We narrowed to 1,336 results for: HRAS
-
Plasmid#61856PurposeExpresses a gRNA plasmid targeting the prion gene's exon 3, negative strandDepositorAvailable SinceFeb. 3, 2015AvailabilityAcademic Institutions and Nonprofits only
-
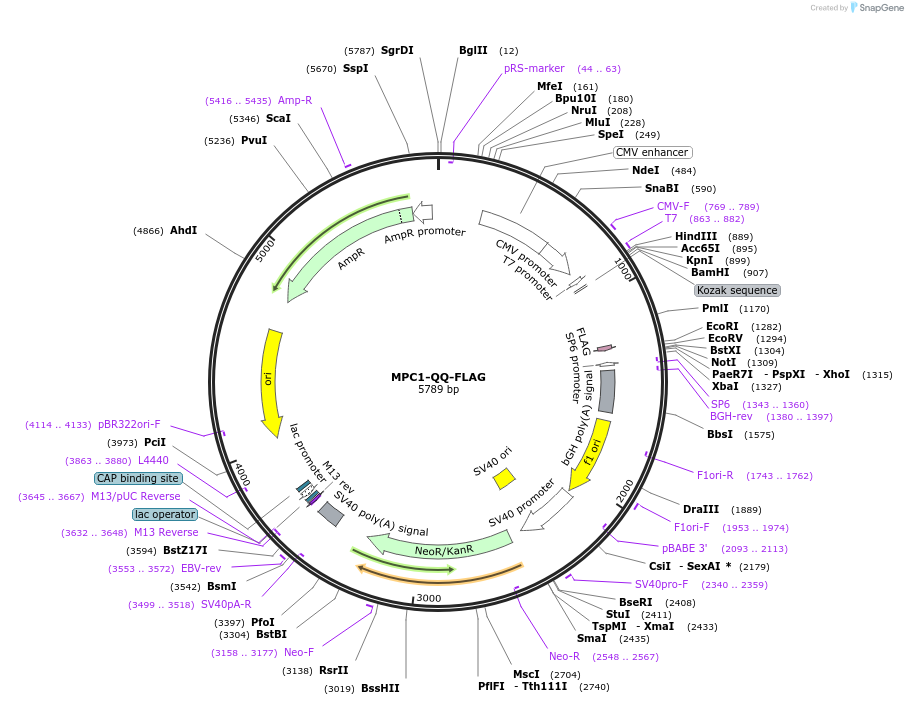
MPC1-QQ-FLAG
Plasmid#228905PurposeExpression of mouse MPC1 in which K45 and K46 motif sites were replaced with glutamine to mimic acetylated lysines with a C terminal FLAG tagDepositorInsertMPC1 (Mpc1 Mouse)
TagsFLAGExpressionMammalianMutationK45 and K46 motif sites were both replaced with g…PromoterCMVAvailable SinceFeb. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
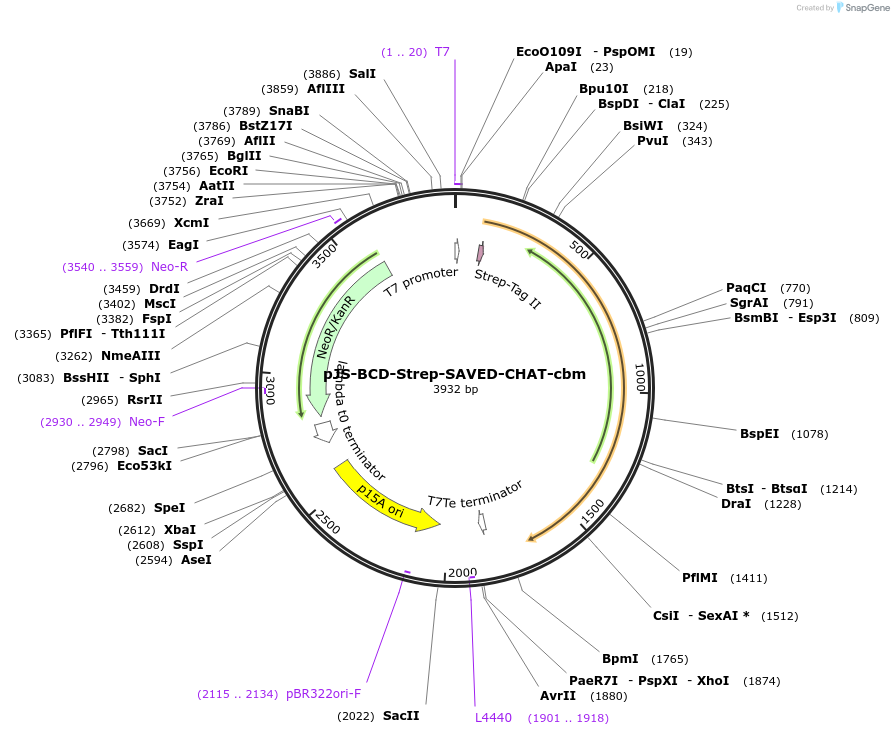
pJS-BCD-Strep-SAVED-CHAT-cbm
