We narrowed to 9,400 results for: Coli
-
Plasmid#67419PurposeExpression of hArl4 with C-term V5 and His6 tag in E. coliDepositorInsertARL4 (ARL4A Human)
UseTagsHis and V5ExpressionBacterialMutationbp 105 A to T; silent mutationPromoterT7/lacOAvailable SinceOct. 5, 2015AvailabilityAcademic Institutions and Nonprofits only -
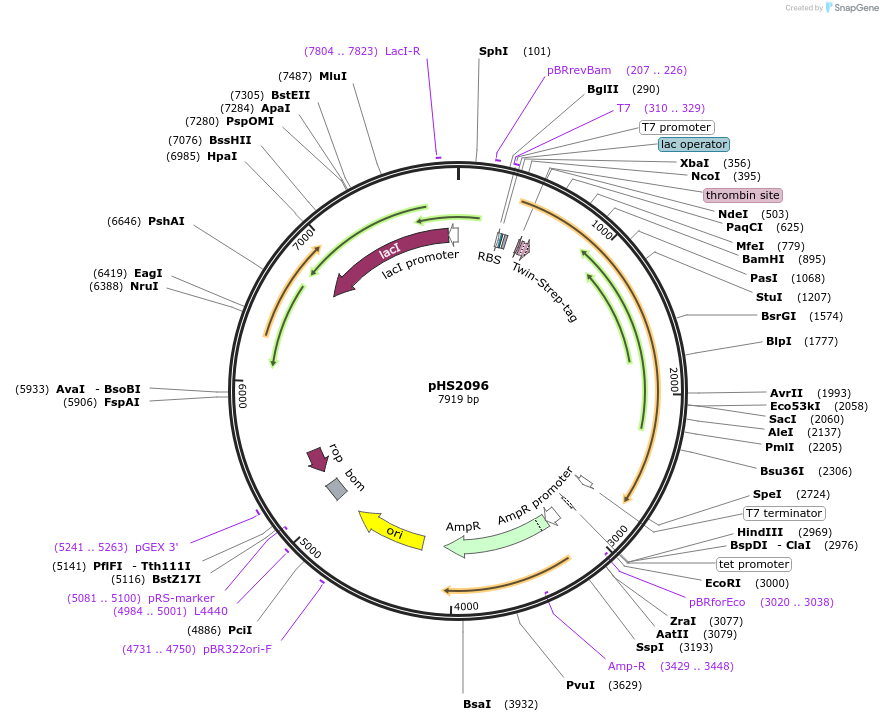
pHS2096
Plasmid#239835PurposeNovaIscB E. coli expression for recombinant purificationDepositorInsertNovaIscB
UseTags14xHis-TwinStrep-bdSUMOExpressionBacterialMutationInsertion of REC domain from NbaCas9-1 and swap o…PromoterT7Available SinceJuly 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
B8-pVTCB052
Plasmid#231302PurposePlasmid used for expression of AI-designed GFP protein B8 in E. coliDepositorInsertB8
UseTagsExpressionBacterialMutationPromoterAvailable SinceMarch 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
pdnCas3-CDA1
Plasmid#229535PurposepMV_hyg encoding dnCas3(H74A+D75A)-CDA1 fusion construct and crRNA expression cassette. Respective crRNA spacer can be introduced via BsmBI restriction cloningDepositorInsertsdnCas3
Cytidine deaminase
Uracil glycosylase inhibitor
UseTagsXTENExpressionBacterial and YeastMutationcodon optimized for S.cerevisiae and codon optimi…PromoterAvailable SinceJan. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
pdnCas3-APOBEC1
Plasmid#229537PurposepMV_hyg encoding dnCas3(H74A+D75A)-rAPOBEC1 fusion construct and crRNA expression cassette. Respective crRNA spacer can be introduced via BsmBI restriction cloningDepositorInsertsrAPOBEC1
dnCas3
Uracil glycosylase inhibitor
UseTagsXTENExpressionBacterial and YeastMutationcodon optimized for S.cerevisiae and codon optimi…PromoterAvailable SinceJan. 23, 2025AvailabilityAcademic Institutions and Nonprofits only -
pQE30-DARPin-E11
Plasmid#226784PurposeExpression of His-tagged DARPin E11 in E. coliDepositorInsertDARPin E11
