We narrowed to 169,365 results for: htt
-
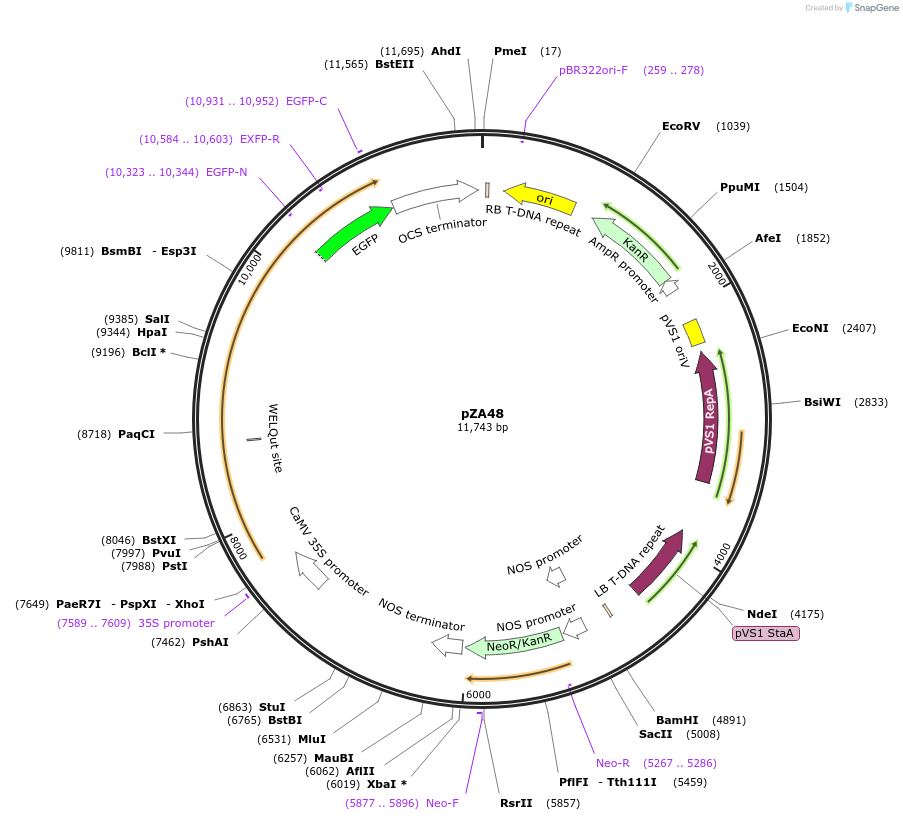
Plasmid#158526PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V/L17E-eGFP. Km plant selection marker.DepositorInsertZAR1-eGFP
TagseGFPExpressionPlantMutationL17E, D481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
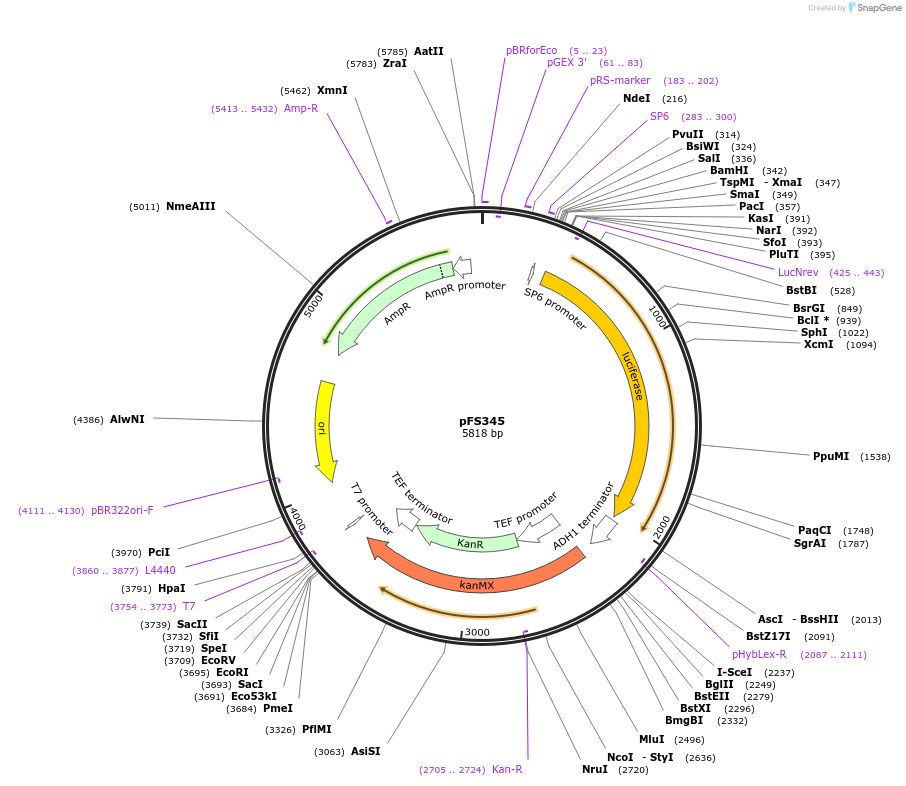
pFS345
Plasmid#89349PurposeBeetle Luciferase C-term tagging vectorDepositorInsertBeetle luciferase
UseC-terminal tagging vectorAvailable SinceJuly 18, 2017AvailabilityAcademic Institutions and Nonprofits only -
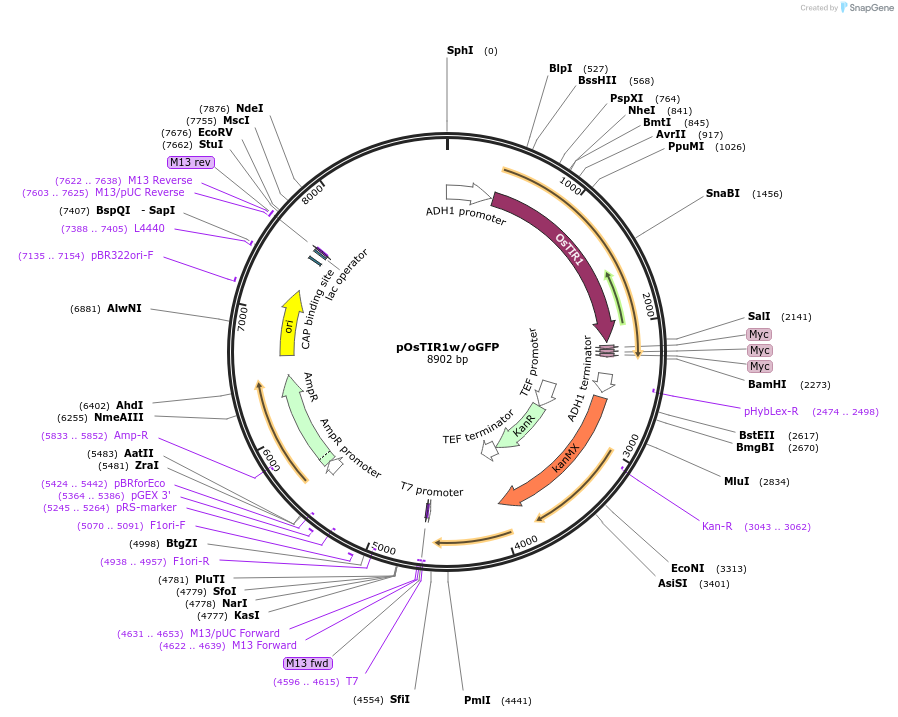
pOsTIR1w/oGFP
Plasmid#102883PurposeExpresses OsTIR1 via the ADH1 promoter in yeast. The cassette is integrated in the HO locus. Assembly and characterization of the auxin-inducible degradation.DepositorInsertOsTIR1
