We narrowed to 5,873 results for: org
-
Plasmid#174769PurposeEntry clone for Golden Gate assemblyDepositorInsertGG_NbZAR1syn-L17E/D481V without STOP codon
UseUnspecifiedMutationL17E/D481VAvailable SinceOct. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
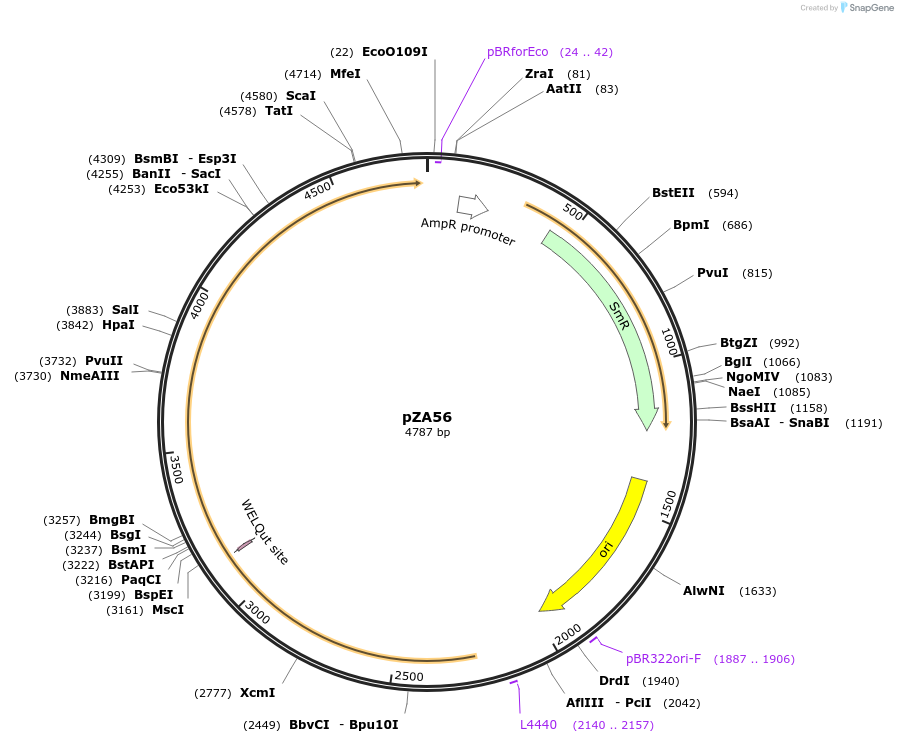
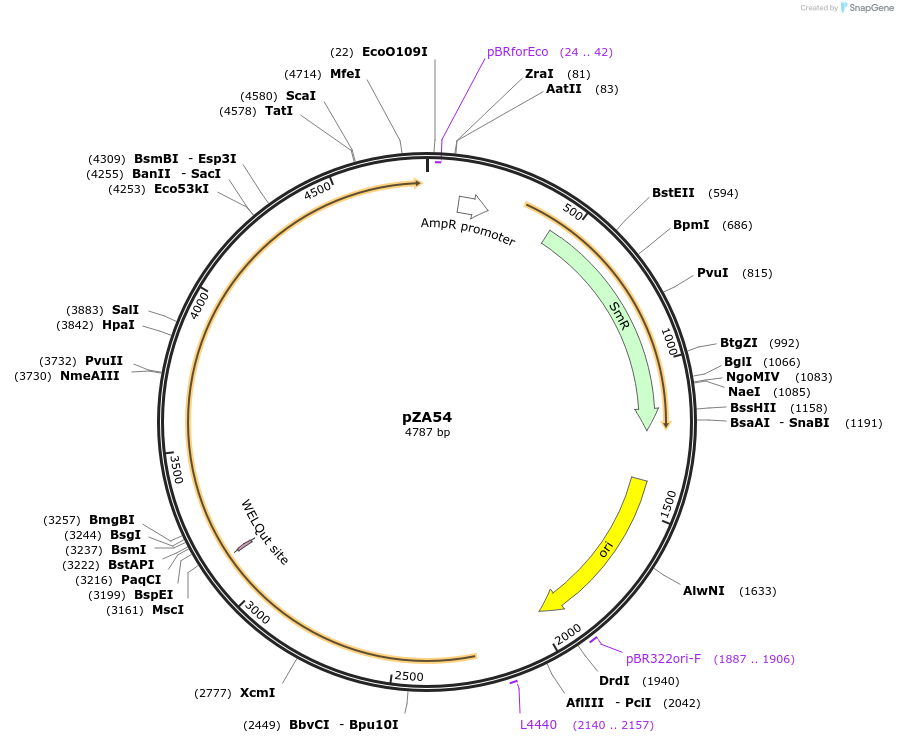
pZA54
Plasmid#174767PurposeEntry clone for Golden Gate assemblyDepositorInsertGG_NbZAR1syn-D481V without STOP codon
UseUnspecifiedMutationD481VAvailable SinceOct. 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
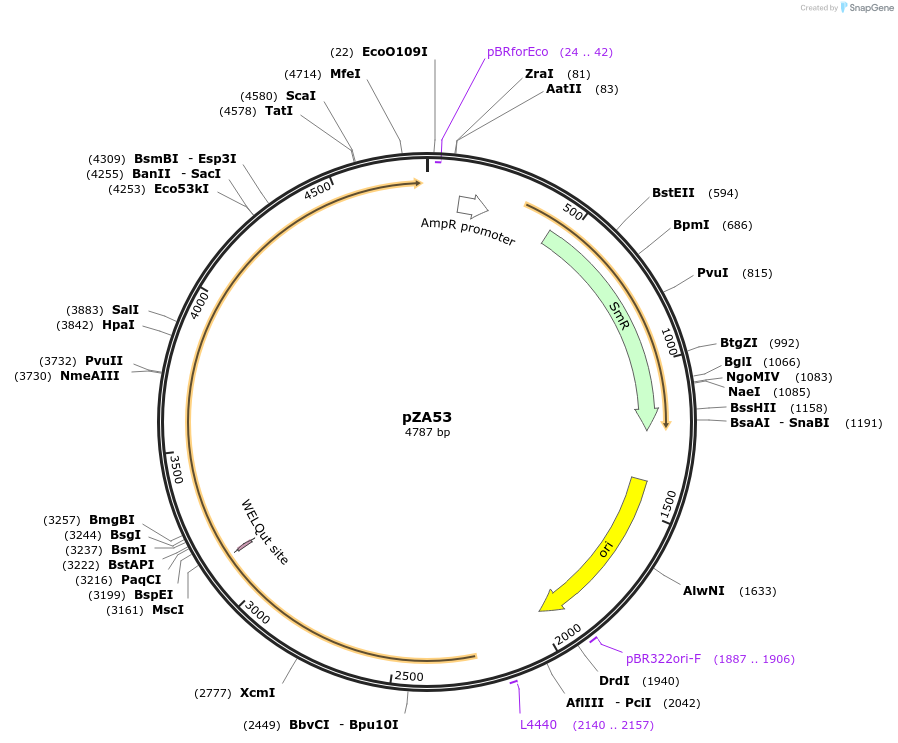
pZA53
Plasmid#174766PurposeEntry clone for Golden Gate assemblyDepositorInsertGG_NbZAR1syn without STOP codon
