We narrowed to 70,205 results for: nin
-
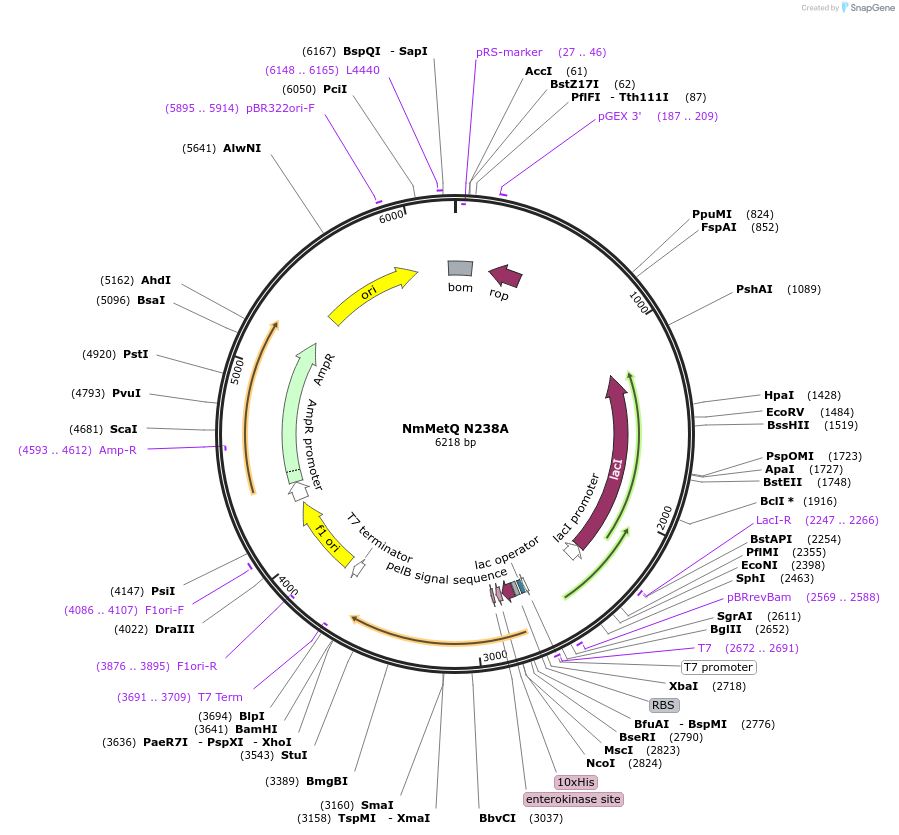
Plasmid#129648PurposeExpresses N238A mutation in the mature metQ gene from Neisseria meningitides, without the signal sequenceDepositorInsertmetQ Gene
Tags10x Histidine Tag, Enterokinase, and pelB signal …ExpressionBacterialMutationChanged Asparagine 238 to AlaninePromoterT7 PromoterAvailable SinceSept. 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
NmMetQ
Plasmid#172111PurposeExpression of Methionine Binding Protein (MetQ) with signal sequenceDepositorInsertMethionine Binding Protein
Tags10x Histidine Tag and N.meningitidis signal seque…ExpressionBacterialMutationChanged Proline 22 to AlaninePromoterT7 PromoterAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