Tags3x mycExpressionYeastPromoterADH1Available SinceJan. 5, 2018AvailabilityAcademic Institutions and Nonprofits only -
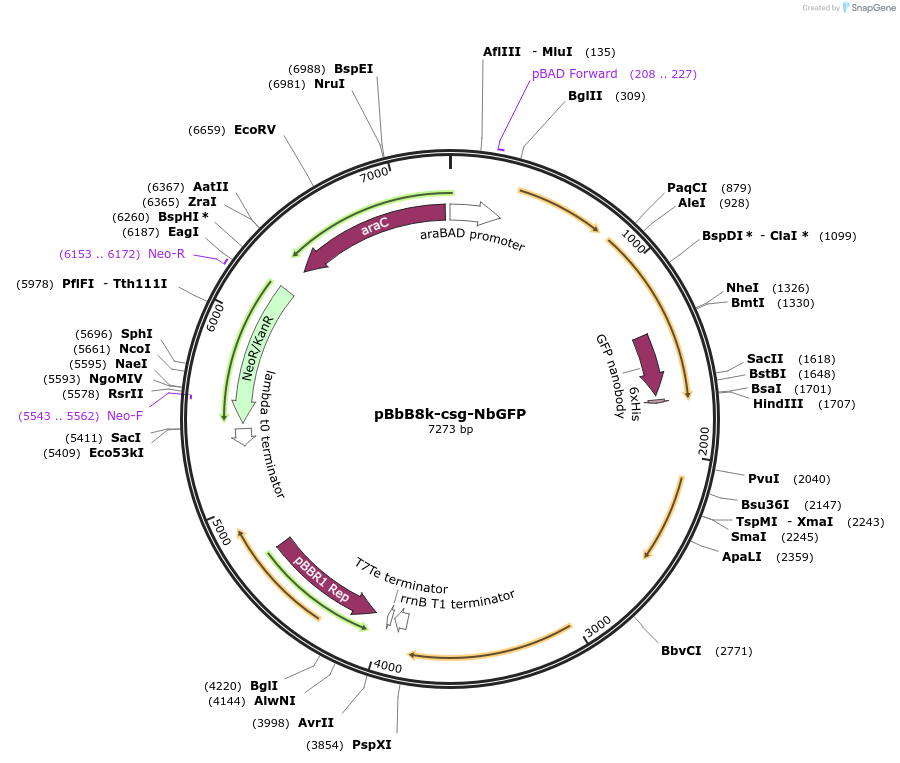
pBbB8k-csg-NbGFP
Plasmid#166858PurposeArabinose-inducible csgBACEFG operon for curli fiber synthesis in which curli subunit CsgA has been fused to a GFP-specific nanobody via a flexible linker. Derived from https://www.addgene.org/35363/.DepositorInsertcsgBACEFG, with GFP-specific nanobody fused to CsgA via flexible linker
UseSynthetic BiologyExpressionBacterialAvailable SinceMarch 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
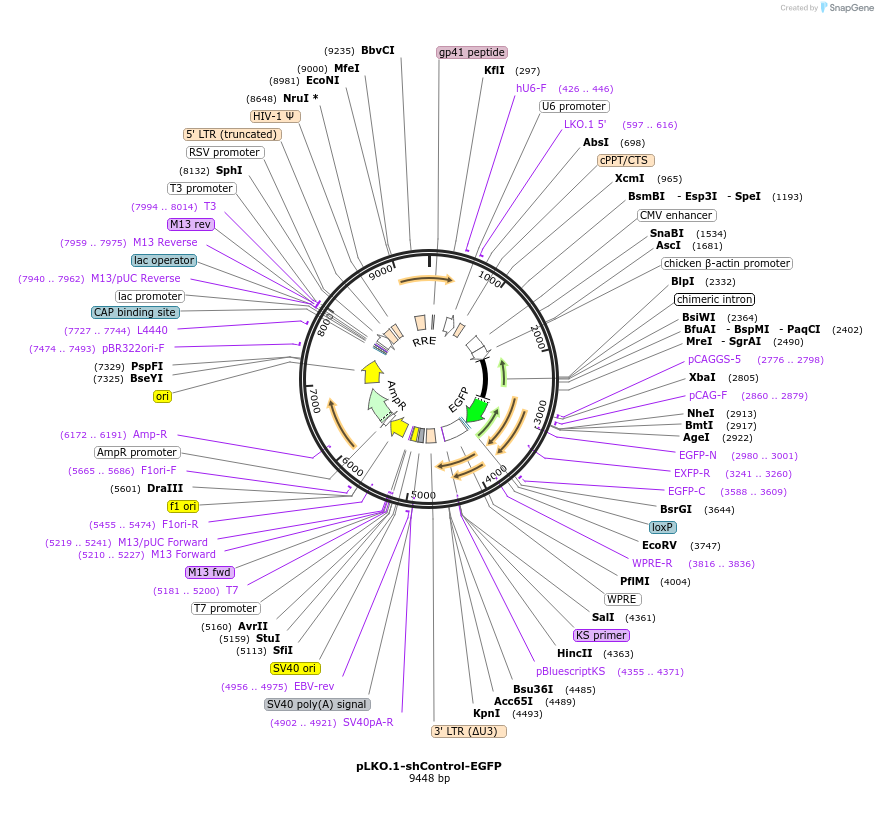
pLKO.1-shControl-EGFP
Plasmid#227685PurposeExpresses EGFP and scramble of shRNA targeting Atg7DepositorInsertscrambled shRNA control
ExpressionMammalianAvailable SinceOct. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
