We narrowed to 26,497 results for: Vit;
-
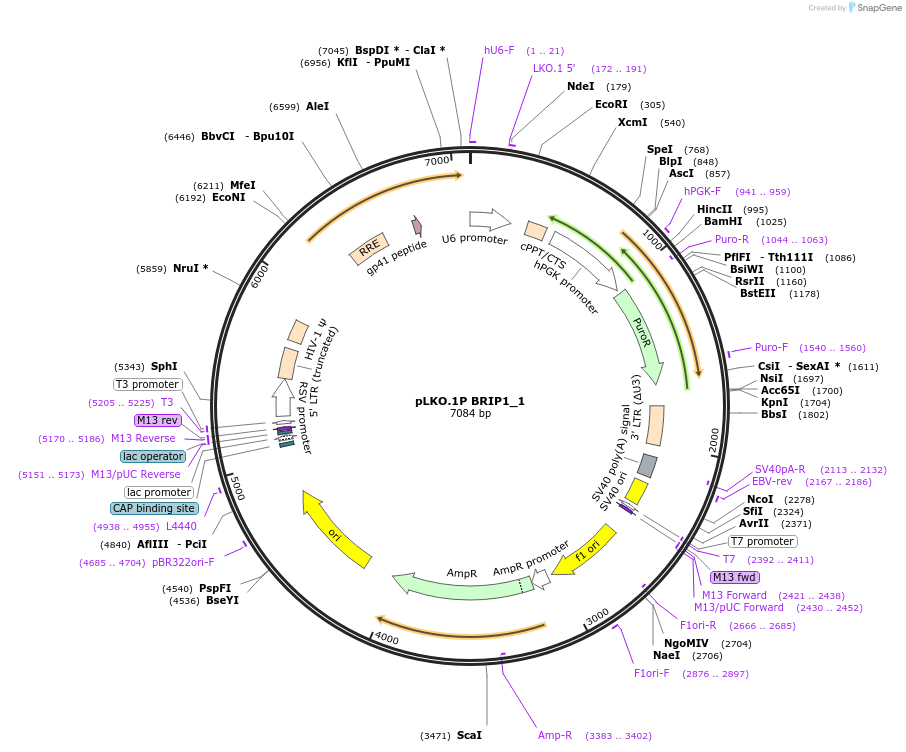
Plasmid#160771PurposeSuppress BRIP1DepositorInsertshBRIP1_1
UseLentiviralAvailable SinceOct. 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
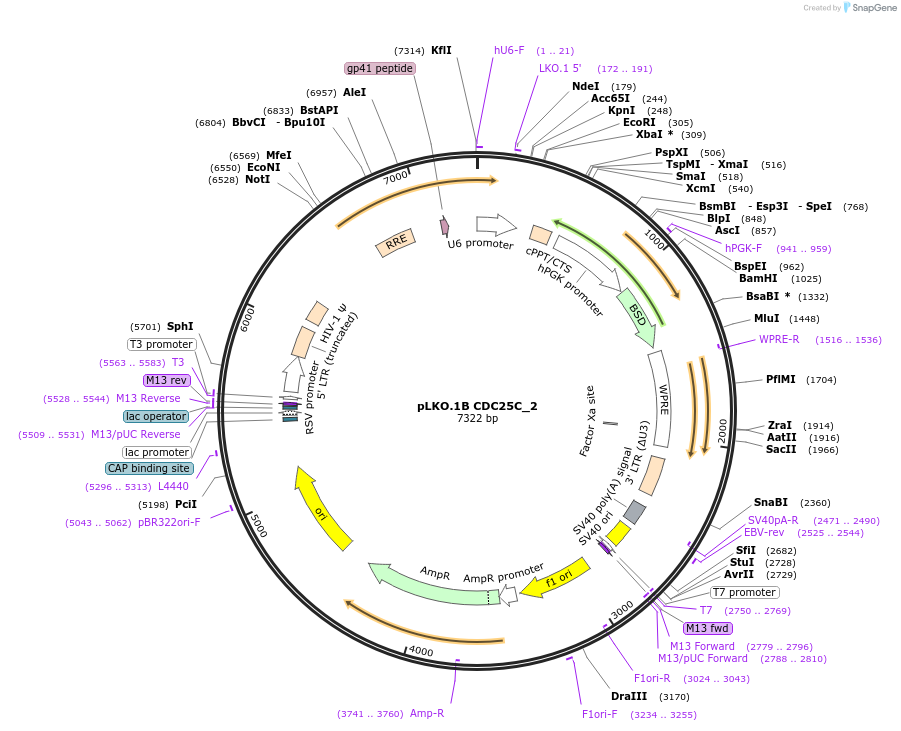
pLKO.1B CDC25C_2
Plasmid#160770PurposeSuppress CDC25CDepositorInsertshCDC25C_2
UseLentiviralAvailable SinceOct. 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
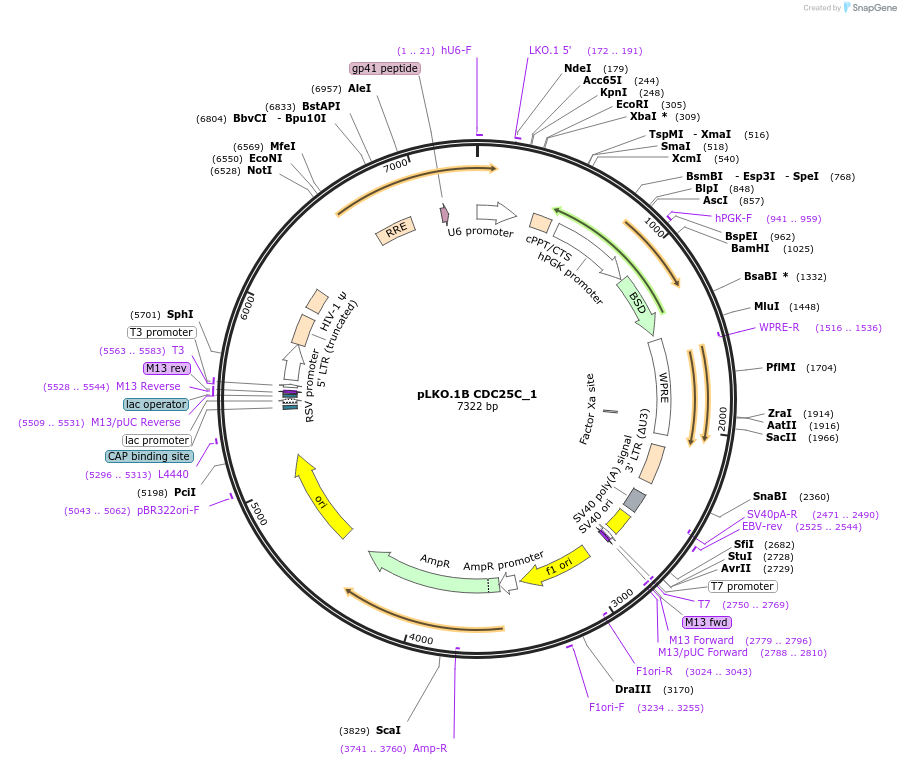
pLKO.1B CDC25C_1
Plasmid#160769PurposeSuppress CDC25CDepositorInsertshCDC25C_1
UseLentiviralAvailable SinceOct. 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
pLKO.1B CDC25A_1
Plasmid#160767PurposeSuppress CDC25ADepositorInsertshCDC25A_1
UseLentiviralAvailable SinceOct. 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
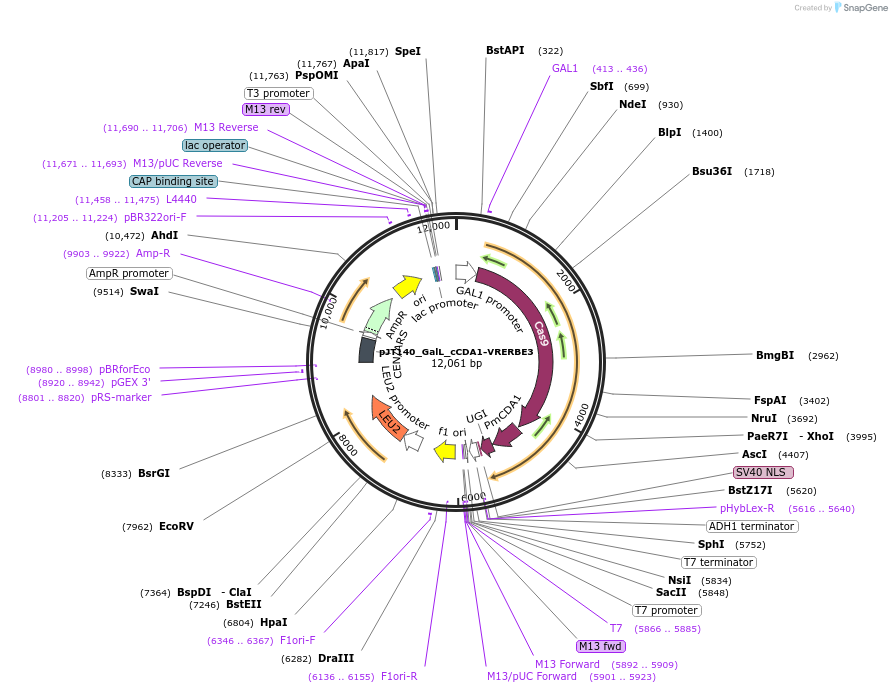