UseUnspecifiedMutationWTAvailable SinceOct. 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
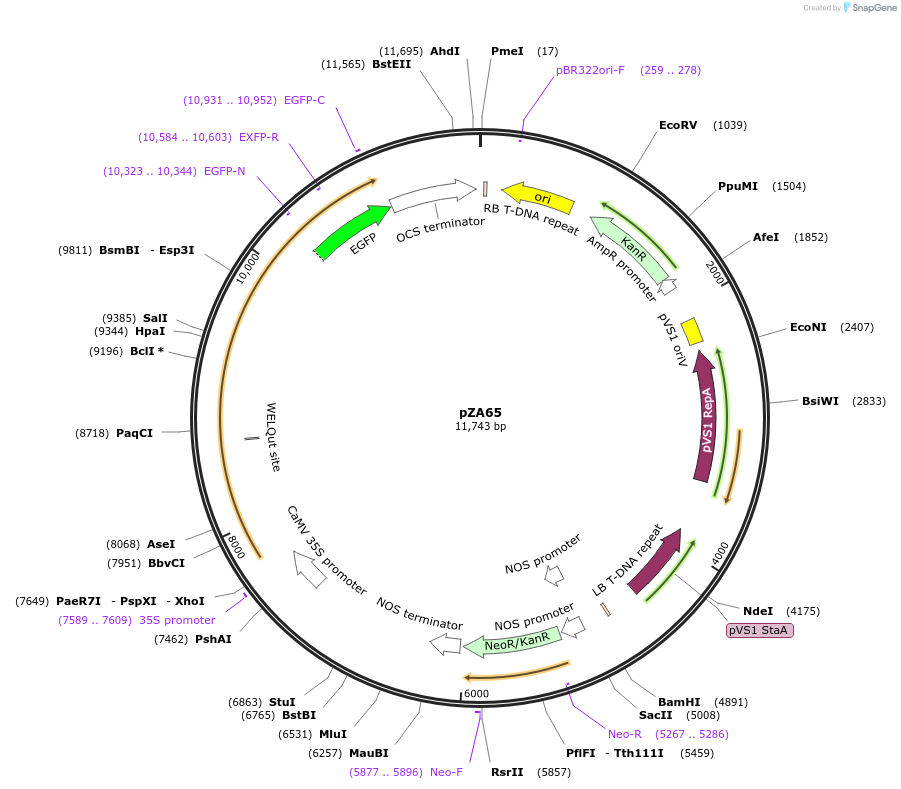
pZA65
Plasmid#174778PurposeIn planta gene expressionDepositorInsertGG_NbZAR1syn-eGFP, Kmr plant selection marker
TagseGFPExpressionPlantMutationWTAvailable SinceOct. 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
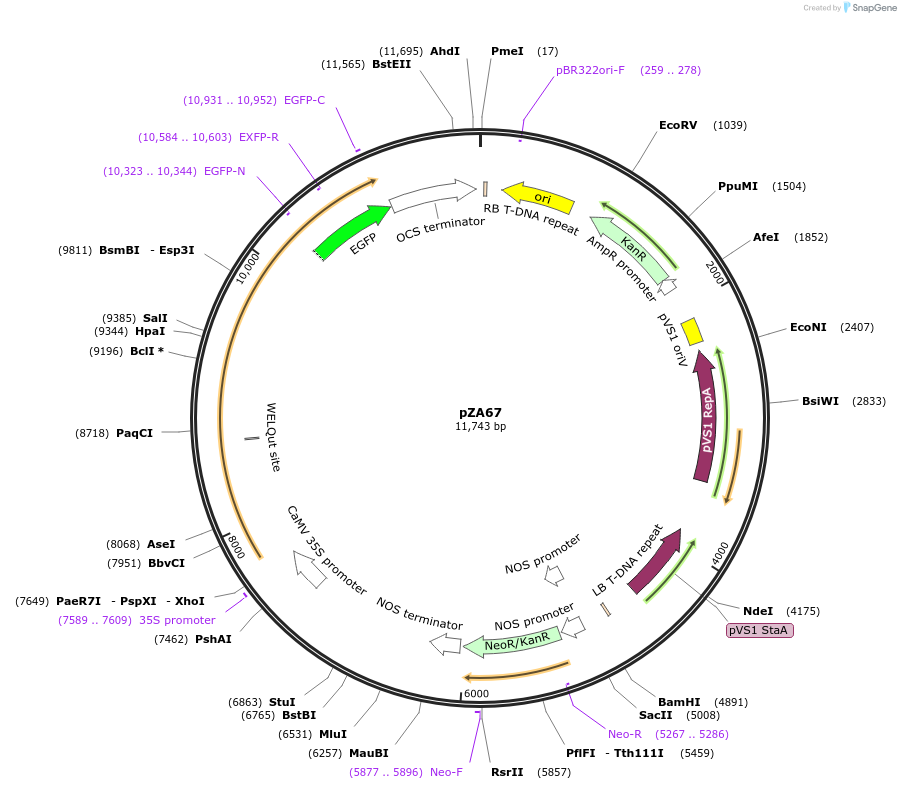
pZA67
Plasmid#174780PurposeIn planta gene expressionDepositorInsertGG_NbZAR1syn-L17E-eGFP, Kmr plant selection marker
TagseGFPExpressionPlantMutationL17EAvailable SinceOct. 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
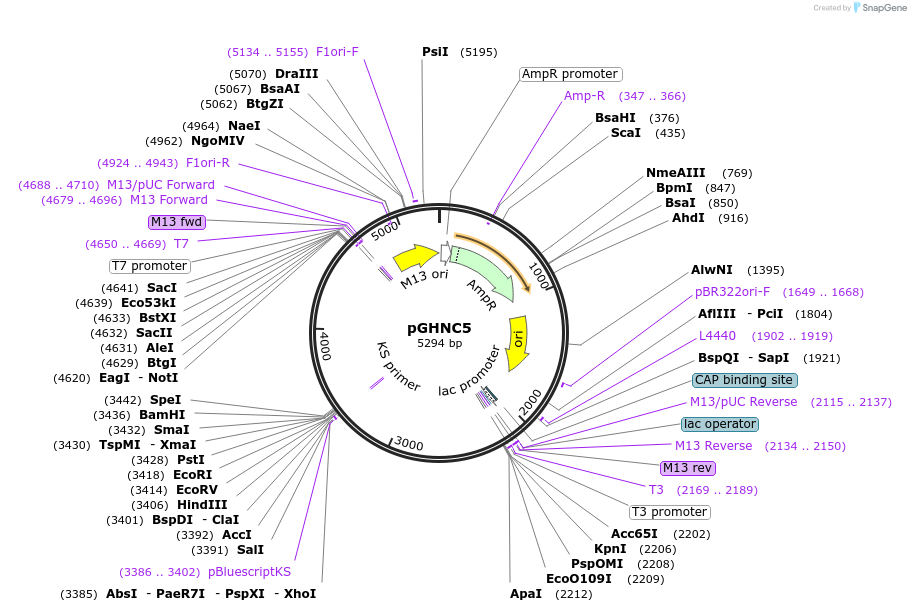
pGHNC5
