We narrowed to 14,239 results for: TIM
-
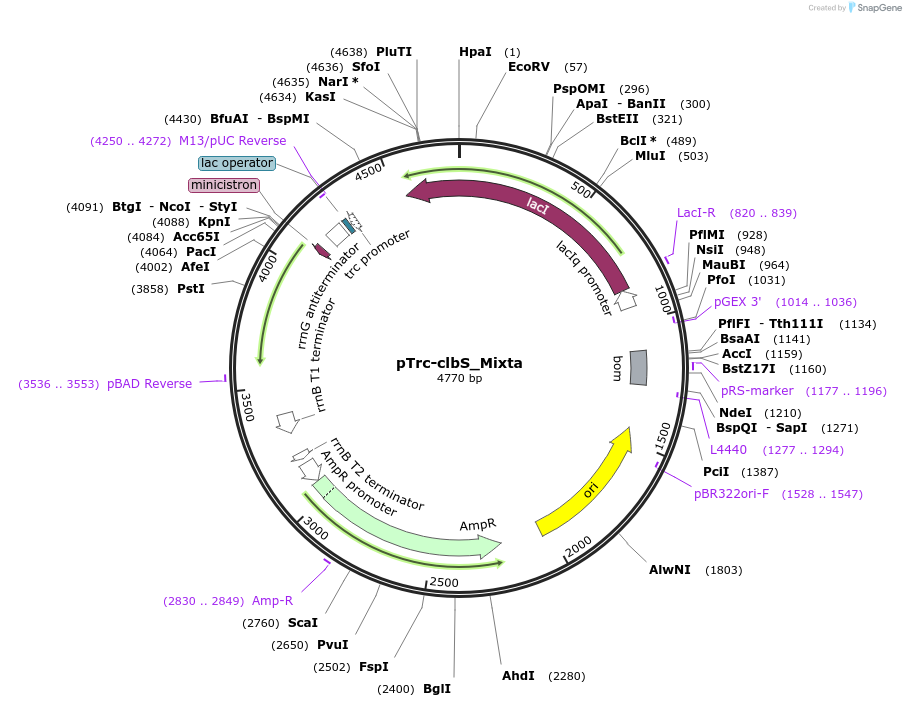
Plasmid#223352PurposeEncoding ClbS from Mixta theicolaDepositorInsertEncoding ClbS from Mixta theicola (Codon optimized for E coli)
ExpressionBacterialPromoterpTrcAvailable SinceNov. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
pTrc-clbS
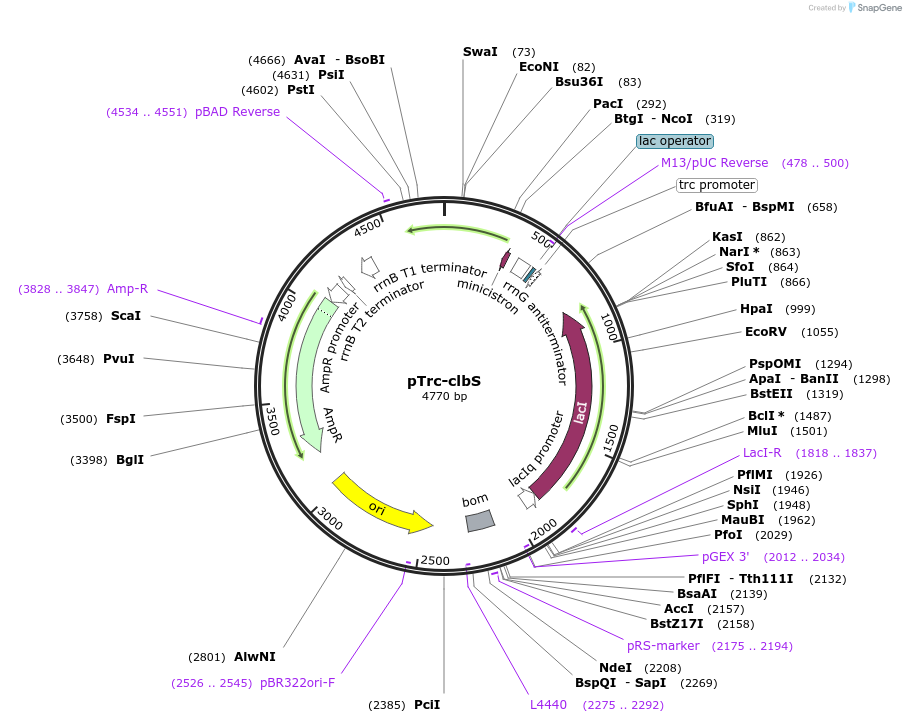
Plasmid#223350PurposeEncoding wild type ClbS from E. coli CFT073DepositorInsertClbS from CFT073 (Codon optimized for E coli)
ExpressionBacterialPromoterpTrcAvailable SinceNov. 8, 2024AvailabilityAcademic Institutions and Nonprofits only -
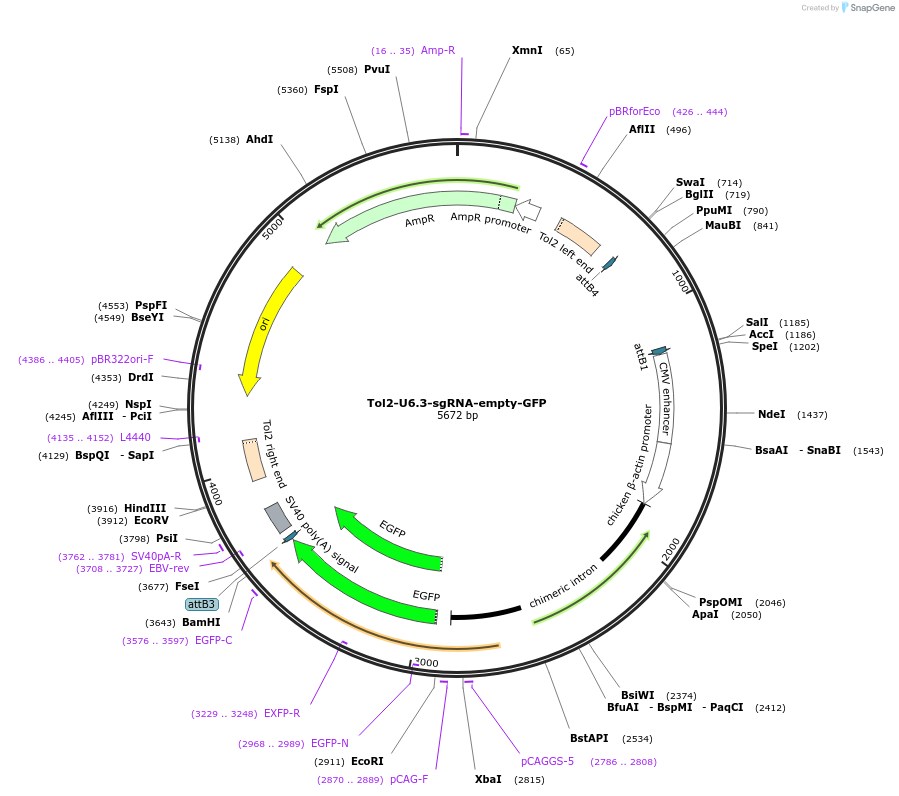
Tol2-U6.3-sgRNA-empty-GFP
Plasmid#221843Purposeempty vector to clone custom sgRNA into BsaI sites to express sgRNA from chick U6.3 promoter expresses GFP reporter from GAGC promoterDepositorInsertEGFP
UseCRISPR; Tol2 transposon optimised for chick expre…ExpressionMammalianPromoterCAGCAvailable SinceJuly 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
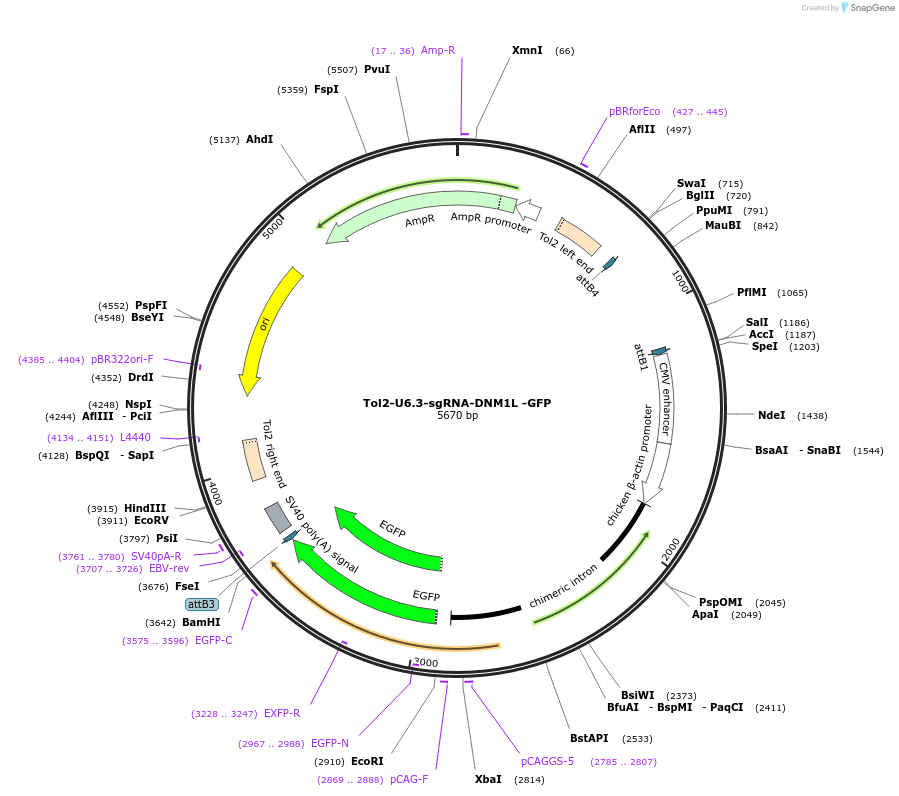
Tol2-U6.3-sgRNA-DNM1L -GFP