Plasmid#214032PurposeBacterial expression plasmid for strep-tagged SAVED-CHAT cA3-binding pocket mutant from Haliangium ochraceumDepositorInsertSAVED-CHAT
TagsStrep-tag IIExpressionBacterialMutationK121E, Q122A, N274A, R276E, Y285A, F286APromoterT7 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
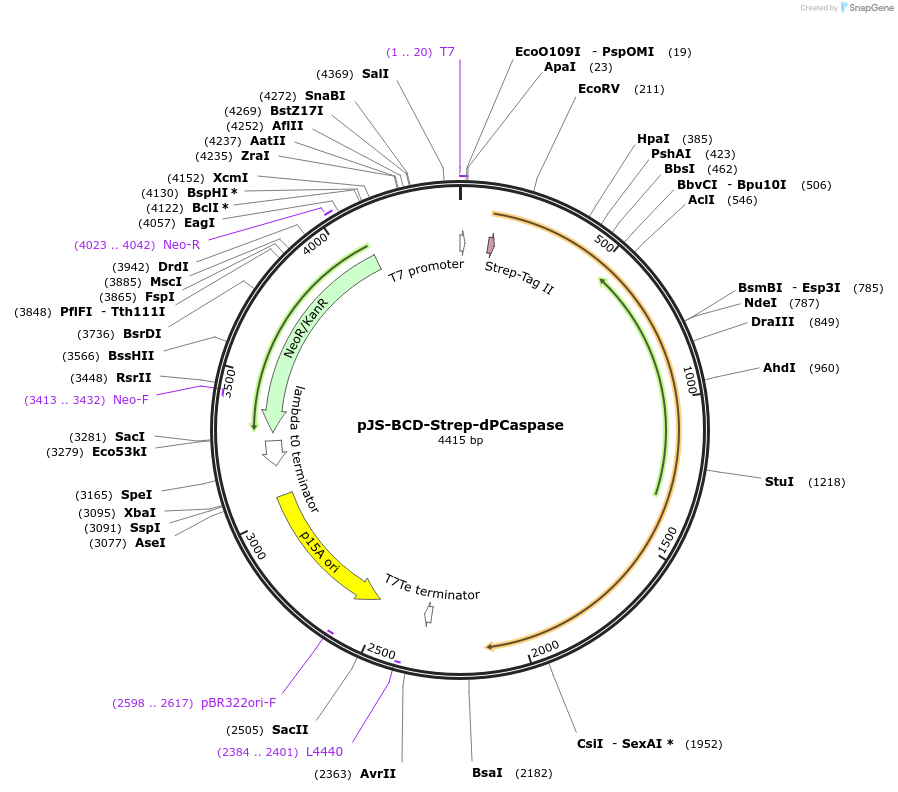
pJS-BCD-Strep-dPCaspase
Plasmid#214035PurposeBacterial expression plasmid for strep-tagged catalytically dead PCaspase (Prokaryotic Caspase) from Haliangium ochraceumDepositorInsertPCaspase
TagsStrep-tag IIExpressionBacterialMutationH79A, C146APromoterT7 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
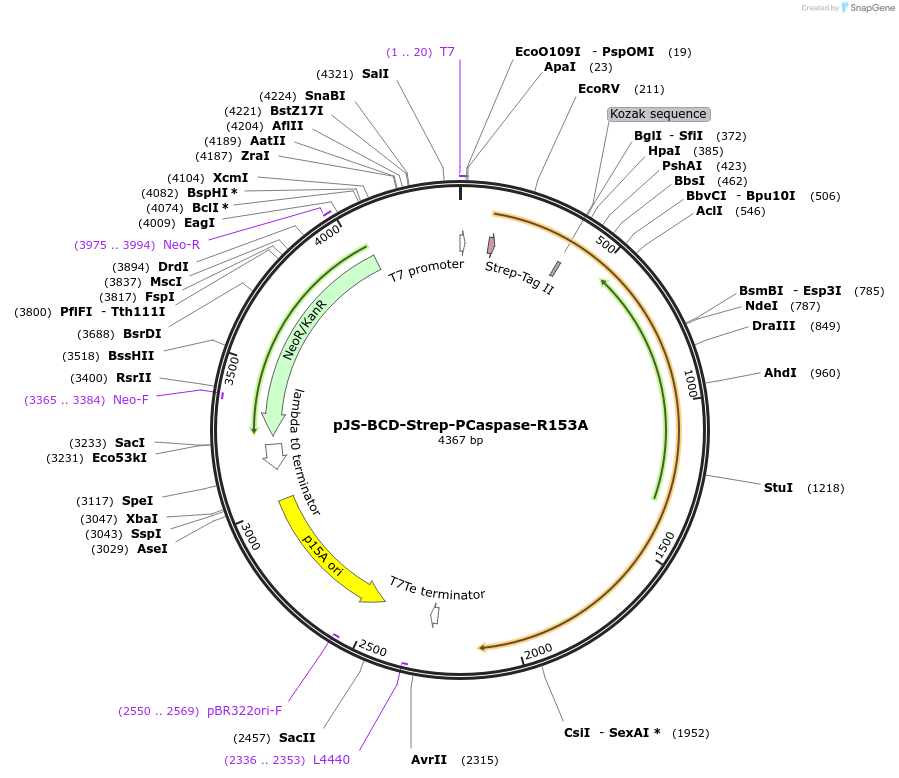
pJS-BCD-Strep-PCaspase-R153A