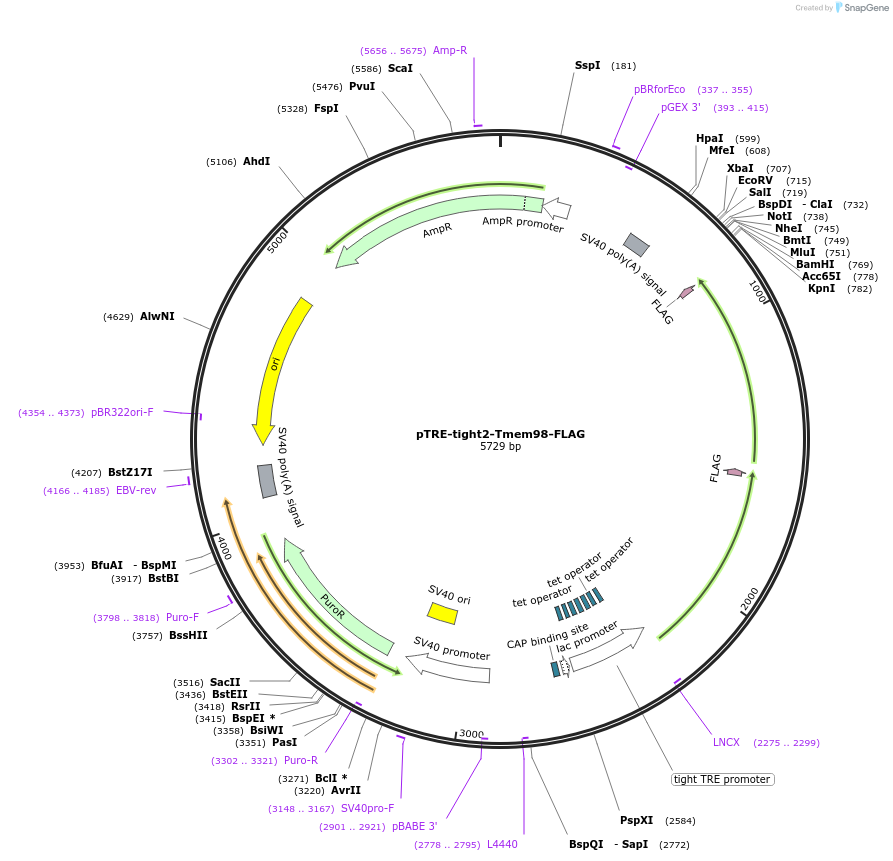
pTRE-tight2-Tmem98-FLAG
Plasmid#124506PurposeExpresses FLAG-tagged mouse TMEM98 in mammalian cells in a doxycycline-dependent manner (C-terminal tag)DepositorAvailable SinceMay 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
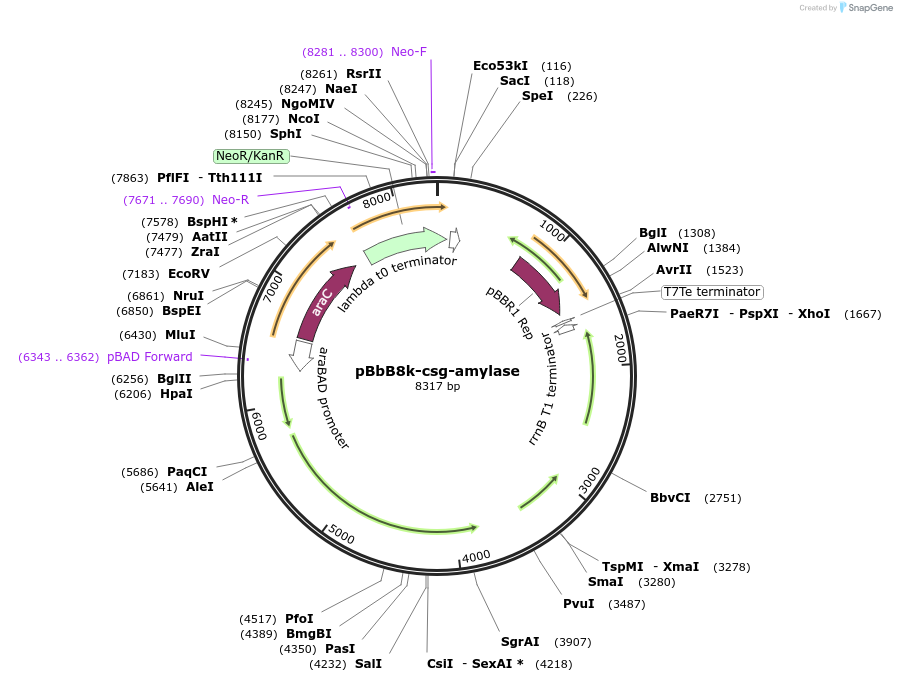
pBbB8k-csg-amylase
Plasmid#166859PurposeArabinose-inducible csgBACEFG operon for curli fiber synthesis in which curli subunit CsgA has been fused to α-amylase via a flexible linker. Derived from https://www.addgene.org/35363.DepositorInsertcsgBACEFG, with α-amylase fused to CsgA via flexible linker
UseSynthetic BiologyExpressionBacterialAvailable SinceApril 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
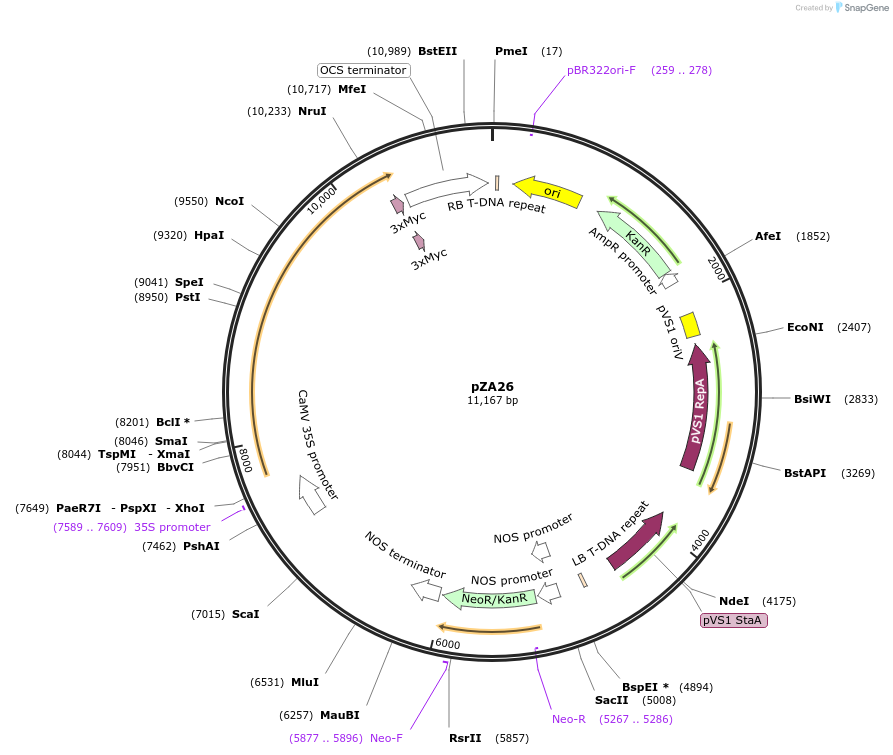
pZA26