Plasmid#221845PurposeTol2 transposon expressing sgRNA targeting chick DNM1L from chick U6.3 promoter expresses GFP reporter from GAGC promoterDepositorInsertsEGFP
DNM1L sgRNA-gTGTTTTCCGACCATCCTCTG
UseCRISPR; Tol2 transposon optimised for chick expre…ExpressionMammalianPromoterCAGC and U6.3 chickAvailable SinceJuly 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
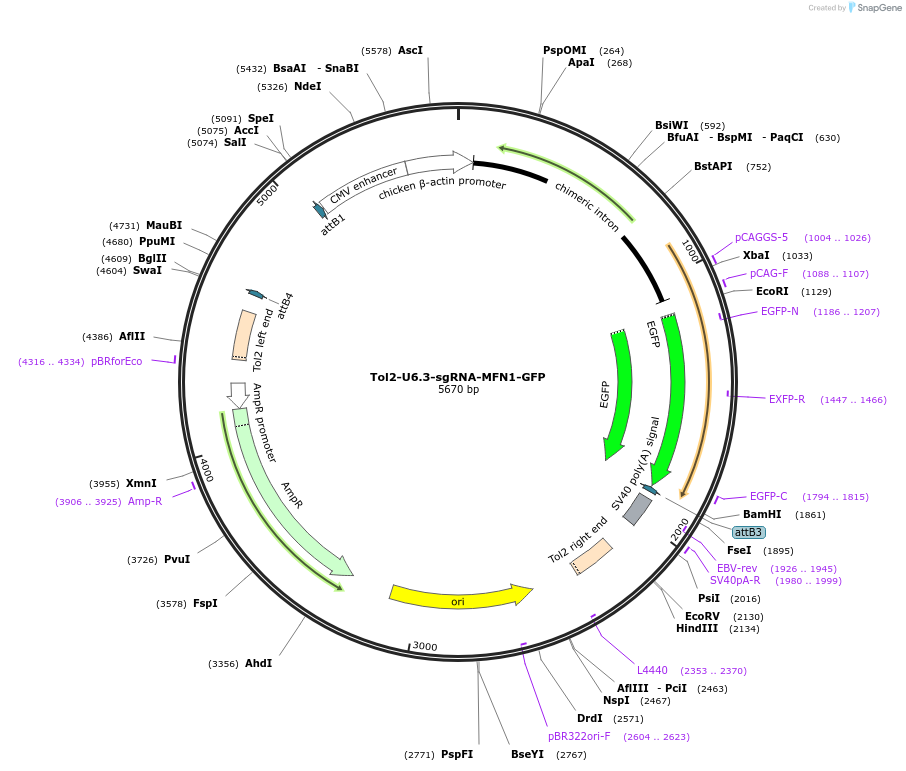
Tol2-U6.3-sgRNA-MFN1-GFP
Plasmid#221846PurposeTol2 transposon expressing sgRNA targeting chick MFN1 from chick U6.3 promoter expresses GFP reporter from GAGC promoteDepositorInsertsEGFP
MFN1 sgRNA-GAGAAGAAGAGCGTCAAGGT
UseCRISPR; Tol2 transposon optimised for chick expre…ExpressionMammalianPromoterCAGC and U6.3 chickAvailable SinceJuly 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
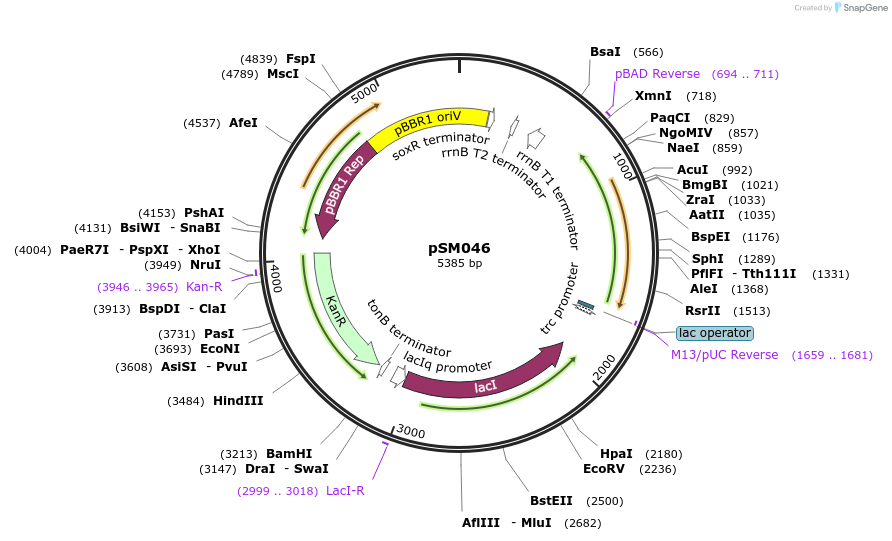
pSM045
Plasmid#217399PurposeKmR, pBTRcK broad host range vector with BTE1(C12-specific fatty acid thioesterase codon-optimised for C. necator) under PtrcDepositorInsertBTE1
ExpressionBacterialPromoterPTrcAvailable SinceJuly 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
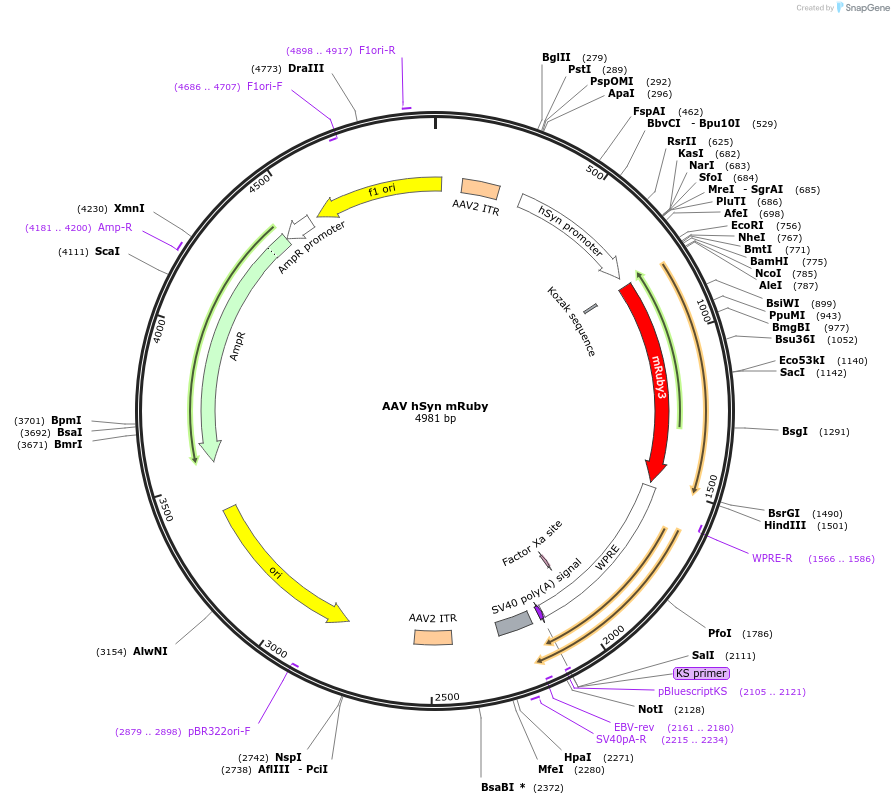
AAV hSyn mRuby
Plasmid#220912PurposeNeuronal specific expression of mRubyDepositorInsertmRuby