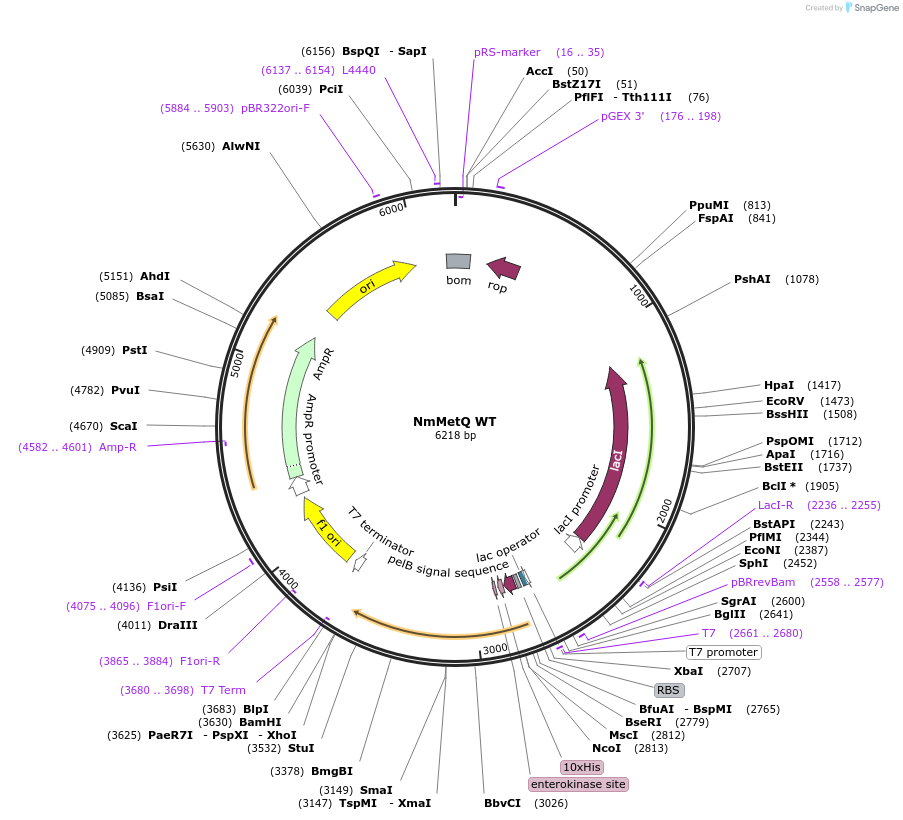
NmMetQ WT
Plasmid#129647PurposeExpresses the mature metQ gene from Neisseria meningitides, without the signal sequenceDepositorInsertmetQ Gene
Tags10x Histidine Tag, Enterokinase, and pelB signal …ExpressionBacterialPromoterT7 PromoterAvailable SinceSept. 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
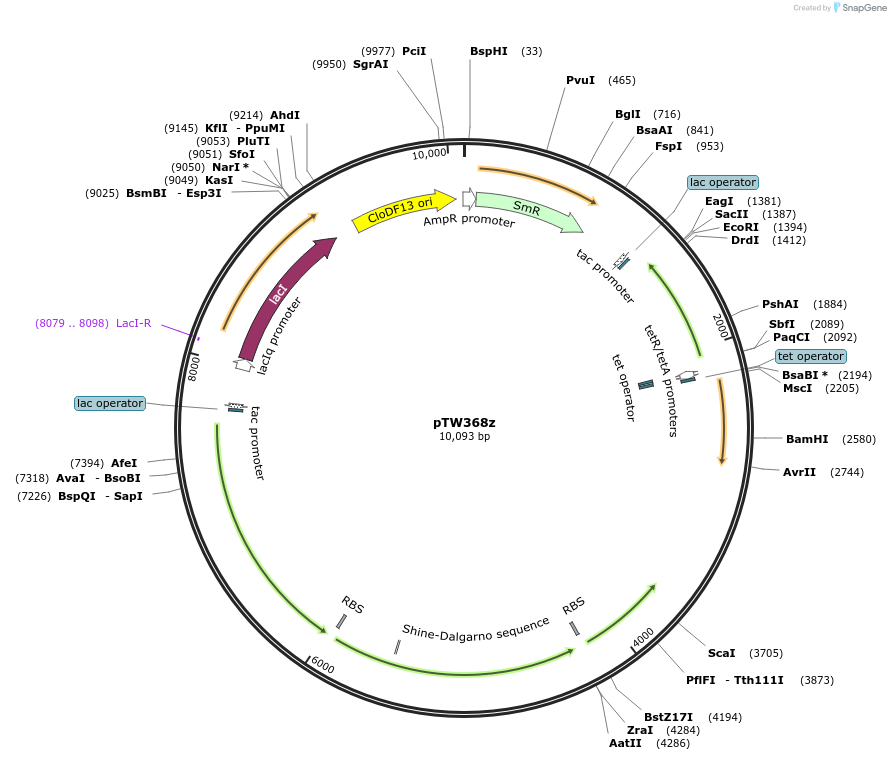
pTW368z
Plasmid#246581PurposeGolden gate acceptor for AtRaf1 library cloningDepositorInsertExpressionBacterialAvailable SinceDec. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
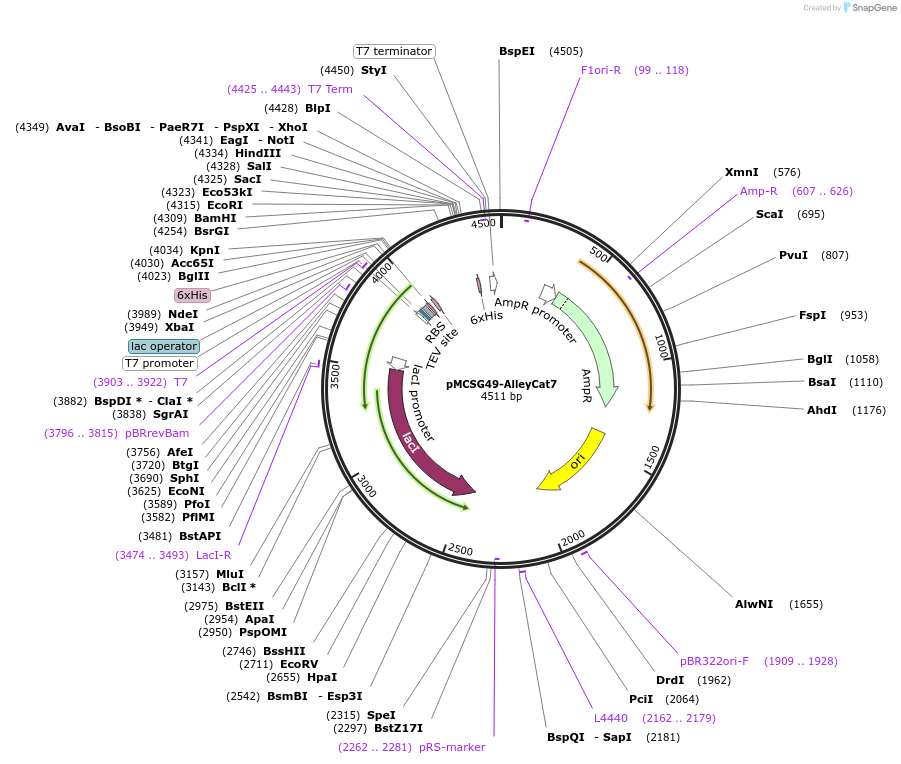
pMCSG49-AlleyCat7
Plasmid#42591DepositorInsertmutated C-terminal portion of calmodulin gene (M76-K148)