UseTagsFLAG and HisExpressionBacterialMutationPromoterAvailable SinceNov. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
pJExpress401-KDAC4 CD H976Y
Plasmid#224250PurposeExpresses human KDAC4 (HDAC4) catalytic domain H976Y in E. coliDepositorInsertKDAC4 (HDAC4 Human)
UseTagsTEV-cleavable His6ExpressionBacterialMutationOnly includes residues 648-1083 (catalytic domain…PromoterT5Available SinceOct. 22, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
pET28a-Fgß-Coh7-ddFLN4-ELP-HIS-ybbR
Plasmid#169135PurposeE. coli expression of Coh7 domain containing Fgß tag, ybbR tag, ELP linker and ddFLN4 fingerprint domain. This construct was designed for AFM-SMFS measurements.DepositorInsertCoh7
UseTags3xELP linker, 6x Histag, Fgβ peptide, ddFLN4 tag,…ExpressionBacterialMutationPromoterAvailable SinceOct. 10, 2024AvailabilityAcademic Institutions and Nonprofits only -
pET28a-SpyCatcher -AVI
Plasmid#223635PurposeProtein purification of SpyCatcher003 (S49C) with AVI and 6xHis tags from E. coliDepositorInsertSpyCatcher003 S49C
UseTagsAVI tag and His6ExpressionBacterialMutationPromoterAvailable SinceSept. 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
pGGI_Myr-YFP-MiniTurboID
Plasmid#222432PurposePotential plasma membrane control.DepositorInsertMiniTurboID (BirA mutant)
UseGolden gate / green gate compatible cloning vectorTagsLinker, Myr, and YFPExpressionMutationaa1-63 deleted; Q65P, I87V, R118S, E140K, Q141R, …PromoterAvailable SinceJuly 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
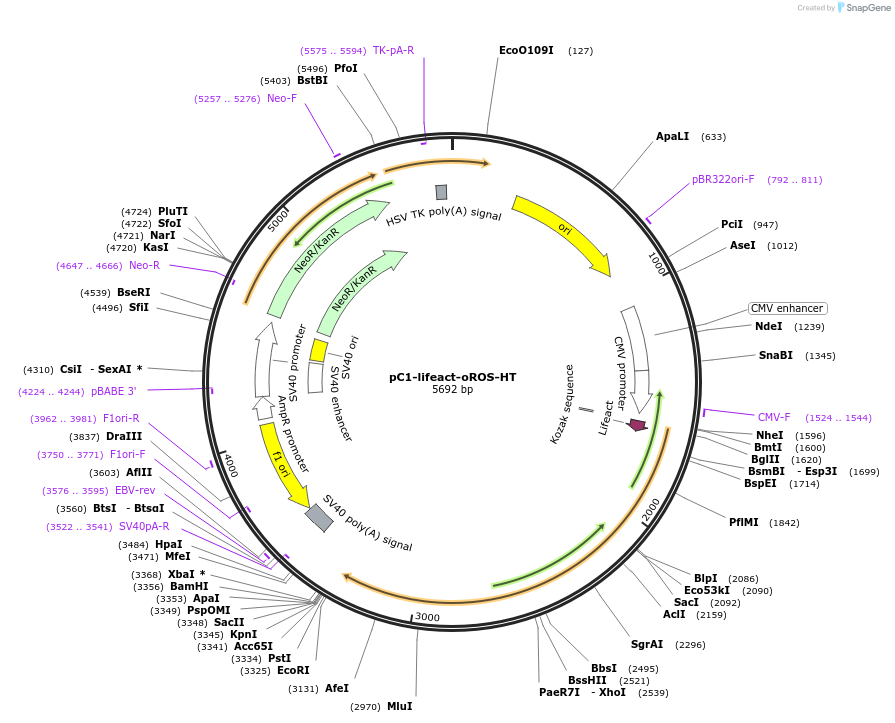
pC1-lifeact-oROS-HT
Plasmid#216420PurposeTargeting of the chemigenetic, fluorescent peroxide sensor oROS-HT to actin filaments.DepositorInsertlifeact-oROS-HT
UseTagsExpressionMammalianMutationPromoterCMVAvailable SinceMay 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
pC3.1_CMV_oROS-G_LF(C199S)
Plasmid#216112PurposeExpresses the loss-of-function mutations C199S of the genetically encoded green fluorescent hydrogen peroxide sensor oROS-G in mamalian cells.DepositorInsertoROS-G_LF(C199S)