Plasmid#214036PurposeBacterial expression plasmid for strep-tagged PCaspase (Prokaryotic Caspase) R153A mutant from Haliangium ochraceumDepositorInsertPCaspase
TagsStrep-tag IIExpressionBacterialMutationR153APromoterT7 promoterAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
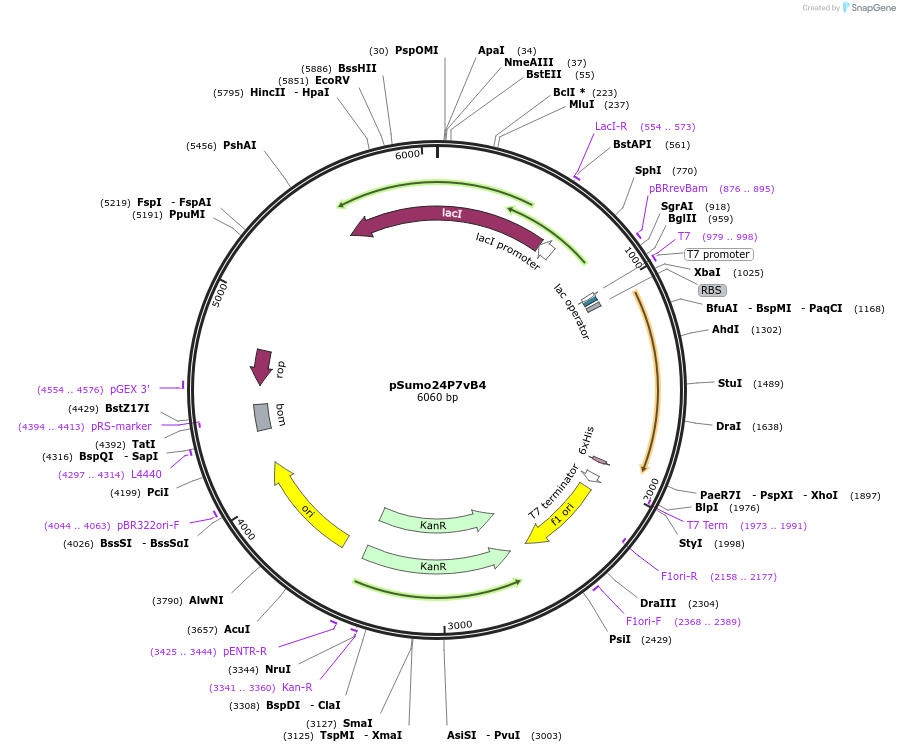
pSumo24P7vB4
Plasmid#113073PurposePlasmid for highly efficient expression of engineered IL24 with binding affinity to cognate receptorsDepositorInsertHuman IL-24 engineered by computational design and fused with N-terminal SUMO tag (IL24 SUMO part (Brachypodium distachyon), Human)
TagsSUMOExpressionBacterialMutationQ60R, K62E, K68E, K77R, M80L, S88D, A89V, Q93R, Q…PromoterT7Available SinceSept. 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