pJT140_GalL_cCDA1-VRERBE3
Plasmid#145103PurposeExpressing base editor cCDA1-VRERBE3 in yeast cellsDepositorInsertcCDA1-VRERBE3
UseCRISPRExpressionYeastMutationspCas9(D10A/D1135V/G1218R/R1335E/T1337R)PromoterGalLAvailable SinceSept. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
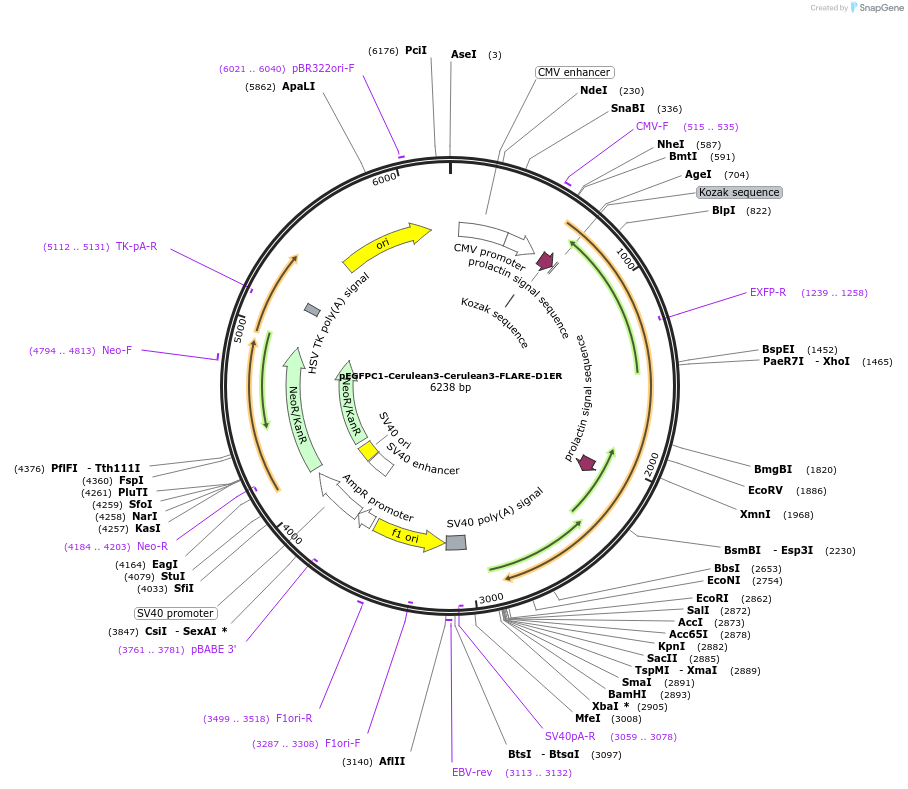
pEGFPC1-Cerulean3-Cerulean3-FLARE-D1ER
Plasmid#123354PurposeCyan-fluorescent homoFRET/anisotropy-based genetically encoded calcium indicator, targeted to endoplasmic reticulum lumen.DepositorInsertcp-moxCerulean3-moxCerulean3-FLARE-D1ER
TagsC-terminal ER retention signal (KDEL), cp-moxCeru…ExpressionMammalianPromoterCMVAvailable SinceSept. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
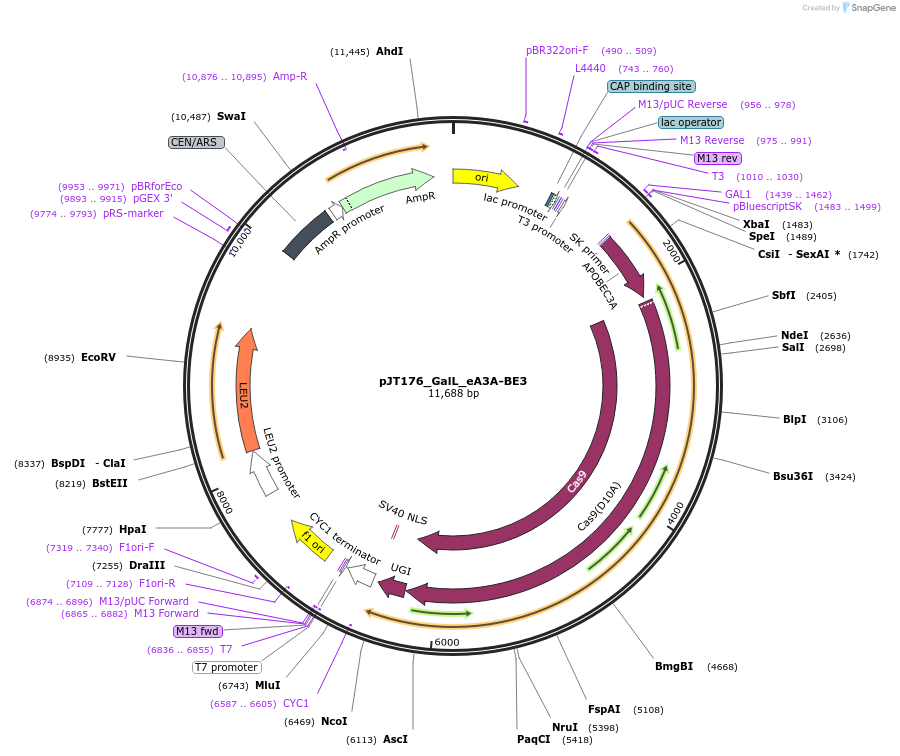
pJT176_GalL_eA3A-BE3
Plasmid#145125PurposeExpressing base editor eA3A-BE3 in yeast cellsDepositorInsertA3A(N57G)-BE3