TagsHis and TEVExpressionBacterialMutationchanged Phenylalanine 92 to Glutamine acid, Isole…PromoterT7Available SinceMarch 21, 2013AvailabilityAcademic Institutions and Nonprofits only -
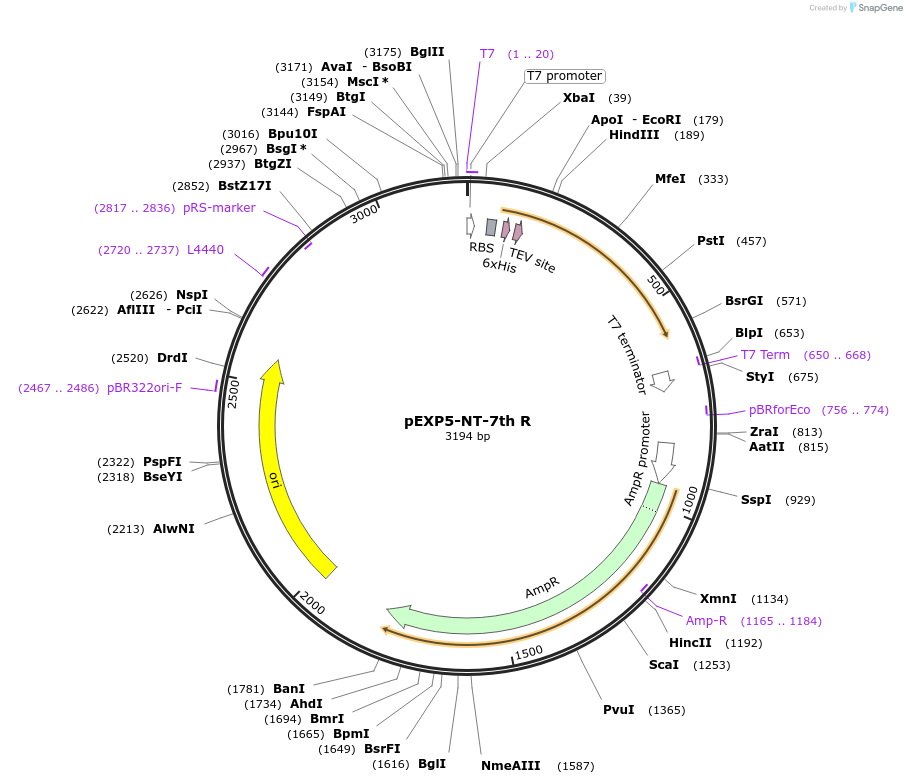
pEXP5-NT-7th R
Plasmid#42573DepositorInsertmutated calmodulin gene
TagsPolyhistidine (6×His) region, TEV recognition sit…ExpressionBacterialMutationchanged Phenylalanine 92 to Glutamine acid, Isole…PromoterT7Available SinceMarch 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
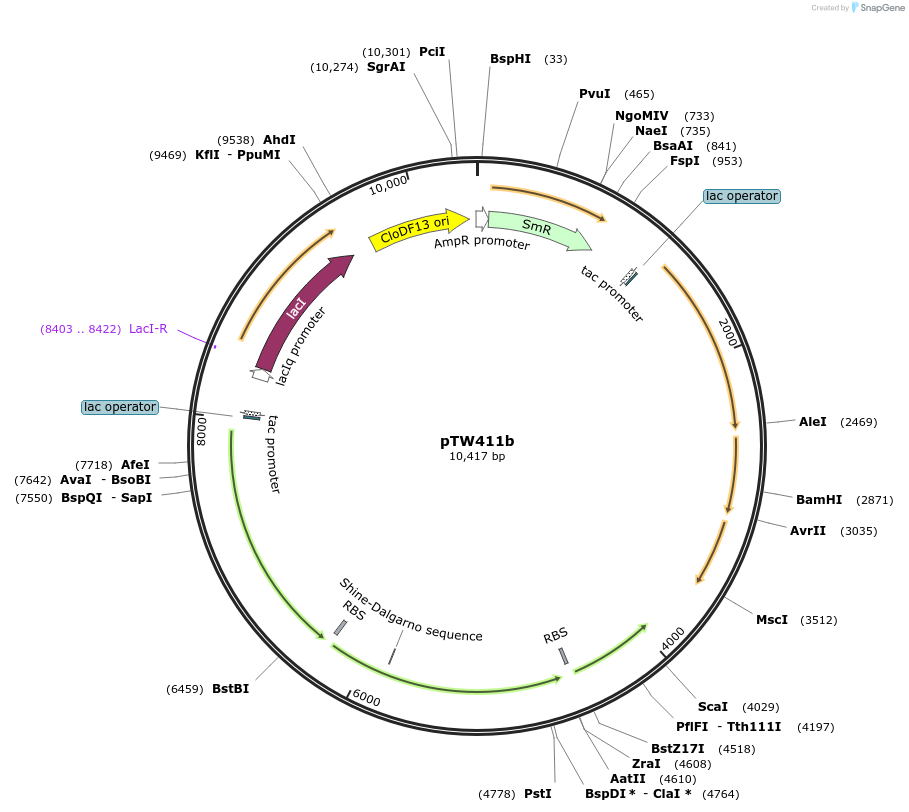
pTW411b
Plasmid#246583Purposedifferent combination of A. thaliana and N. tabacum Rubisco assembly factorsDepositorExpressionBacterialAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
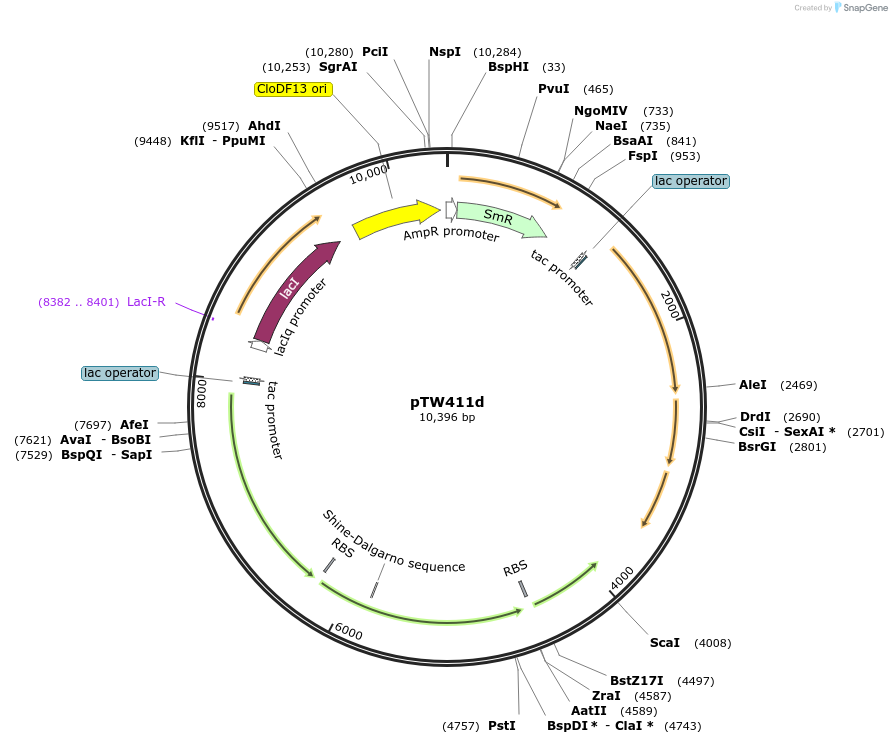
pTW411d