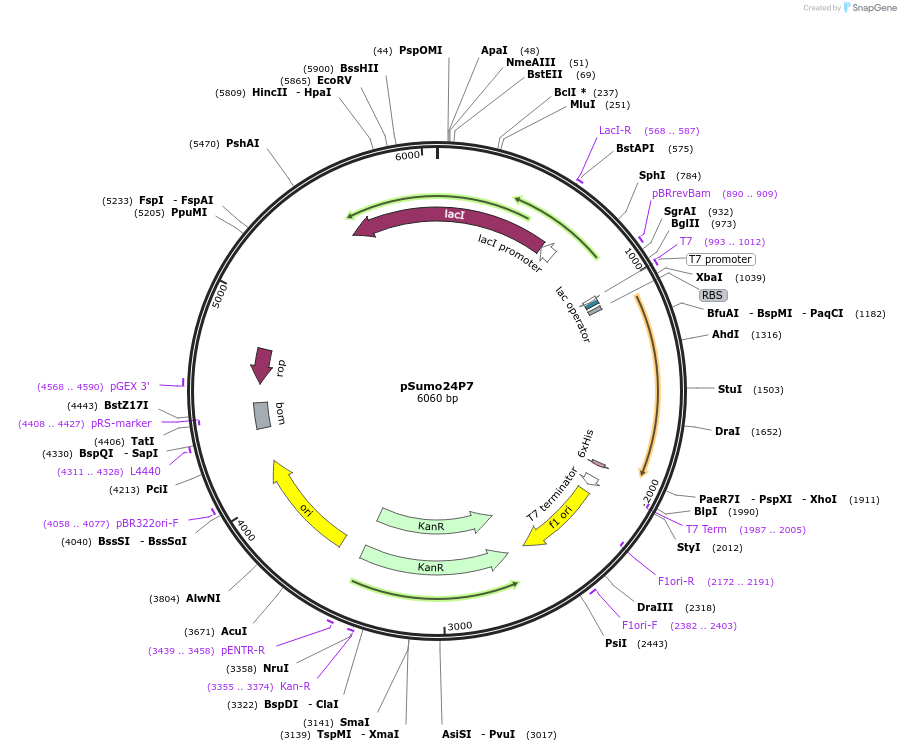
pSumo24P7
Plasmid#113072PurposePlasmid for highly efficient expression of engineered IL24 with mutated binding sitesDepositorInsertHuman IL-24 engineered by computational design and fused with N-terminal SUMO tag (IL24 SUMO part (Brachypodium distachyon), Human)
TagsSUMOExpressionBacterialMutationQ60R, K62E, K68E, K77R, M80L, S88D, A89V, Q93R, Q…PromoterT7Available SinceSept. 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
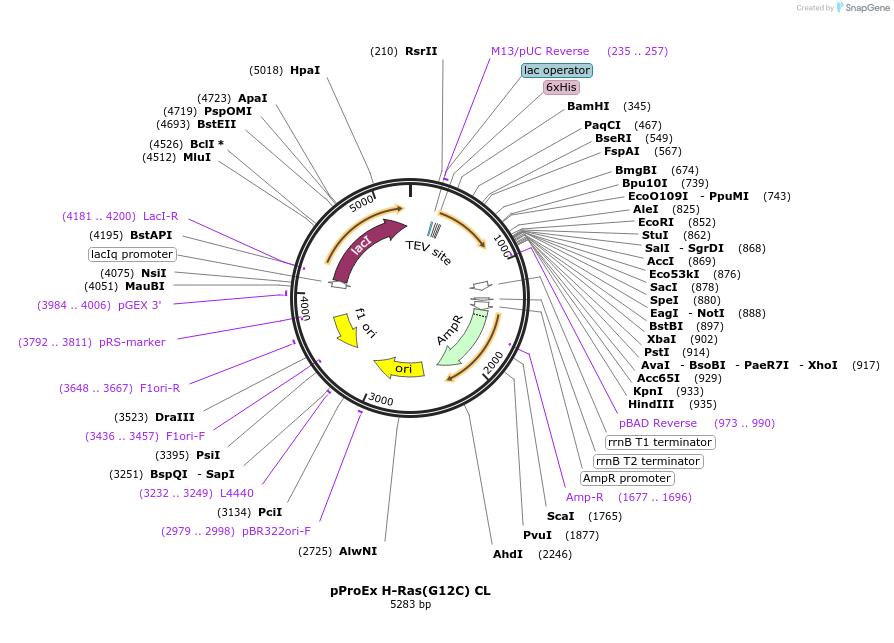
pProEx H-Ras(G12C) CL
Plasmid#224261PurposeProtein purification of H-Ras (G12C)DepositorAvailable SinceSept. 18, 2024AvailabilityAcademic Institutions and Nonprofits only -
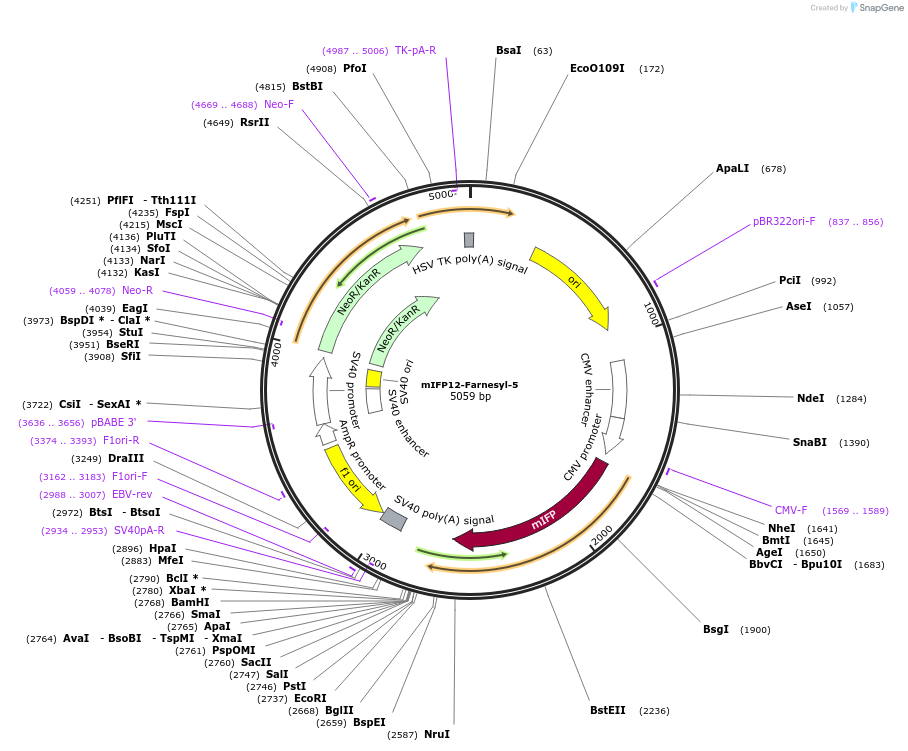
mIFP12-Farnesyl-5