UseAAVExpressionMammalianPromoterhSynAvailable SinceJune 14, 2024AvailabilityAcademic Institutions and Nonprofits only -
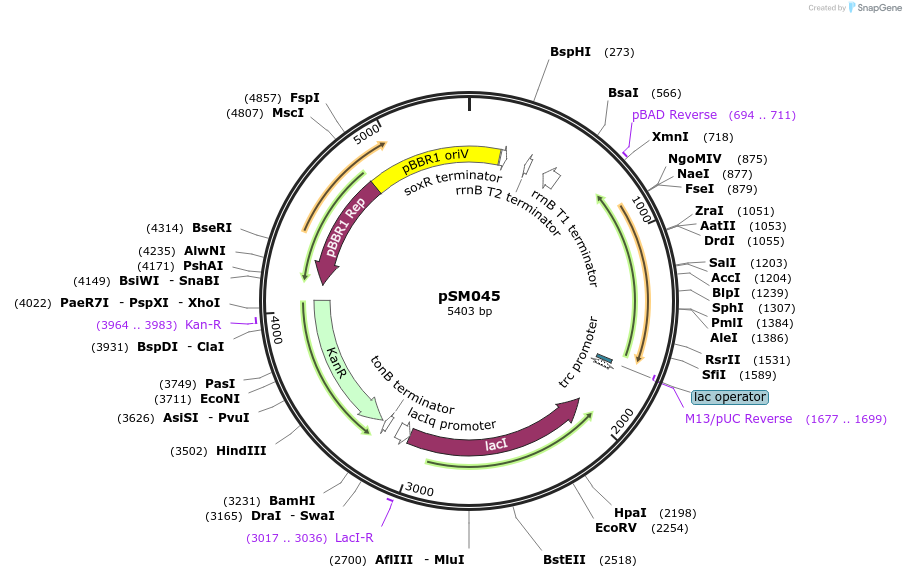
pSM046
Plasmid#217400PurposeKmR, pBTRcK broad host range vector with BTE2(C12-specific fatty acid thioesterase codon-optimised for C. necator) under PtrcDepositorInsertBTE2
UseSynthetic BiologyExpressionBacterialPromoterPTrcAvailable SinceJune 4, 2024AvailabilityAcademic Institutions and Nonprofits only -
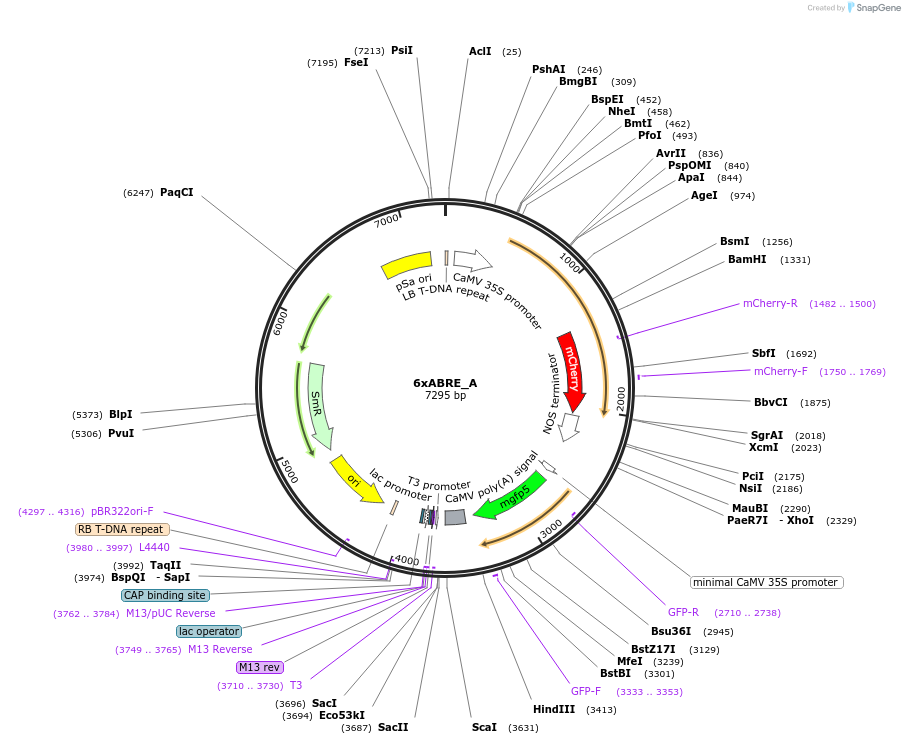
6xABRE_A
Plasmid#185803PurposeA synthetic ABA signaling reporter designed by multimerization of ABRE (7bp) and its flanking sequences from the ABI1 promoterDepositorInsert6xABRE(ABI1)+-86MP+erGFP
ExpressionPlantAvailable SinceFeb. 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
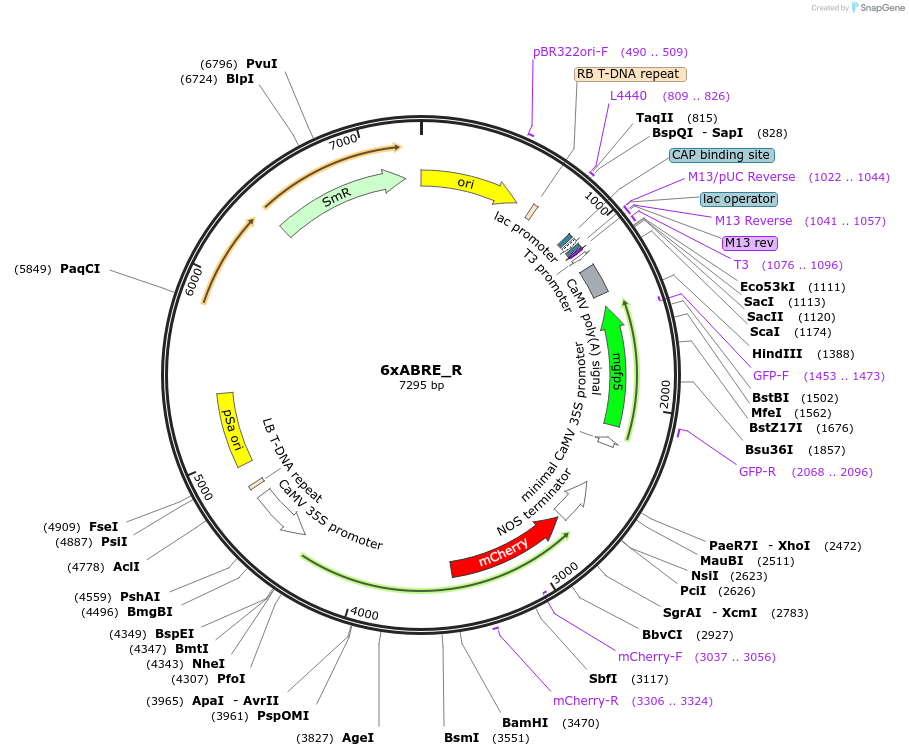
6xABRE_R
Plasmid#185804Purposea synthetic ABA signaling reporter designed by multimerization of ABRE (7bp) and its flanking sequences from the RD29A promoterDepositorInsert6xABRE(RD29A)+-86MP+erGFP
ExpressionPlantAvailable SinceJan. 24, 2024AvailabilityAcademic Institutions and Nonprofits only -