Plasmid#158504PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 D498V-4xMyc. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
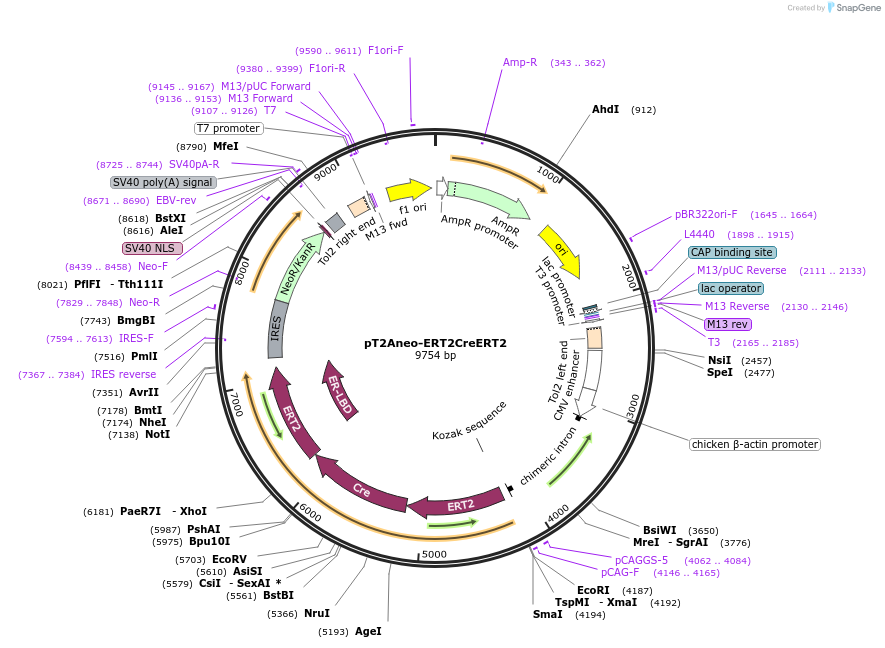
pT2Aneo-ERT2CreERT2
Plasmid#173850PurposeEncoding Cre recombinase (activated in response to tamoxifen)DepositorInsertERT2CreERT2
ExpressionMammalianAvailable SinceOct. 21, 2021AvailabilityAcademic Institutions and Nonprofits only -
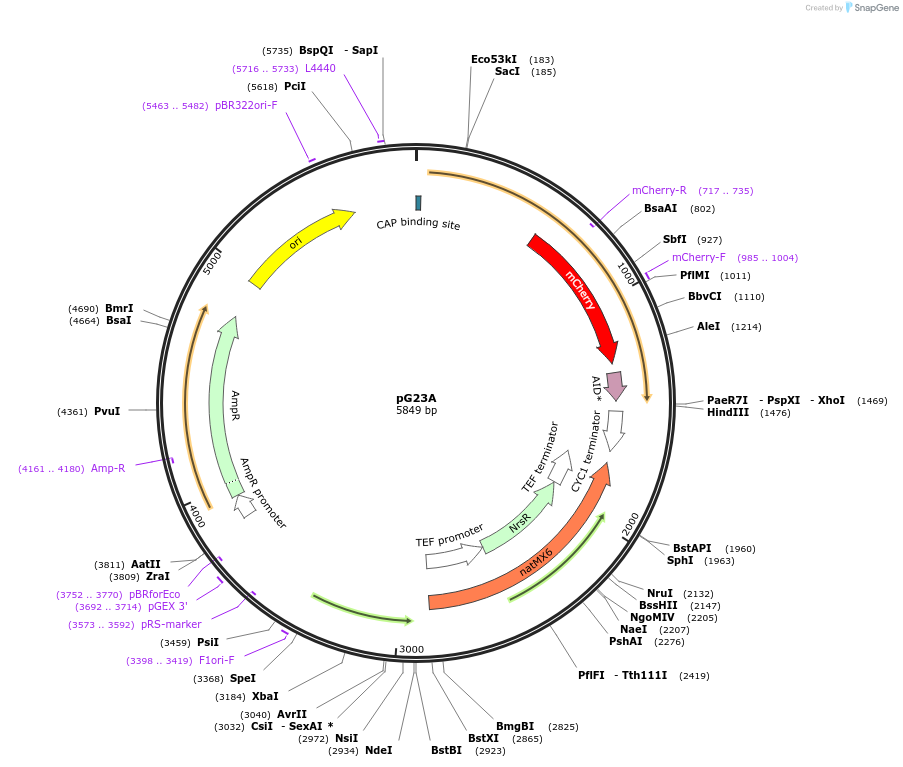
pG23A
Plasmid#102884PurposePlasmid used for the C-terminal tagging of Cdc14 with mCherry and the auxin-inducible degron in yeast.DepositorInsertCdc14/mCherry-AID(71-114)-NatMX (CDC14 Budding Yeast)
TagsAID(71-114) and mCherryExpressionYeastPromoterNative promoterAvailable SinceJan. 5, 2018AvailabilityAcademic Institutions and Nonprofits only -
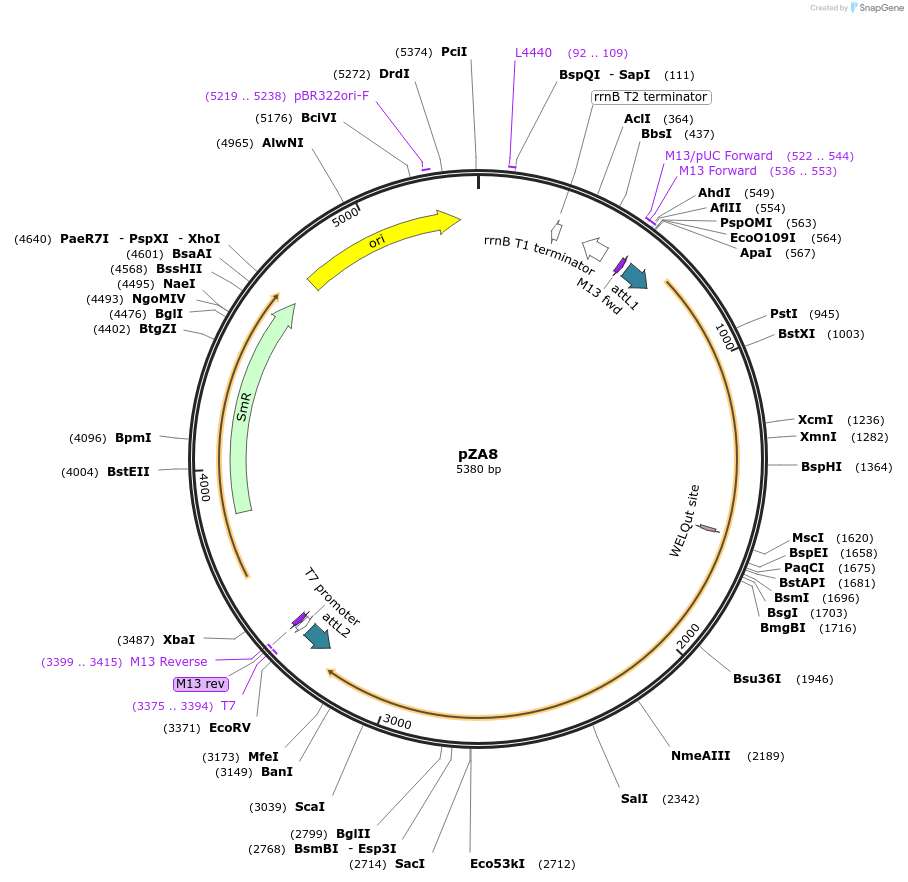
pZA8