Plasmid#246585Purposedifferent combination of A. thaliana and N. tabacum Rubisco assembly factorsDepositorInsertExpressionBacterialAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
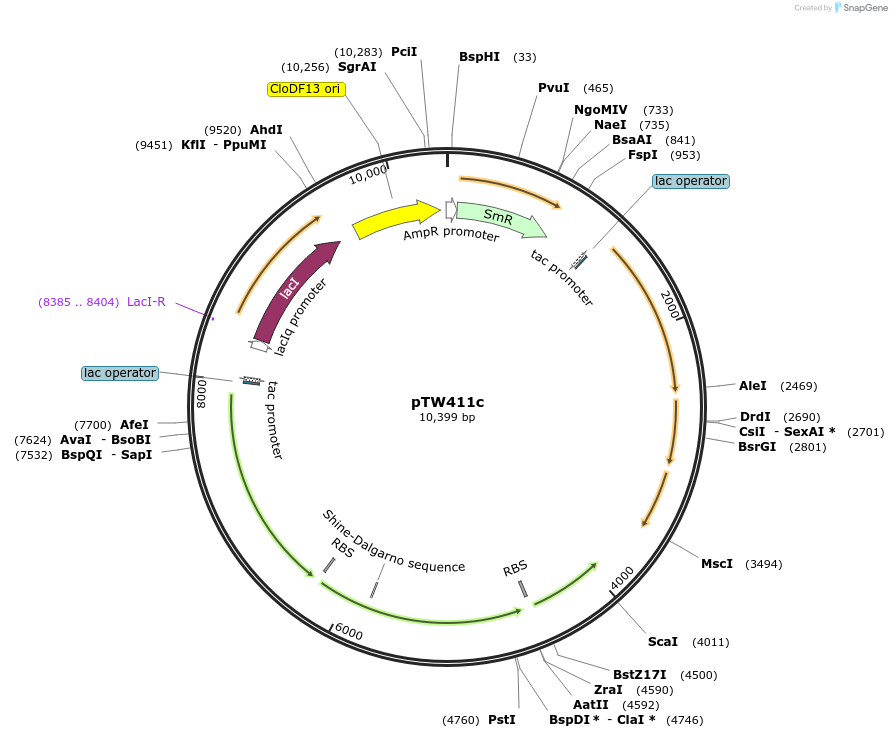
pTW411c
Plasmid#246584Purposedifferent combination of A. thaliana and N. tabacum Rubisco assembly factorsDepositorInsertExpressionBacterialAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
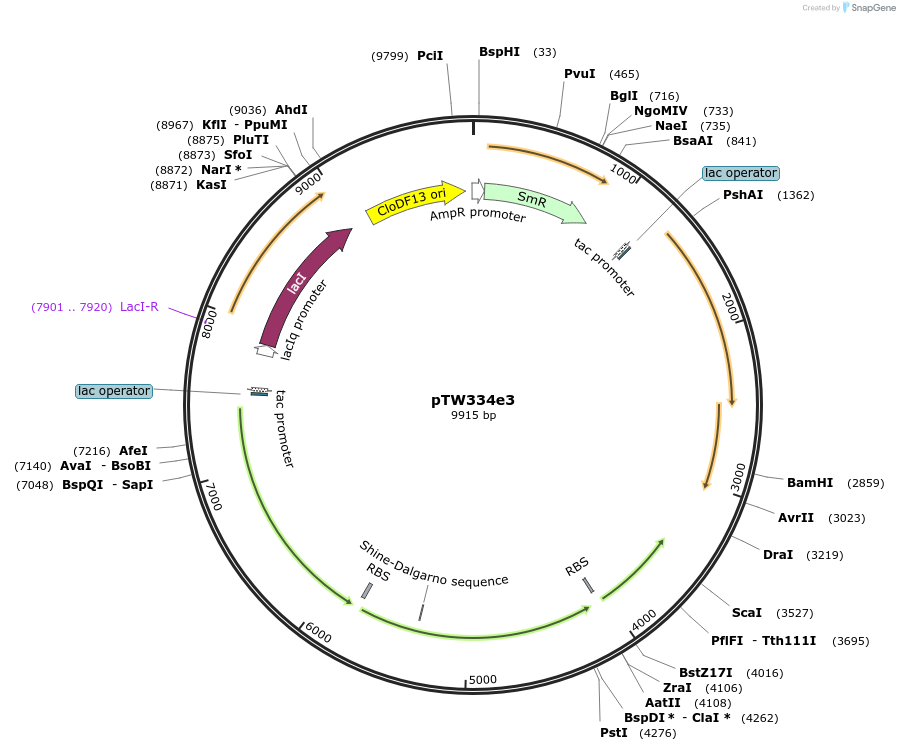
pTW334e3
Plasmid#246588Purposesequential knock out of A. thaliana Rubisco assembly factorsDepositorInsertExpressionBacterialAvailable SinceNov. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
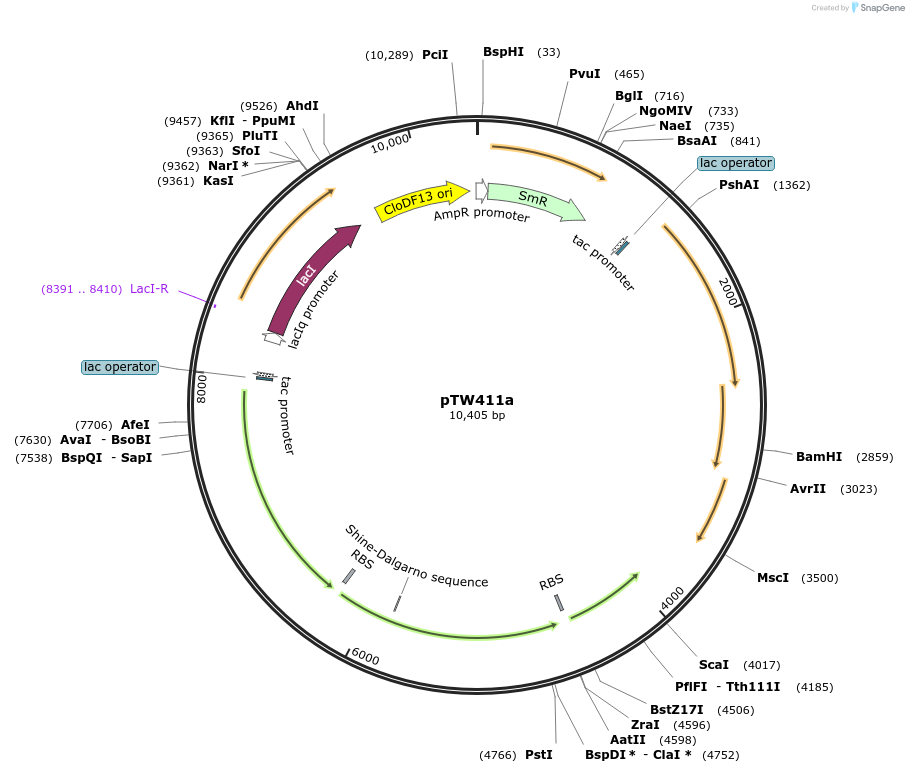
pTW411a
Plasmid#246582Purposedifferent combination of A. thaliana and N. tabacum Rubisco assembly factorsDepositorInsertExpressionBacterialAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