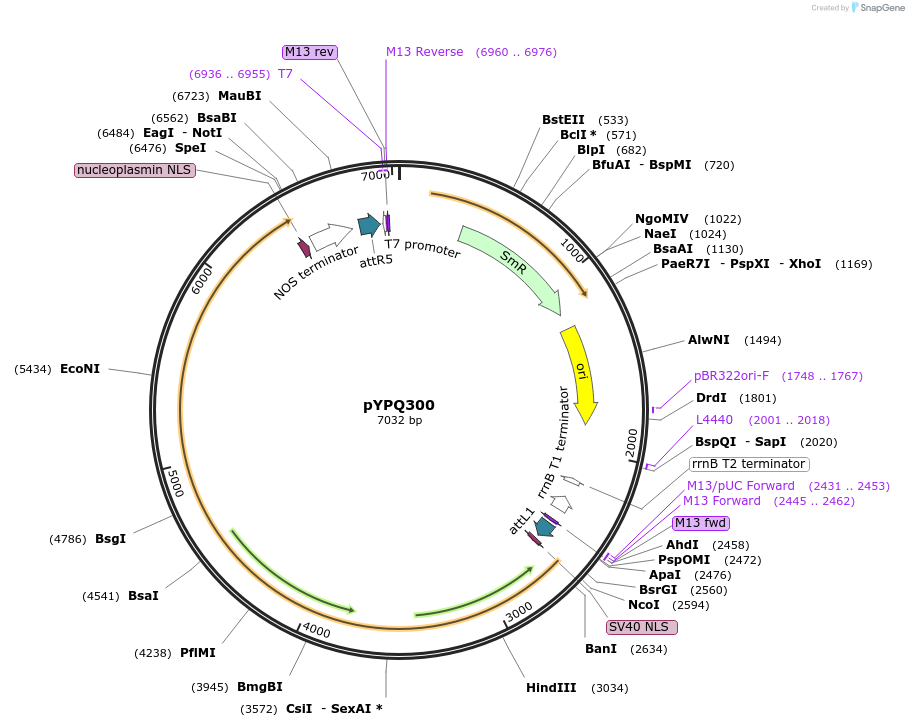
pYPQ300
Plasmid#205414PurposeGateway entry plasmid (attL1 & attR5) expressing rice codon optimized Ev1Cas12a without promoterDepositorInsertEv1Cas12a
UseCRISPR; Gateway compatible cas12a entry cloneTagsNLS (N terminal on insert) NLS (C terminal on ins…ExpressionPlantAvailable SinceNov. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
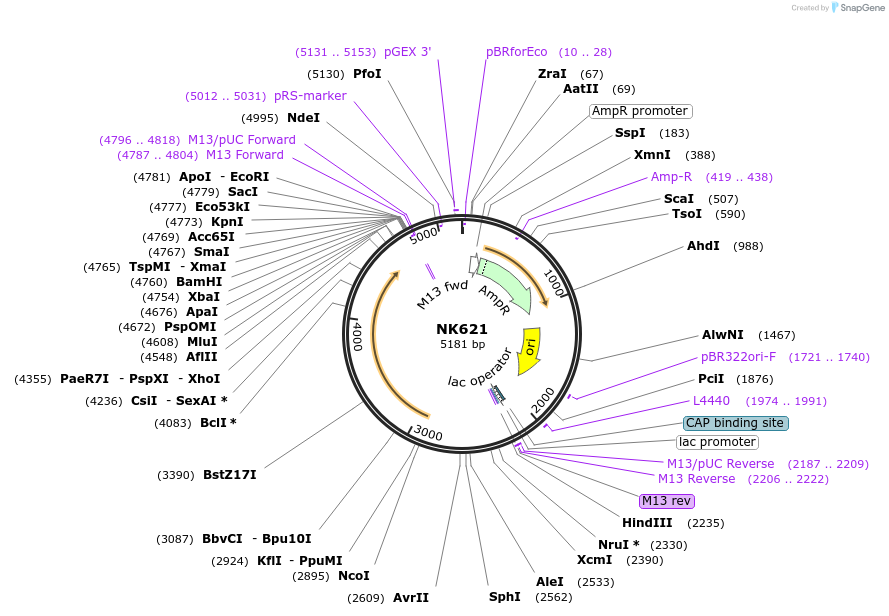
NK621
Plasmid#196405PurposeStrong expression of firefly luciferase codon-optimized for S. rosettaDepositorInsertSro5actin-firefly-Sro3actin
UseLuciferaseMutationN/A (synthetic)Available SinceAug. 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
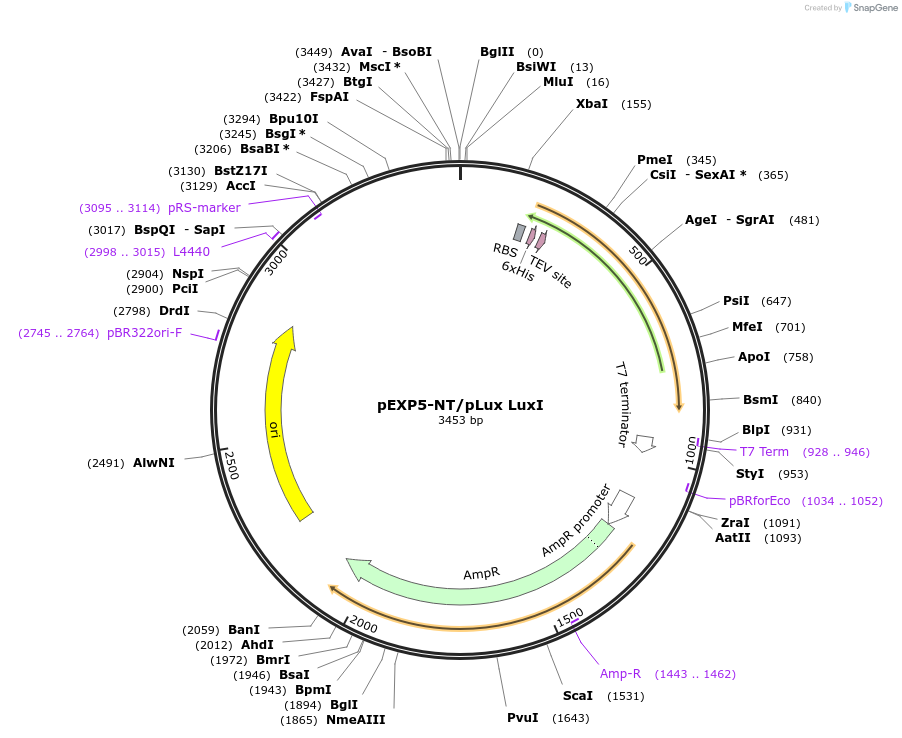
pEXP5-NT/pLux LuxI
Plasmid#193627PurposeInducible expression of LuxI by the transcription factor complex LuxR-3OC6 homoserine lactone (HSL). LuxI is codon optimized for E. coli.DepositorInsertLuxI
Tags6xHisExpressionBacterialPromoterLux promoterAvailable SinceJuly 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
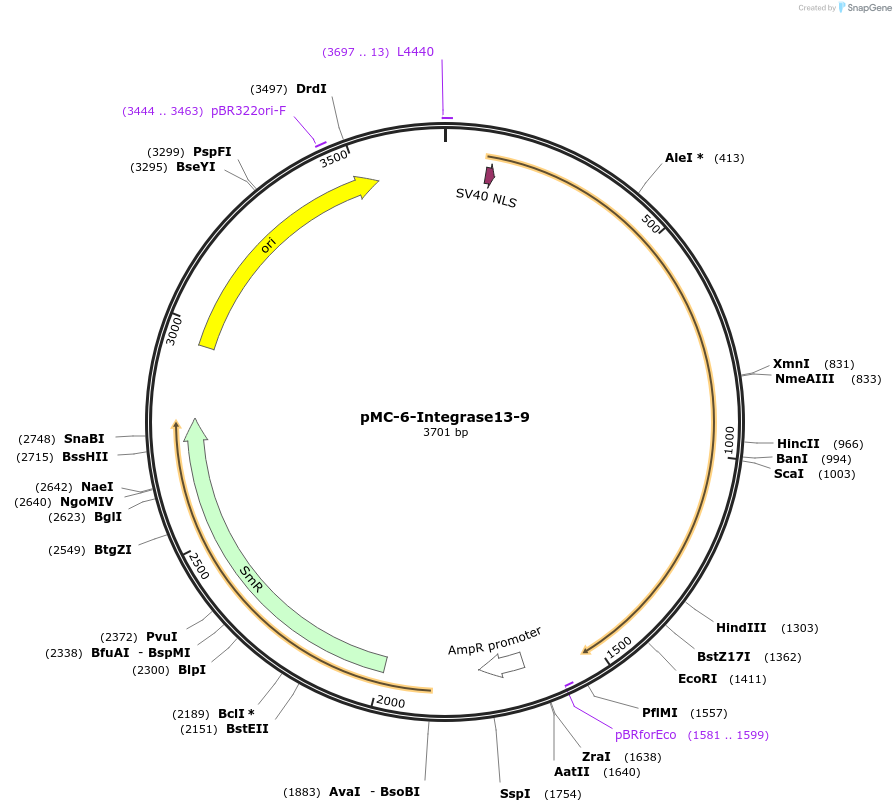
pMC-6-Integrase13-9
