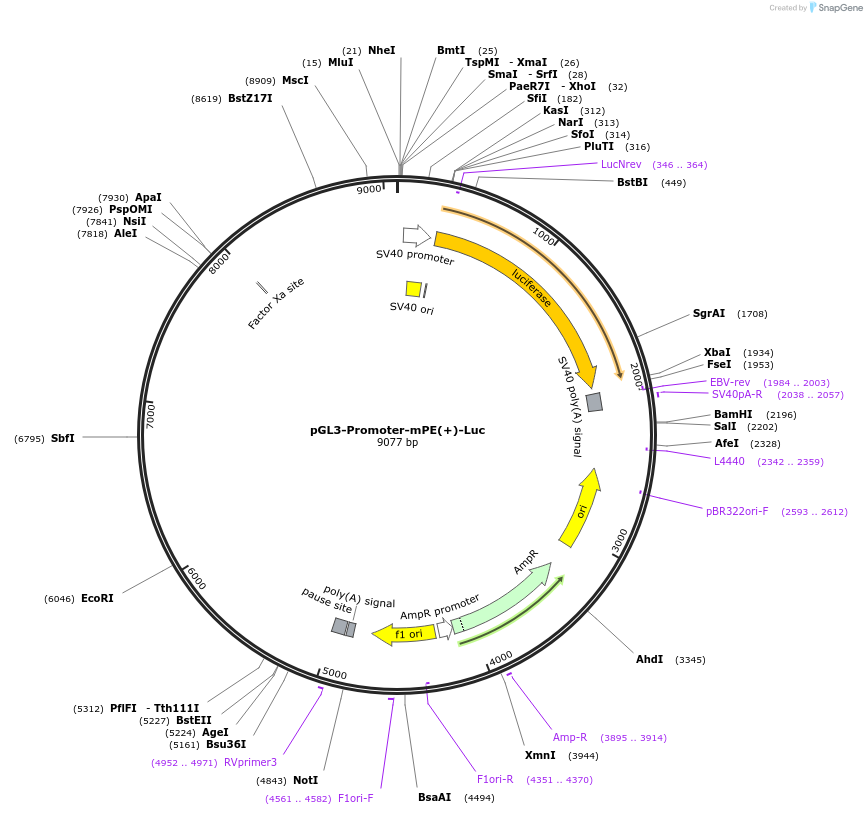
pGL3-Promoter-mPE(+)-Luc
Plasmid#172005PurposeLuciferase Reporter for Mouse PTEN Enhancer PE - Forward OrientationDepositorInsertMouse PE sequence (mPE)
UseLuciferaseExpressionBacterial and MammalianPromoterSV40 PromoterAvailable SinceJuly 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
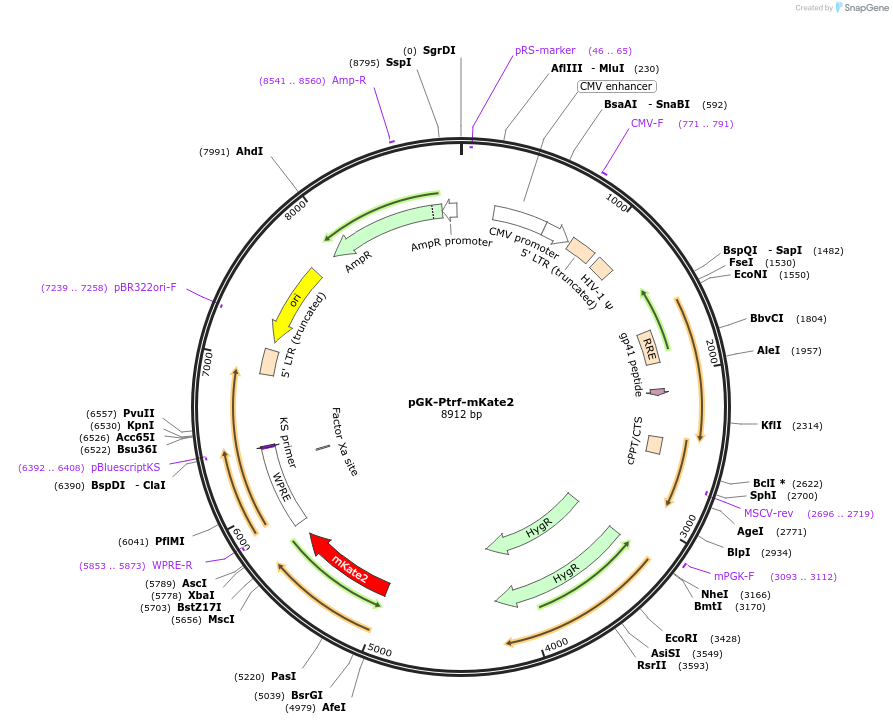
pGK-Ptrf-mKate2
Plasmid#128765PurposeFluorescent mKate reporter for PtrfDepositorAvailable SinceNov. 5, 2020AvailabilityAcademic Institutions and Nonprofits only -
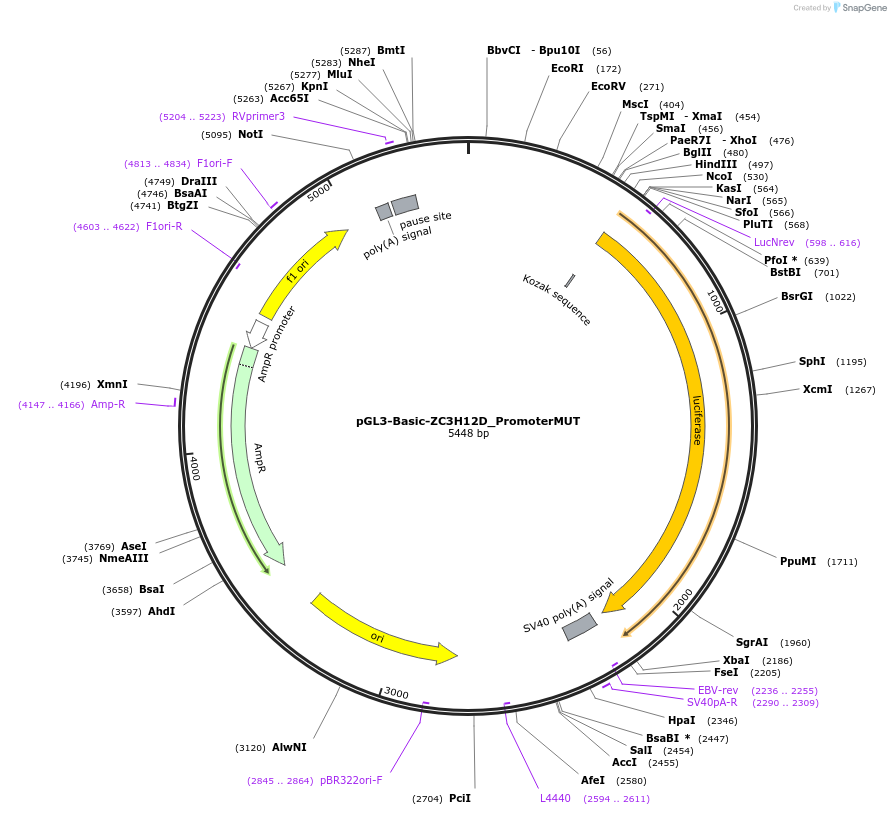
pGL3-Basic-ZC3H12D_PromoterMUT