UseCRISPRExpressionYeastMutationA3A(N57G); spCas9(D10A)PromoterGalLAvailable SinceJuly 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
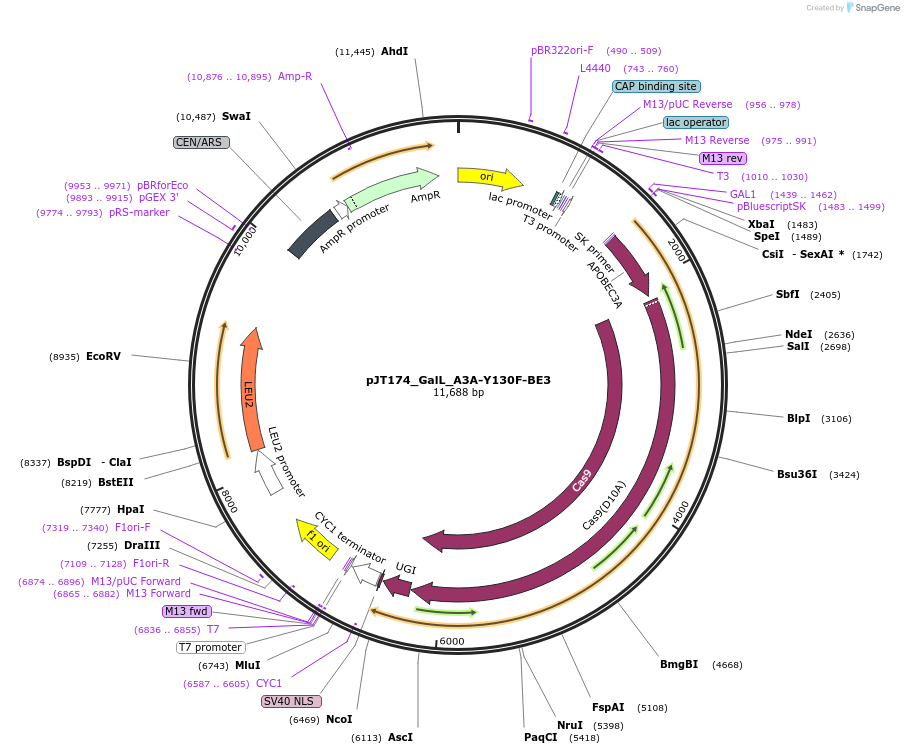
pJT174_GalL_A3A-Y130F-BE3
Plasmid#145123PurposeExpressing base editor A3A-Y130F-BE3 in yeast cellsDepositorInsertA3A(Y130F)-BE3
UseCRISPRExpressionYeastMutationA3A(Y130F); spCas9(D10A)PromoterGalLAvailable SinceJuly 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
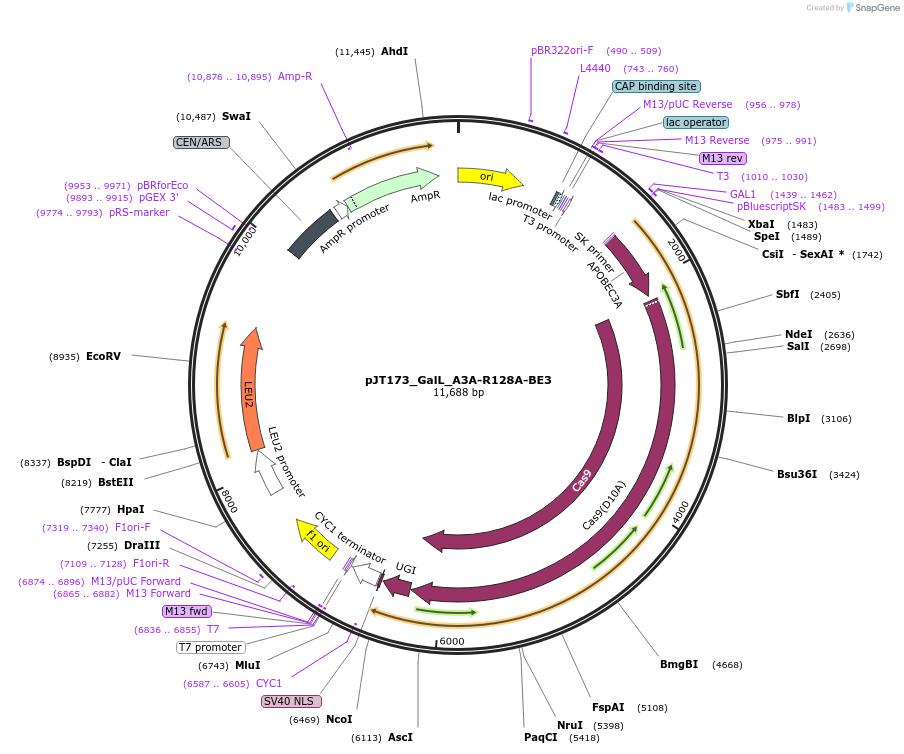
pJT173_GalL_A3A-R128A-BE3
Plasmid#145122PurposeExpressing base editor A3A-R128A-BE3 in yeast cellsDepositorInsertA3A(R128A)-BE3
UseCRISPRExpressionYeastMutationA3A(R128A); spCas9(D10A)PromoterGalLAvailable SinceJuly 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
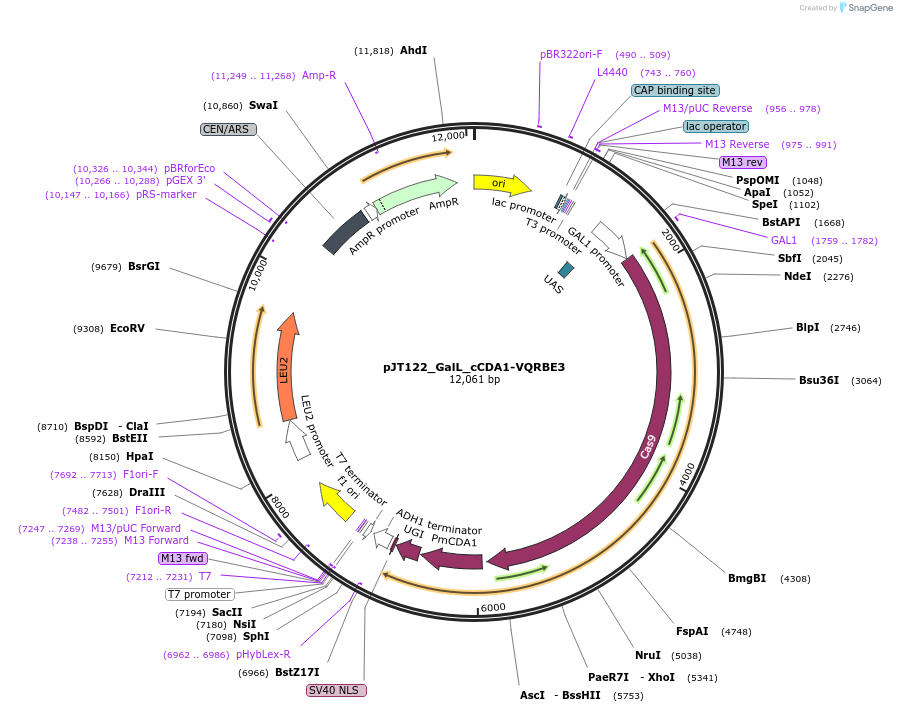