Plasmid#56251PurposeLocalization: Plasma Membrane, Excitation: 683, Emission: 704DepositorInsertFarnesyl (HRAS Human)
TagsmIFP12ExpressionMammalianMutationencodes final 20 aa of NM_001130442.1PromoterCMVAvailable SinceApril 28, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
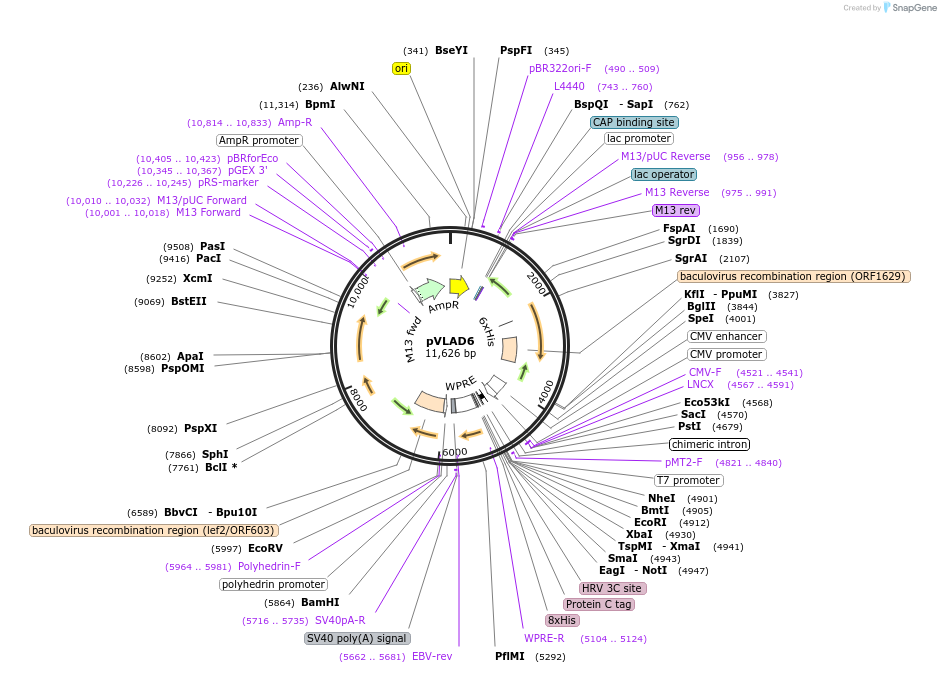
pVLAD6
Plasmid#41850DepositorTypeEmpty backboneExpressionMammalianAvailable SinceFeb. 14, 2013AvailabilityAcademic Institutions and Nonprofits only -
pVLAD8L
Plasmid#41853DepositorTypeEmpty backboneExpressionMammalianAvailable SinceFeb. 14, 2013AvailabilityAcademic Institutions and Nonprofits only -
pVLAD8
Plasmid#41852DepositorTypeEmpty backboneExpressionMammalianAvailable SinceFeb. 14, 2013AvailabilityAcademic Institutions and Nonprofits only -