Plasmid#158486PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 L17E/D481V with STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationL17E, D481V, STOP codonAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
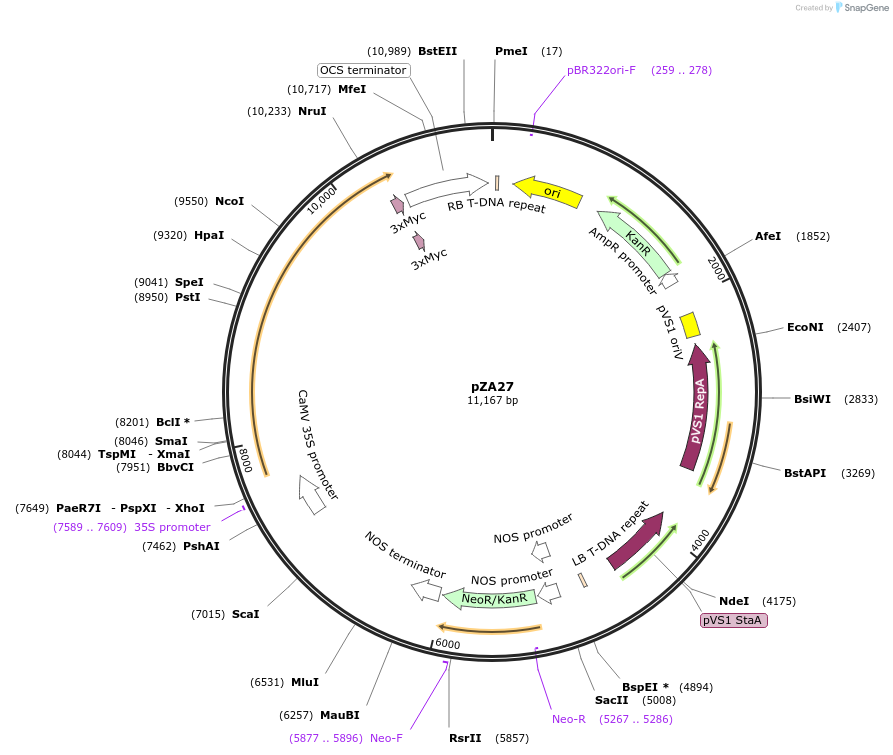
pZA27
Plasmid#158505PurposeIn planta gene expression of Golden gate compatible A. thaliana ZAR1 L17E-4xMyc. Km plant selection marker.DepositorAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
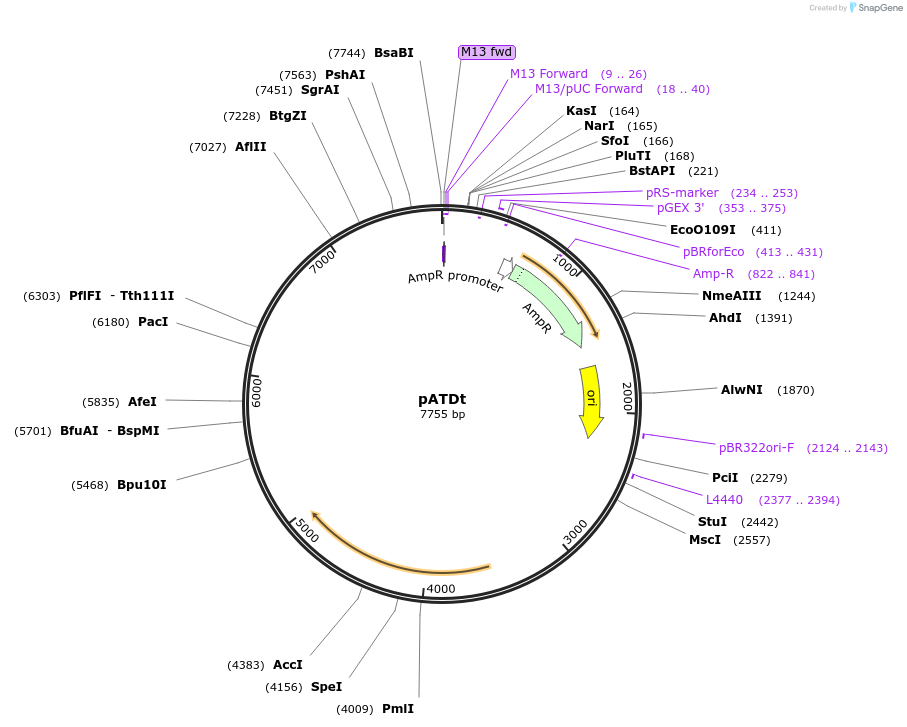
pATDt
Plasmid#249499PurposeExpresses tdTomato – Monodirectional chloroplast expression vector (psbH Rescue) in Chlamydomonas Reinhardtii photosynthetically deficient strain CC4388DepositorInserttdTomato (red fluorescence)
UseChlamydomonas reinhardtii chloroplast genomeExpressionBacterialMutationtdTomato results from the fusion of two copies of…PromoterThe psbD/ 5′ UTR, a light-activated chloroplast p…Available SinceJan. 21, 2026AvailabilityAcademic Institutions and Nonprofits only -