Plasmid#24701DepositorInsertDirect Repeats of Tobacco Rb7-6 MARs
UseE. coli phagemidAvailable SinceJuly 8, 2010AvailabilityAcademic Institutions and Nonprofits only -
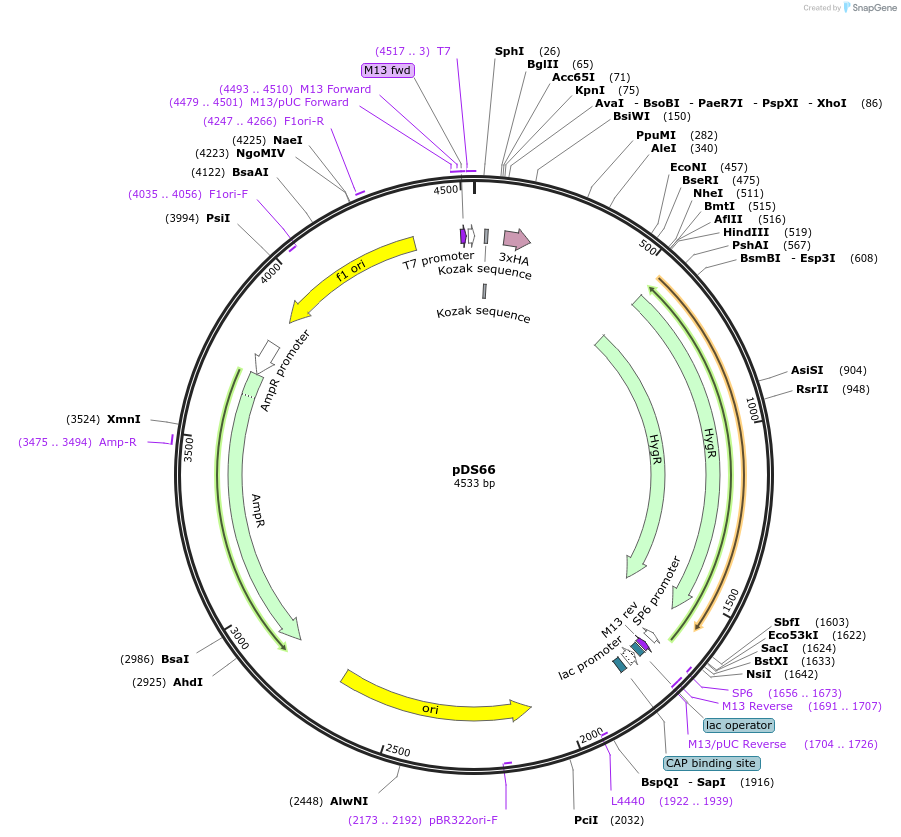
pDS66
Plasmid#48364Purpose3XHA-intergenic region of TUB-HYG (modified from PMID 16269191)DepositorInsert3XHA-intergenic region of TUB-HYG (modified from PMID 16269191)
UseCre/Lox; Knockout in t. bruceiAvailable SinceOct. 17, 2013AvailabilityAcademic Institutions and Nonprofits only -
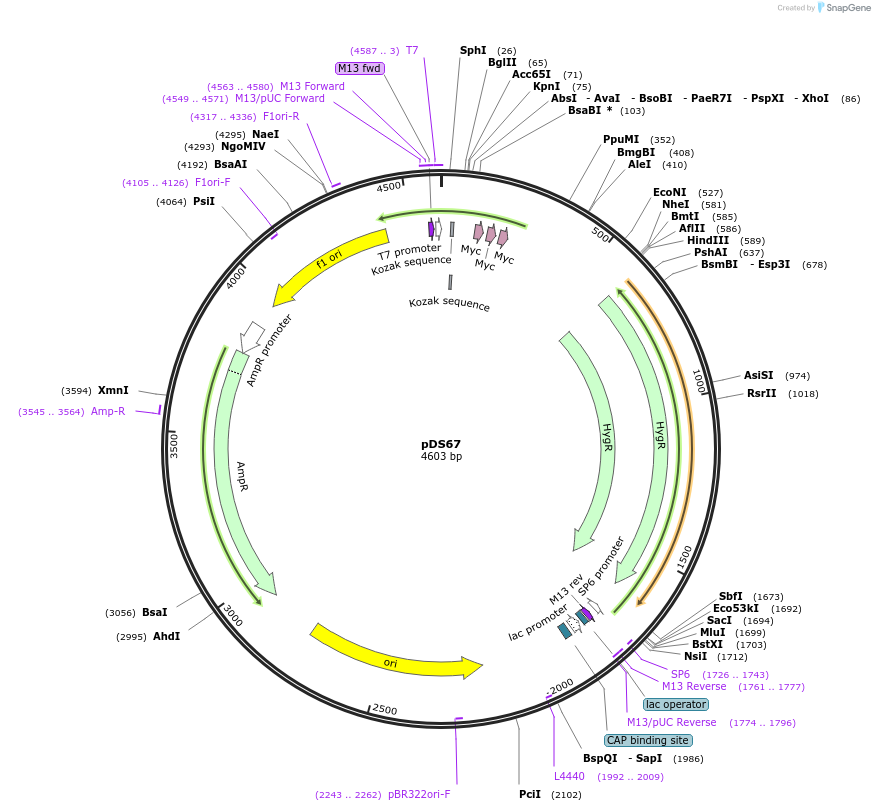
pDS67
Plasmid#48365Purpose3XMYC-intergenic region of TUB-HYG (modified from PMID 16269191)DepositorInsert3XMYC-intergenic region of TUB-HYG (modified from PMID 16269191)