Plasmid#153067PurposeZC3H12D promoter region carrying mutations in BHLHE40 binding sites, cloned upstream of luciferase reporterDepositorInsertZC3H12D promoter region mutated in BHLHE40 binding sites (ZC3H12D Human)
UseLuciferaseExpressionMammalianMutationMutagenesis to disrupt 3 BHLHE4040 binding sites.…PromoterNoneAvailable SinceSept. 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
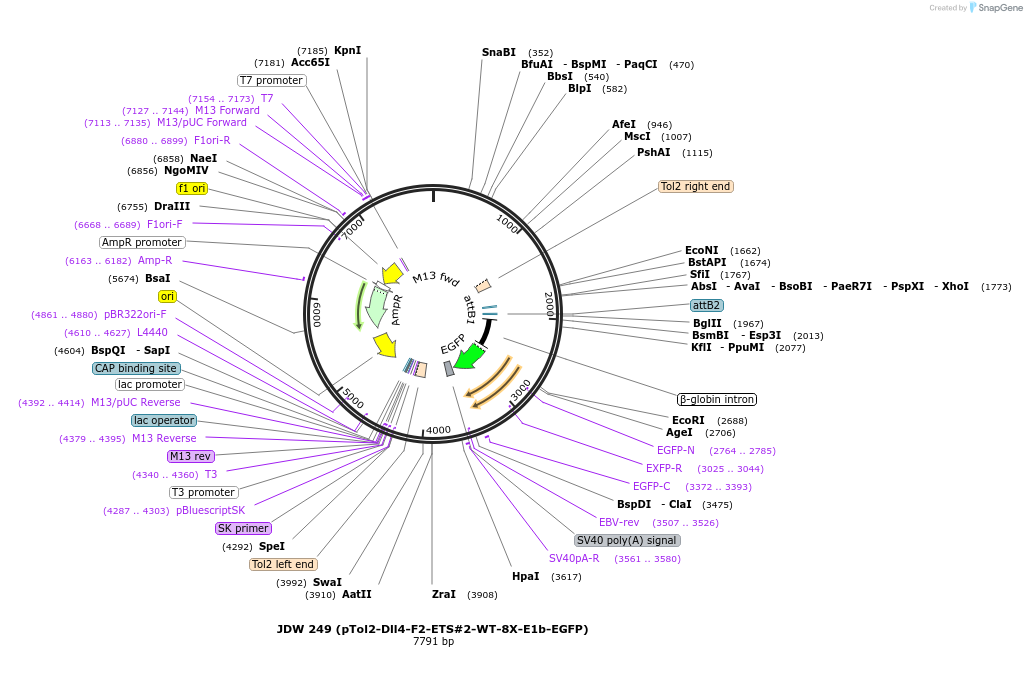
JDW 249 (pTol2-Dll4-F2-ETS#2-WT-8X-E1b-EGFP)
Plasmid#156417Purpose8X copies of the ETS#2 site of the murine Dll4 intronic enhancer and a minimal E1b reporter driving expression of EGFP flanked by Tol2 sitesDepositorInsertmurine Dll4 F2-6/F8 ETS site B/ site #2, 8X