pJT122_GalL_cCDA1-VQRBE3
Plasmid#145087PurposeExpressing base editor cCDA1-VQRBE3 in yeast cellsDepositorInsertcCDA1-VQRBE3
UseCRISPRExpressionYeastMutationspCas9(D10A, D1135V, R1335Q, T1337R)PromoterGalLAvailable SinceJuly 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
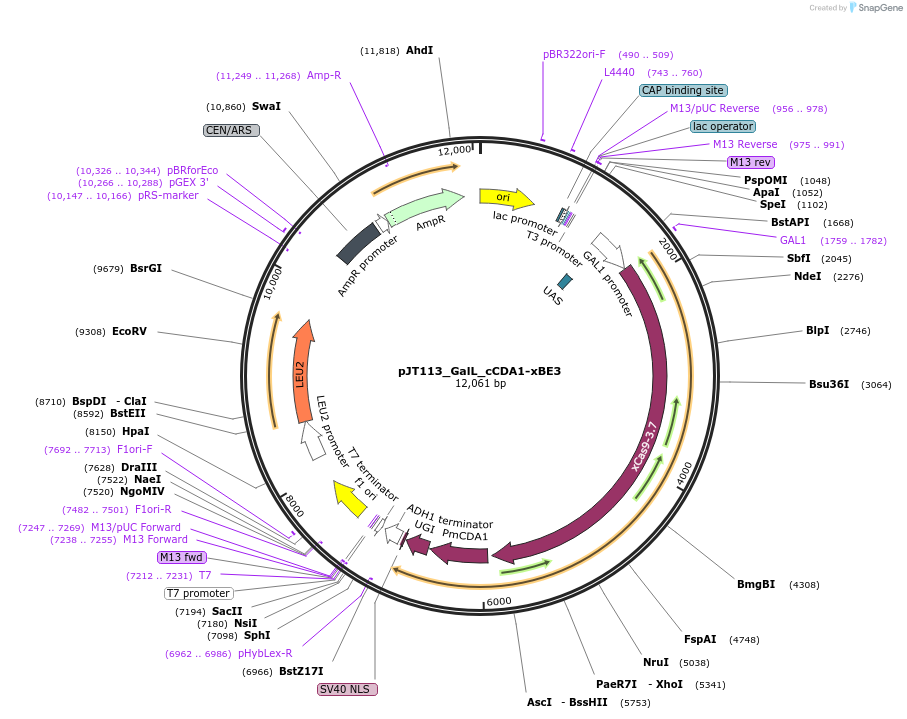
pJT113_GalL_cCDA1-xBE3
Plasmid#145079PurposeExpressing base editor cCDA1-xBE3 in yeast cellsDepositorInsertcCDA1-xBE3
UseCRISPRExpressionYeastMutationspCas9(D10A, A262T, R324L, S409I, E480K, E543D, M…PromoterGalLAvailable SinceJuly 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
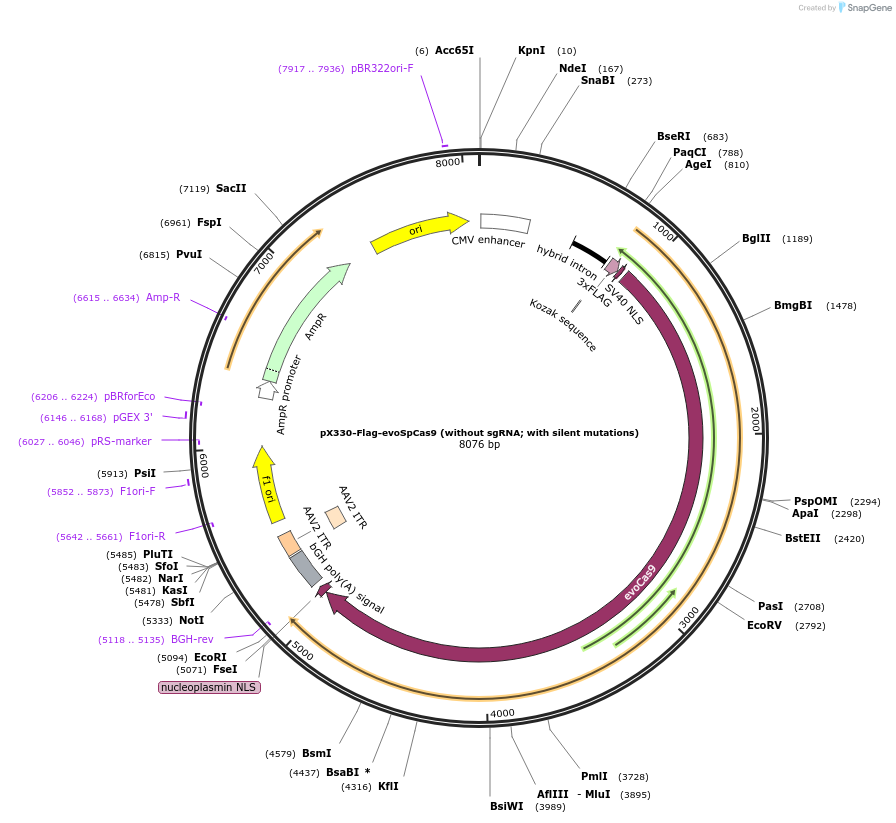
pX330-Flag-evoSpCas9 (without sgRNA; with silent mutations)
Plasmid#126758PurposeExpression plasmid for human codon-optimized increased fidelity evoSpCas9 (without U6-sgRNA coding sequence, with silent mutations)DepositorInsertevoSpCas9