UseCre/Lox; Knockout in t. bruceiAvailable SinceOct. 17, 2013AvailabilityAcademic Institutions and Nonprofits only -
pET23a-MmUBB
Plasmid#69556PurposeBacterial expression; in vitro transcription/translation (T7)DepositorAvailable SinceDec. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
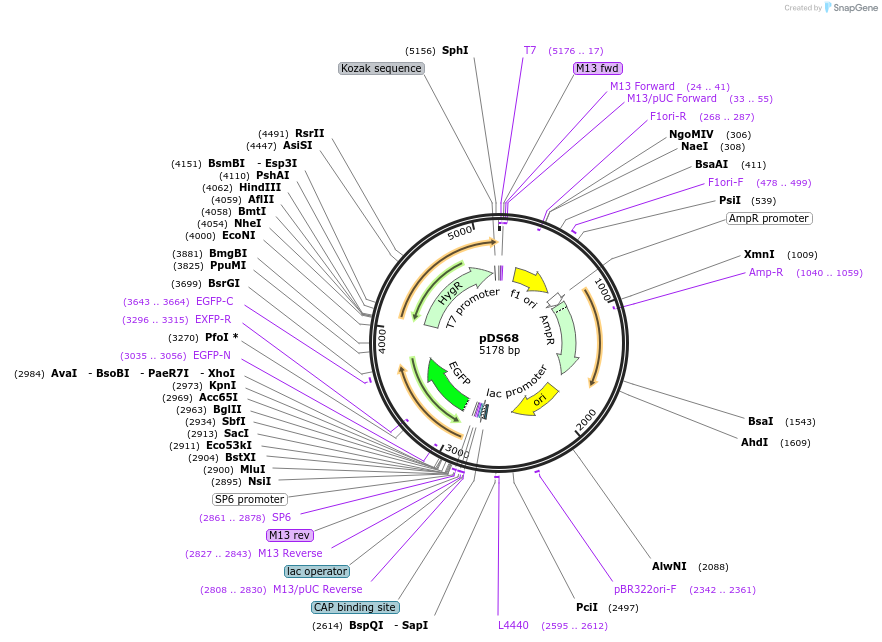
pDS68
Plasmid#48366PurposeGFP-intergenic region of TUB-HYG (modified from PMID 16269191)DepositorInsertGFP-intergenic region of TUB-HYG (modified from PMID 16269191)
UseCre/Lox; Knockout in t. bruceiAvailable SinceJan. 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
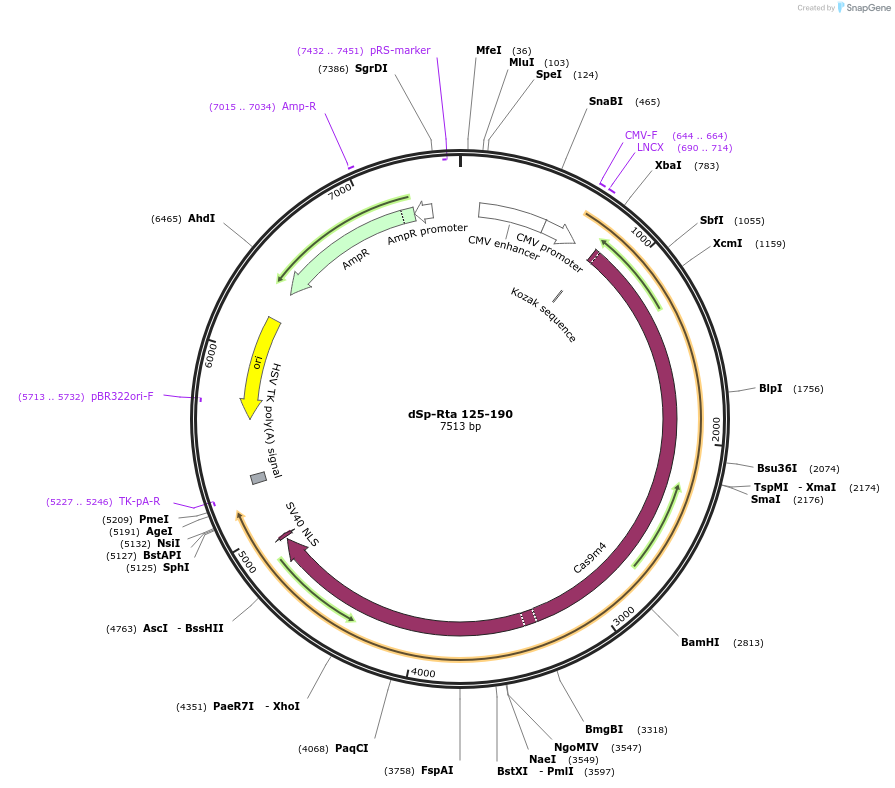
dSp-Rta 125-190