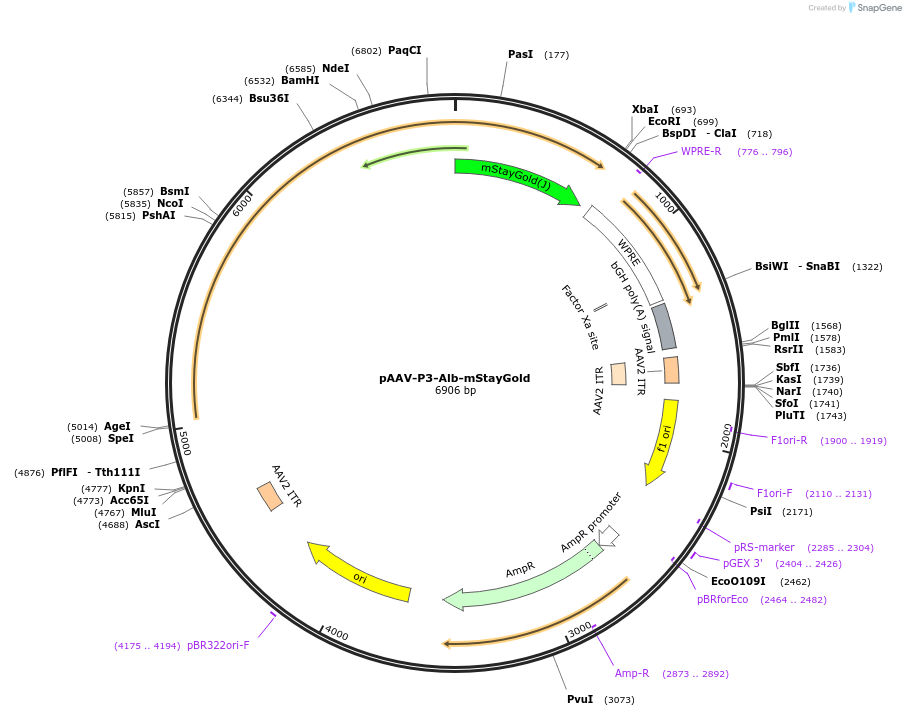
pAAV-P3-Alb-mStayGold
Plasmid#234549PurposeTo produce AAV with the P3 promoter to express Alb(mouse)-mStayGold(Ando) fusion proteinDepositorInsertAlb-mStayGold
UseAAVMutationNonePromoterP3Available SinceOct. 22, 2025AvailabilityAcademic Institutions and Nonprofits only -
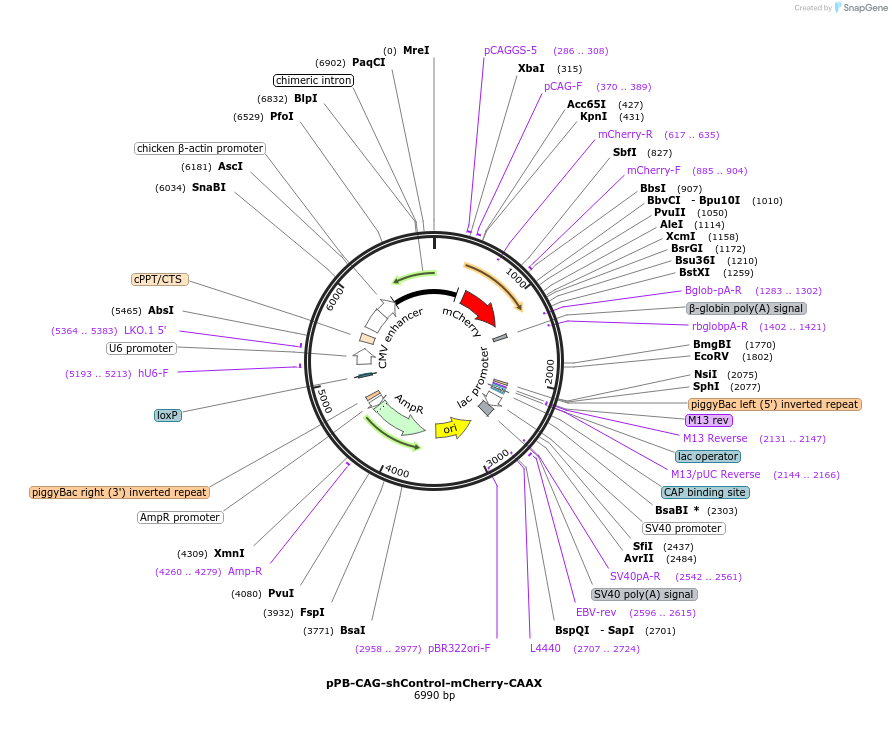
pPB-CAG-shControl-mCherry-CAAX
Plasmid#227689PurposeMembrane tethered mCherry and scramble of shRNA targeting Atg7DepositorInsertscrambled shRNA control
ExpressionMammalianAvailable SinceOct. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
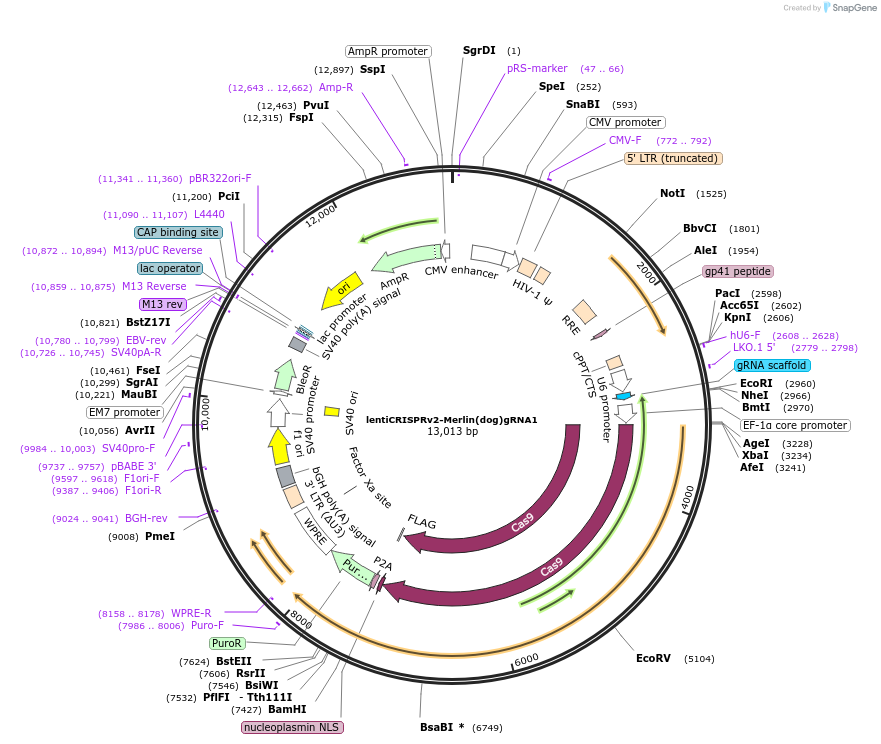
lentiCRISPRv2-Merlin(dog)gRNA1