UseTagsExpressionMammalianMutationPromoterCMVAvailable SinceMay 28, 2024AvailabilityAcademic Institutions and Nonprofits only -
pET151-GyrA_F164R238
Plasmid#216230PurposeS. aureus USA300 GyrA mutant F164 R238 residues codon-optimized for E. coliDepositorInsertDNA gyrase
UseTags6xHisExpressionBacterialMutationChanged arginine 238 to serine, phenylalanine 168…PromoterT7Available SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
pET151-GyrB_A467
Plasmid#216234PurposeS. aureus USA300 GyrB mutant A467 residue codon-optimized for E. coliDepositorInsertDNA gyrase
UseTags6xHisExpressionBacterialMutationChanged alanine 467 to valinePromoterT7Available SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
pET151-GyrB_T522
Plasmid#216235PurposeS. aureus USA300 GyrB mutant T522 residue codon-optimized for E. coliDepositorInsertDNA gyrase
UseTags6xHisExpressionBacterialMutationChanged threonine 522 to isoleucinePromoterT7Available SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
pGEX-4T1-MBP-TEV-NAP1_delta-NDP52 (S37K/A44E)
Plasmid#208872PurposeFor expression in E. coli of NAP1 (delta-NDP52)DepositorInsertNAP1 (AZI2 Human)
UseTagsMBP-TEVExpressionBacterialMutationS37K/A44E (disrupts NDP52 binding)PromoterAvailable SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
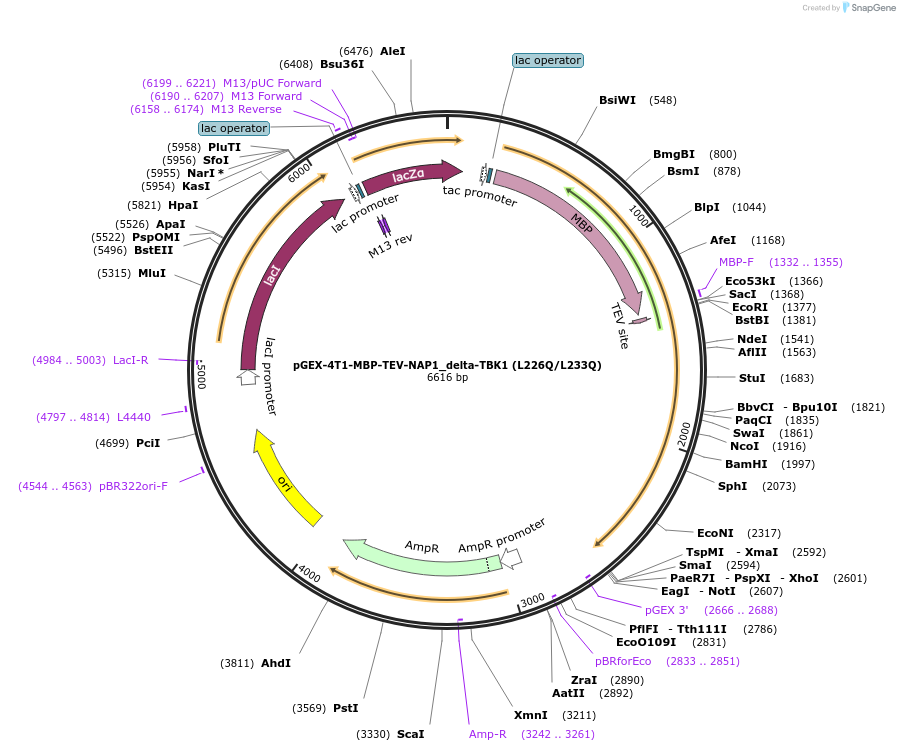
pGEX-4T1-MBP-TEV-NAP1_delta-TBK1 (L226Q/L233Q)
Plasmid#208873PurposeFor expression in E. coli of NAP1 (delta-TBK1)DepositorInsertNAP1 (AZI2 Human)
UseTagsMBP-TEVExpressionBacterialMutationL226Q/L233Q (disrupts TBK1 binding)PromoterAvailable SinceApril 19, 2024AvailabilityAcademic Institutions and Nonprofits only -
pCgVchCAST-crtYf
Plasmid#203816PurposeExpression of CRISPR-associated transposases, shuttle vector for Corynebacterium glutamicum / E. coliDepositorInsertVchTniQ, VchCas5/8, VchCas7, VchCas6, VchTnsABC, CRISPR(crtYf)
UseCRISPRTagsExpressionBacterialMutationPromoterAvailable SinceAug. 7, 2023AvailabilityAcademic Institutions and Nonprofits only -