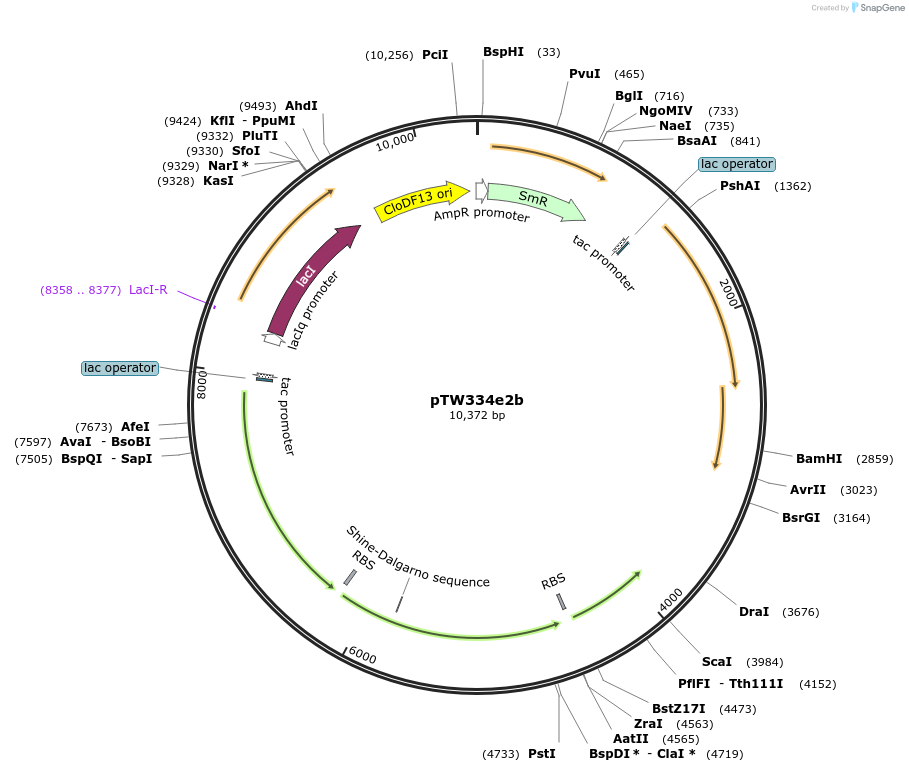
pTW334e2b
Plasmid#246587Purposesequential knock out of A. thaliana Rubisco assembly factorsDepositorInsertExpressionBacterialMutationAtBSD2W108A/L109EAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
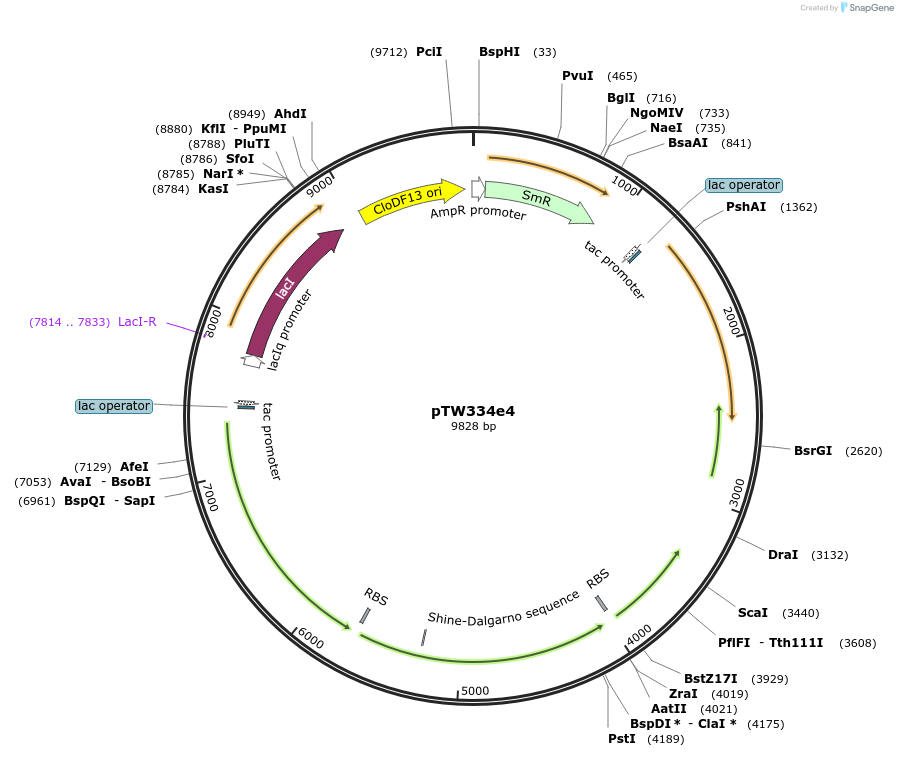
pTW334e4
Plasmid#246589Purposesequential knock out of A. thaliana Rubisco assembly factorsDepositorInsertExpressionBacterialAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
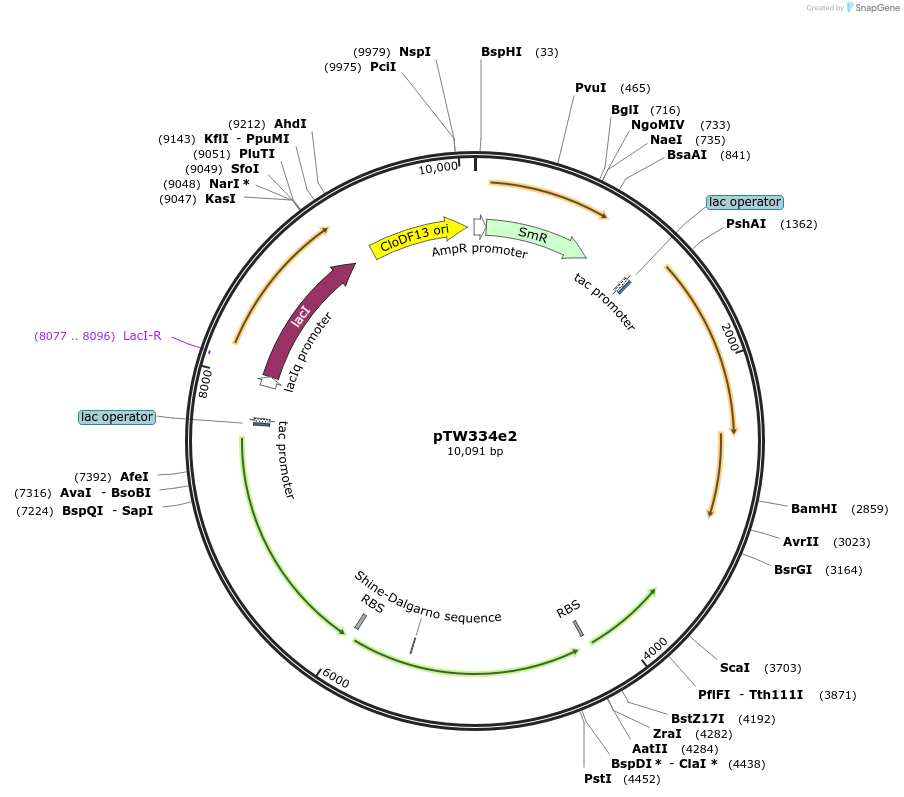
pTW334e2
Plasmid#246586Purposesequential knock out of A. thaliana Rubisco assembly factorsDepositorInsertExpressionBacterialAvailable SinceOct. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