Plasmid#99664PurposeExpresses dSp Cas9 fused to truncated RTA activation domainDepositorArticleInsertdCas9
Tagstruncated RTA activation domain ( 125-190)ExpressionMammalianMutationdead Cas9Available SinceNov. 27, 2018AvailabilityAcademic Institutions and Nonprofits only -
pRB7-6
Plasmid#24700DepositorInsertTobacco Rb7 MAR (SAR)
UseE. coli phagemidAvailable SinceJuly 8, 2010AvailabilityAcademic Institutions and Nonprofits only -
pKlab676-HBEGF-2a-mWasabi-WPRE
Plasmid#249029PurposeThis plasmid expresses the mWasabi flourescent tag for integration into mammalian cell linesDepositorInsertmWasabi
UseCRISPRAvailable SinceJan. 21, 2026AvailabilityAcademic Institutions and Nonprofits only -
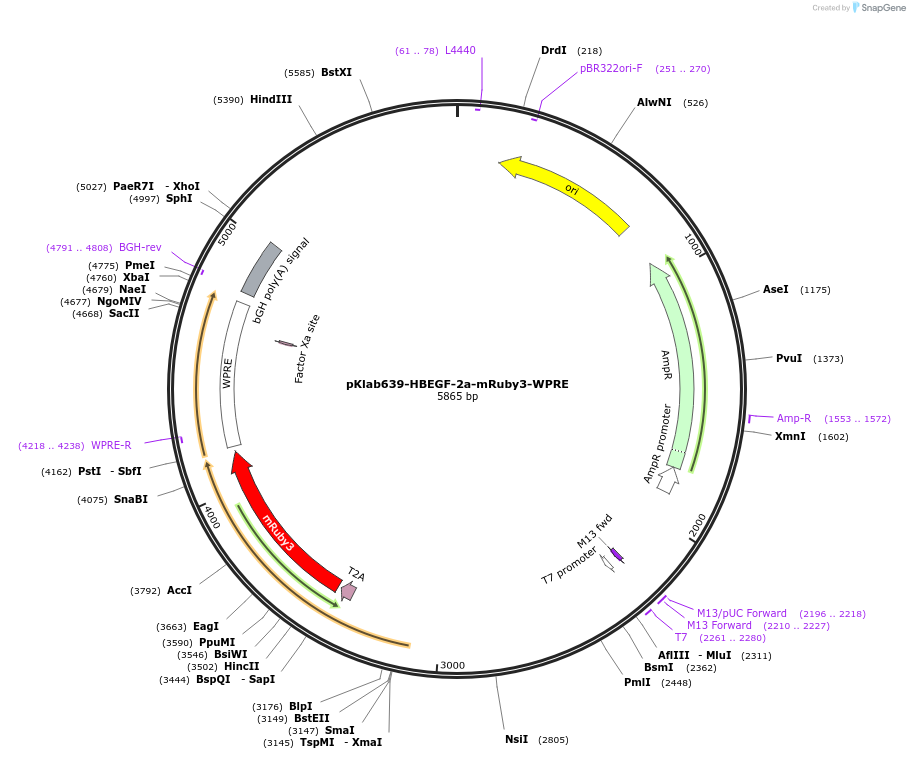
pKlab639-HBEGF-2a-mRuby3-WPRE