His SUMO TEV R354H SPOP 28-359
Plasmid#197011PurposeE. coli expression vectorDepositorAvailable SinceMay 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
pnEA-NpM-HsNot4_275-575-del377-402-GB1His6_AG
Plasmid#148785PurposeE. coli of HsNot4_275-575-del307-402DepositorAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
pnEA-NpM-HsNot4_275-575-del400-428-GB1His6_AG
Plasmid#148787PurposeE. coli of HsNot4_275-575-del400-428DepositorAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
pXD70DAmt1-DadR/DadS-Pdadh-DadhABC(R506S, A2)
Plasmid#191631PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), native promoter-dopamine dehydroxylase from dopamine-metabolizing E. lenta A2 strain (R506S mutation) with transcriptional regulators DadR/DadSDepositorInsertDopamine dehydroxylase (R506S mutation, inactive form)
UseTagsExpressionBacterialMutationPromoterAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
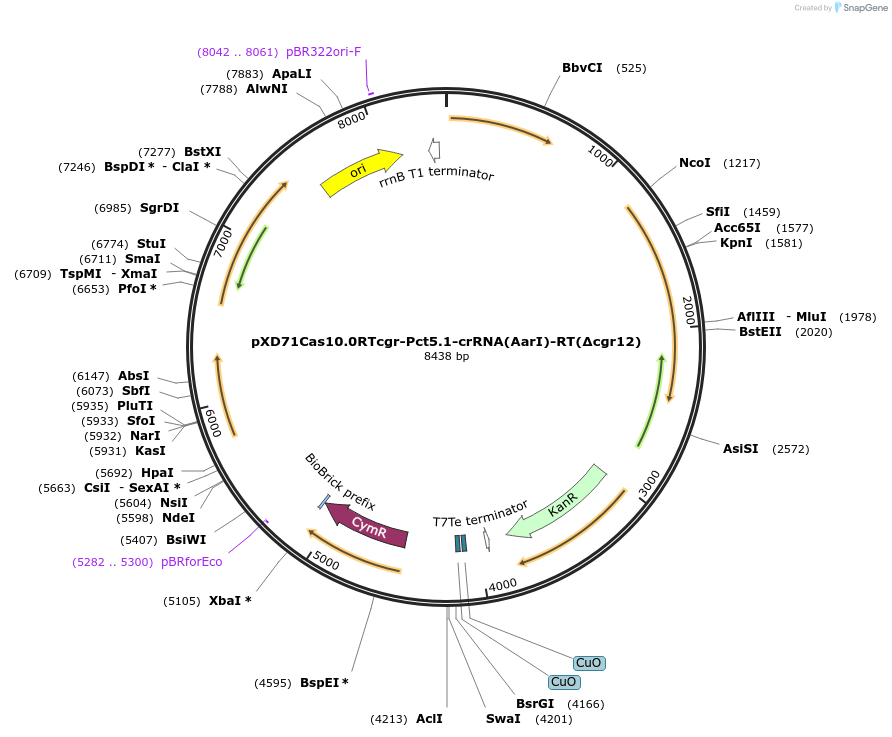
pXD71Cas10.0RTcgr-Pct5.1-crRNA(AarI)-RT(Δcgr12)
Plasmid#191651PurposeE. coli - Eggerthella lenta shuttle plasmid (KanR), cumate-inducible promoter Pct5.1-crRNA(nontargeting spacer) cgr1/cgr2-deleting repair template, used as negative control for pXD71Cas10.4RT4DepositorInsertCymR
UseCRISPRTagsExpressionBacterialMutationPromoterAvailable SinceJan. 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
KTK_113
Plasmid#180540PurposeEntry vector containing Signal peptide region of CsgA including SecB-N22 sequencedDepositorInsertSignal peptide region of CsgA including SecB-N22 sequenced
UseTagsExpressionBacterialMutationPromoterAvailable SinceAug. 17, 2022AvailabilityAcademic Institutions and Nonprofits only -
pBICI_high ori
Plasmid#179273PurposeExpresses CuCBP and BaSP in E. coli BL21(DE3)DepositorInsertsSucrose Phosphorylase from Bifidobacterium adolescentis (BAD_RS00415 Bifidobacterium adolescentis)
Cellobiose phosphorylase from Cellulomonas uda