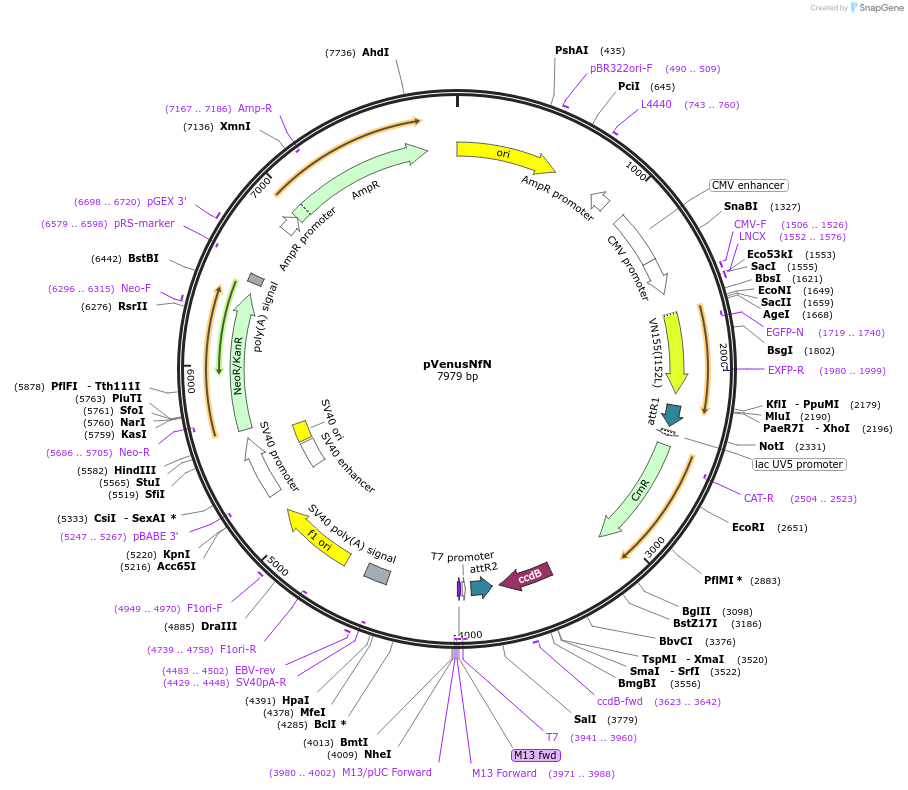
pVenusNfN
Plasmid#128564PurposeProtein interaction screening tool with Gateway-compatible BiFC vectors comprising of a set of 4 vectors permiting N or C fusions of candidate genes to the N or C fragment of Split-Venus.DepositorInsertVenus
ExpressionMammalianAvailable SinceJan. 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
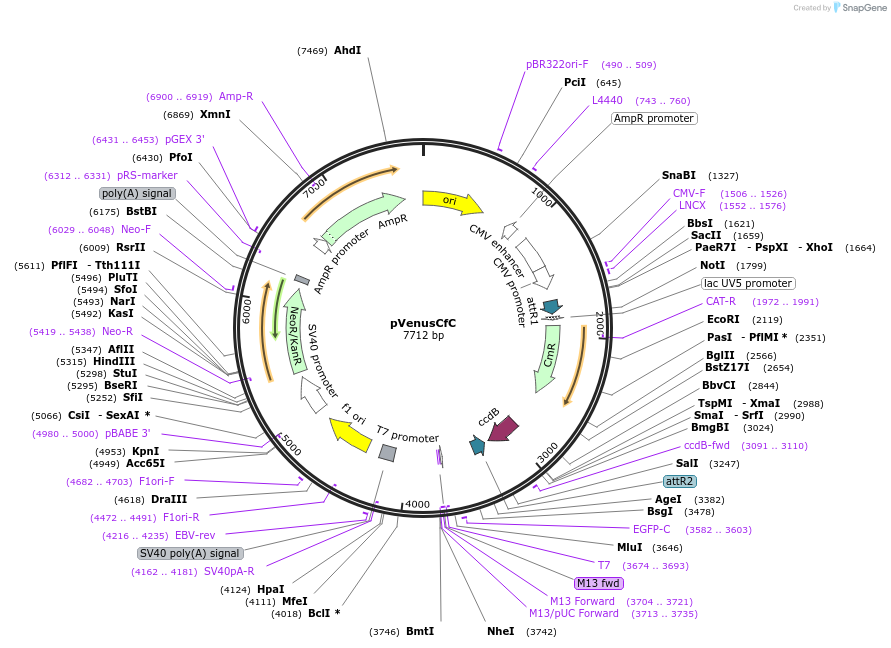
pVenusCfC
Plasmid#128567PurposeProtein interaction screening tool with Gateway-compatible BiFC vectors comprising of a set of 4 vectors permiting N or C fusions of candidate genes to the N or C fragment of Split-Venus.DepositorInsertVenus
ExpressionMammalianAvailable SinceJan. 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