Plasmid#214823PurposeA knockout vector for dog Merlin.DepositorInsertA gRNA targeting the dog NF2(Merlin) gene and the cDNA of CRISPR-Cas9
UseCRISPR and LentiviralExpressionMammalianAvailable SinceJuly 22, 2025AvailabilityAcademic Institutions and Nonprofits only -
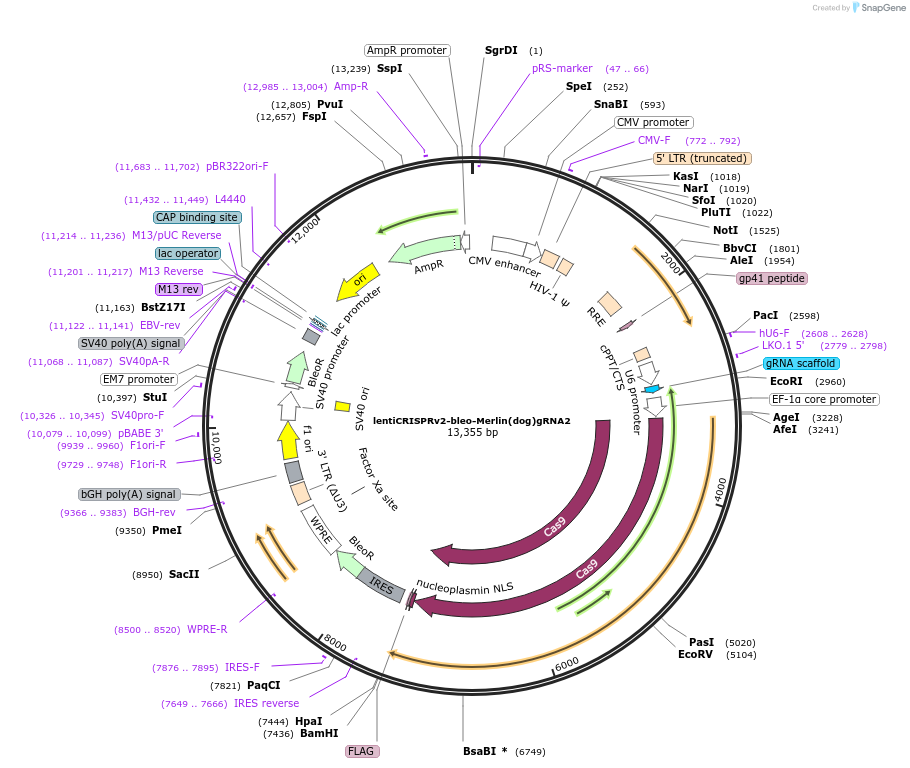
lentiCRISPRv2-bleo-Merlin(dog)gRNA2
Plasmid#214824PurposeA knockout vector for dog Merlin.DepositorInsertA gRNA targeting the dog NF2(Merlin) gene and the cDNA of CRISPR-Cas9
UseCRISPR and LentiviralExpressionMammalianAvailable SinceJuly 22, 2025AvailabilityAcademic Institutions and Nonprofits only -
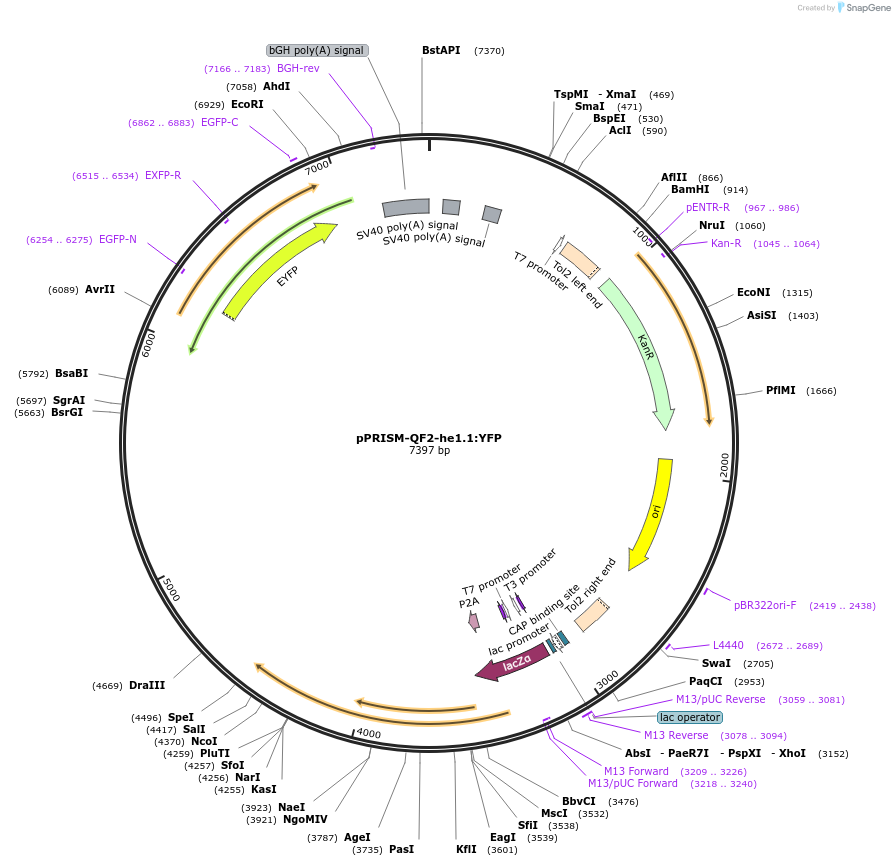
pPRISM-QF2-he1.1:YFP
Plasmid#184819PurposeModified version of GeneWeld method from Wierson et al., 2020 (https://doi.org/10.7554/eLife.53968). Backbone into which homology arms are cloned for CRISPR/Cas9-mediated integration of QF2.DepositorInsertQF2-pA-he1.1:YFP