UseZebrafish transgenesisPromoterE1b min pro/b-globin intronAvailable SinceAug. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
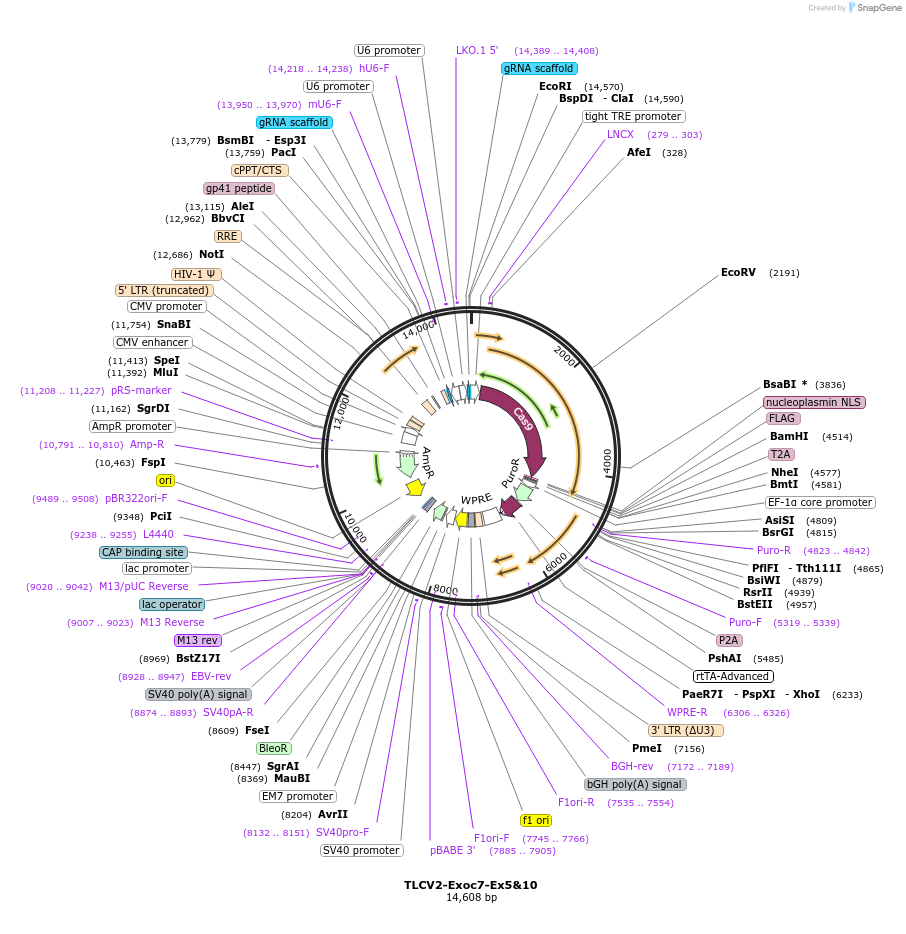
TLCV2-Exoc7-Ex5&10
Plasmid#133303PurposeEGFP reporter sequence was removed from TLCV2 vector and two gRNAs targeting mouse Exoc7 gene were cloned into the modified TLCV2 vector.DepositorInsertExocyst complex component 7 (Exoc7 Mouse)
UseCRISPR and LentiviralExpressionMammalianPromotermU6 promoterAvailable SinceNov. 15, 2019AvailabilityAcademic Institutions and Nonprofits only -
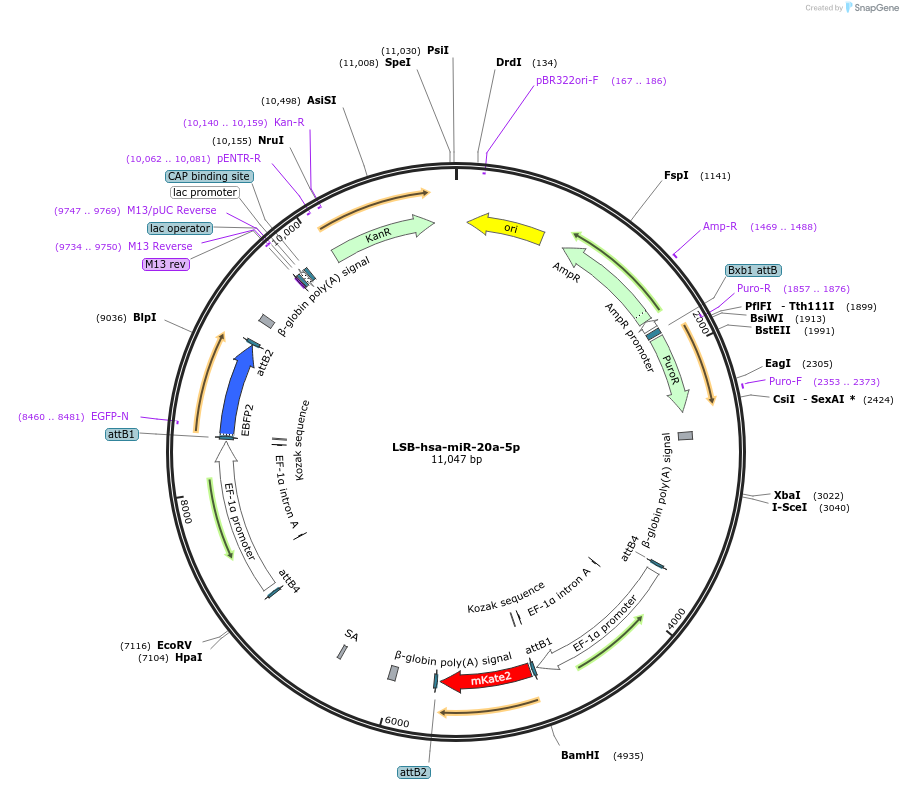