pVLAD6L
Plasmid#41851DepositorTypeEmpty backboneExpressionMammalianAvailable SinceFeb. 14, 2013AvailabilityAcademic Institutions and Nonprofits only -
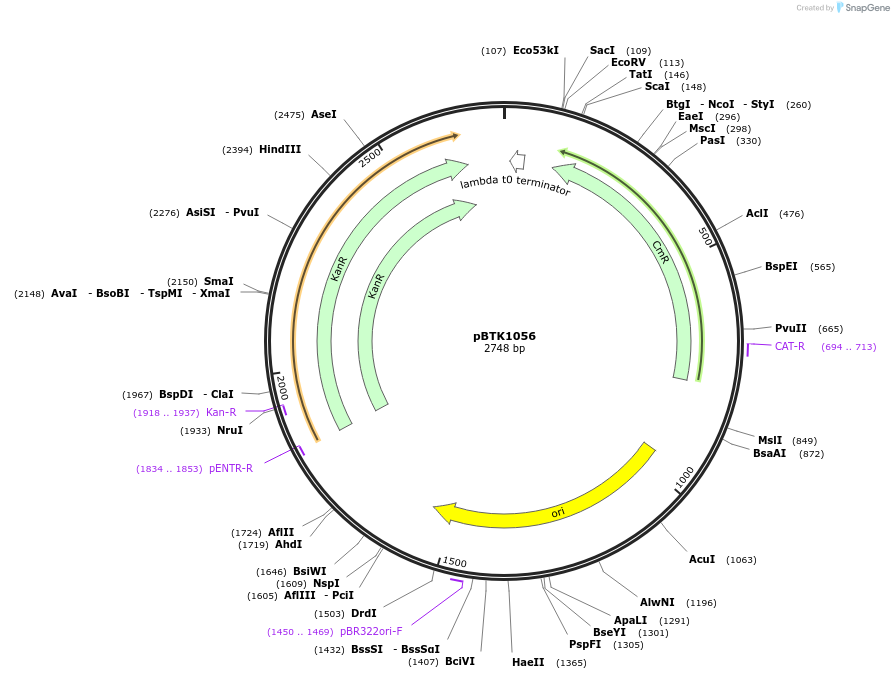
pBTK1056
Plasmid#209546PurposeBTK Type 4 antibiotic resistance cassette for Golden Gate assemblyDepositorInsertType 4 Kanamycin resistance cassette
UseSynthetic BiologyAvailable SinceJan. 12, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
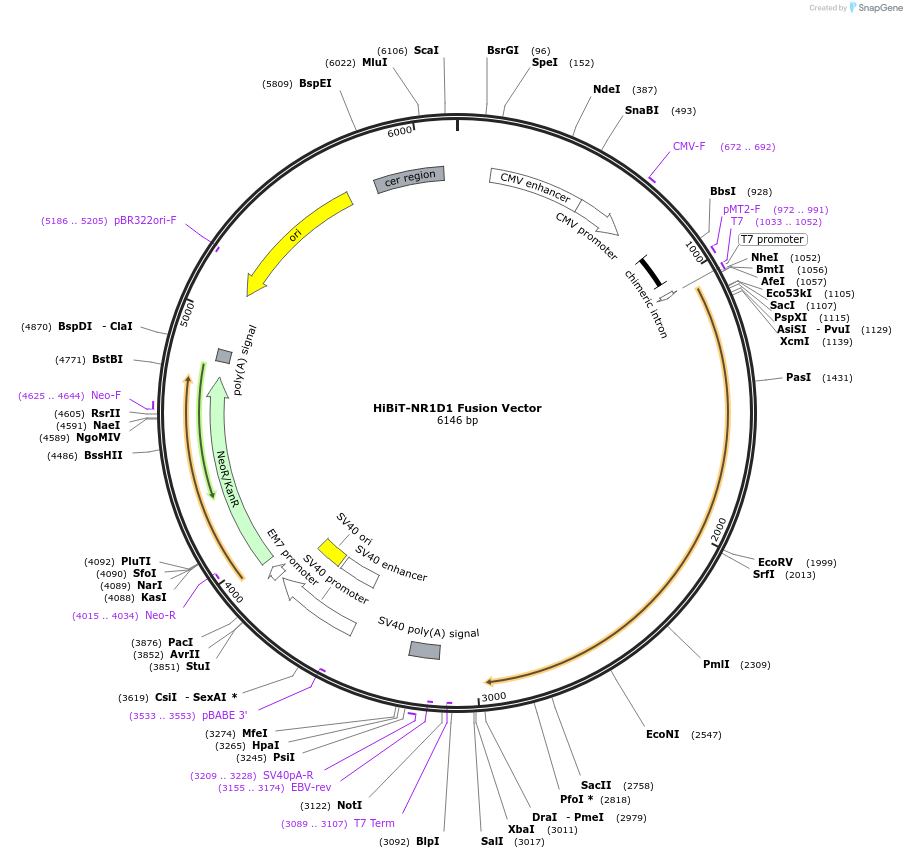
HiBiT-NR1D1 Fusion Vector
Plasmid#238609PurposeExpressHiBiT-NR1D1 Fusion Protein in Mammalian Cells under a CMV promoterDepositorHas ServiceDNAInsertNR1D1 (NR1D1 Human)
TagsHiBiTExpressionMammalianMutationNonePromoterCMVAvailable SinceSept. 14, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