Plasmid#197728PurposePhytobrick (MoClo) Level 0 PartDepositorInsertIntegrase13 (tomato optimized)
ExpressionPlantAvailable SinceJune 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
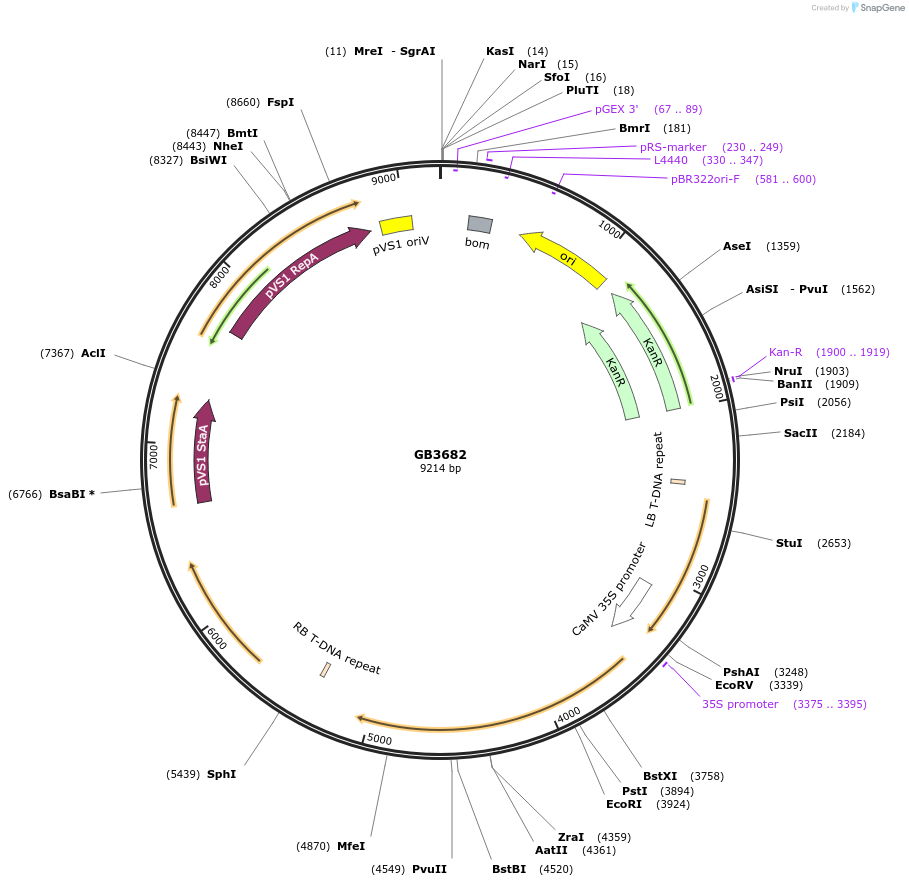
GB3682
Plasmid#187807PurposeTranscriptional unit for expression of alcohol O-acetyltransferase from Saccharomyces pastorianus strain CBS 1483 chromosome SeVIII-SeXV, codon optimized for Nicotiana.DepositorInsertSpATF1-2
ExpressionPlantAvailable SinceJune 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
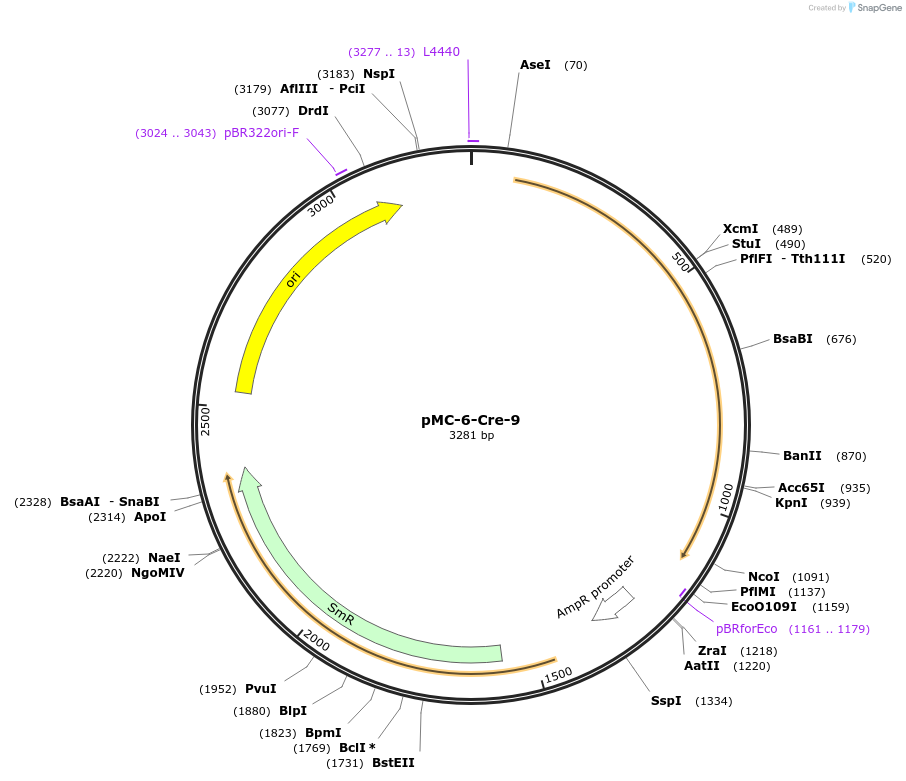
pMC-6-Cre-9
Plasmid#197721PurposePhytobrick (MoClo) Level 0 PartDepositorInsertCre recombinase (maize optimized)
ExpressionPlantAvailable SinceMay 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
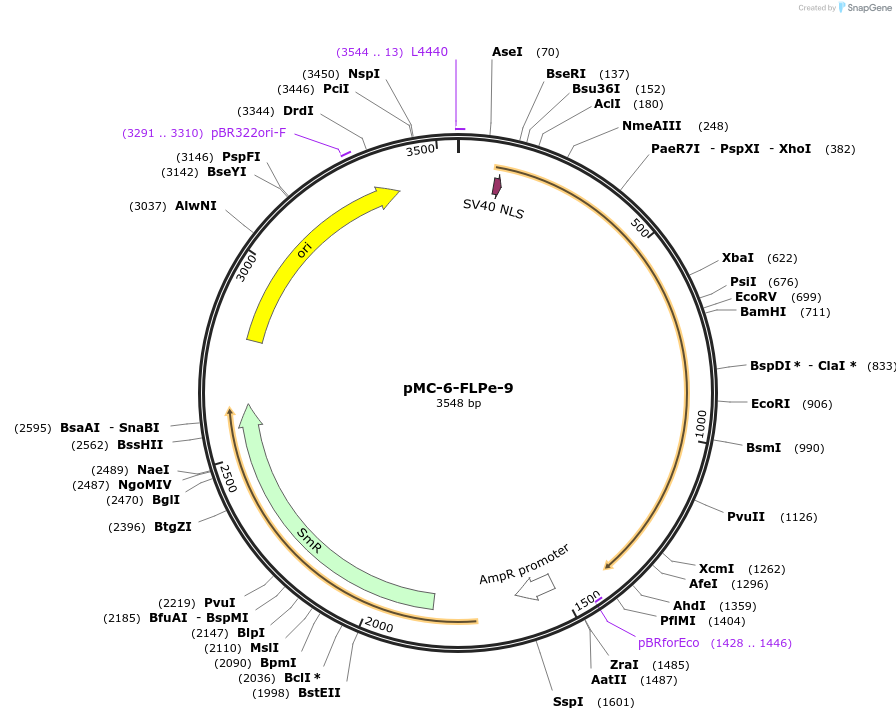
pMC-6-FLPe-9
Plasmid#197724PurposePhytobrick (MoClo) Level 0 PartDepositorInsertFLPe recombinase (tomato optimized)
ExpressionPlantAvailable SinceMay 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
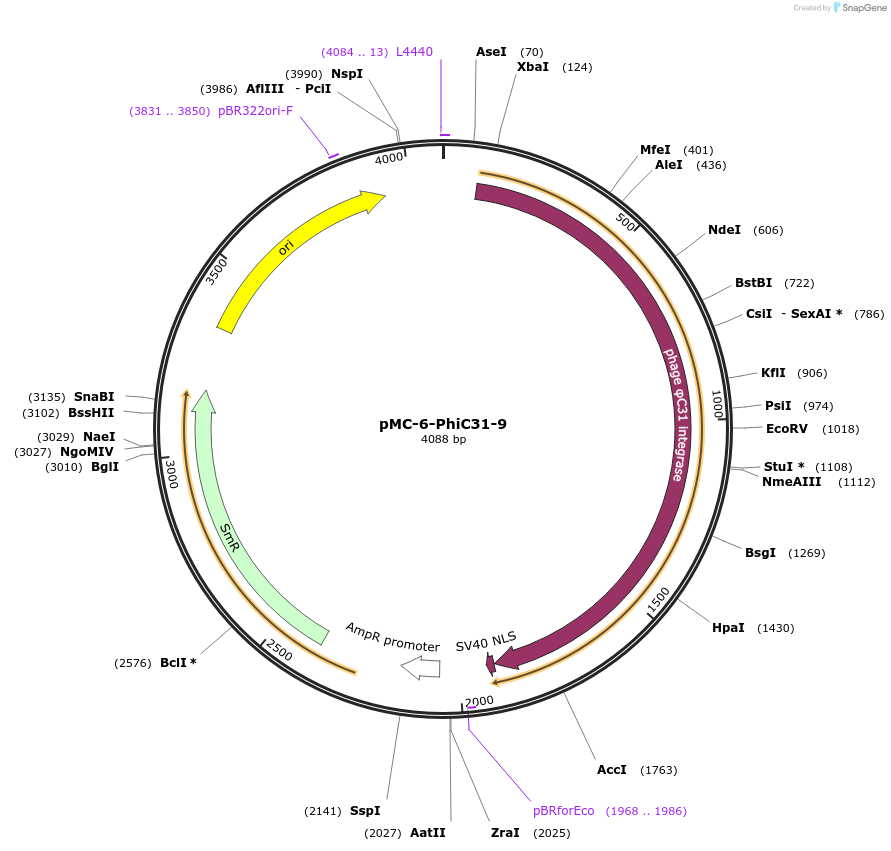
pMC-6-PhiC31-9
Plasmid#197725PurposePhytobrick (MoClo) Level 0 PartDepositorInsertPhiC31 recombinase (tomato optimized)