UseCRISPRTags3xFLAG and NLSExpressionMammalianMutationM495V, Y515N, K526E, R661QPromoterCBhAvailable SinceMarch 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
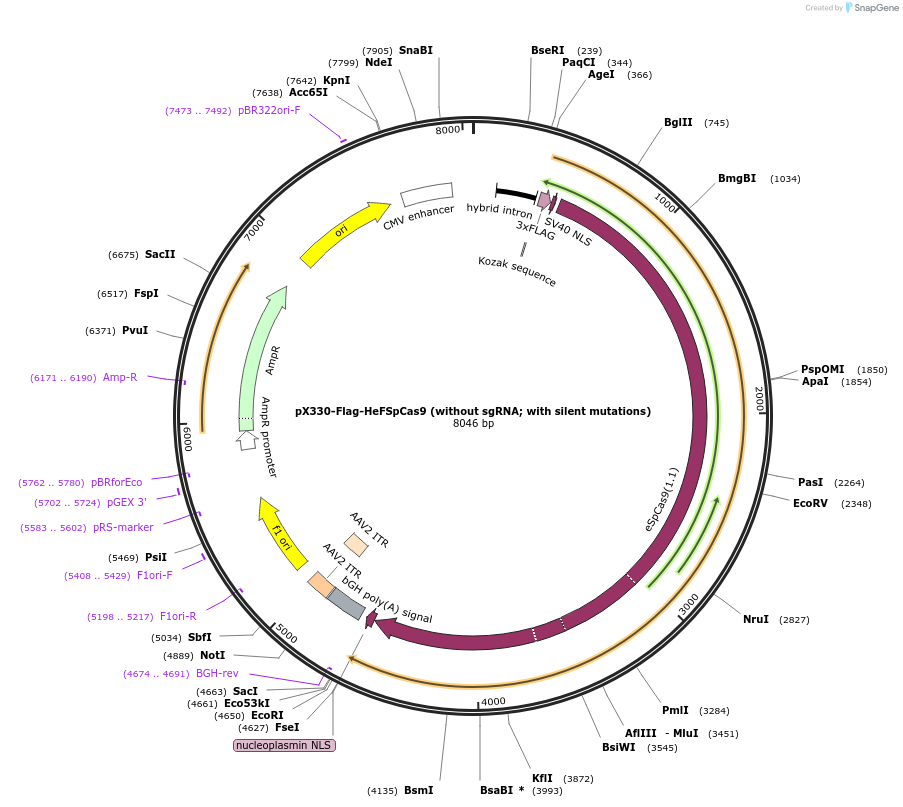
pX330-Flag-HeFSpCas9 (without sgRNA; with silent mutations)
Plasmid#126759PurposeExpression plasmid for human codon-optimized increased fidelity HeFSpCas9 (without U6-sgRNA coding sequence, with silent mutations)DepositorInsertHeFSpCas9
UseCRISPRTags3xFLAG and NLSExpressionMammalianMutationN497A, R661A, Q695A, K848A, Q926A, K1003A, R1060APromoterCBhAvailable SinceMarch 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
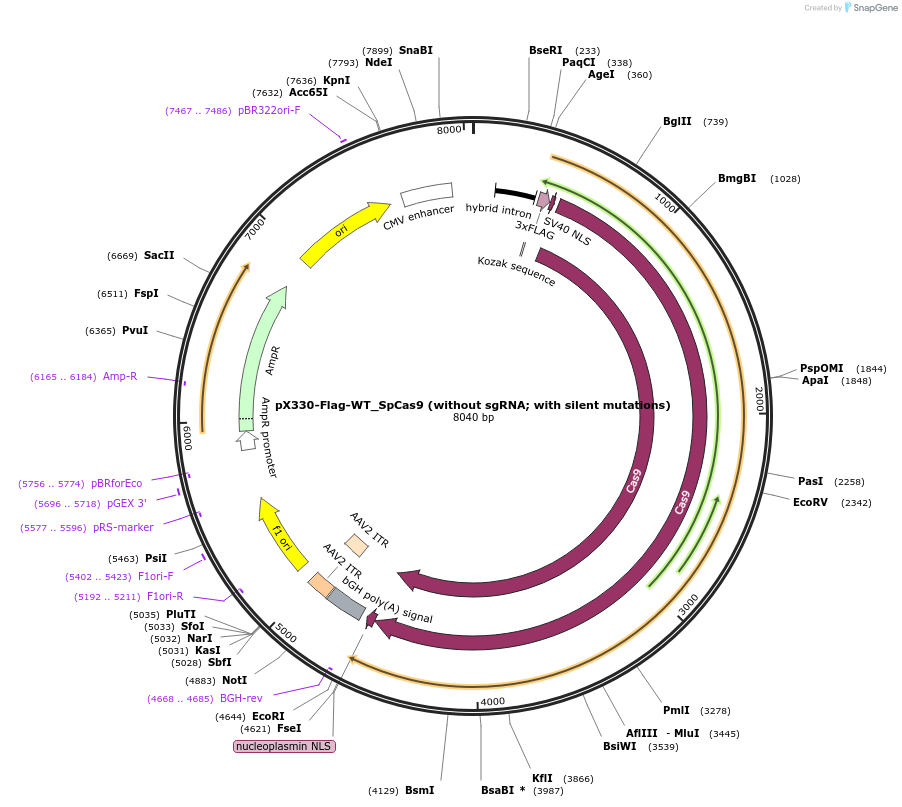
pX330-Flag-WT_SpCas9 (without sgRNA; with silent mutations)
Plasmid#126753PurposeExpression plasmid for human codon-optimized WT SpCas9 (without U6-sgRNA coding sequence, with silent mutations)DepositorInsertWT SpCas9