UseSynthetic BiologyTagsHis Tag and Strep - Tag IIExpressionBacterialMutationPromoterT7 lacOAvailable SinceApril 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
pBtPIPLC_N168C His
Plasmid#173809PurposeExpression and purification of the mature form of Bacillus thuringiensis phosphaatidyl inositol specific phospholipase C (PI-PLC), UniProt ID P08954 from E. coli BL21 CodonPlus (DE3) RIL cellsDepositorInsertBacillus thuringiensis 1-phosphatidylinositol phosphodiesterase
UseTags6XHisExpressionBacterialMutationAsn168Cys for labelingPromoterT7Available SinceAug. 25, 2021AvailabilityAcademic Institutions and Nonprofits only -
pET51b-His-TEV-SNAP-tag_dead_mt
Plasmid#167270PurposeOverexpression of the dead variant (C145A) of SNAP-tag in E. coliDepositorInsertSNAP-tag
UseTagsHis x10ExpressionBacterialMutationC145APromoterT7Available SinceApril 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
pET51b-His-TEV-SNAP-tag_fast_dead_mt
Plasmid#167272PurposeOverexpression of the fast (E30R) dead variant (C145A) of SNAP-tag in E. coliDepositorInsertSNAPf-tag
UseTagsHis x10ExpressionBacterialMutationE30R-C145APromoterT7Available SinceApril 14, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGEX-optimized E6AP F727D
Plasmid#165101Purposeecoli optimized expression of a GST fusion of Human E6AP F727DDepositorInsertE6AP F727D (UBE3A Human)
UseTagsGST from plasmid.ExpressionBacterialMutationcodon optimized for E. coli expressionPromoterAvailable SinceMarch 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
pET28a-ybbr-XMod-Doc (BMA-KO, S199AzF)-HIS
Plasmid#153444PurposeE. coli expression (amber suppression) of Rc. XDocB binding mode A knock-out mutant with serine at position 199 replaced with amber codon.DepositorInsertRc.XDocB
UseTags6xHis and ybbr tagExpressionBacterialMutationSerine 199 was mutated to amber codon (TAG). Argi…PromoterT7 promoterAvailable SinceJuly 23, 2020AvailabilityAcademic Institutions and Nonprofits only