LSB-hsa-miR-20a-5p
Plasmid#103342PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-20a-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-20a-5p target (MIR20A Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
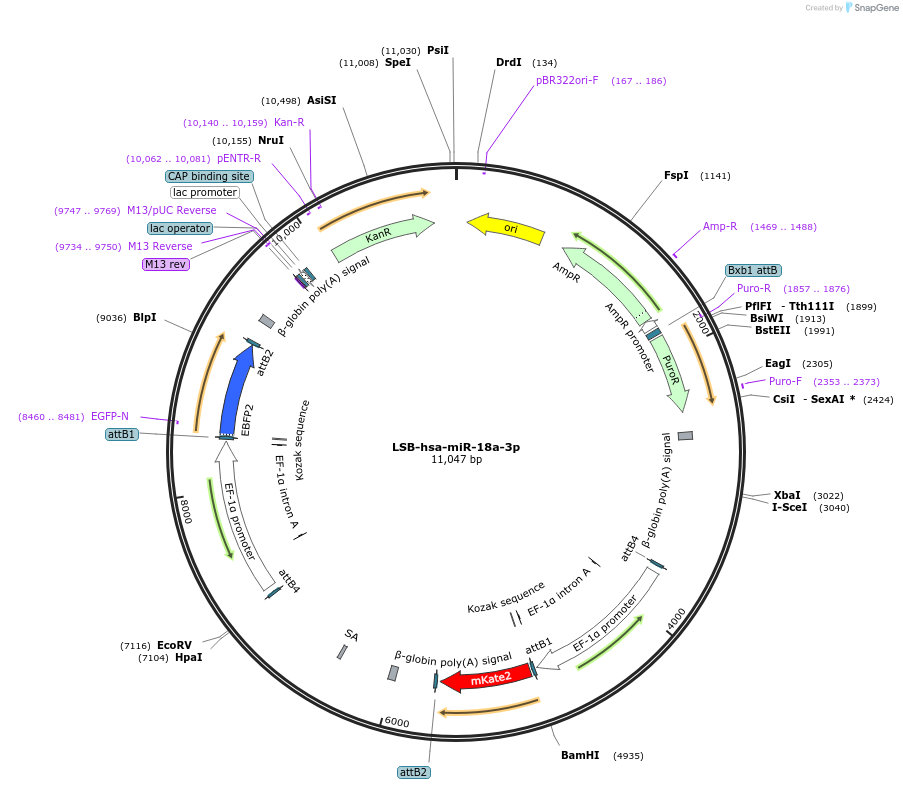
LSB-hsa-miR-18a-3p
Plasmid#103292PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-18a-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-18a-3p target (MIR18A Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
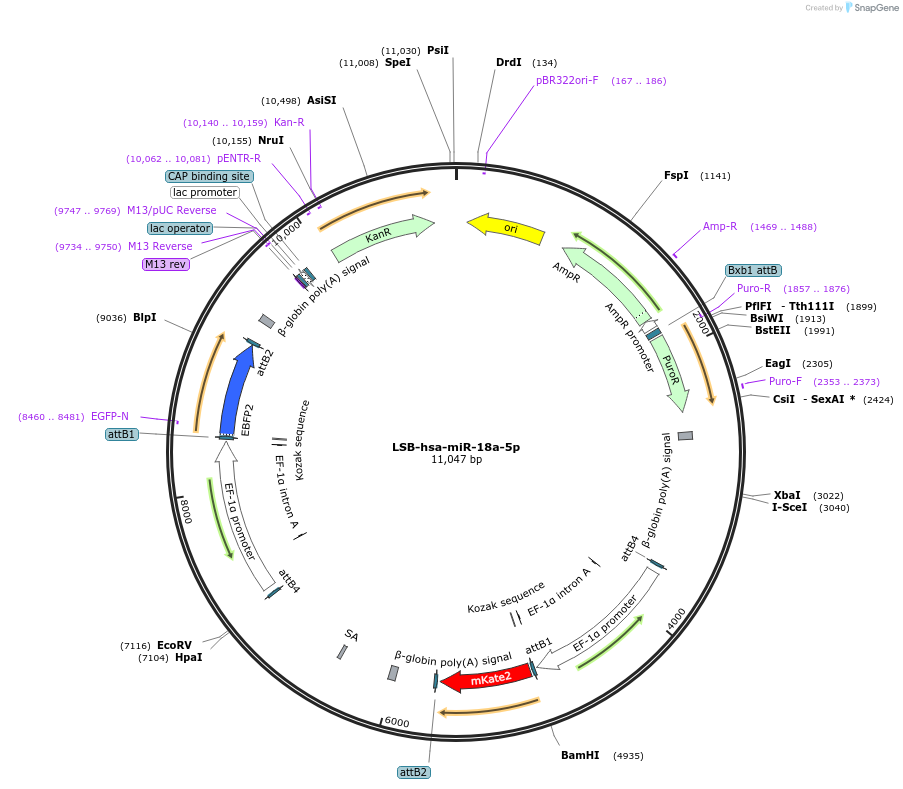
LSB-hsa-miR-18a-5p
Plasmid#103293PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-18a-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-18a-5p target (MIR18A Human)