UseCRISPRTags3xFLAG and NLSExpressionMammalianPromoterCBhAvailable SinceMarch 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
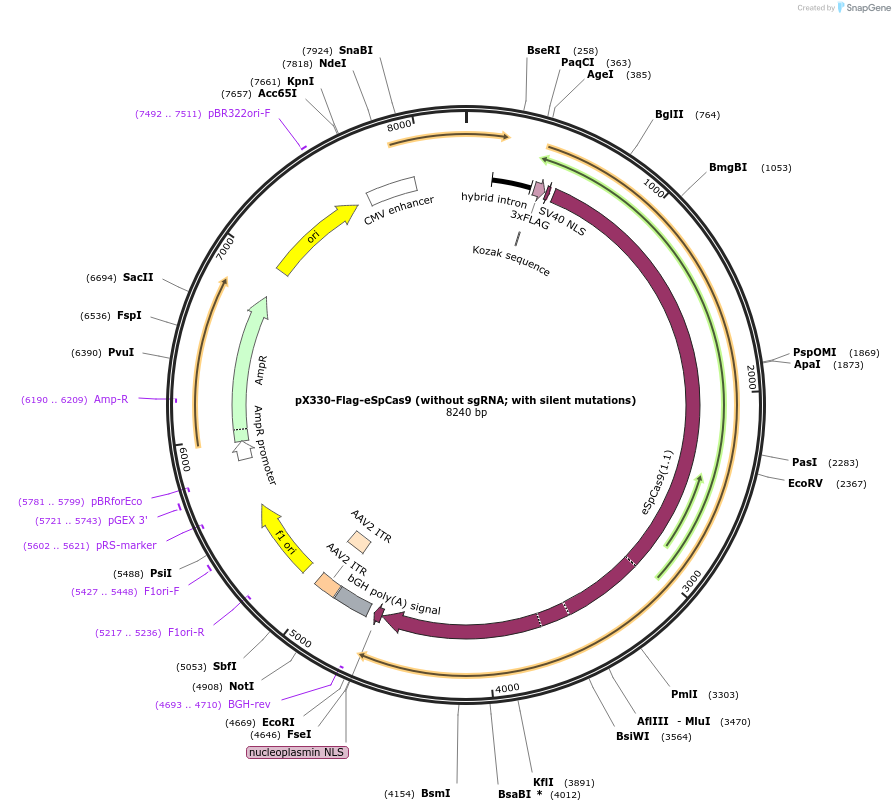
pX330-Flag-eSpCas9 (without sgRNA; with silent mutations)
Plasmid#126754PurposeExpression plasmid for human codon-optimized increased fidelity eSpCas9 (without U6-sgRNA coding sequence, with silent mutations)DepositorInserteSpCas9
UseCRISPRTags3xFLAG and NLSExpressionMammalianMutationK848A, K1003A, R1060APromoterCBhAvailable SinceMarch 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
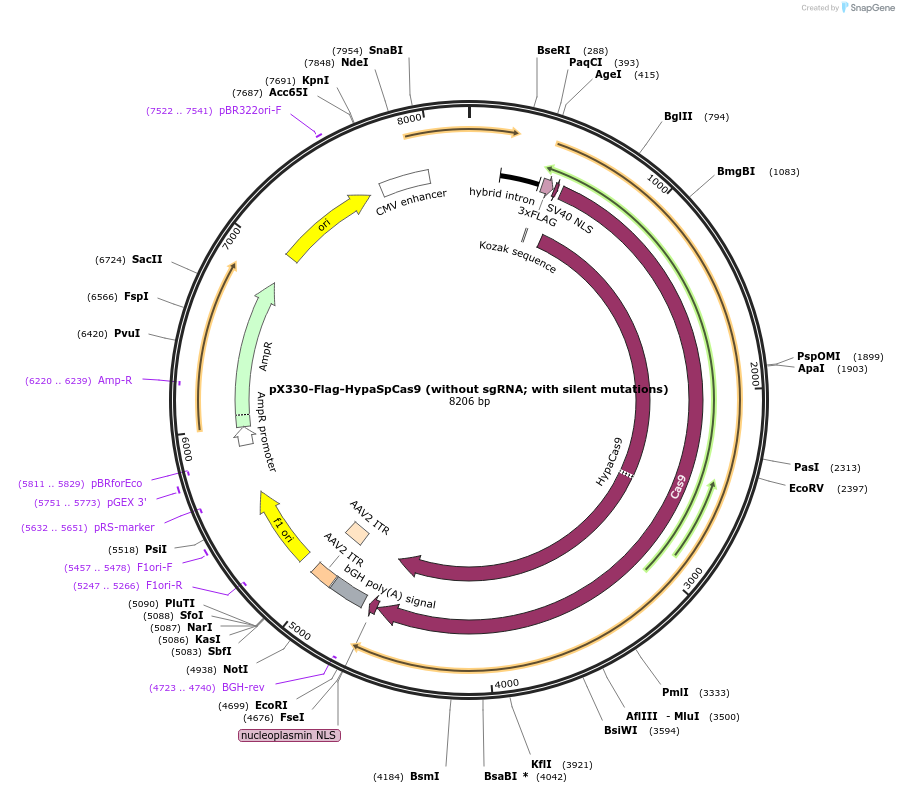
pX330-Flag-HypaSpCas9 (without sgRNA; with silent mutations)
Plasmid#126756PurposeExpression plasmid for human codon-optimized increased fidelity HypaSpCas9 (without U6-sgRNA coding sequence, with silent mutations)DepositorInsertHypaSpCas9