ExpressionPlantAvailable SinceMay 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
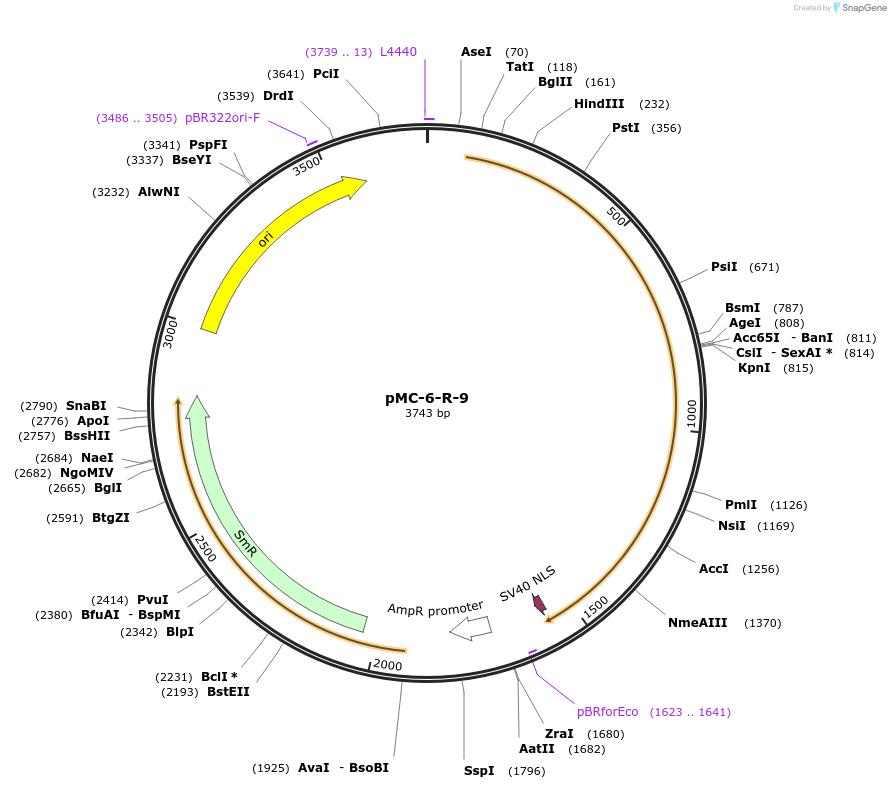
pMC-6-R-9
Plasmid#197726PurposePhytobrick (MoClo) Level 0 PartDepositorInsertR recombinase (tomato optimized)
ExpressionPlantAvailable SinceMay 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
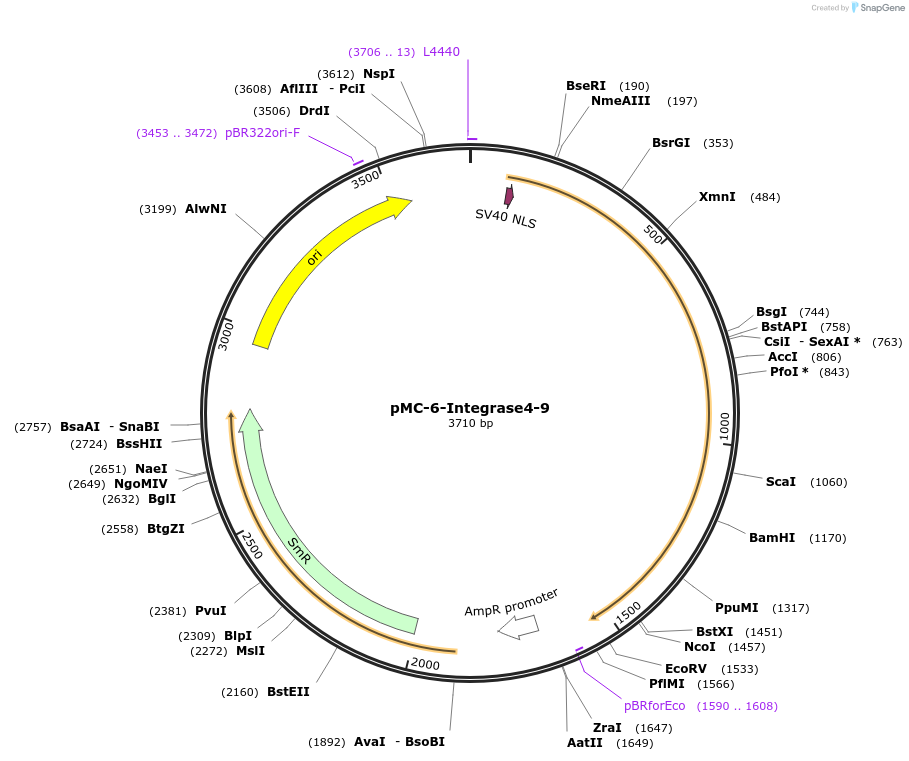
pMC-6-Integrase4-9
Plasmid#197727PurposePhytobrick (MoClo) Level 0 PartDepositorInsertIntegrase4 (tomato optimized)
ExpressionPlantAvailable SinceMay 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
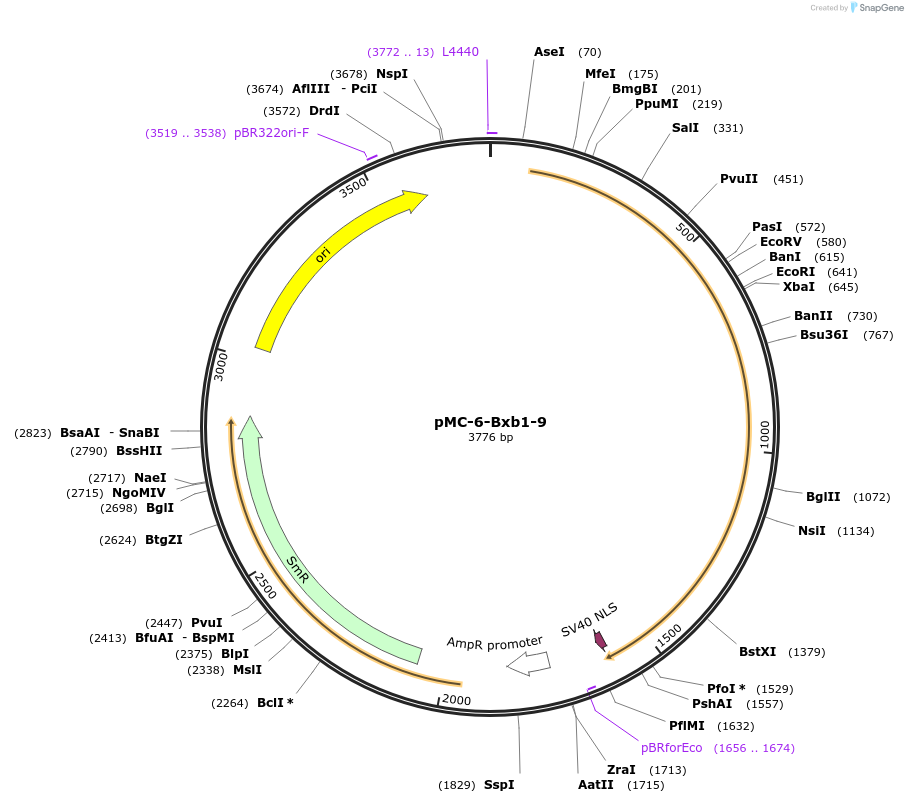
pMC-6-Bxb1-9
Plasmid#197730PurposePhytobrick (MoClo) Level 0 PartDepositorInsertBxb1 recombinase (tomato optimized)
ExpressionPlantAvailable SinceMay 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
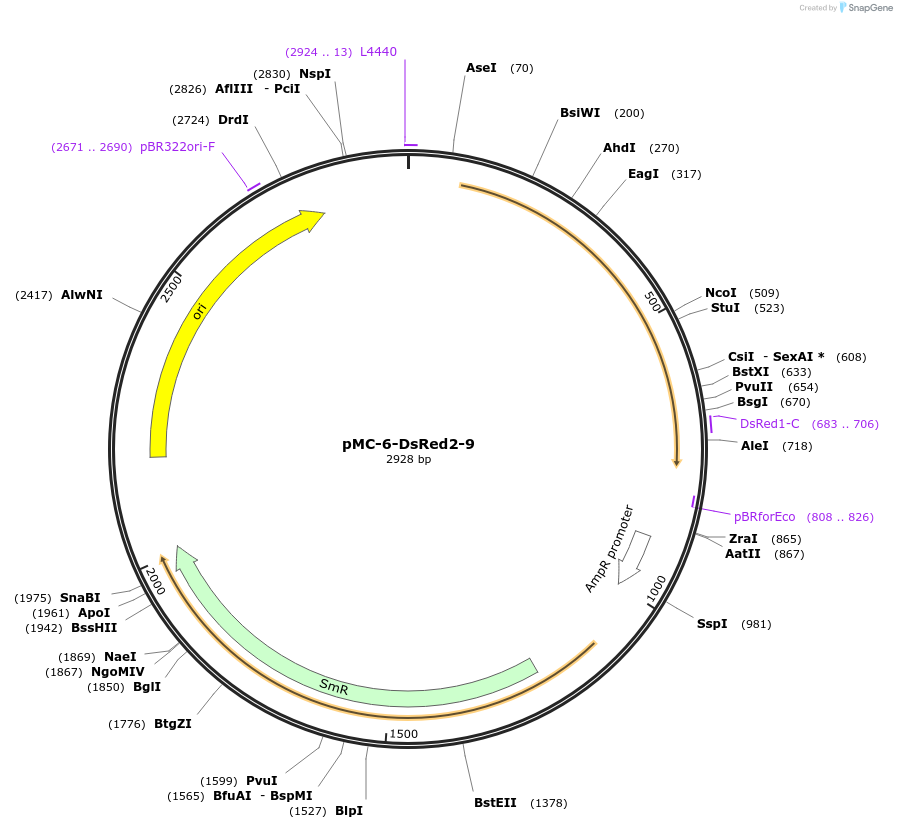
pMC-6-DsRed2-9
Plasmid#197735PurposePhytobrick (MoClo) Level 0 PartDepositorInsertDsRed2 (maize optimized)
ExpressionPlantAvailable SinceMay 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
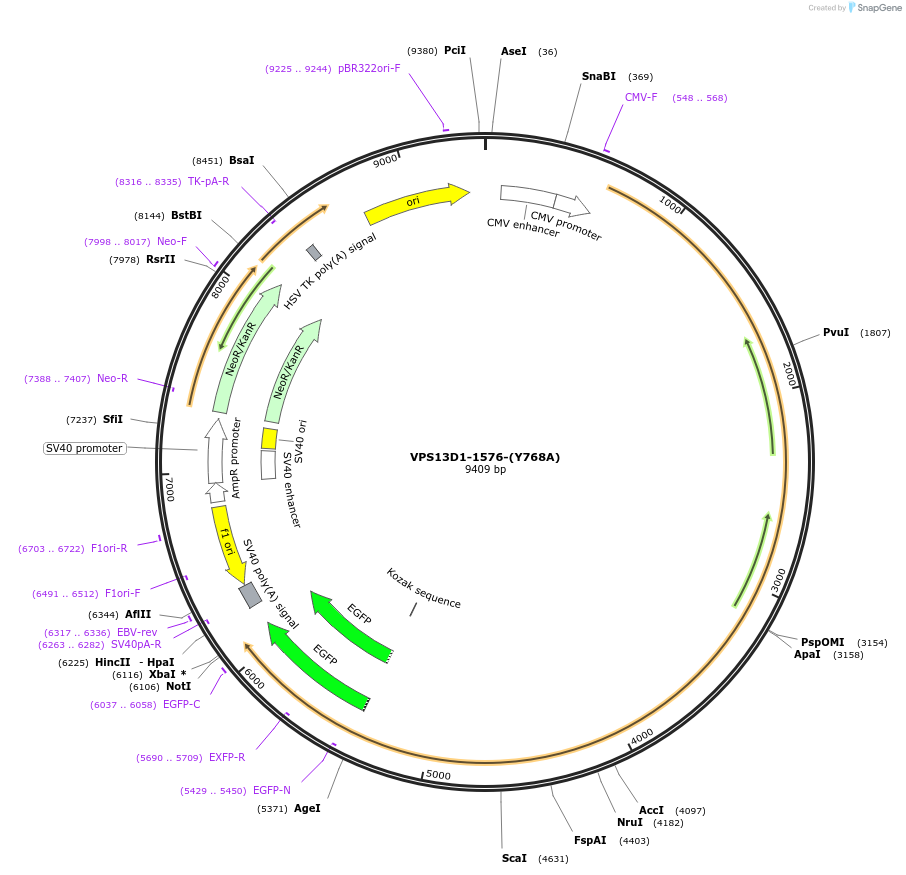
VPS13D1-1576-(Y768A)