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
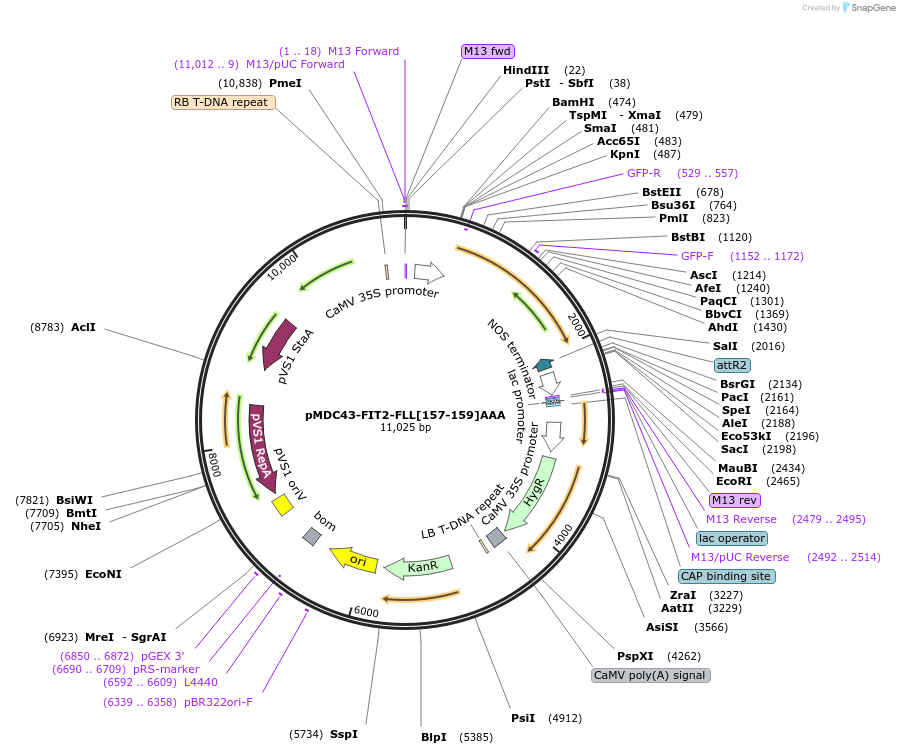
pMDC43-FIT2-FLL[157-159]AAA
Plasmid#96994PurposeExpress GFP-tagged mutated mouse FIT2 gene in plants.DepositorInsertMmFIT2 (Fitm2 Mouse)
TagsGFPExpressionPlantMutationAmino acid residues 157-159 were mutated (FLL[157…Promoter35SAvailable SinceAug. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
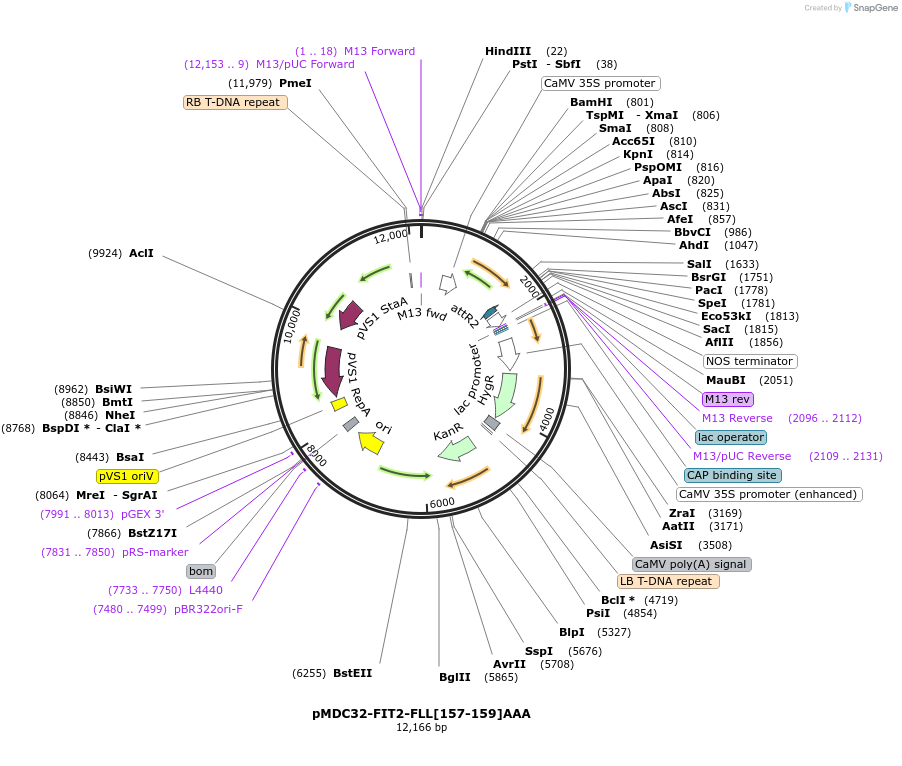
pMDC32-FIT2-FLL[157-159]AAA
Plasmid#96991PurposeExpress mouse FIT2 gene in plants.DepositorInsertMmFIT2 (Fitm2 Mouse)