UseCRISPRTags3xFLAG and NLSExpressionMammalianMutationN692A, M694A, Q695A, H698APromoterCBhAvailable SinceMarch 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
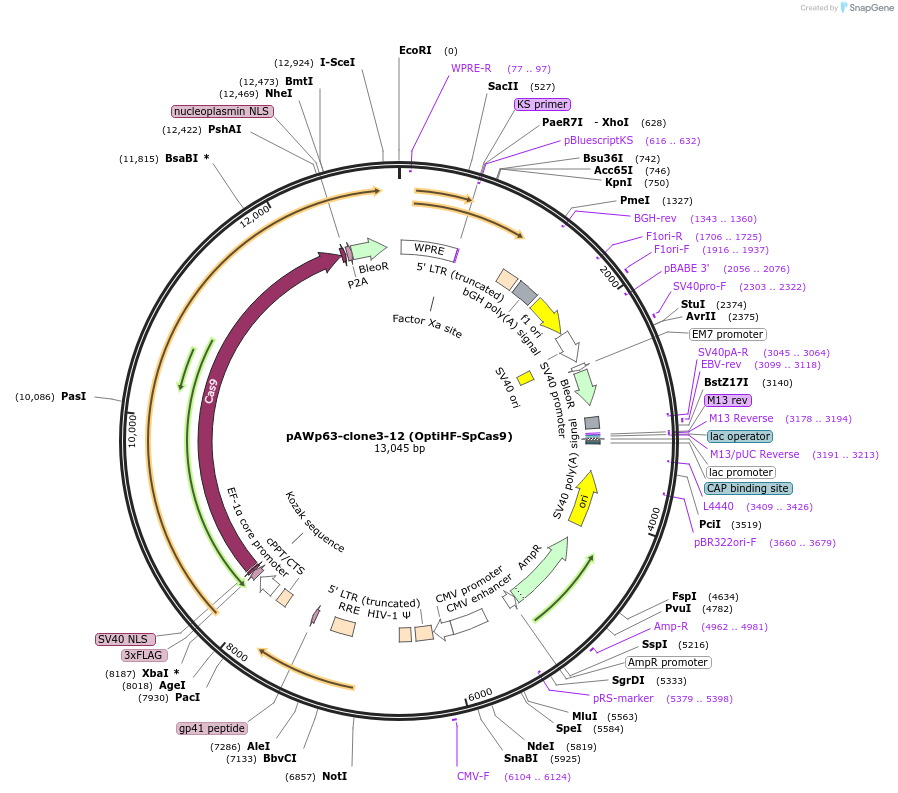
pAWp63-clone3-12 (OptiHF-SpCas9)
Plasmid#131737PurposeExpresses OptiHF-SpCas9 in mammalian cellsDepositorInsertOptiHF-SpCas9
UseLentiviralTagsFlagMutationQ695A+K848A+E923M+T924V+Q926APromoterEFSAvailable SinceDec. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
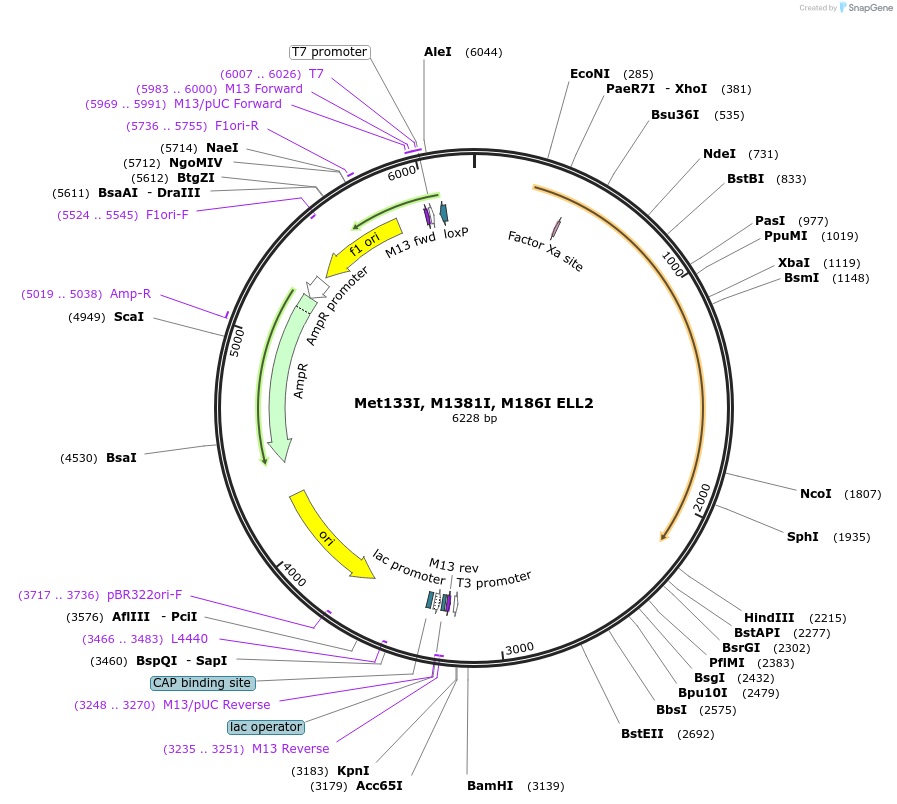
Met133I, M1381I, M186I ELL2
Plasmid#127263PurposeFor in vitro translation of human ELL2 with Met133I, M1381I, M186IDepositorInsertELL2 (Met133ILeu, M1381ILeu, M186ILeu) (ELL2 Human)