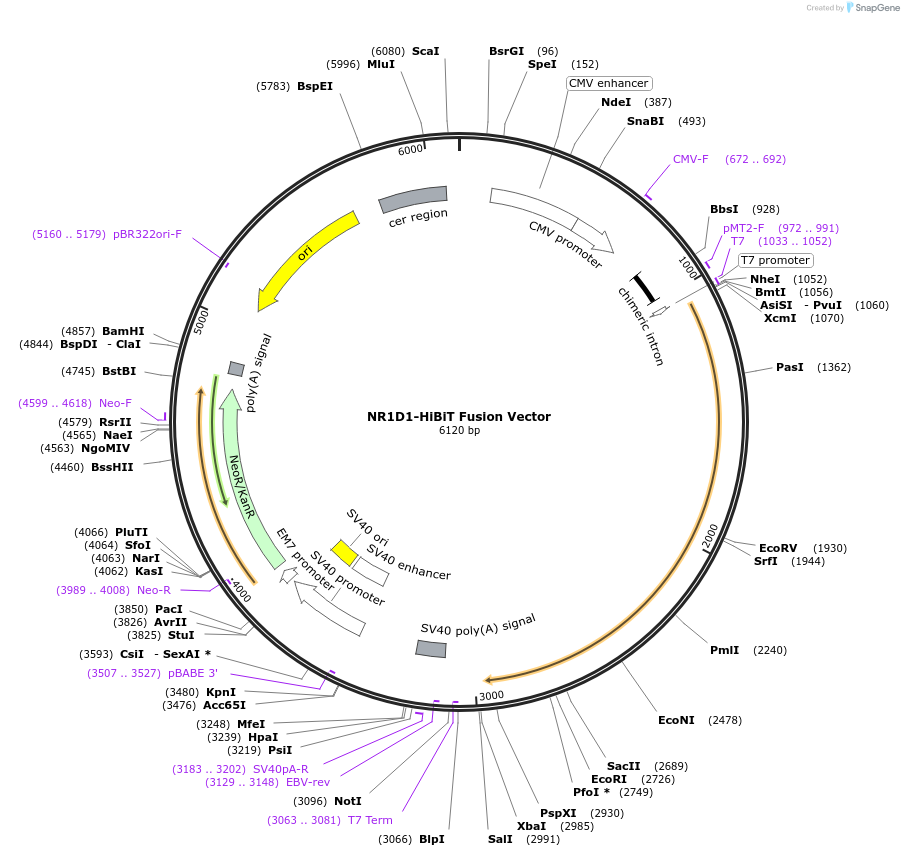
NR1D1-HiBiT Fusion Vector
Plasmid#238610PurposeExpress NR1D1-HiBiT Fusion Protein in Mammalian Cells under a CMV promoterDepositorHas ServiceDNAInsertNR1D1 (NR1D1 Human)
TagsHiBiTExpressionMammalianMutationNonePromoterCMVAvailable SinceSept. 14, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
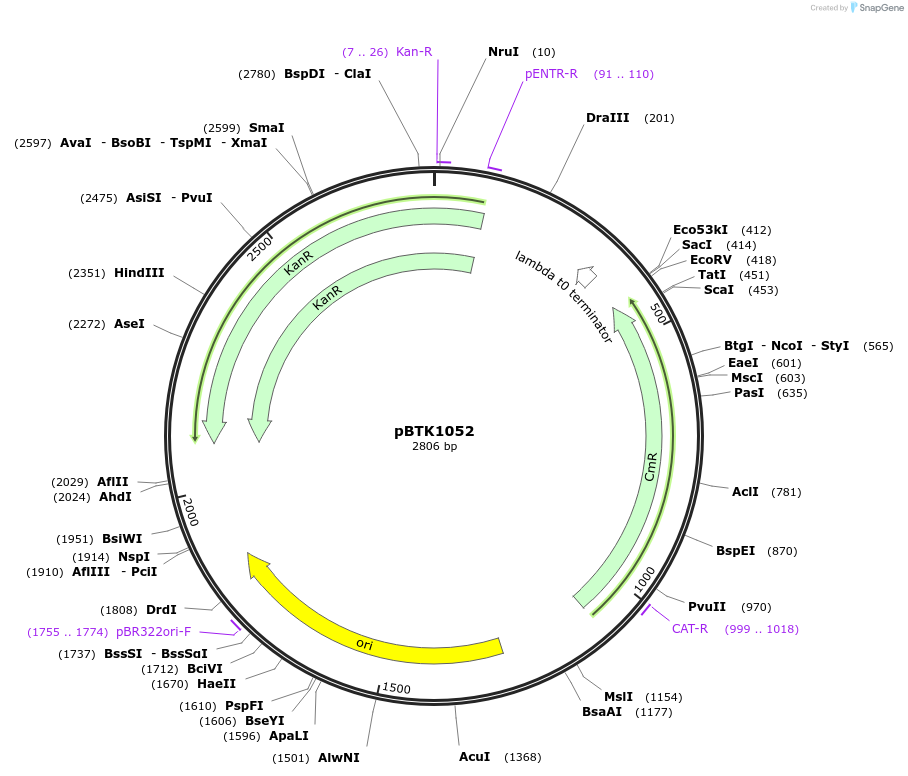
pBTK1052
Plasmid#209542PurposeBTK Type 2 antibiotic resistance cassette for Golden Gate assemblyDepositorInsertType 2 Kanamycin resistance cassette
UseSynthetic BiologyAvailable SinceMay 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
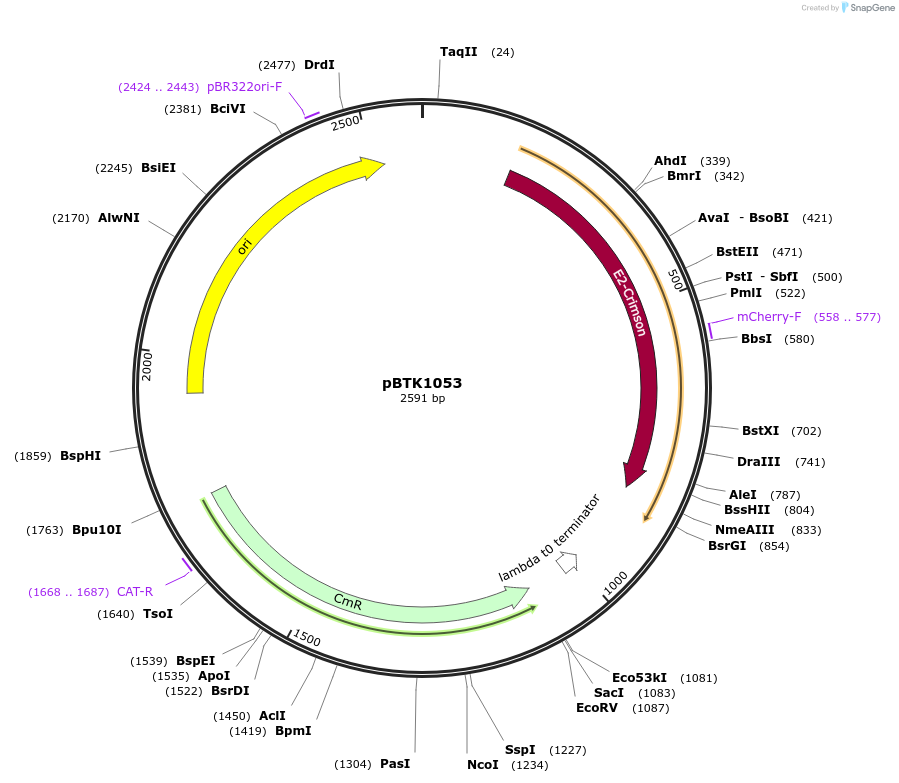
pBTK1053