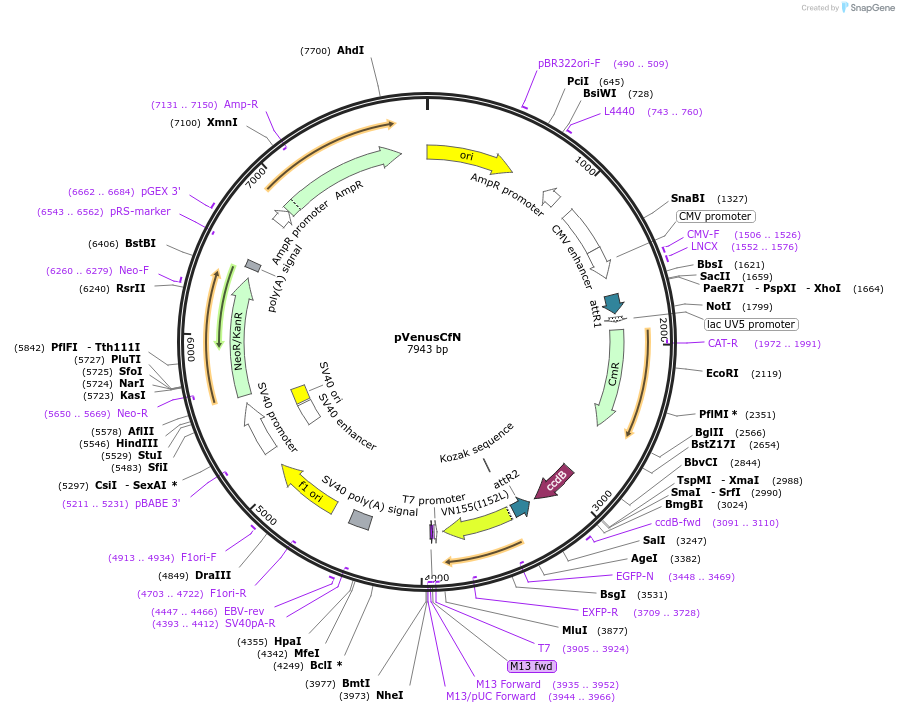
pVenusCfN
Plasmid#128566PurposeProtein interaction screening tool with Gateway-compatible BiFC vectors comprising of a set of 4 vectors permiting N or C fusions of candidate genes to the N or C fragment of Split-Venus.DepositorInsertVenus
ExpressionMammalianAvailable SinceJan. 15, 2021AvailabilityAcademic Institutions and Nonprofits only -
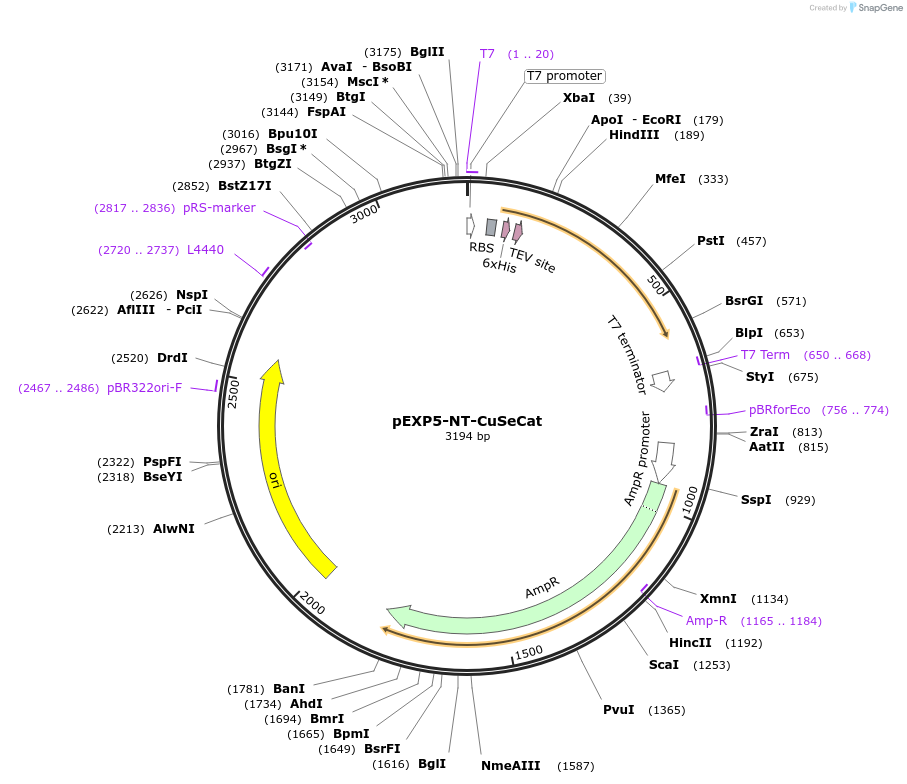
pEXP5-NT-CuSeCat
Plasmid#42568DepositorInsertmutated calmodulin gene
TagsPolyhistidine (6×His) region, TEV recognition sit…ExpressionBacterialMutationchanged Phenylalanine 92 to Glutamine acid, Valin…PromoterT7Available SinceMarch 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
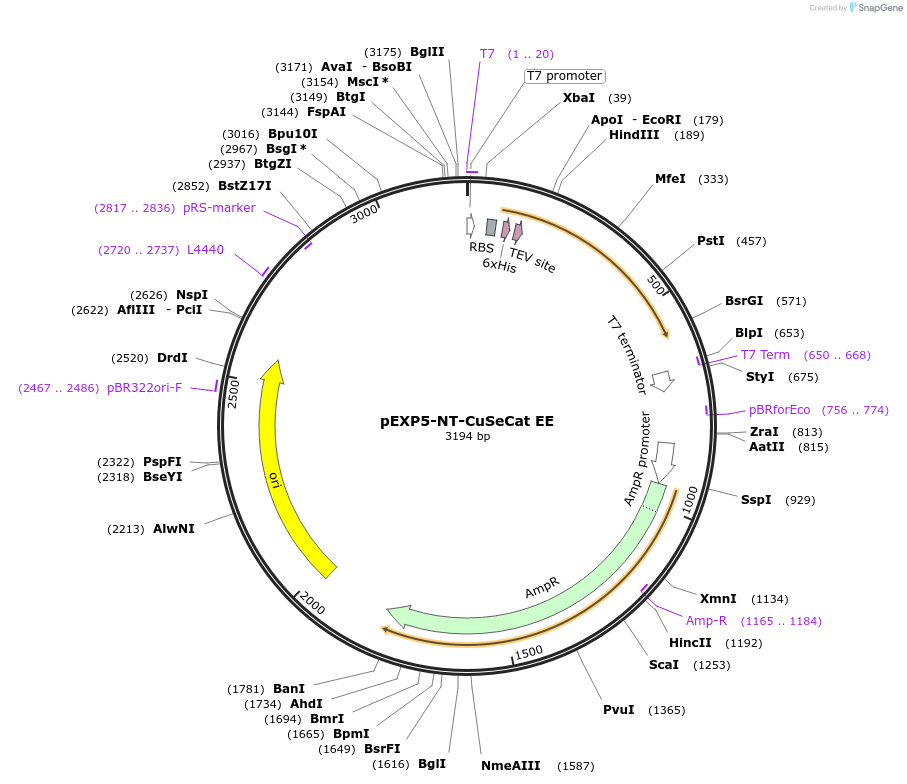
pEXP5-NT-CuSeCat EE