Plasmid#249030PurposeThis plasmid expresses the mRuby3 flourescent tag for integration into mammalian cell linesDepositorInsertmRuby3
UseCRISPRAvailable SinceJan. 21, 2026AvailabilityAcademic Institutions and Nonprofits only -
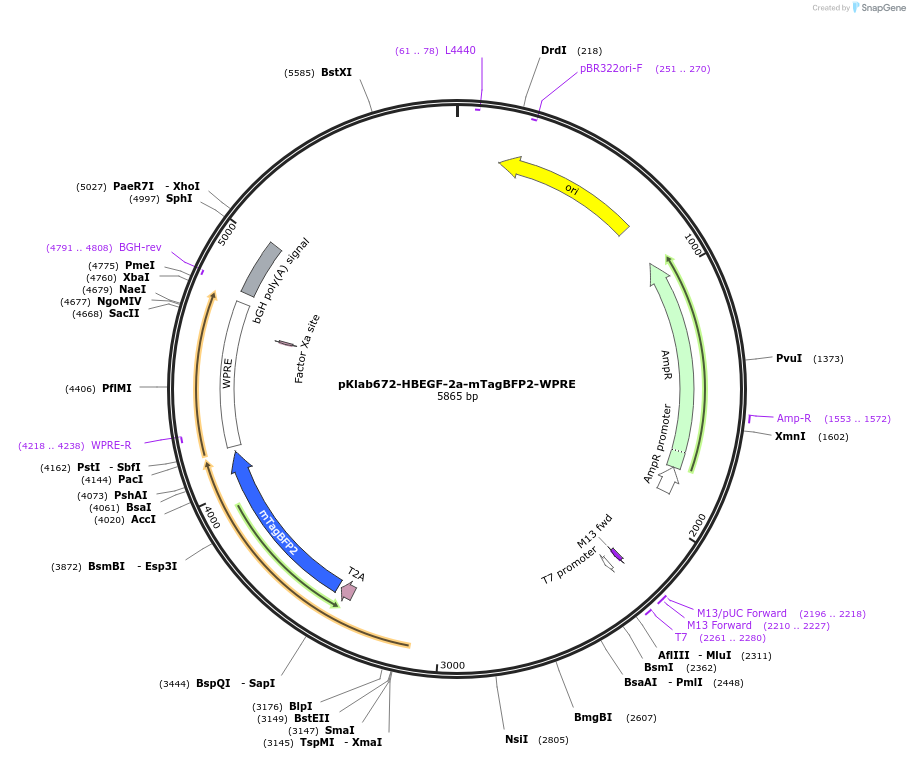
pKlab672-HBEGF-2a-mTagBFP2-WPRE
Plasmid#249031PurposeThis plasmid expresses the mTagBFP2 flourescent tag for integration into mammalian cell linesDepositorInsertmTagBFP2
UseCRISPRAvailable SinceJan. 21, 2026AvailabilityAcademic Institutions and Nonprofits only -
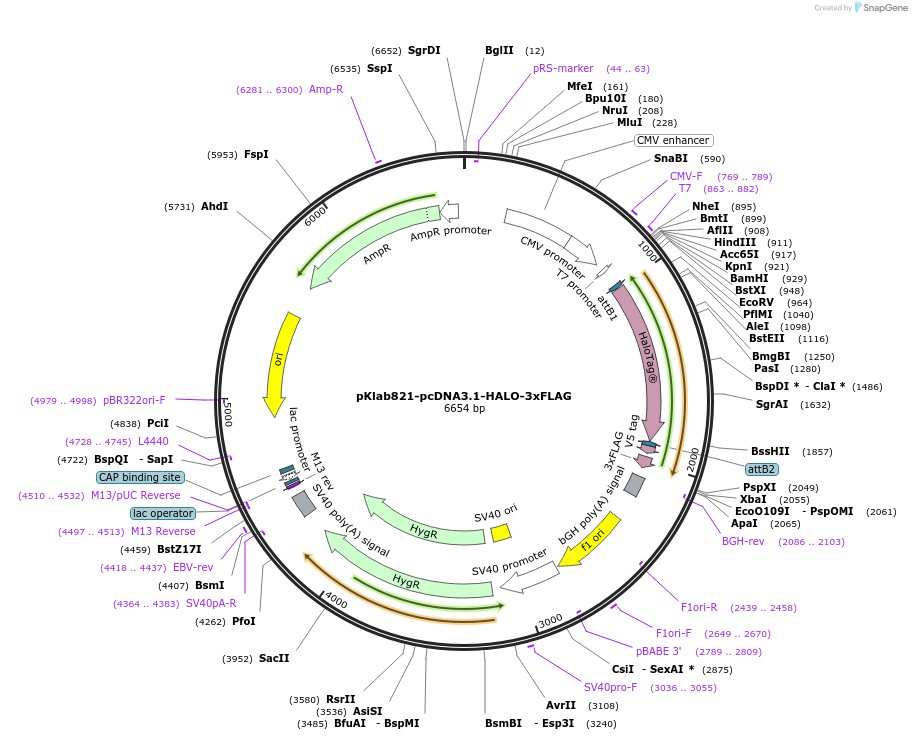
pKlab821-pcDNA3.1-HALO-3xFLAG
Plasmid#249007PurposeThis plasmid expresses the C terminally 3x-FLAG tagged HALO protein sequences for pulldown assaysDepositorInsertHALO