UseCRISPR; Backbone for cloning of homology arms (fo…Available SinceMay 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
pPRISM-QF2-he1.1:YFP-rln3a-HA
Plasmid#184820PurposeModified version of GeneWeld method from Wierson et al., 2020 (https://doi.org/10.7554/eLife.53968). Contains homology arm inserts for integration of QF2 into rln3a gene by CRISPR/Cas9 integration.DepositorInsertrln3a-HA
UseCRISPR; For integration of qf2 by homology-direct…Available SinceMay 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
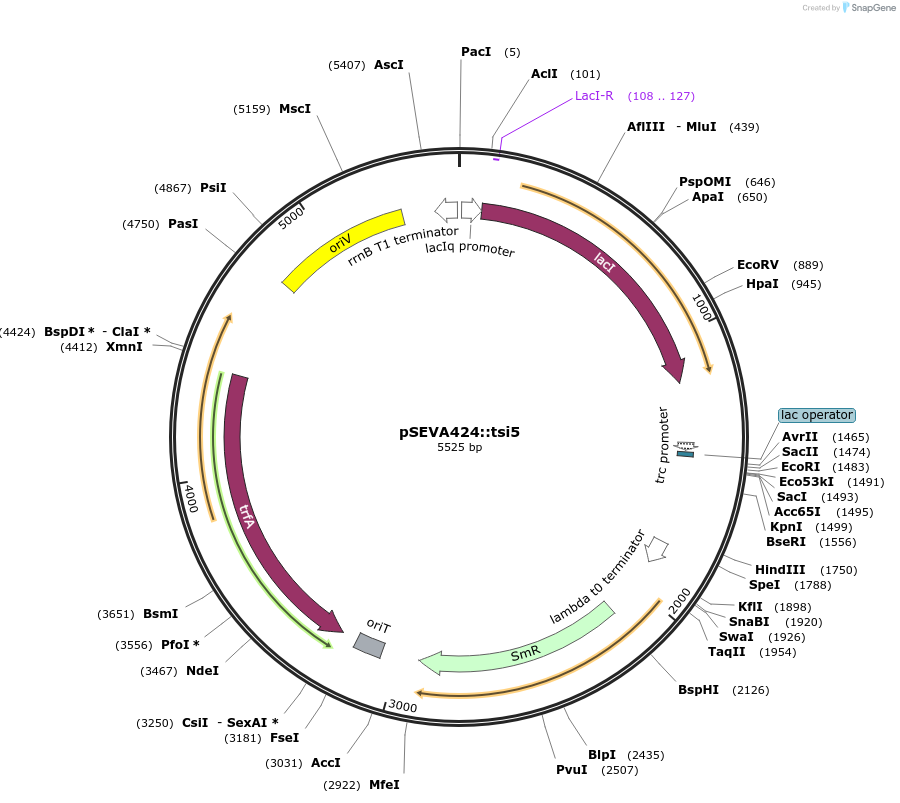
pSEVA424::tsi5
Plasmid#192960PurposeExpresses the protein Tsi51-76 for biological function studies in a broad-spectrum of organismsDepositorInserttsi5
ExpressionBacterialAvailable SinceFeb. 3, 2023AvailabilityAcademic Institutions and Nonprofits only