Plasmid#194006PurposeY to A mutation in VPS13D geneDepositorAvailable SinceApril 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
VPS13D1-1576-(Y1253S)
Plasmid#194005PurposeY to S mutation in VPS13D geneDepositorAvailable SinceApril 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
pLKO.1 - TRC mTagBFP2-Synapsin1a
Plasmid#191567PurposeExpresses an shRNA and mTagBFP2-synapsin1aDepositorTypeEmpty backboneUseLentiviral and RNAiExpressionMammalianPromoterU6 for shRNAAvailable SinceOct. 17, 2022AvailabilityAcademic Institutions and Nonprofits only -
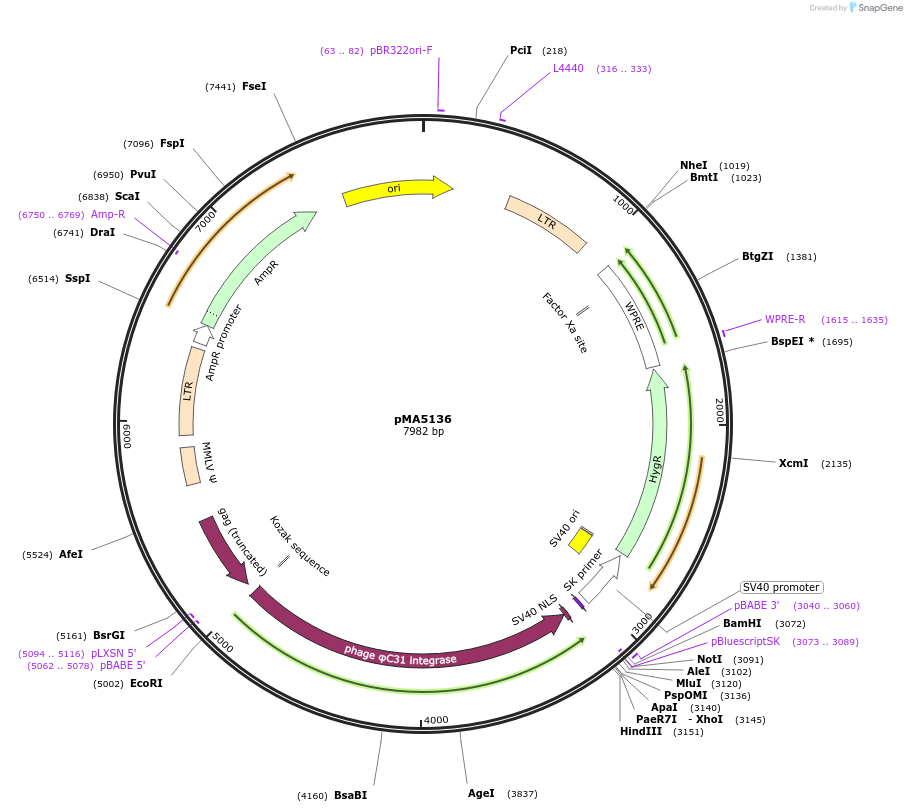
pMA5136
Plasmid#184853PurposeA retroviral vector encoding hygromycin resistance and PhiC31 recombinaseDepositorInsertPhiC31o recombinase
UseRetroviral; Phic31 attp-attb recombination/excisi…ExpressionMammalianMutationCodon-optimized for E. coli expressionPromoterLTRAvailable SinceAug. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
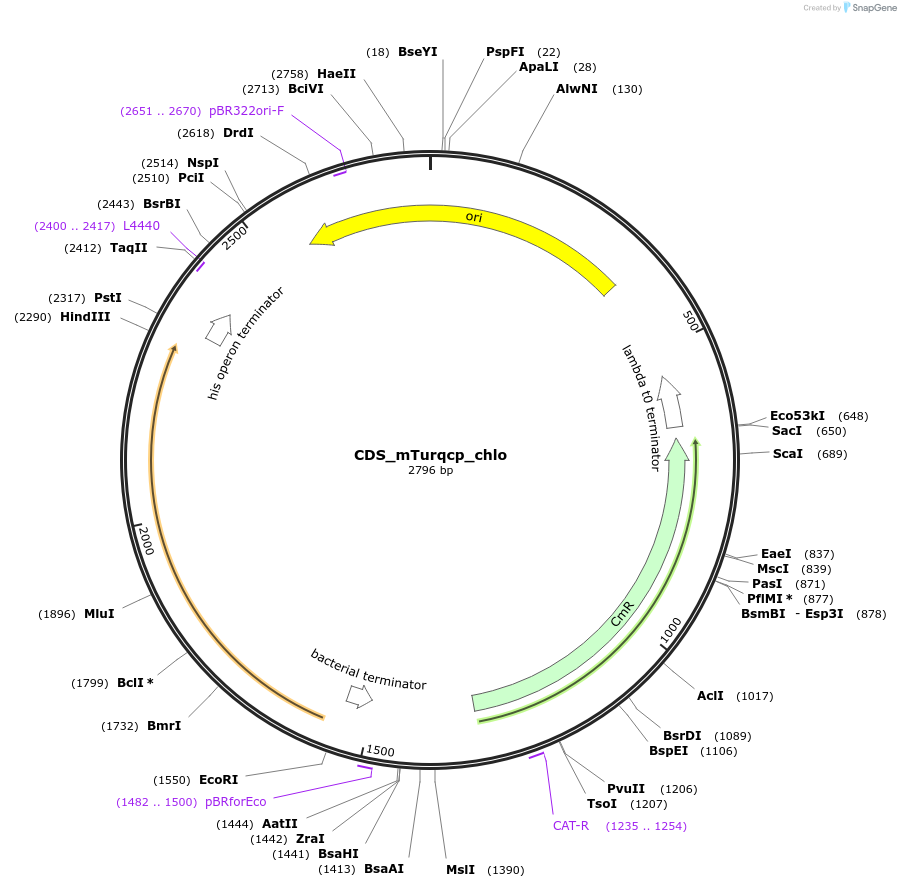
CDS_mTurqcp_chlo
Plasmid#163936PurposeL0 part - CDSDepositorInsertFP optimised for Marchantia polymorpha chloroplast expression, from Genbank ID KT364744