UseIn vitro translationMutationThree Mets (133, 138, 186) to IleuPromoterT7Available SinceNov. 14, 2019AvailabilityAcademic Institutions and Nonprofits only -
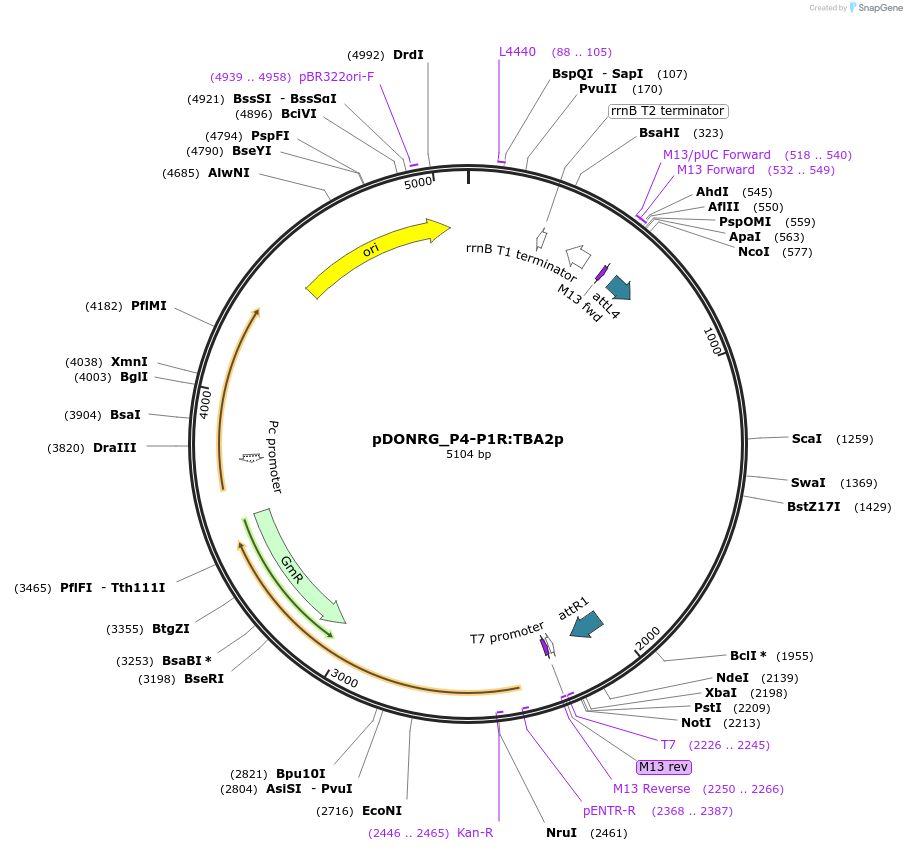
pDONRG_P4-P1R:TBA2p
Plasmid#128429PurposeGateway (Invitrogen) promoter clone (pDONRG_P4-P1R) containing the TBA2 (At1g62060) promoter sequence (1346 base pairs), for use in Three-way Gateway cloning.DepositorInsertTESTA ABUNDANT 2 (AT1G62060 Mustard Weed)
UseGateway promoter entry clonePromoterTESTA ABUNDANT 2Available SinceOct. 4, 2019AvailabilityIndustry, Academic Institutions, and Nonprofits -
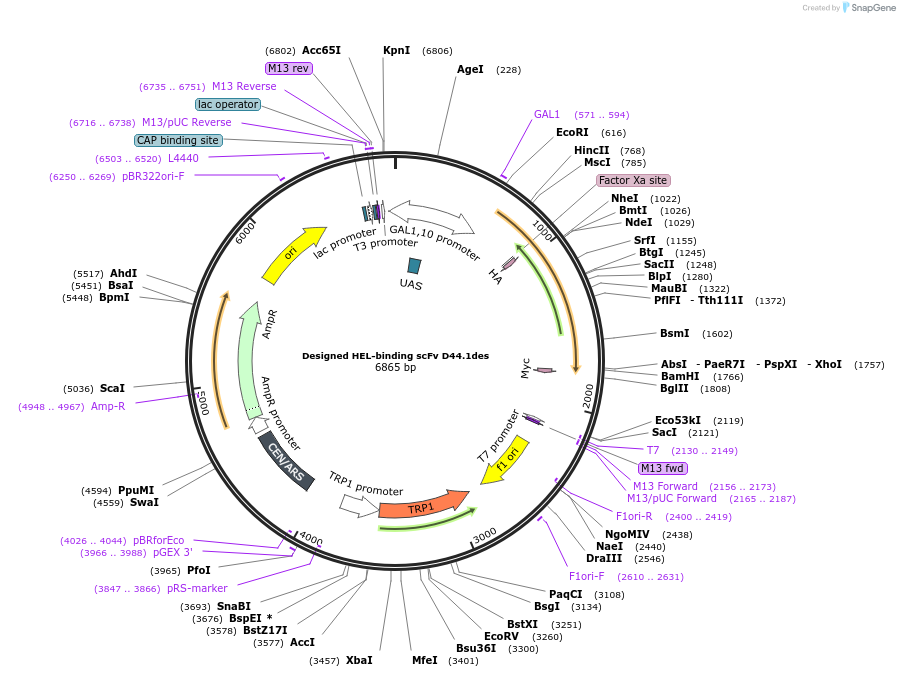
Designed HEL-binding scFv D44.1des
Plasmid#111719PurposeYeast Surface Display of D44.1desDepositorInsertD44.1 des
Tagscmyc tagExpressionYeastMutationL Q89G, L S55V, L S43P, H V37M, H H43G, H W47Y, H…Available SinceJuly 22, 2019AvailabilityAcademic Institutions and Nonprofits only