Plasmid#42569DepositorInsertmutated calmodulin gene
TagsPolyhistidine (6×His) region, TEV recognition sit…ExpressionBacterialMutationchanged Phenylalanine 92 to Glutamine acid, Valin…PromoterT7Available SinceMarch 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
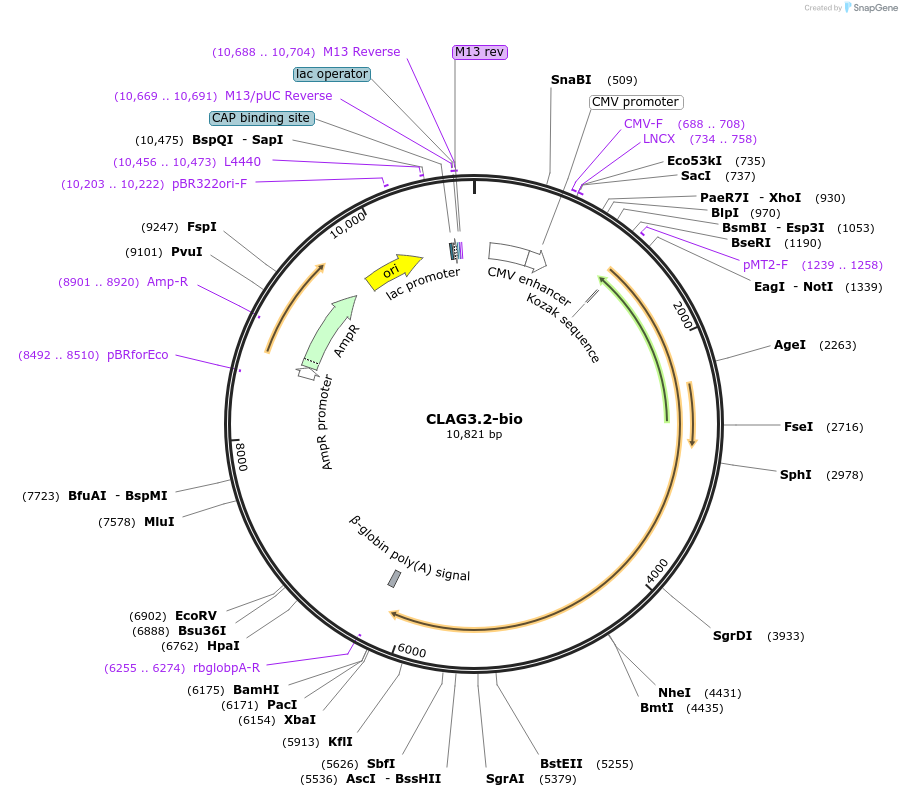
CLAG3.2-bio
Plasmid#47797PurposeExpresses enzymatically monobiotinylated full-length CLAG3.2 with no N-linked glycans upon cotransfection with BirA in mammalian cells. Contains a C-terminal rat Cd4 d3+4 tag.DepositorInsertCodon-optimised CLAG3.2
Tagsenzymatic biotinylation sequence and rat Cd4 d3+4ExpressionMammalianMutationExogenous signal peptide of mouse origin, changed…PromoterCMVAvailable SinceFeb. 4, 2014AvailabilityAcademic Institutions and Nonprofits only