ExpressionBacterialAvailable SinceNov. 30, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
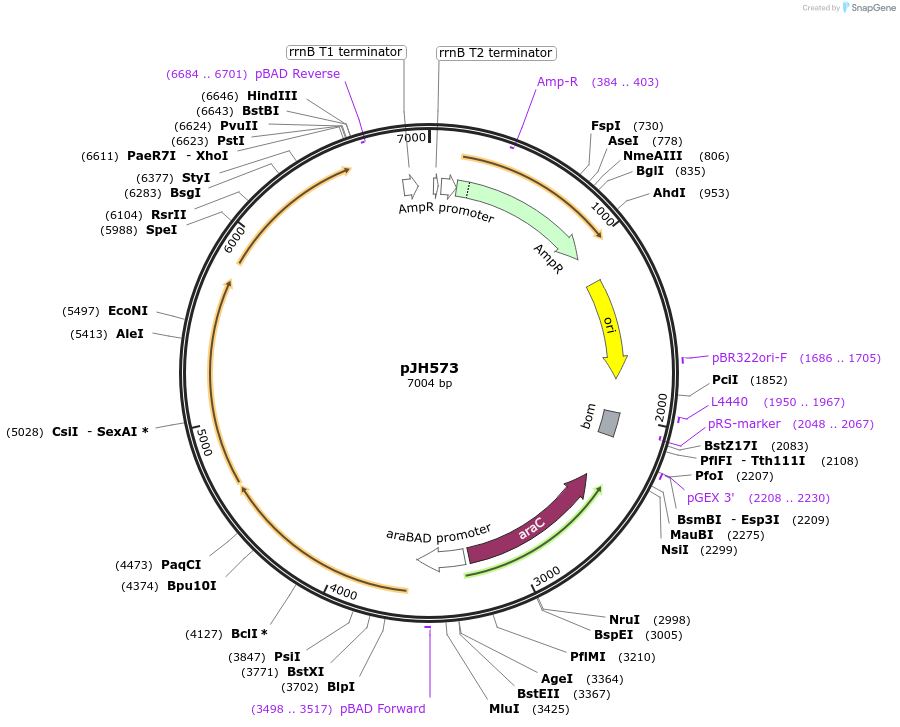
pJH573
Plasmid#172157PurposepylBCD variant in pBAD backboneDepositorInsertPara (Untagged pylBCD); AraC
ExpressionBacterialMutationCodon-optimized Methanosarcina acetivorans pylBCDAvailable SinceSept. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
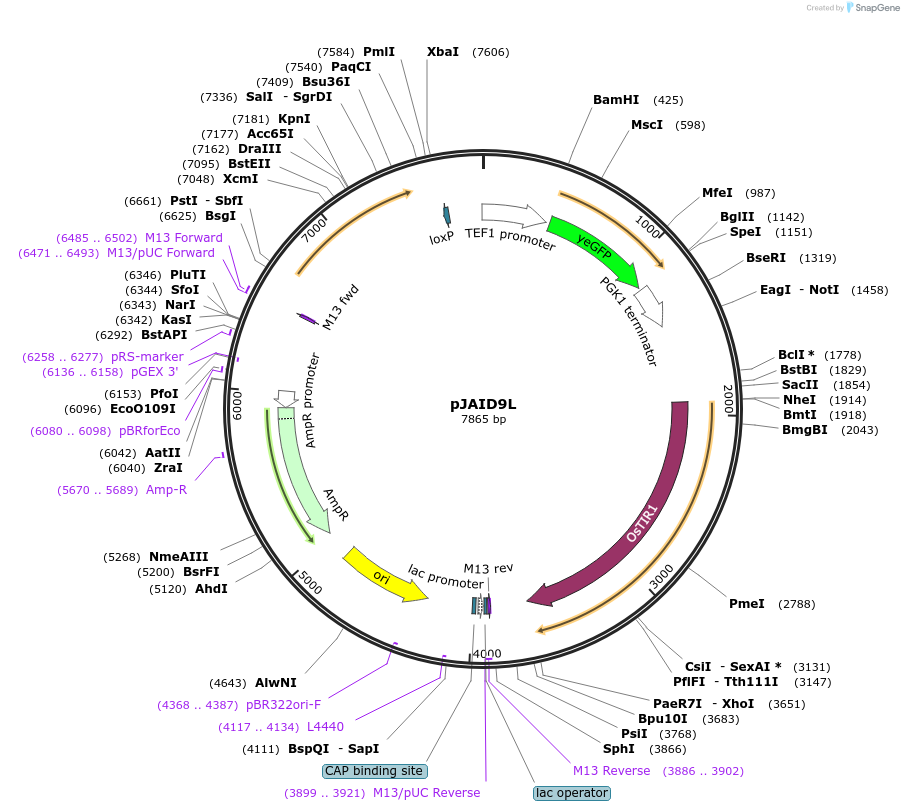
pJAID9L
Plasmid#165052PurposeIntegrative expression of yEGFP and rice auxin receptor gene OsTIR1. OsTIR1 is codon-optimised for expression in S. cerevisiaeDepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>yEGFP>T-PGK1-P-ACS2>OsTIR1opt>T-URA3
ExpressionYeastMutationWTAvailable SinceMarch 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
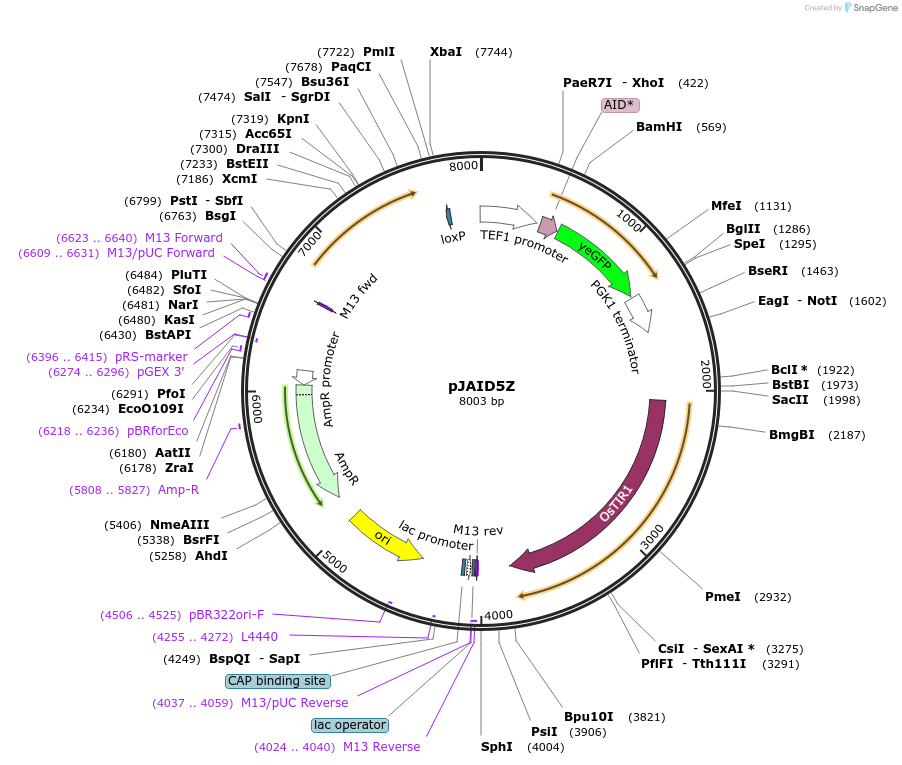
pJAID5Z
Plasmid#165047PurposeIntegrative expression of AID* (mini auxin inducible degron)-yEGFP and rice auxin receptor gene OsTIR1.DepositorInsertP-URA3>KlURA3>T-AgTEF1-P-TEF1>AID*>yEGFP>T-PGK1-P-ACS2>OsTIR1opt>T-URA3
ExpressionYeastMutationOsTIR1 is codon-optimised for expression in S. ce…Available SinceMarch 12, 2021AvailabilityAcademic Institutions and Nonprofits only