ExpressionPlantMutationAmino acid residues 157-159 were mutated (FLL[157…Promoter35SAvailable SinceAug. 17, 2017AvailabilityAcademic Institutions and Nonprofits only -
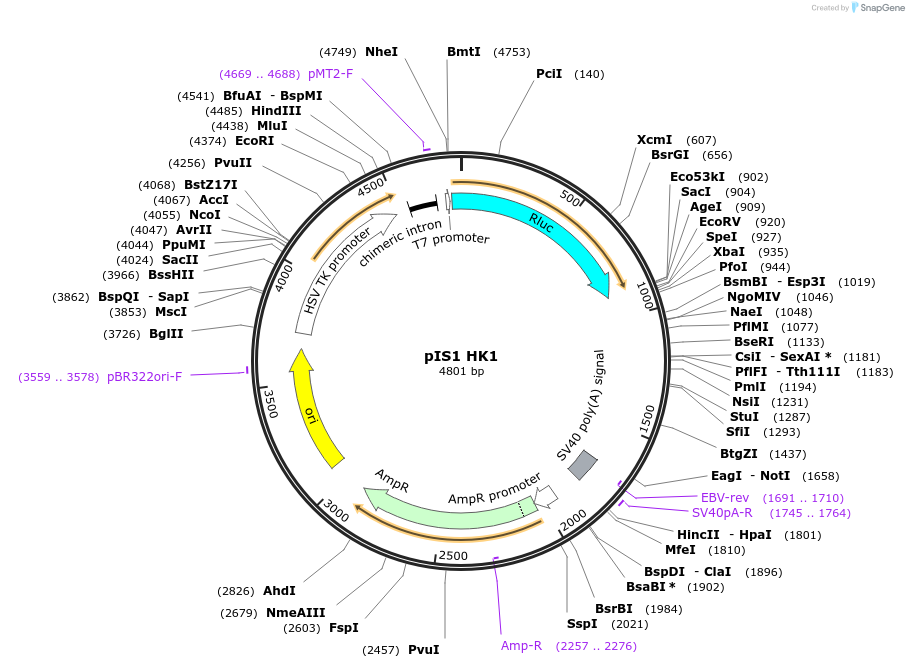
pIS1 HK1
Plasmid#60794PurposeLuciferase reporter to measure miR-155 mediated differential repression. Contains HK1 3' UTR and wild-type miR-155 sitesDepositorInsertHK1 3'UTR and wild-type miR-155 binding site (HK1 Human)
UseLuciferaseExpressionMammalianPromoterthymidine kinase (HSV-TK promoter)Available SinceOct. 21, 2014AvailabilityAcademic Institutions and Nonprofits only -
pIS1 mHK1
Plasmid#60795PurposeLuciferase reporter to measure miR-155 mediated differential repression. Contains HK1 3' UTR and mutated miR-155 sitesDepositorInsertHK1 3'UTR and mutated miR-155 binding site (HK1 Human)
UseLuciferaseExpressionMammalianMutationmutated miR-155 binding sitePromoterthymidine kinase (HSV-TK promoter)Available SinceOct. 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
pMiR-CIP2A-stop-mut
Plasmid#53713PurposeLuciferase reporter assay for CIP2A with mutated firefly stop codonDepositorInsertCIP2A (CIP2A Human)
UseLuciferaseMutationMutation on the stop codon of firefly luciferase …Available SinceJuly 2, 2014AvailabilityAcademic Institutions and Nonprofits only