Tags3x-FLAG-V5ExpressionMammalianPromoterCMV PromoterAvailable SinceJan. 21, 2026AvailabilityAcademic Institutions and Nonprofits only -
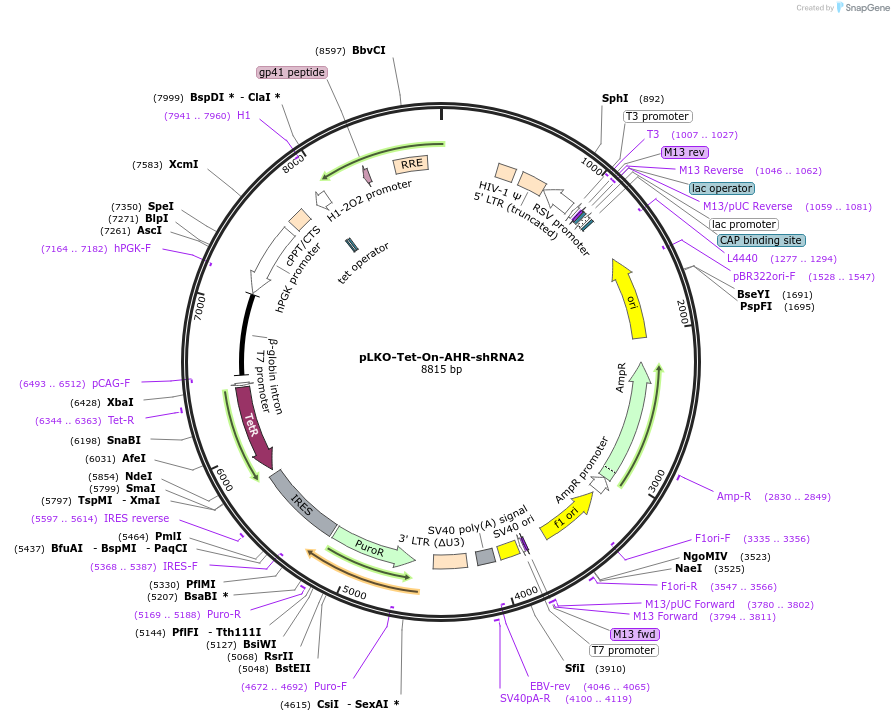
pLKO-Tet-On-AHR-shRNA2
Plasmid#166908PurposeLentivirus for inducible knockdown of human AHRDepositorInsertAHR shRNA
UseLentiviralExpressionMammalianAvailable SinceOct. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
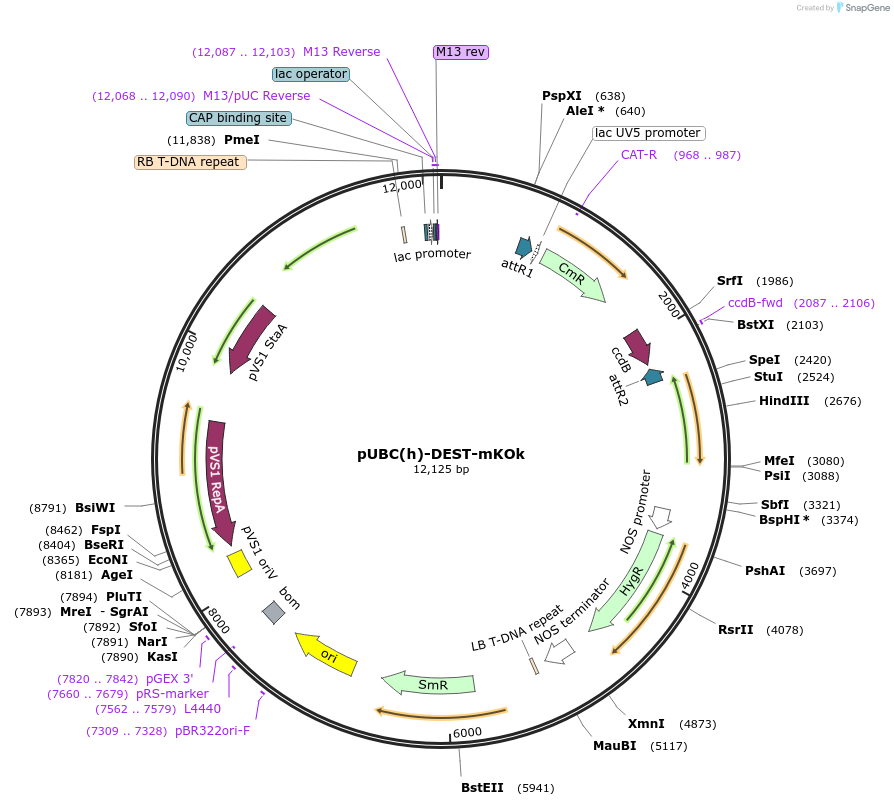
pUBC(h)-DEST-mKOk
Plasmid#224830PurposeEmpty Gateway DEST vector for expressing C-terminal tagged proteins with mKusabiraOrangekand hygromycin selection in plantsDepositorTypeEmpty backboneTagsmKusabiraOrangekExpressionPlantAvailable SinceJuly 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