Plasmid#209543PurposeBTK Type 3-4 fluorescent protein cassette for Golden Gate assemblyDepositorInsertType 3-4 E2-Crimson cassette
UseSynthetic BiologyPromoterCP25Available SinceApril 30, 2025AvailabilityAcademic Institutions and Nonprofits only -
MSCV.IRES.hCD4
Plasmid#35712DepositorInsertsIRES
hCD4
UseRetroviralExpressionMammalianAvailable SinceApril 19, 2012AvailabilityAcademic Institutions and Nonprofits only -
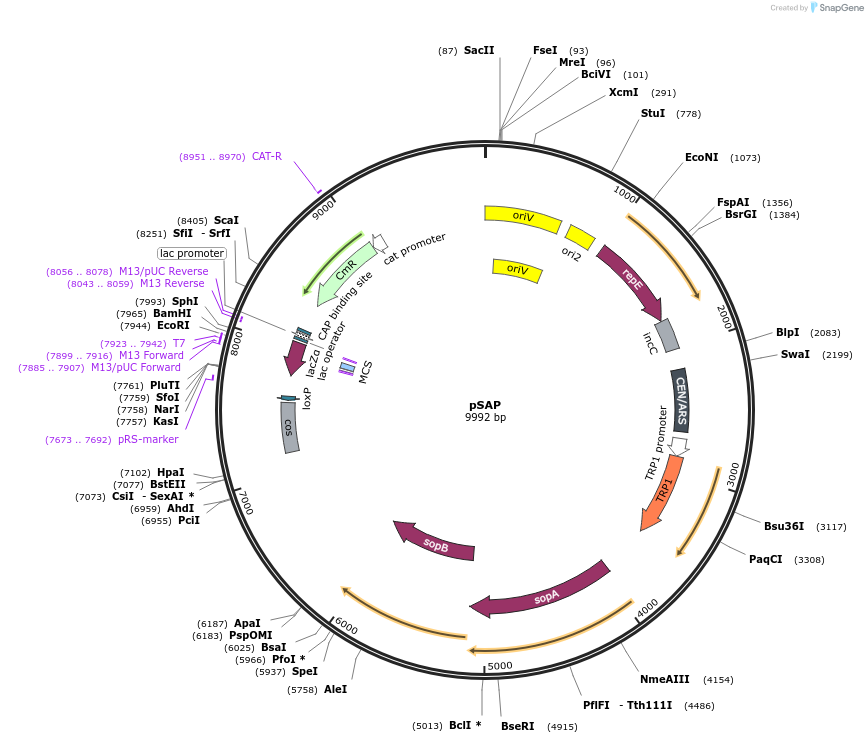
pSAP
Plasmid#206429PurposeA vector backbone domesticated for SapI (pCC1BAC-based), contains elements for selection, maintenance, and propagation in S. cerevisiae (TRP1) and E. coli (cat).DepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceJan. 19, 2024AvailabilityAcademic Institutions and Nonprofits only