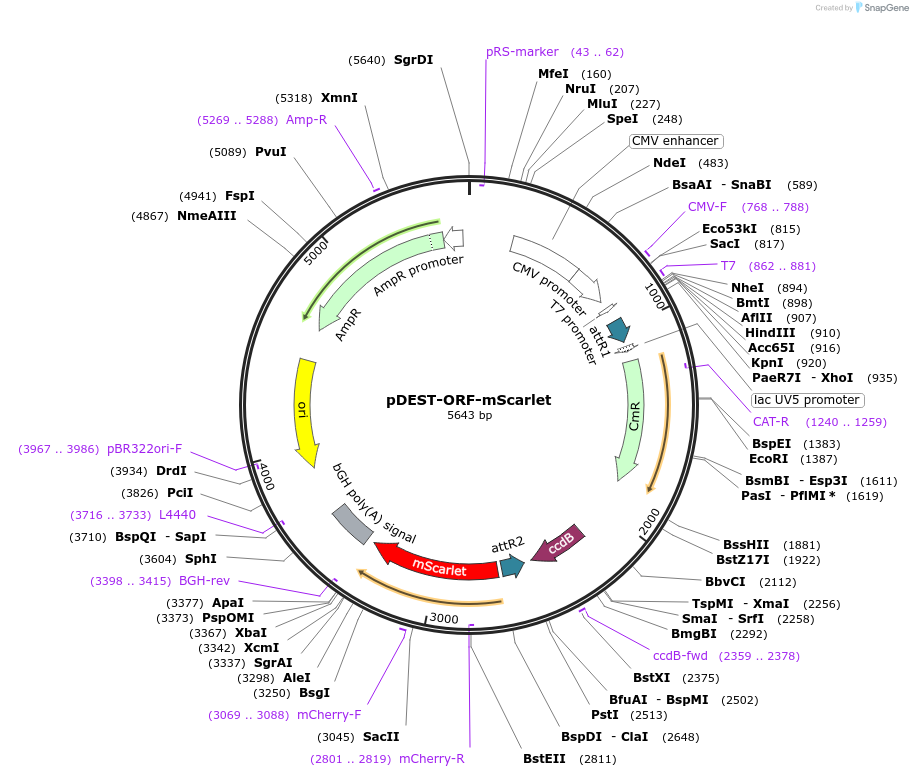
pDEST-ORF-mScarlet
Plasmid#217453PurposeGateway destination vector with C-terminal mScarletDepositorTypeEmpty backboneUseGateway destination vectorTagsmScarletExpressionMammalianPromoterCMVAvailable SinceMay 29, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
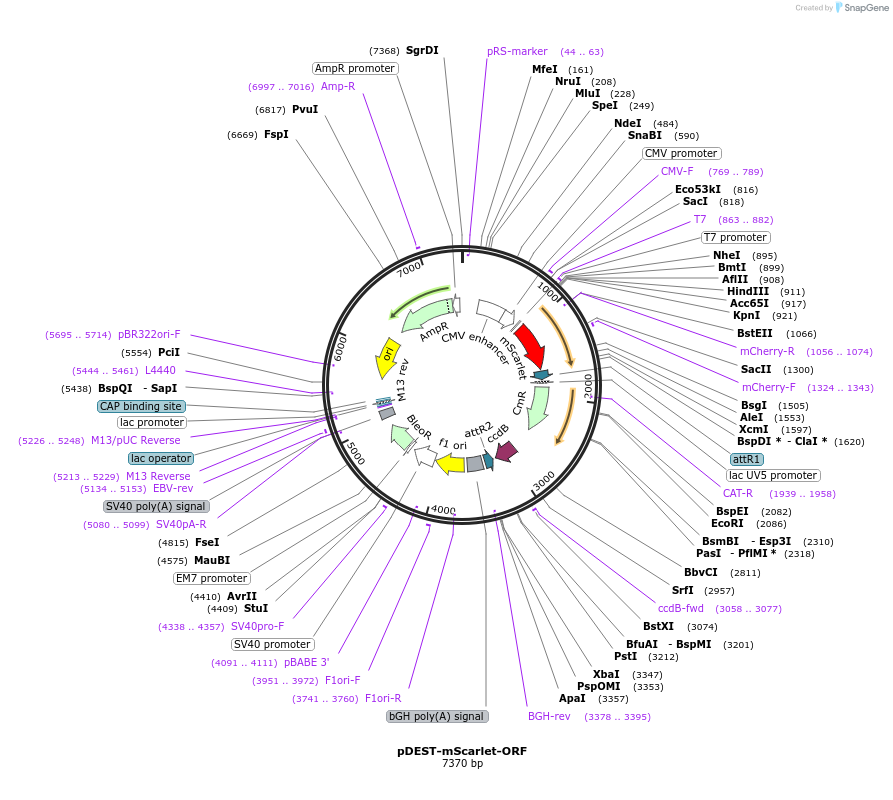
pDEST-mScarlet-ORF
Plasmid#217454PurposeGateway destination vector with N-terminal mScarletDepositorTypeEmpty backboneUseGateway destination vectorTagsmScarletExpressionMammalianPromoterCMVAvailable SinceMay 29, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits